CD11cloB220+ interferon-producing killer dendritic cells are activated natural killer cells (original) (raw)
Abstract
Interferon-producing killer dendritic cells (IKDCs) are a recently described subset of CD11cloB220+ cells that share phenotypic and functional properties of DCs and natural killer (NK) cells (Chan, C.W., E. Crafton, H.N. Fan, J. Flook, K. Yoshimura, M. Skarica, D. Brockstedt, T.W. Dubensky, M.F. Stins, L.L. Lanier, et al. 2006. Nat. Med. 12:207–213; Taieb, J., N. Chaput, C. Menard, L. Apetoh, E. Ullrich, M. Bonmort, M. Pequignot, N. Casares, M. Terme, C. Flament, et al. 2006. Nat. Med. 12:214–219). IKDC development appears unusual in that cytokines using the interleukin (IL)-2 receptor β (IL-2Rβ) chain but not those using the common γ chain (γc) are necessary for their generation. By directly comparing Rag2−/−γc −/y, Rag2−/−IL-2Rβ−/−, Rag2−/−IL-15−/−, and Rag2−/−IL-2−/− mice, we demonstrate that IKDC development parallels NK cell development in its strict IL-15 dependence. Moreover, IKDCs uniformly express NK-specific Ncr-1 transcripts (encoding NKp46), whereas NKp46+ cells are absent in Ncr1gfp/+γc −/y mice. Distinguishing features of IKDCs (CD11cloB220+MHC-II+) were carefully examined on developing NK cells in the bone marrow and on peripheral NK cells. As B220 expression was heterogeneous, defining B220lo versus B220hi NK1.1+ NK cells could be considered as arbitrary, and few phenotypic differences were noted between NK1.1+ NK cells bearing different levels of B220. CD11c expression did not correlate with B220 or major histocompatibility complex (MHC) class II (MHC-II) expression, and most MHC-II+ NK1.1+ cells did not express B220 and were thus not IKDCs. Finally, CD11c, MHC-II, and B220 levels were up-regulated on NK1.1+ cells upon activation in vitro or in vivo in a proliferation-dependent fashion. Our data suggest that the majority of CD11cloB220+ “IKDC-like” cells represent activated NK cells.
During innate and adaptive immune responses, NK cells and DCs appear to play distinct, complementary, and synergistic roles. NK cells, by virtue of their capacity to respond without prior sensitization, can directly eliminate infected or transformed target cells and elaborate soluble factors (cytokines and chemokines) that recruit and amplify the inflammatory response. DCs, in contrast, are the sentinels that, once activated, take up antigen, mature, and migrate to secondary lymphoid organs, where they present the processed antigen to naive T cells to initiate the adaptive immune response. In addition, DCs and NK cells can interact in a reciprocal fashion that results in the mutual activation of both types of cells (for review see reference 1). Mature DCs can prime NK cells to enhance their functional capacities (2), whereas activated NK cells secrete IFN-γ that up-regulates antigen presentation in DCs and can “edit” DC responses through the elimination of immature DCs (3, 4). In this way, NK cells and DCs represent two highly specialized cell types that interact in a complementary fashion. This cellular strategy could provide an additional level of control, because complete immune responses would not be efficiently achieved by activation of either cell type in isolation.
Recently, the functional dichotomy of DCs and NK cells has been challenged with the characterization of a hybrid cell subset called IFN-producing killer DCs (IKDCs) (5, 6). IKDCs were characterized as CD3−CD19− cells with a CD11cloB220+ phenotype and were clearly distinguished from plasmacytoid DCs (pDCs) because of the absence of Ly6C (Gr-1) expression (5). CD11cloB220+Gr-1− cells expressed NK1.1 and CD49b in addition to other NK cell surface markers (NKG2D and Ly49 family receptors), and like DCs, they expressed co-stimulatory molecules (CD80 and CD86) and MHC class II (MHC-II) (5, 6). Moreover, initial reports demonstrated that IKDCs possess NK cell–like cytotoxicity and inducible IFN-γ production, as well as a DC-like ability to secrete IFN-α and to stimulate transgenic OVA-specific T cells (5, 6). Based on these observations, IKDCs appear to represent a hybrid effector cell that can act both as the “assassin” (NK cells) and as the “messenger” (DCs).
Although the evidence for dual IKDC function was clearly demonstrated at the population level (i.e., in the CD11cloB220+ subset), it could not be rigorously ruled out that IKDC activities were simply the additive property of two distinct cell types that shared a CD11cloB220+ phenotype. Neither CD11c nor B220 are DC-specific markers, as B cells express B220 and activated B cells and CD8 T cells, and most NK cells were shown to express CD11c (7–10). Only a minor fraction of IKDCs was able to produce type I IFN upon stimulation in vitro (5), and direct evidence that the same cell can produce both IFN-α and IFN-γ is still lacking. Moreover, recent reports have challenged the idea that IKDCs secrete IFN-α upon stimulation (11, 12), suggesting that minor pDC contamination in IKDC purification may have accounted for the type I IFN detected in a previous report (5).
Concerning their development from hematopoietic precursors, IKDCs appeared to have a truly unique cytokine dependency, because they were not detected in mice lacking the IL-2Rβ chain (required for IL-2 and IL-15 receptor function), whereas they were present in mice lacking the common γ chain (γc; required for IL-2, -4, -7, -9, -15, and -21 receptors) (5, 6), suggesting the existence of a new IL-2Rβ–dependent, γc-independent signaling pathway. Alternatively, IKDCs could also be IL-2Rβ independent but might be missing from IL-2Rβ−/− mice secondary to the intense inflammatory syndrome that these mice develop (13). Lastly, genetic background differences in the mice analyzed could explain the divergent results.
Considering their curious developmental, phenotypic, and functional qualities, we have analyzed the development of “IKDC-like” cells (the CD11cloB220+ subset) using a series of C57BL/6 mice deficient in selected cytokines and their receptors. Using mice that report expression of the NK cell–specific NKp46 molecule (Ncr-1gfp/+ mice) (14), we demonstrate that IKDC-like cells are developmentally related to NK cells and share their IL-15 dependence. Moreover, many of the distinguishing features of IKDC-like cells (B220, CD11c, and MHC-II expression) are inducible in NK cells. Our results challenge the view that IKDC-like cells belong to a separate DC lineage but provide evidence that the majority of these cells resemble activated NK cells.
RESULTS AND DISCUSSION
Cells expressing the IKDC-like profile are bona fide NK cells
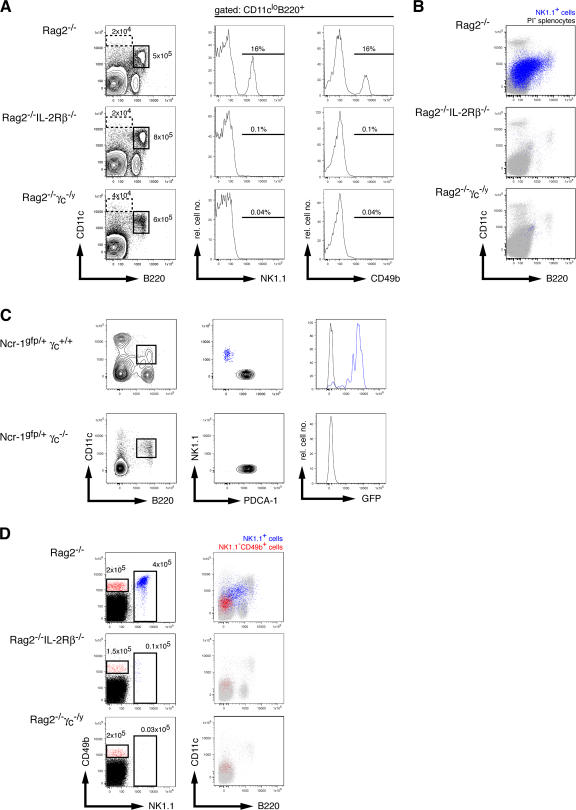
Cells with the CD11cloB220+ phenotype are heterogeneous, as both CD11c and B220 are expressed by B, T, and NK lymphocytes and by diverse DC subsets. By excluding CD3+ and CD19+ cells, the CD11cloB220+ subset has been shown to contain pDCs (NK1.1−Ly6C+) and a novel NK1.1+CD49b+ subset called IKDCs (5, 6). Previous reports detected IKDCs in Rag2−/−γc −/y mice but not in IL-2Rβ−/− mice (5, 6). These surprising results suggested the existence of a novel _il2rg_-dependent but _il2rb_-dependent signaling pathway that promoted IKDC development. Alternatively, the absence of IKDCs in IL-2Rβ−/− mice could have resulted from the inflammatory syndrome that these mice develop with age (13). To address these possibilities, we examined the CD11cloB220+ subset in Rag2−/− mice that were deficient or not in the γc or IL-2Rβ chains (Fig. 1 A). All mice were backcrossed onto the C57BL/6 background and carried the Nkrp1c B6 allele (allowing detection by NK1.1). We found that all mice had similar absolute numbers of BM CD11cloB220+ cells but that the deficiency in γc or IL-2Rβ completely ablated development of NK1.1+ or CD49b+ cells within the CD11cloB220+ subset (Fig. 1 A). Analysis of splenocytes from these mice yielded similar results (Fig. 1 B). To determine which IL-2Rβ–dependent ligand was driving IKDC development, we further analyzed Rag2−/−IL-2−/− and Rag2−/−IL-15−/− mice. The CD11cloB220+NK1.1+ subset was present in the former but not the latter (Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20071451/DC1), confirming the essential role for IL-15 in the development of all NK1.1+ cells (15).
Figure 1.
NK1.1+CD11cloB220+ (IKDC-like) cells are NK cells. (A) Analysis of BM cells from Rag2−/− (top row), Rag2−/−IL-2Rβ−/− (middle row), and Rag2−/−γc −/y (bottom row) mice. CD11c versus B220 profiles of BM cells from the indicated mice (left) with gates for cDCs (dotted line) and pDCs (continuous line) with their absolute cell numbers are given. (middle and right) NK1.1 and CD49b histograms, respectively, of the gated CD11cloB220+ cells from the left. The percentages of NK1.1+ (middle) and CD49b+ (right) cells among gated CD11cloB220+ cells are indicated. (B) Analysis of splenocytes from the same mice as in A. Shown are population overlays of viable cells (gray) with NK1.1+CD49b+ cells (blue). (C) Analysis of ncr1gfp/+γc+/+ and ncr1gfp/+γc−/y mice. (left) CD11c versus B220 profile of gated CD3−CD19− splenocytes of the indicated mice. (middle) PDCA-1 versus NK1.1 profiles on gated CD11cloB220+ cells. (right) GFP fluorescence (NKp46 expression) on gated NK1.1+ and PDCA-1+ cells. (D) NK1.1 versus CD49b profile of BM cells from the same mice as in A. (left) The gates indicate CD49b+NK1.1− (red) and NK1.1+ (blue) cells, respectively, and the absolute cell numbers are indicated. (right) A population overlay of the gated populations for CD11c versus B220 expression. The results shown are representative of two to three animals per genotype.
NKp46 is an NK cell–specific receptor expressed in human, bovine, and mouse NK cells encoded by the Ncr-1 gene (14, 16–18). Mice expressing GFP under Ncr-1 transcriptional control elements show strong GFP fluorescence in CD3−NK1.1+ cells (14, 18). Using Ncr-1gfp/+ mice (14), we found that two cell types were present in the CD3−CD19−CD11cloB220+ subset: the first consisted of pDCs that expressed the pDC antigen 1 (PDCA-1) marker, whereas the second included the putative IKDCs expressing NK1.1 (Fig. 1 C). Remarkably, the CD11cloB220+NK1.1+ subset highly expressed NKp46 (also confirmed by cell-surface staining; unpublished data), indicating that IKDC-like cells were more closely related to NK cells than previously anticipated. Moreover, NKp46 expression was completely eliminated when Ncr-1gfp/+ mice were made deficient in γc (Fig. 1 C). NKp46 expression was not detected in pDCs (Fig. 1 C) or conventional DCs (cDCs; not depicted) under any condition.
A recent report indicated that IKDCs in γc-deficient mice differed from IKDCs in C57BL/6 mice in that they were NK1.1− but expressed CD49b (19). We therefore studied the NK1.1−CD49b+ cells that were equally present in the BM of Rag2−/−, Rag2−/−IL-2Rβ−/−, and Rag2−/−γc −/y mice (Fig. 1 D). Compared with NK1.1+CD49+ cells (of which a fraction expressed the CD11cloB220+ phenotype), NK1.1−CD49b+ cells lacked CD11c and B220 expression (Fig. 1 C). Thus, NK1.1+ cells with the CD11cloB220+ phenotype (IKDC-like cells) require IL-15 for their development and express NKp46; these are characteristics of bone fide NK cells.
B220 expression on developing and mature NK cells
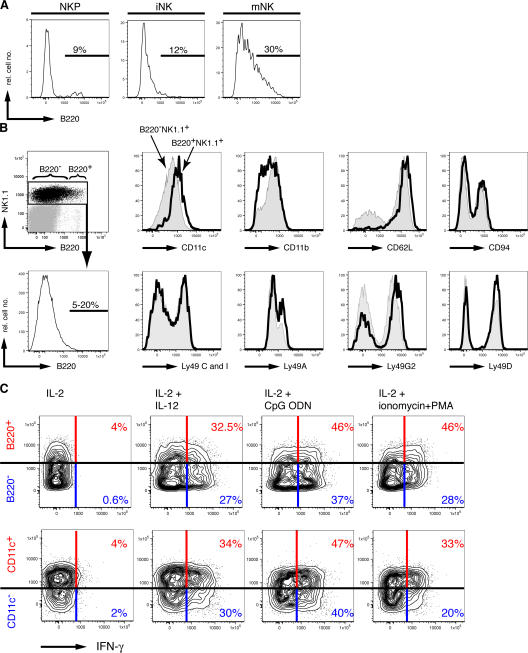
We studied further the expression of B220 by cells of the NK cell lineage. NK cell precursors were essentially B220−, whereas a minor population of immature NK cells were B220lo (Fig. 2 A). In contrast, ∼10-30% of mature NK1.1+CD49b+ NK cells in the BM and spleen clearly expressed B220 in a rather homogeneous fashion (Fig. 2, A and B). Because B220 expression on peripheral NK cells was not bimodal, we consider it arbitrary to designate B220− versus B220+ NK cell subsets, as others have done (5, 6). Examining the phenotypes of NK1.1+ cells bearing low versus high levels of B220 (Fig. 2 B), the frequency of NK cells expressing Ly49 receptors (A, C/I, G2, or D) or CD94 was not substantially different. B220+ NK cells were uniformly CD62Lhi and had slightly higher expression of CD11c (Fig. 2 B), although CD11c expression was not bimodal. In contrast, the overall density of CD11b was reduced on B220+ NK cells. No differences in the expression of NKG2D, CD49b, and CD44 were found between B220− and B220+ NK cells (unpublished data).
Figure 2.
Phenotype and function of B220− and B220+ NK cells. (A) B220 expression on BM NK cell precursors (CD122+NK1.1−CD49b−; left), immature NK cells (CD122+NK1.1+CD49b−; middle), and mature NK cells (CD122+NK1.1+CD49b+; right) from Rag2−/− mice. (B) Splenocytes from Rag2−/− mice analyzed for NK1.1 and B220 expression. (top left) The distribution of NK1.1+ cells expressing B220 and the gating for B220− and B220+ NK cells. (bottom left) Histogram shows the B220 expression on the same gated NK1.1+ cells. The range of percentages of B220+ cells among total NK1.1+ cells is given (n = 10). The remaining histograms show overlays of CD11c, CD11b, CD62L, and CD94 (top, from left to right) and Ly49C/I, Ly49A, Ly49G2, and Ly49D (bottom, from left to right) expressed by B220+NK1.1+ cells (continuous line) and B220−NK1.1+ cells (shaded) from Rag2−/− splenocytes. (C) Expression of IFN-γ in NK1.1+ cells in relation to B220 expression (top) and CD11c expression (bottom). NK cells above the horizontal bar are B220+ (top) or CD11c+ (bottom), and the red numbers indicate the percentage of IFN-γ+ cells among total B220+ or CD11c+ NK1.1+ cells, respectively. The cells below the horizontal bar are B220− (top) or CD11c− (bottom), and the blue numbers indicate the percentage of IFN-γ+ cells in total B220− or CD11c− NK1.1+ cells, respectively. The results shown are representative of two to three experiments. iNK, immature NK cell; mNK, mature NK cell; NKP, NK cell precursor.
Previous reports have demonstrated that B220+ NK cells have enhanced functional activities compared with B220− NK cells (5, 6). We found that both B220− and B220+ NK cells were similarly capable of IFN-γ production after stimulation in vitro with IL-2 and IL-12 or IL-2 and CpG oligodeoxynucleotide (ODN; Fig. 2 C). Moreover, IFN-γ production did not correlate with CD11c expression (Fig. 2 C). However, when stimulated with pharmacological agents (PMA plus ionomycin), we found that CD11c+ or B220+ NK cells produced somewhat more IFN-γ than CD11c− or B220− NK cells (Fig. 2 C). Collectively, these results fail to support the notion that B220hi cells represent a distinct phenotypic and functional NK1.1+ NK cell subset.
Activation-induced changes in B220, CD11c, and MHC-II expression in NK cells
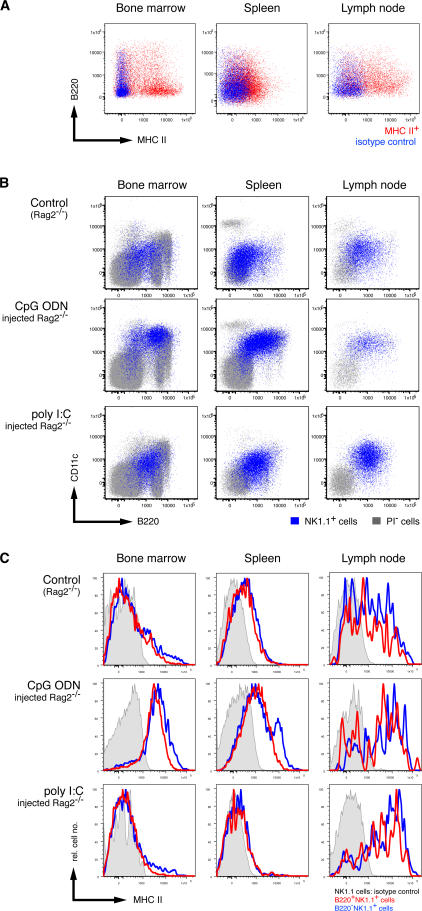
MHC-II expression has been proposed as a defining characteristic for CD11cloB220+ IKDC-like cells (5). Consistent with a previous report (5), few splenic NK cells (CD3−NK1.1+) from C57BL/6 mice expressed MHC-II molecules on their cell surface under steady-state conditions (Fig. 3 A). The highest proportion of MHC-II+ cells among CD3−NK1.1+ cells could be detected in the lymph nodes (10–50%), although a subset of MHC-II+ NK cells in the BM was detected (Fig. 3 A). Surprisingly, only a minority of MHC-II+ NK cells coexpressed B220, and in the tissues examined, MHC-II and B220 expression on NK cells was not correlated in an obvious fashion (Fig. 3 A). Similar results were obtained when examining coexpression of B220 and CD86 on NK cells (unpublished data). These results argue against the existence of a widely distributed IKDC-like NK1.1+ cell that coexpresses B220 and MHC-II (5, 6).
Figure 3.
Activation-induced expression of B220, CD11c, and MHC-II by NK1.1+ cells. (A) MHC-II versus B220 expression (red) of gated CD3−CD19−NK1.1+ BM (left), spleen (middle), and lymph node (right) cells from C57BL/6 mice. The blue staining represents the isotype control for the anti–MHC-II antibodies. (B) Population overlays of NK1.1+ cells (blue) on CD11c versus B220 dot plots of total cells (gray) from the BM (left), spleen (middle), and lymph nodes (right) harvested from Rag2−/− mice injected with PBS (top row), CpG ODN 1668 (middle row), or poly I:C (bottom row). (C) Expression of MHC-II molecules by B220+NK1.1+ (red) and B220−NK1.1+ (blue) cells from the same mice and organs analyzed in B. Shaded histograms represent isotype control staining by NK1.1+ cells from the respective mice and organs. The results shown are representative of three to five experiments.
The low frequency of MHC-II+ NK cells might reflect the steady-state, noninflammatory situation in normal, healthy mice. We next analyzed whether inflammatory stimuli would modify the NK cell phenotype in vivo. Mice were injected with either poly I:C (a Toll-like receptor [TLR] 3 ligand) or CpG ODN (a TLR9 ligand), and 3 d later, CD3−NK1.1+ cells were analyzed for CD11c, MHC-II, and B220. CD11c levels increased dramatically on NK cells from the BM, spleen, and lymph nodes after TLR activation (Fig. 3 B). Concurrently, most NK cells up-regulated B220 expression. As a result, ∼30–50% of NK cells from poly I:C– or CpG ODN–treated mice became IKDC-like (Fig. 3 B). The increase in CD11c+B220+ NK cells was likely caused by up-regulation of these markers rather than by preferential proliferation of CD11cloB220+NK1.1+ cells, because Ki67 expression (a marker of cells undergoing cell division) was predominantly found in NK1.1+B220neg cells (unpublished data). Interestingly, MHC-II expression was increased on all NK cells in mice treated with CpG ODN but only in lymph node NK cells in mice receiving poly I:C treatment (Fig. 3 C), suggesting a differential threshold for induction of MHC-II on NK cells after activation in this tissue. As for the case with resting NK cells (Fig. 3 A), no direct correlation could be made with inducible MHC-II expression in NK cells expressing different levels of B220 (not depicted). These results suggested that CD11c, B220, and MHC-II behave as classical activation markers on NK cells, although their inducible expression is differentially controlled. Finally, increased CD86 expression paralleled inducible MHC-II expression (unpublished data). Collectively, these results show that mouse NK cells appear to closely resemble human NK cells in their inducible expression of MHC-II and co-stimulatory molecules (20).
NK cell proliferation increases B220 expression in vitro and in vivo
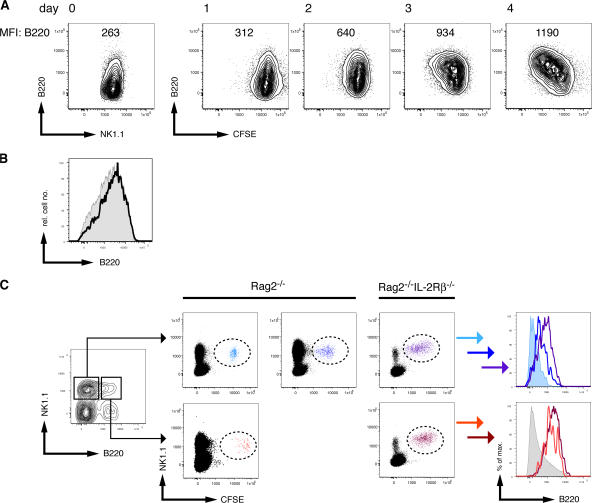
To understand the mechanisms and dynamics of inducible B220 expression by NK1.1+ cells, we labeled total splenocytes from Rag2−/− mice with CFSE and cultured them in IL-2. B220 expression on NK1.1+ cells was analyzed daily in relation to cytokine-induced proliferation. During the first 2 d, most NK cells divided asynchronously, as indicated by the even dilution of CFSE in the bulk NK cell population, which was accompanied by an increase in B220 expression (a twofold increase in mean fluorescence intensity). By days 3 and 4, a clear fraction of NK cells had further proliferated, and their B220 expression was approximately fivefold higher than that observed on NK cells at the start of the culture. Similar results were obtained in IL-2–supplemented cultures of lymph node– or thymus-derived NK cells (Fig. 4 A and not depicted). All IL-2–cultured NK cells demonstrated increased expression of NK1.1, CD11c, and B220 (unpublished data), suggesting that cytokine-induced proliferation can generate an IKDC-like phenotype from resting NK cells.
Figure 4.
Dynamic B220 expression by NK1.1+ cells. (A) Total CFSE-labeled splenocytes from Rag2−/− mice were cultured with IL-2. At the indicated times, cells were harvested and analyzed for B220 expression. The values of mean fluorescence intensity (MFI) indicate the expression level of B220 at the cell surface of the NK1.1+ cells analyzed. (B) Expression of B220 by sorted B220+ (shaded) or B220− (continuous line) NK1.1+CD49b+ cells from Rag2−/− mice after 7 d of culture in IL-2. The results shown are representative of three experiments. (C, left) The dot plot indicates the gates used to sort B220−NK1.1+ cells (left gate) and B220+NK1.1+ cells (right gate) from Rag2−/− mice. The sorted cells were adoptively transferred into NK cell–proficient (Rag2−/−) or NK cell–deficient (Rag2−/−IL-2Rβ−/−) hosts. Some of the NK cell–proficient hosts receiving B220−NK1.1+ cells were injected with poly I:C at the day of transfer. (middle) CFSE dilution of NK1.1+ cells 3 d after transfer of B220− (top) and B220+ (bottom) cells into the indicated hosts. (right) The histograms show the expression levels of B220 by the transferred cells 3 d after the transfer, as indicated. The colors in the histograms match the populations defined by the dashed ovals in the middle panels. The results shown are representative of two to five animals per group from two to three experiments.
To study whether B220+ NK cells can up-regulate this protein after activation, we sorted splenic B220+ and B220− NK cells (NK1.1+CD49b+) from Rag2−/− mice and cultured them with IL-2. At day 7, we found that all NK cells from the sorted B220− population now expressed B220 at the cell surface at levels similar to the sorted B220+ population (Fig. 4 B).
To further investigate the proliferation-induced B220 expression on NK cells, we purified splenic B220− and B220+NK1.1+ cells from Rag2−/− mice, labeled them with CFSE, and adoptively transferred them into NK cell–deficient Rag2−/−IL-2Rβ−/− recipient mice. The analysis of CD11c expression by the sorted B220− and B220+ NK cells confirmed that both populations expressed CD11c, with B220+ NK cells expressing slightly higher CD11c levels (Fig. S2, available at http://www.jem.org/cgi/content/full/jem.20071451/DC1). Both NK cell populations underwent “homeostatic” proliferation (21–23) based on their CFSE-dilution profiles. Although B220+ NK cells maintained B220 expression, the B220− NK cells became B220+ (Fig. 4 C). We next assessed whether B220 induction on B220− NK cells was proliferation dependent by transferring CFSE-labeled B220− and B220+ NK cells into NK cell–proficient (B−, T−, and NK+) Rag2−/− recipients. Most transferred NK cells did not divide in these hosts (Fig. 4 C), likely because of competition with endogenous NK cells for cellular resources such as IL-15 (22). Under these conditions, B220− NK cells remained B220−, whereas B220+ NK cells maintained B220 expression (Fig. 4 C). Lastly, we assessed the effects of TLR engagement on the fate of adoptively transferred B220− NK cells in Rag2−/− recipients. In contrast to untreated Rag2−/− mice, B220− NK cells proliferated in poly I:C–treated Rag2−/− recipients, with concomitant up-regulation of B220 expression (Fig. 4 C). Thus, B220 expression is up-regulated on proliferating NK cells in vivo. These results suggest that TLR ligands may modify NK cell activation states through stimulation of NK cell proliferation.
T cell stimulatory activity of B220+ and B220− NK cells
Peptide-pulsed CD11cloB220+NK1.1+ cells were reported to stimulate antigen-specific T cells (5), suggesting that IKDCs could function as APCs to stimulate adaptive immune responses. We next assessed whether T cell stimulatory activity was a unique property of B220+ NK cells. “Marilyn” TCR-transgenic cells specific for the male antigen Dby presented by I-Ab (24) were used as responder cells. Female Rag2−/− mice were injected with CpG ODN, and 3 d later, B220+ NK cells, B220− NK cells, pDCs, and cDCs were isolated. Cells were pulsed with the Dby peptide, and pulsed and nonpulsed cells were cultured with sorted, CFSE-labeled Marilyn T cells. After 5 d, T cell proliferation was assessed by CFSE dilution. Both B220+ and B220− NK cells were able to stimulate division of ∼30% of Marilyn CD4+ T cells (Fig. 5). Nevertheless, CpG-activated NK cells were clearly less efficient than either pDCs or cDCs in their T cell stimulatory capacity. This result contrasts with a previous study that demonstrated B220+ NK cells to be more efficient than pDCs in T cell stimulation (5). The difference may lie in the choice of a TCR-transgenic mouse model (OT-II vs. Marilyn) that may differ in activation threshold.
Figure 5.
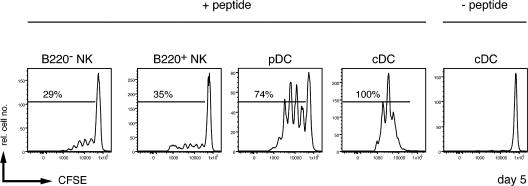
T cell stimulatory potential of NK1.1+ NK cells is independent of B220 expression. Sorted B220−NK1.1+ cells, B220+NK1.1+ cells, pDCs, and cDCs from CpG ODN 1668−treated mice (3 d before the sort) were incubated with the MHC-II–restricted H-Y peptide Dby and co-cultured with sorted, CFSE-labeled Vβ6+CD4+ T cells from Marilyn TCR transgenic mice. 3 d later, cells were harvested and analyzed for the CFSE dilution. The percentages of cells having undergone one or more cell divisions are indicated. Sorted B220−NK1.1+ cells, B220+NK1.1+ cells, pDCs, and cDCs that were not incubated with the Dby peptide failed to stimulate proliferation of Marilyn T cells (not depicted). The results shown are representative of two animals per group.
Conclusions
In this report, we provide evidence that NK1.1+ cells with the CD11cloB220+ phenotype (IKDC-like cells) are bona fide NK cells. All CD3−NK1.1+ cells share the same developmental requirements (γc, IL-2Rβ, and IL-15) and express the same NK cell–specific genes (NKp46). These features clearly distinguish NK cells from diverse DC subsets.
We also demonstrate inducible B220 and CD11c expression after NK cell activation in vitro and in vivo. The capacity of NK cells to express high levels of CD11c after activation in vivo might explain the increased frequency of intratumoral CD11c+NK1.1+ cells detected in patients treated with imatinib mesylate and IL-2, as reported by Taieb et al. (6). Similarly, their activated status would explain why B220+NK1.1+ cells isolated from these patients exhibit an increased functional competence in vitro and in vivo (6). Other markers suggested to be specific for CD11cloB220+NK1.1+ cells, such as MHC-II and CD86, were clearly not confined to these cells but were also expressed by B220lo/− NK cells in naive mice. Some of these activation-induced phenotypic changes (acquisition of B220) are closely linked to cellular proliferation. Up-regulation of B220 and CD11c has been previously detected on activated NK T cells and CD8+ T cells (10, 25). As such, these “atypical” markers may signal recent cytokine stimulation, thereby representing a conserved mechanism that operates in cytotoxic cells.
MATERIALS AND METHODS
Mouse strains.
Mice deficient in Rag2 (Rag2−/−) and its derivatives deficient in γc (Rag2−/−γc−/y), in the IL-2Rβ chain (Rag2−/−IL-2Rβ−/−; provided by S. Koyasu, Keio University, Tokyo, Japan), in IL-2 (Rag2−/−IL-2−/−), or in IL-15 (Rag2−/−IL-15−/−) on the C57BL/6 background have been previously described (15, 26). Mice carrying a GFP reporter in the NK cell–specific Ncr-1 locus (Ncr1gfp/+) have been previously described (14), and male Ncr1gfp/+ mice were crossed to female Rag2−/−γc−/− mice to create γc-deficient Ncr1gfp/+ male mice. C57BL/6 mice were obtained from Charles River Laboratories. Mice were maintained in specific pathogen-free conditions at the Institut Pasteur and were used at 4–10 wk of age. Mouse experiments were approved by an institutional committee at the Institut Pasteur and validated by the French Ministry of Agriculture.
Antibodies and reagents.
mAbs directly conjugated to FITC, PE, PE-Cy7, PerCP-Cy5.5, PE-Cy5.5, allophycocyanin, allophycocyanin-Cy7, or biotin specific for the following antigens (clone name in parenthesis) were used: B220 (RA3-6B2), CD3 (145-2C11), CD11b (M1/70), CD11c (N418 and HL3, respectively), CD19 (1D3), CD49b (DX5), CD62L (MEL14), CD86 (GL1), CD94 (18d3), CD122 (TM-β1), I-A/I-E (M5/1 14.15.2), Ly49A (JR9), Ly49G2 (4D11), Ly49C/I (5E6), NK1.1 (PK136), PDCA-1 (PDCA-1), and IFN-γ (XMG1.2). Antibodies were purchased from Becton Dickinson, eBioscience, Biolegend, and Miltenyi Biotec. The polyclonal goat anti–mouse anti-NKp46 serum was purchased from R&D Systems. FCS and PBS were purchased from Invitrogen.
Isolation of lymphoid cells and flow cytometric analysis.
Single-cell suspensions from the BM, spleen, lymph nodes, and thymus and FACS analysis were performed using a flow cytometer (FACSCanto or FACSCanto II; Becton Dickinson) with FACSDiva software (5.02; Becton Dickinson), as previously described (27). Datasets were analyzed using FlowJo software (Tree Star, Inc.). Cell sorting was performed on a cell sorter (FACSAria, BD Biosciences; access was provided by the Flow Cytometry Platform at the Institut Pasteur).
Adoptive transfer of sorted NK cell subpopulations.
Splenic cell suspensions from Rag2−/− mice were sorted as NK1.1+CD49b+B220+ or B220− cells and injected i.v. into alymphoid recipients. At the given times after transfer (as shown in the figures), splenic lymphocytes were prepared, and their phenotype was determined. Alternatively, cells were labeled with CFSE after sorting, injected i.v. into Rag2−/− or Rag2−/−IL-2Rβ−/− mice, and analyzed 3 d later.
In vitro cultures.
Splenic cell suspensions from Rag2−/− mice were labeled with 5 μm CFSE, and 1–2 × 105 cells per 200 μl were cultured in flat-bottom microtiter plates in 1,000 U/ml IL-2 (PeproTech). For co-cultures of Marilyn T cells with NK cells or IKDC-like cells, we sorted NK1.1+B220− and NK1.1+B220+ cells from female Rag2−/− mice and incubated half of the sorted cells with 100 nM Dby peptide. 2 × 104 of the sorted cells were cultured with CFSE-labeled Vβ6+CD4+ T cells from Marilyn Rag2−/− transgenic mice (28) at a 1:1 ratio. As controls, we co-cultured Marilyn T cells with sorted NK1.1+B220− and NK1.1+B220+ cells that had not been incubated with peptide, and with sorted pDCs and cDCs from the same female Rag2−/− mice. CpG ODN–activated cells were obtained from Rag2−/− mice that received 35 μg CpG ODN (i.p.) 3 d earlier.
In vivo activation.
Rag2−/− or C57BL/6 mice were injected i.p. with either CpG ODN 1668 (S-oligo; Sigma-Proligo) or poly I:C (Sigma-Aldrich). At the time points indicated in the figures, cells from the indicated organs were isolated and analyzed.
Online supplemental material.
Fig. S1 shows an overlay of NK1.1+CD49b+ NK cells on a CD11c versus B220 dot plot of viable BM cells derived from Rag2−/−, Rag2−/−IL-2−/−, and Rag2−/−IL-15−/− mice. Fig. S2 shows the expression of B220 and CD11c by sorted B220+ and B220−NK1.1+CD49b+ splenocytes from Rag2−/− mice. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20071451/DC1.
Supplemental Material
[Supplemental Material Index]
Acknowledgments
We thank Shigeo Koyasu for providing Rag2−/−IL-2Rβ−/− mice, and Veronique Braud, Béatrice Breart, Nicholas Huntington, and Agathe Burgess for stimulating discussions.
This work was supported by grants from the Institut Pasteur and the Institut National de la Santé et de la Recherche Médicale, and as an Equipe Labelisé of the Ligue National Contre le Cancer.
The authors have no conflicting financial interests.
References
- 1.Degli-Esposti, M.A., and M.J. Smyth. 2005. Close encounters of different kinds: dendritic cells and NK cells take centre stage. Nat. Rev. Immunol. 5:112–124. [DOI] [PubMed] [Google Scholar]
- 2.Lucas, M., W. Schachterle, K. Oberle, P. Aichele, and A. Diefenbach. 2007. Dendritic cells prime natural killer cells by trans-presenting interleukin 15. Immunity. 26:503–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferlazzo, G., M.L. Tsang, L. Moretta, G. Melioli, R.M. Steinman, and C. Munz. 2002. Human dendritic cells activate resting natural killer (NK) cells and are recognized via the NKp30 receptor by activated NK cells. J. Exp. Med. 195:343–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Piccioli, D., S. Sbrana, E. Melandri, and N.M. Valiante. 2002. Contact-dependent stimulation and inhibition of dendritic cells by natural killer cells. J. Exp. Med. 195:335–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan, C.W., E. Crafton, H.N. Fan, J. Flook, K. Yoshimura, M. Skarica, D. Brockstedt, T.W. Dubensky, M.F. Stins, L.L. Lanier, et al. 2006. Interferon-producing killer dendritic cells provide a link between innate and adaptive immunity. Nat. Med. 12:207–213. [DOI] [PubMed] [Google Scholar]
- 6.Taieb, J., N. Chaput, C. Menard, L. Apetoh, E. Ullrich, M. Bonmort, M. Pequignot, N. Casares, M. Terme, C. Flament, et al. 2006. A novel dendritic cell subset involved in tumor immunosurveillance. Nat. Med. 12:214–219. [DOI] [PubMed] [Google Scholar]
- 7.Laouar, Y., F.S. Sutterwala, L. Gorelik, and R.A. Flavell. 2005. Transforming growth factor-beta controls T helper type 1 cell development through regulation of natural killer cell interferon-gamma. Nat. Immunol. 6:600–607. [DOI] [PubMed] [Google Scholar]
- 8.Postigo, A.A., A.L. Corbi, F. Sanchez-Madrid, and M.O. de Landazuri. 1991. Regulated expression and function of CD11c/CD18 integrin on human B lymphocytes. Relation between attachment to fibrinogen and triggering of proliferation through CD11c/CD18. J. Exp. Med. 174:1313–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schleicher, U., A. Hesse, and C. Bogdan. 2005. Minute numbers of contaminant CD8+ T cells or CD11b+CD11c+ NK cells are the source of IFN-gamma in IL-12/IL-18-stimulated mouse macrophage populations. Blood. 105:1319–1328. [DOI] [PubMed] [Google Scholar]
- 10.Huleatt, J.W., and L. Lefrancois. 1995. Antigen-driven induction of CD11c on intestinal intraepithelial lymphocytes and CD8+ T cells in vivo. J. Immunol. 154:5684–5693. [PubMed] [Google Scholar]
- 11.Vremec, D., M. O'Keeffe, H. Hochrein, M. Fuchsberger, I. Caminschi, M. Lahoud, and K. Shortman. 2007. Production of interferons by dendritic cells, plasmacytoid cells, natural killer cells, and interferon-producing killer dendritic cells. Blood. 109:1165–1173. [DOI] [PubMed] [Google Scholar]
- 12.Welner, R.S., R. Pelayo, K.P. Garrett, X. Chen, S.S. Perry, X.H. Sun, B.L. Kee, and P.W. Kincade. 2007. Interferon-producing killer dendritic cells (IKDCs) arise via a unique differentiation pathway from primitive c-kitHiCD62L+ lymphoid progenitors. Blood. 109:4825–4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suzuki, H., T.M. Kundig, C. Furlonger, A. Wakeham, E. Timms, T. Matsuyama, R. Schmits, J.J. Simard, P.S. Ohashi, H. Griesser, et al. 1995. Deregulated T cell activation and autoimmunity in mice lacking interleukin-2 receptor beta. Science. 268:1472–1476. [DOI] [PubMed] [Google Scholar]
- 14.Gazit, R., R. Gruda, M. Elboim, T.I. Arnon, G. Katz, H. Achdout, J. Hanna, U. Qimron, G. Landau, E. Greenbaum, et al. 2006. Lethal influenza infection in the absence of the natural killer cell receptor gene Ncr1. Nat. Immunol. 7:517–523. [DOI] [PubMed] [Google Scholar]
- 15.Vosshenrich, C.A., T. Ranson, S.I. Samson, E. Corcuff, F. Colucci, E.E. Rosmaraki, and J.P. Di Santo. 2005. Roles for common cytokine receptor gamma-chain-dependent cytokines in the generation, differentiation, and maturation of NK cell precursors and peripheral NK cells in vivo. J. Immunol. 174:1213–1221. [DOI] [PubMed] [Google Scholar]
- 16.Sivori, S., M. Vitale, L. Morelli, L. Sanseverino, R. Augugliaro, C. Bottino, L. Moretta, and A. Moretta. 1997. p46, a novel natural killer cell–specific surface molecule that mediates cell activation. J. Exp. Med. 186:1129–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Storset, A.K., S. Kulberg, I. Berg, P. Boysen, J.C. Hope, and E. Dissen. 2004. NKp46 defines a subset of bovine leukocytes with natural killer cell characteristics. Eur. J. Immunol. 34:669–676. [DOI] [PubMed] [Google Scholar]
- 18.Walzer, T., M. Blery, J. Chaix, N. Fuseri, L. Chasson, S.H. Robbins, S. Jaeger, P. Andre, L. Gauthier, L. Daniel, et al. 2007. Identification, activation, and selective in vivo ablation of mouse NK cells via NKp46. Proc. Natl. Acad. Sci. USA. 104:3384–3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ullrich, E., M. Bonmort, G. Mignot, N. Chaput, J. Taieb, C. Menard, S. Viaud, T. Tursz, G. Kroemer, and L. Zitvogel. 2007. Therapy-induced tumor immunosurveillance involves IFN-producing killer dendritic cells. Cancer Res. 67:851–853. [DOI] [PubMed] [Google Scholar]
- 20.Hanna, J., T. Gonen-Gross, J. Fitchett, T. Rowe, M. Daniels, T.I. Arnon, R. Gazit, A. Joseph, K.W. Schjetne, A. Steinle, et al. 2004. Novel APC-like properties of human NK cells directly regulate T cell activation. J. Clin. Invest. 114:1612–1623. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Cooper, M.A., J.E. Bush, T.A. Fehniger, J.B. VanDeusen, R.E. Waite, Y. Liu, H.L. Aguila, and M.A. Caligiuri. 2002. In vivo evidence for a dependence on interleukin 15 for survival of natural killer cells. Blood. 100:3633–3638. [DOI] [PubMed] [Google Scholar]
- 22.Ranson, T., C.A. Vosshenrich, E. Corcuff, O. Richard, W. Muller, and J.P. Di Santo. 2003. IL-15 is an essential mediator of peripheral NK-cell homeostasis. Blood. 101:4887–4893. [DOI] [PubMed] [Google Scholar]
- 23.Prlic, M., B.R. Blazar, M.A. Farrar, and S.C. Jameson. 2003. In vivo survival and homeostatic proliferation of natural killer cells. J. Exp. Med. 197:967–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scott, D., C. Addey, P. Ellis, E. James, M.J. Mitchell, N. Saut, S. Jurcevic, and E. Simpson. 2000. Dendritic cells permit identification of genes encoding MHC class II-restricted epitopes of transplantation antigens. Immunity. 12:711–720. [DOI] [PubMed] [Google Scholar]
- 25.Koyasu, S. 1994. CD3+CD16+NK1.1+B220+ large granular lymphocytes arise from both α-βTCR+CD4−CD8− and γ-δTCR+CD4−CD8− cells. J. Exp. Med. 179:1957–1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuwajima, S., T. Sato, K. Ishida, H. Tada, H. Tezuka, and T. Ohteki. 2006. Interleukin 15-dependent crosstalk between conventional and plasmacytoid dendritic cells is essential for CpG-induced immune activation. Nat. Immunol. 7:740–746. [DOI] [PubMed] [Google Scholar]
- 27.Vosshenrich, C.A., M.E. Garcia-Ojeda, S.I. Samson-Villeger, V. Pasqualetto, L. Enault, O. Richard-Le Goff, E. Corcuff, D. Guy-Grand, B. Rocha, A. Cumano, et al. 2006. A thymic pathway of mouse natural killer cell development characterized by expression of GATA-3 and CD127. Nat. Immunol. 7:1217–1224. [DOI] [PubMed] [Google Scholar]
- 28.Lantz, O., I. Grandjean, P. Matzinger, and J.P. Di Santo. 2000. Gamma chain required for naive CD4+ T cell survival but not for antigen proliferation. Nat. Immunol. 1:54–58. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental Material Index]