Protein-DNA chimeras for single molecule mechanical folding studies with the optical tweezers (original) (raw)
. Author manuscript; available in PMC: 2010 May 4.
Published in final edited form as: Eur Biophys J. 2008 Jan 9;37(6):729–738. doi: 10.1007/s00249-007-0247-y
Abstract
Here we report on a method that extends the study of the mechanical behavior of single proteins to the low force regime of optical tweezers. This experimental approach relies on the use of DNA handles to specifically attach the protein to polystyrene beads and minimize the non-specific interactions between the tethering surfaces. The handles can be attached to any exposed pair of cysteine residues. Handles of different lengths were employed to mechanically manipulate both monomeric and polymeric proteins. The low spring constant of the optical tweezers enabled us to monitor directly refolding events and fluctuations between different molecular structures in quasi-equilibrium conditions. This approach, which has already yielded important results on the refolding process of the protein RNase H (Cecconi et al. in Science 309: 2057–2060–2005), appears robust and widely applicable to any protein engineered to contain a pair of reactive cysteine residues. It represents a new strategy to study protein folding at the single molecule level, and should be applicable to a range of problems requiring tethering of protein molecules.
Keywords: Laser tweezers, DNA handles, Protein-DNA chimeras, Single molecule mechanical manipulation, Protein folding
Introduction
Single molecule force spectroscopy has emerged as a new and powerful technique to study protein folding, allowing a host of new questions to be addressed. The method allows one to investigate areas previously inaccessible to study such as: (1) the response of individual molecules to a denaturant (force) along a well-defined reaction coordinate, defined by the molecular end-to-end distance, (2) rare unfolding and refolding events, (3) the magnitude and range of the forces responsible for maintaining the tertiary structure of molecules, (4) alternative regions of the energy landscape not accessible with thermal or chemical denaturation. To date, the primary tool for protein single molecule force spectroscopy has been the atomic force microscope (AFM). Recently, however, we reported on the mechanical un/refolding of E. coli ribonuclease HI (RNase H) using the optical tweezers (Cecconi et al. 2005) by employing the use of covalently attached double strand DNA “handles”. Here we present the development of such DNA/protein chimeras as a general method to investigate the mechanical stability of single proteins across any axis using the optical tweezers.
For mechanical folding studies using the AFM, the protein of interest is tethered between a cantilever and a substrate (Rounsevell et al. 2004). A crucial issue in these studies is to maintain a large separation between the relatively large tethering surfaces. This requirement is easy to fulfill when manipulating micrometer-long polymers, such as titin (Rief et al. 1997), but it becomes an experimental challenge when working with small globular proteins whose ends are only a few nanometers apart. One solution to this problem is to create polyproteins similar to titin, where the molecule consists of a linear array of globular domains. Such polyproteins are usually synthesized through recombinant DNA techniques—creating a gene that will express the entire protein polymer (Carrion-Vazquez et al. 1999; Best et al. 2001). The synthesis of such polyproteins can be very laborious and is often complicated by the bacterial recombination machinery, which tends to rearrange such repeated proteins (Graham and Maio 1992, and our unpublished data), and by solubility problems (Steward et al. 2002). In addition, the repeating units in these polyproteins are connected via their N- and C-termini, thus preventing the application of force on the protein along alternative axes. The ability to apply force across different domains of a molecule allows one to explore the anisotropic nature of the energy landscape of proteins (Brockwell et al. 2003; Carrion-Vazquez et al. 2003; Dietz et al. 2006a; Dietz and Rief 2006).
To date, AFM studies have focused on characterizing the mechanical unfolding of proteins in the high force regime. Under these circumstances, it is difficult to study the refolding process, due to the high spring constants of commercially available cantilevers (typically in the 10–100 pN/nm range) and the correspondingly high loading rates (Berkemeier et al. 2006; Lee et al. 2006; Walther et al. 2007). Moreover, these experiments are almost always carried out far from equilibrium, and as such, it is difficult to follow the folding–unfolding reaction reversibly. Many of these limitations can be overcome by using the force-measuring optical tweezers (Smith et al. 2003).
The low mechanical stiffness of the optical tweezer technique enables us to collect force–extension data at close to equilibrium conditions and directly monitor fluctuations between different conformations. Like the AFM, samples for the optical tweezer are also tethered between solid supports—in this case two polystyrene beads. Our approach relies on the use of molecular handles that function as spacers between the protein and the beads making it possible to manipulate single protein domains. The two molecular handles (each a ~500 bp dsDNA) can be attached via disulfides to any cysteine residues exposed on the surface of the protein, so that various pulling geometries can be used. In addition to our previously reported studies on the protein RNase H (Cecconi et al. 2005), we have successfully used this new method to manipulate variants of several globular proteins, as well as to pull on polyproteins synthesized from monomers post-translationally. For the proteins examined, we find that the DNA handles do not significantly alter the structure and folding/refolding properties of the proteins.
Materials and methods
Protein preparation
Cysteine-modified proteins were generated using the Quik-Change Site-Directed mutagenesis kit from Stratagene. All genes were cloned into high copy plasmids under a T7 promoter (pAED4 or pET27). The proteins were purified using their respective published protocols with the difference that 1 mM DTT was present during all purification steps (Dabora and Marqusee 1994; Llinas and Marqusee 1998; Robic et al. 2002).
Gel electrophoresis
Linear polyacrylamide SDS gels were prepared using a Bio-Rad apparatus. Gradient polyacrylamide SDS gels, 4–20%, were purchased from Bio-Rad.
The fluorescent dyes SYPRO Red (specific for proteins) and SYBR Green II (specific for RNA and DNA) were purchased from Molecular Probes. Fluorescently stained gels were scanned and visualized using a Typhoon 8600, from Molecular Dynamics. After imaging processing with Photoshop, the protein bands that stained with SYPRO Red appear red, while DNA bands dyed with SYBR Green II appear green. When both protein and DNA were present, the bands appear yellow.
Atomic force microscopy
AFM was performed in air with a Digital Instrument Nanoscope III, in tapping mode, using tips from Nanosensors (pointprobes, type NCH-100). Molecular constructs were diluted to a final concentration of 2 nM in deposition buffer (10 mM HEPES, pH 7.5, 10 mM NaCl, 2 mM MgCl2). Twenty microliters of this solution were deposited onto freshly cleaved mica (Asheville-Schoonmaker mica company #472X8/10XSF) and allowed to adsorb onto the surface for 1 min. The sample was then gently washed with doubly distilled water and dried with a stream of nitrogen.
DTDP activation of cysteine-modified proteins
Protein molecules in buffer A (0.1 M NaH2PO4/Na2HPO4, pH 7.0) were typically reduced with 10 mM DTT for ~1 h at RT, and then buffer exchanged into 0.1 M NaH2PO4/Na2HPO4, pH 5.5 using size-exclusion columns, either Sephadex G-25 (Pierce) or HiTrap desalting columns (Amersham Biosciences). The resulting protein was reacted with a concentrated stock of DTDP (4.5 or 10 mM in 0.1 M NaH2PO4/Na2HPO4, pH 5.5, 15% acetonitrile) such that DTDP was in 5- to 25-fold molar excess of the protein, and allowed to react for 2–24 h at RT. The excess of DTDP was then removed using size-exclusion columns equilibrated with buffer A.
Attachment of DNA handles to proteins
Typically, a 30–40 μM DNA handle solution in 15 mM NaH2PO4/Na2HPO4, pH 7.0 was reduced with an excess of DTT (40:1 molar ratio) at RT for 1 h, and then buffer exchanged into 0.1 M NaH2PO4/Na2HPO4, pH 8.0 using three Micro Bio-Spin 6 chromatography columns (Bio-Rad). The resulting DNA molecules were immediately reacted with a thiol-pyridine activated protein solution. After the first step of the sequential attachment of handles to proteins, the protein-handle complexes were isolated from the unreacted proteins by gel filtration, using a Tos-oHaas G2000SWXL 5-mm column (7.8 × 300 mm) on a Shimadzu LC10A series HPLC system.
Generation of DNA handles
The 558 bp DNA handles were generated in large quantities by PCR using Taq DNA polymerase and pGEMEX 1 plasmid DNA from Promega as template. Usually 400–500 μg of handles were generated at a time using 9 ml of PCR reaction. The two types of handles were generated using the primer 5′ thiol-GCT-ACC-GTA-ATT-GAG-ACC-AC together with either the primer 5′ biotin-CAA-AAA-ACC-CCT-CAA-GAC-CC or the primer 5′ digoxigenin-CAA-AAA-ACC-CCT-CAA-GAC-CC. The PCR products were purified using HiSpeed Plasmid Maxi Kit, from QIAGEN. Short handles (20 and 40 bp) were generated by annealing complementary oligonucleotides.
Attachment of protein-DNA chimeras to polystyrene beads
Protein-DNA constructs were first reacted with polystyrene beads coated with anti-digoxigenin antibodies (dig-beads) for 5–15 min at room temperature (RT). Dig-beads were generated by coupling anti-digoxigenin antibodies (Roche) to 3.18 μm proteinG-coated beads (Spherotech). After reacting with the protein-DNA construct, a dig-bead was placed in the optical trap and then brought in close proximity to a 2.10 μm streptavidin-coated bead (Spherotech) which was held in place at the end of a pipette, until a tether between the two beads was attained.
Circular dichroism experiments
CD data were taken using an Aviv 62 DS spectropolarimeter. Temperature melts were equilibrated for 3 min at every temperature. Blank sample values (buffer only) were subtracted from all data.
Results
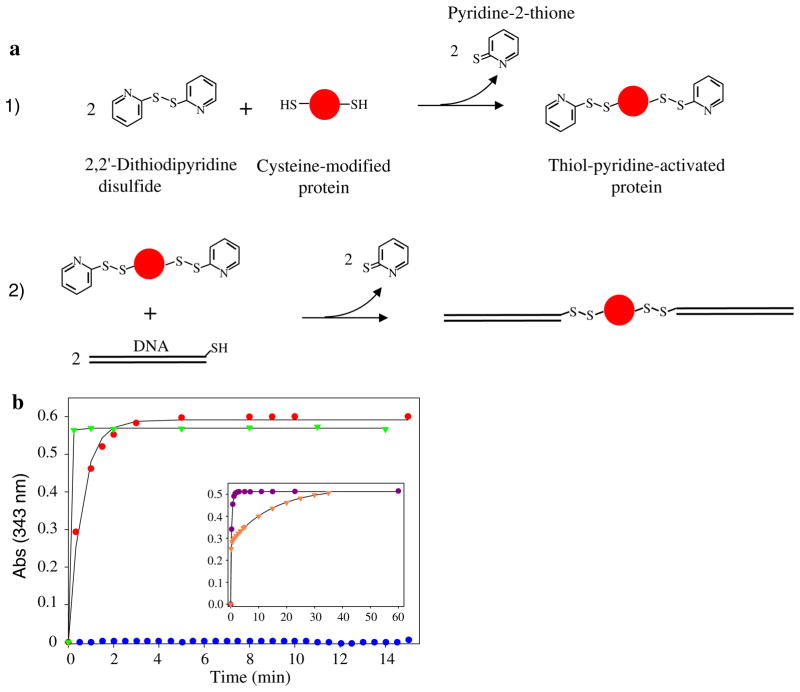
DNA handles, both short (20 or 40 bp) and long (558 bp), were attached to cysteine-modified proteins via disulfide bond formation. To speed up the oxidation reaction as well as to allow direct monitoring of the reaction, the thiol groups were activated with 2,2′-dithiodipyridine (DTDP) (Grassetti and Murray 1967; Pedersen and Jacobsen 1980; Riener et al. 2002) (Fig. 1a). In theory, DTDP can be used to activate the thiol groups of either DNA or proteins. In practice, however, we almost exclusively used the second strategy as it allowed the use of mass spectrometry to follow the reaction, and the activated proteins could then be used as the starting material to synthesize polyproteins (see below).
Fig. 1.
Attachment of DNA handles to protein molecules. a Schematic of the reactions used to: (1) activate protein’s cysteine residues with DTDP, and (2) attach DNA molecules to activated protein thiols. b Time course of the release of pyridine-2-thione during the activation of RNase H*Q4C/V155C (red dots), T4L*K16C/D159C (green triangles), and of the cysteine-free variant RNase H* (blue dots). Inset, activation of T4L*T21C/K124C with (purple dots) and without 3M GdmCl (orange triangles)
Attachment of DNA handles to a single protein domain
DTDP activation of cysteine-modified proteins
Two specific cysteine residues were engineered into otherwise cysteine-free proteins. The proteins were purified in the presence of 1 mM dithiothreitol (DTT) and then further reduced with a 10- to 25-fold molar excess of DTT (pH 5.5) for approximately 2–24 h at RT. DTT was then removed by size-exclusion chromatography and the protein immediately reacted with an excess of DTDP. The time course of the DTDP activation was monitored spectrophotometrically at 343 nm via the release of pyridine-2-thione (Fig. 1b). The reaction often reached completion in a few minutes, and the experimental data could be well-fit by a single exponential, indicating equal reactivity of the two cysteines. Occasionally, however, a double-exponential fit was required (Fig. 1b inset), suggesting differential accessibility of one of the two thiols. In this case, addition of the denaturant guanidinium chloride (GdmCl) abolished the differential reactivity. Mass spectrometry was used to confirm that the protein was activated with two thiol pyridines, and the activated protein could then be stably stored at 4°C for months, Table 1.
Table 1.
Mass spectrometry measurements of the molecular weight of different proteins before and after DTDP-activations
| Protein | Calculated M.W. (Da) | Measured M.W. (Da) |
|---|---|---|
| RNase H* Q4C/V155C | 17,479.80 | 17,480.39 ± 0.82 |
| DTDP-activated RNase H* Q4C/V155C | 17,698.10 | 17,698.41 ± 1.68 |
| T4L*K16C/D159C | 18,565.3 | 18,566.1 ± 1.05 |
| DTDP-activated T4L* K16C/D159C | 18,783.3 | 18,783.3 ± 1.07 |
| T4L* D61C,D159C | 18,577.4 | 18,578.8 ± 0.7 |
| DTDP-activated T4L* D61C/D159C | 18,795.4 | 18,796.7 ± 1.23 |
In cases where differential reactivity of the thiols is seen it should be possible to selective modify one of the two cysteines. While this is not important in our application because the two DNA handles are essentially equivalent, selective modification could be important in other applications such as the selective incorporation of dyes suitable for fluorescence energy transfer experiments.
Attachment of DNA molecules to single protein domains
In order to obtain samples with the ability to bind differentially to two functionally diverse polystyrene beads (specifically, a bead coated with anti-digoxigenin antibodies and another coated with streptavidin), two distinctly modified 558 bp dsDNA handles were attached to the protein molecules. One handle contained a 5′ thiol group and a 5′ biotin moiety; the other handle cotained a 5′ thiol group and a 5′ digoxigenin moiety. The attachment of the two handles was carried out as described below, either sequentially or through a one-step chemical reaction.
Sequential attachment
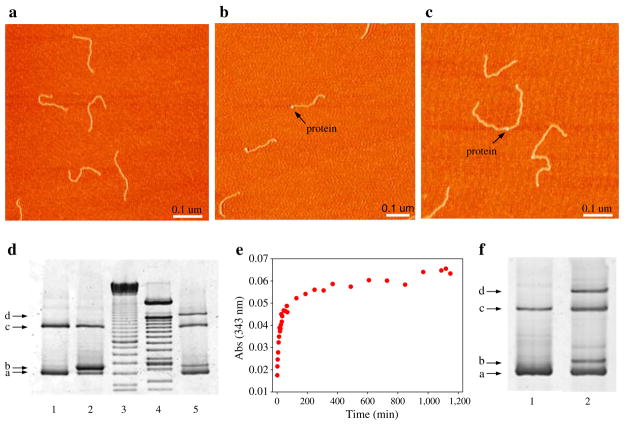
To ensure the maximum likelihood of two different handles attaching to each protein, the handles can be attached sequentially. First a five to tenfold excess of derivatized protein was added to one handle (2–4 h at RT); this reaction usually reached completion in 20–30 min as determined by the time course of the release of pyridine-2-thione (data not shown). Unreacted protein was then removed by size-exclusion chromatography, and a five to tenfold molar excess of the second handle was added (RT for 24–72 h) to complete the reaction. The attachment of the second handle to the protein is typically much slower than the attachment of the first one; this reaction usually takes days to reach completion. This slower kinetics is likely the result of the electrostatic repulsion between the DNA molecule already attached to the protein and the incoming one. The product of the reaction was analyzed both by SDS-PAGE and AFM (Fig. 2).
Fig. 2.
Attachment of 558 bp DNA handles to protein molecules. a–c Show atomic force microscopy images of DNA handles alone, RNase H bound to one handle and RNase H bound to two handles, respectively. d SDS-PAGE analysis of the sequential attachment of DNA handles to T4L*T21C/K124C. This 4% gel shows handles alone (lane 1, position a and c; the latter band is the result of the reaction between two DNA handles through their thiol groups), T4 lysozyme bound to one handle (lanes 2 and 5, positions b), and T4 lysozyme bound to two DNA handles (lane 5, position d). Lanes 3 and 4 show two different DNA ladders. As expected, the bands in position b and d are not present when the sample is treated with proteinase K (data not shown). When the sample is run on a native gel, the molecules electroeluted from band d all display the same structure of the two DNA-protein-DNA complexes shown in c (data not shown). The gel was stained with SYBR Green II and is shown in gray scale. e Time course of the release of pyridine-2-thione during the one-step attachment of both DNA handles to RNase H. The kinetics of the reaction is biphasic. f Four percent SDS-PAGE gel of handles alone (lane 1), and of the product of the onestep attachment (lane 2). The bands corresponding to RNase H bound to one or two handles are visible in position b and d, respectively
One-step attachment
Handles can also be attached to proteins through a simpler, more direct and less time-consuming, one-step reaction. In this case, a thiol-pyridine activated protein was reacted with a two to fourfold excess of a 1:1 mixture of the two handles at RT for 12–24 h. The final result is a mixture of DNA-modified proteins where only 50% of the molecules have the required configuration of one biotin- and one digoxigenin-labeled handle. As expected, the time course of this reaction is biphasic; a fast phase occurring during the first 20–30 min, probably corresponding to the attachment of the protein to one handle, is followed by a much slower phase that likely corresponds to the attachment of the second DNA molecule (Fig. 2e). This method provides a good yield of correctly labeled proteins and has been our method of choice to prepare samples for the optical tweezer. The one-step attachment protocol does not require any further purification, as only correctly labeled proteins will function in the laser tweezer experiments. Other species present in the sample, such as protein without/with one handle or protein flanked by identical handles, are unable to bind simultaneously to the two differently functionalized beads and therefore do not complicate our data. Sporadically, force–extension curves lacking any discontinuity were observed; these traces likely come from the stretching of DNA–DNA dimers. The same one-step attachment reaction was also successfully used to attach small handles (20–40 bp long) to single globular proteins (data not shown).
Synthesis of polyproteins and their attachment to DNA handles
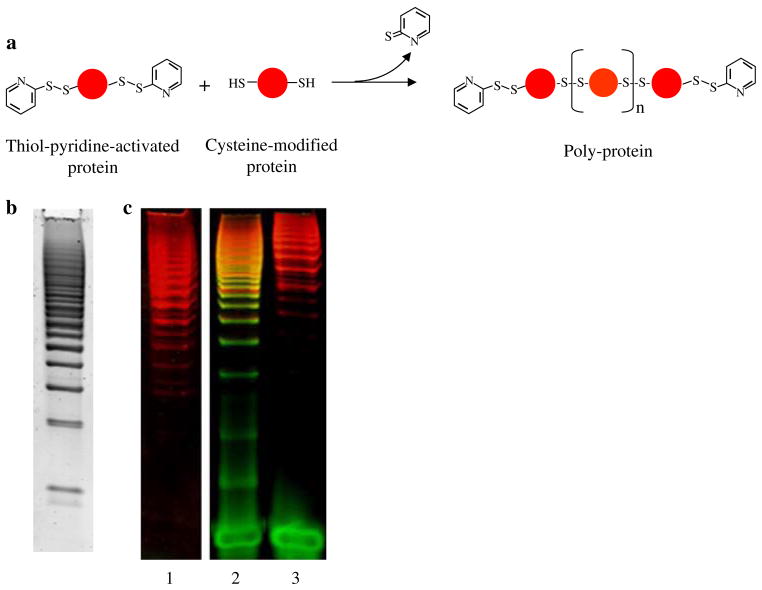
Polymers of proteins for use in single-molecule studies were constructed using the same DTDP-mediated reaction. After engineering two solvent-exposed cysteine residues, the polymerization was carried out in two steps (Fig. 3a). First, thiol-pyridine activated proteins were reacted with a twofold molar excess of unactivated molecules at RT overnight, and then more activated proteins (usually ¼ of the amount used in the first reaction) were added to the reaction to ensure that most of the polymer ends were capped with thiol-pyridine activated cysteines. The extent of the polymerization reaction can be assessed by SDS-PAGE (Fig. 3b, c); polymers as long as 20–30 monomeric units can be reproducibly synthesized with this method. After the reaction, HPLC size-exclusion chromatography was used to eliminate the excess of protein monomer, as well as to select the longest polymers (Fig. 3c, lane 1). This approach presents several advantages over the traditional polymerization methods (Dietz et al. 2006b): (1) it does not require the engineering of tandem repeats of a protein gene and the expression of polyproteins, (2) the connections between adjacent domains along the polymers are created through cysteine residues that can be placed anywhere on the surface of the protein, allowing the protein structure to be pulled along different axes of force application, and (3) polymers longer than ten monomers can easily be obtained.
Fig. 3.
Synthesis of protein polymers and their attachment to DNA handles. a Schematic of the reaction used to polymerize proteins. b A 4–20% gradient SDS-PAGE gel of the polymerization reaction of RNase H stained with SYPRO Red and shown in gray scale. c A 4–20% gradient SDS-PAGE gel of poly-RNase H alone (lane 1), poly-RNase H after reaction with 40 bp DNA handles bearing a 5′-thiol group (lane 2), and poly-RNase H after reaction with 40 bp DNA handle (lane 3). The gel was stained both with SYBR Green II and SYPRO Red
DNA handles were attached to these polyproteins through the same one-step attachment method described above. In this case, shorter polymers were generated during the attachment reaction, probably as a result of the DNA thiols attacking the polyprotein’s internal disulfide bonds (compare lanes 1 and 2 in Fig. 3c).
Characterization of protein-DNA chimeras
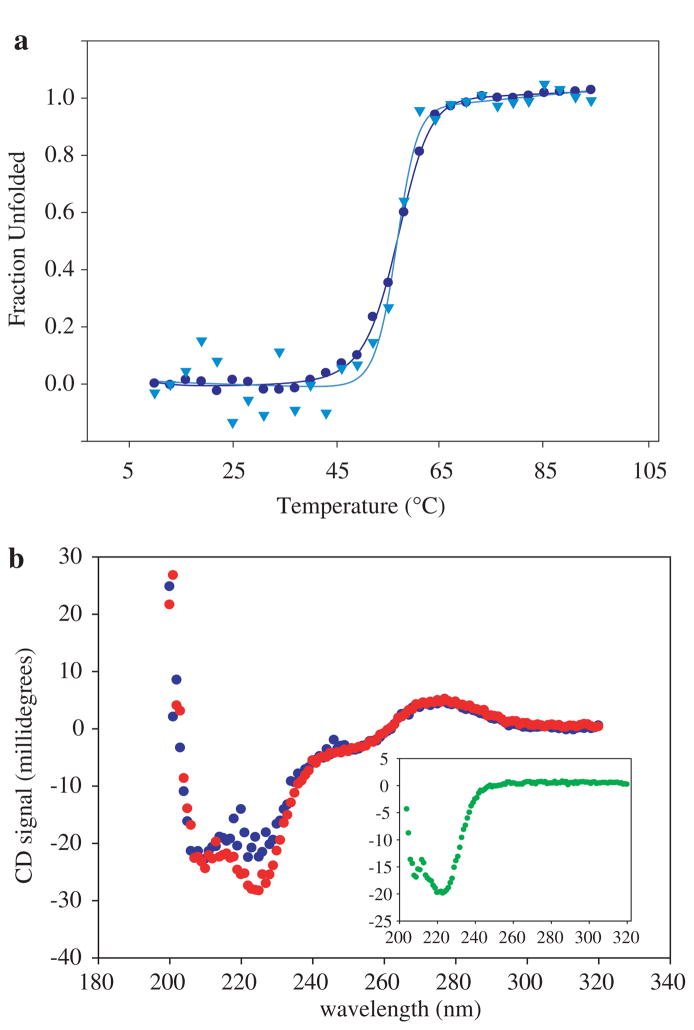
A series of traditional bulk experiments can be carried out to assess the effect of the attached handles on the protein’s structure, stability, and folding. Circular dichroism (CD) studies were carried out on protein bound to small handles (20 or 40 bp DNA) as it is technically difficult to obtain enough sample with long handles for CD studies. For the proteins studied to date, the CD spectra suggest that the proteins maintain their overall fold, and thermal denaturation studies indicate that the handles do not have a significant effect on the thermal stability (e.g., Fig. 4). In addition, RNase H modified with long DNA handles retains enzymatic activity (Cecconi et al. 2005). Of course, the lack of a notable perturbation will be protein specific and needs to be verified for any new sample.
Fig. 4.
Effect of DNA handles on protein stability. a Temperature melts of T4L*D61C/D159C alone (blue dots) and attached to two single stranded 20 base DNAs (light blue triangles). The data were normalized to show the fraction of protein unfolded at each temperature. In both cases, the two-state fits to the data yield a _T_m of 57°C. b CD spectra of T4L*D61C/D159C attached to two single stranded 20 base DNAs, before (red dots) and after (blue dots) thermal unfolding. The CD spectrum of the protein alone is shown in the inset (green dots)
Mechanical manipulation of DNA-modified proteins using the optical tweezers
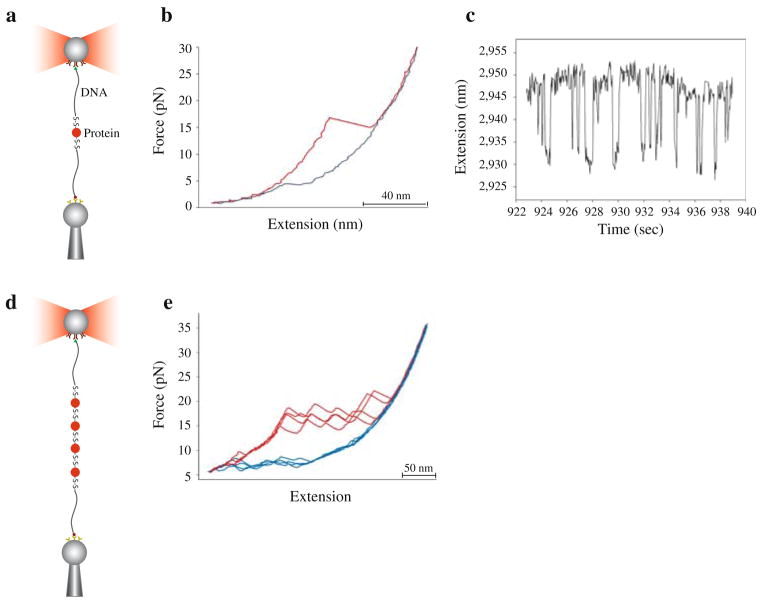
Globular proteins coupled to long DNA handles were manipulated using the optical tweezer set-up depicted in Fig. 5. The protein of interest is tethered between two polystyrene beads using DNA handles. One polystyrene bead is held in place at the end of a pipette by suction, while the other is held in the optical trap. During the experiment, the protein is stretched and relaxed by moving the pipette relative to the optical trap, and the forces applied on the molecule are determined by measuring the change in momentum of the light beams leaving the trap (Smith et al. 2003). The force–extension curves for DNA have been well characterized (Smith et al. 1992, 1996; Seol et al. 2007) therefore the contribution of the DNA handles to the overall signal can be easily distinguished. Handle fluctuations can sometimes give rise to small transitions in the recorded traces. These transitions however can be carefully characterized with control experiments, where handles alone are pulled, and easily distinguished in the overall signal. Figure 5b shows a typical force–extension curve obtained by pulling on a single globular protein. The unfolding of a protein is usually a very cooperative process, characterized by a sudden increment in the molecular end-to-end distance and a drop in force. Conversely, the refolding process gives rise to upward transitions in the relaxation traces that usually restore the original length of the molecule. If the protein unfolds and refolds reversibly, fluctuations between different molecular states can be monitored by keeping the tension on the molecule at a constant preset value using a force feedback mode of the optical tweezers instrument (Fig. 5c, Cecconi et al. 2005).
Fig. 5.
Experimental set-up and force–extension curves. a Schematic of a single globular protein attached to polystyrene beads through DNA handles. One handle is derivatized with a 5′ biotin moiety, which interacts with a streptavidin-coated bead held in place at the end of a pipette by suction. The other handle is derivatized with a 5′ digoxigenin moiety, which interacts with an antibody-coated bead held in a laser trap. b Force–extension cycles obtained by stretching and relaxing a single RNase H molecule (see also Cecconi et al. 2005). c Extension versus time trace of an RNase H molecule held at constant force, F = 6.0 pN. d Schematic of a protein polymer attached to polystyrene beads through DNA handles. e Force–extension cycles obtained by stretching and relaxing a tetramer of RNase H molecules multiple times using long handles. f Force–extension cycles obtained by stretching and relaxing a polymer of RNase H molecules (likely a heptamer) multiple times using short handles
Polyproteins were mechanically manipulated through either short (40 bp) or long (558 bp) handles (Fig. 5e). Short handles are easier to synthesize and attach to proteins; however, they are sensitive to force and melt at forces above 40 pN (S. Smith, personal communication). Long handles keep the attachment points further away from each other, thereby reducing the interactions between the tethering surfaces, and can be overstretched to check for multiple molecular attachments.
The stretching and relaxation of polyproteins yielded force–extension curves characterized by saw-tooth-like patterns (Fig. 5e), as expected from the unfolding (Bustamante et al. 1997; Forman and Clarke 2007) and refolding of tandem repeats of protein domains. The number of peaks observed with different molecular tethers varied because of the heterogeneity in the number of domains; however typically 4–10 peaks were observed. We have not thoroughly investigated the differences due to the handle length; qualitatively, however, the saw-tooth-like patterns obtained with short and long handles were similar. A tethered molecule could be pulled and relaxed several times before it broke or detached from the beads. Dissimilarities between successive unfolding–refolding cycles reflect the stochastic nature of the thermally facilitated denaturation and renaturation processes.
Discussion
Advances in single molecule manipulation techniques through the use of AFM and optical tweezers have made it possible to study the behavior of proteins under mechanical stress (Forman and Clarke 2007). Proteins are subject to mechanical stress in a variety of biological processes (Bustamante et al. 2004) and the response of proteins to mechanical stimuli is largely dependent on their particular mechanical properties and varies among different systems. Extracellular matrix proteins, for example, such as fibronectin and tenascin, as well as intracellular cytoskeletal proteins, such as spectrin and _α_-actinin, resist mechanical stress to help retain the shape of tissues and cells. Other proteins yield to mechanical perturbation to allow their translocation from one cellular compartment to another. For example, many mitochondrial proteins synthesized in the cytosol must be mechanically unfolded for translocation into mitochondria (Matouschek 2003). Mechanosensors respond to force with subtle structural deformations that mediate the transduction of mechanical stimuli into cellular processes (Janmey and Weitz 2004; Ingber 2006; Vogel 2006). Understanding the basic principles that govern the response of different proteins to force will provide insight about the mechanisms by which fundamental biological processes in the cell are influenced and regulated by direct mechanical interactions.
Over the last decade AFM has been the technique of choice for most of these studies. The results have yielded important insights into the mechanical properties of several molecules and have started to tease out the biophysical basis of a protein’s response to force. Thanks to these experiments, we now have a better understanding of: (1) the magnitude of the forces that hold together protein structures, (2) the relationship between topology and mechanical resistance, (3) the energy profiles of mechanical denaturation trajectories, and (4) the dependence of the tensile strength on the direction of the applied force vector. These studies, however, have been limited mostly to the characterization of mechanical behavior of protein molecules in the high force regime. Forces below 15–20 pN are very difficult to measure with probes whose spring constant is of the order of 10–100 pN/nm. Moreover, the extrapolation of the high-force unfolding data to lower forces might not always lead to a correct description of the mechanical behaviors of the molecules at the lower tensions. Indeed, some experiments suggest that lower loading rates may reveal very different mechanical properties (Williams et al. 2003).
Optical tweezers are much better suited to study the behavior of molecules in the low-force regime. The low spring constant of optical traps (~0.1 pN/nm) permits better control of the force applied to the molecules and the study of their mechanical properties under much lower loading rates. The engineered polyproteins used in AFM experiments (usually 8–10 monomers long), however are not suited for use in the optical tweezers. The polystyrene beads used in the tweezers are much larger (radius of curvature ~500 nm or bigger) than an typical AFM tip (radius of curvature ~10 nm) and therefore more space between the ends of the sample is required to avoid non-specific interactions between the tethering surfaces. Until the approach reported here (and used in Cecconi et al. 2005), the only proteins amenable to study with the optical tweezers have been micrometer-long molecules naturally organized in linear arrays of hundreds of globular domains, such as titin (Tskhovrebova et al. 1997; Kellermayer et al. 2000). These experiments have revealed important aspects of the overall mechanical properties of these molecules, but failed to provide information on specific domain behavior due to heterogeneity of these naturally occurring polymers.
The use of protein/DNA chimeras is widely applicable to any protein that can be engineered to have two exposed cysteine residues. The axis of force application on the molecule can be changed at will by altering the location of the protein’s cysteines. The response to force of different polyprotein domains along different pulling axes has already been investigated with AFM by several groups, using diverse experimental strategies (Brockwell et al. 2003; Carrion-Vazquez et al. 2003; Dietz et al. 2006a; Dietz and Rief 2006). Particularly noteworthy to this regard is the cysteine-based methodology developed by Rief group to polymerize GFP and study the response to force of this protein along five different pulling axes (Dietz et al. 2006a, b). The method described here is unique in its ability to study the anisotropy of the energy landscape of individual protein monomers, while avoiding the protein-specific limitations of previously described alternative tethering methods (Brockwell et al. 2003; Carrion-Vazquez et al. 2003).
To successfully generate DNA-protein constructs for use in laser tweezer experiments large concentrations (μM) of both protein and DNA are required in the coupling reaction. This entails the expression and purification of large amounts of protein and the generation of substantial quantities of DNA, with the resulting necessity of purchasing large amounts of expensive material, such as derivatized primers. Once generated, however, these protein-DNA complexes are stable for months at 4°, and the amount of sample generated is sufficient to perform hundreds of experiments.
Recently Garcia-Manyes et al. (2007) used a very complimentary approach to study the behavior of individual protein molecules in the high force regime of the AFM. In this AFM study, single protein monomers were flanked by small peptide handles, consisting of 12 amino acid residues at the C-terminus and four amino acid residues at the N-terminus. Inspite of the very short handles, and the fact that the tethering of the protein relies on the non-specific adsorption of the molecule on the AFM probe, the authors were able to successfully capture unfolding and refolding events of individual protein monomers and characterize their kinetics.
The specific effect of the DNA handles on the energy landscape of a tethered protein has not been investigated yet. In the case of a tethered RNA hairpin, however, variations in the length of the handles from 1 to 10 kbp resulted in changes of less than 1 kT in the energy landscape (Manosa et al. 2007). Based on these data, we expect the effects on the protein landscape to be fairly small.
In sum, the optical tweezers’ low spring constant permits the manipulation of individual molecules near equilibrium and the direct monitoring of fluctuations between different molecular structures, making it possible to follow in real-time the interconversion of molecules between alternative structures and to characterize the kinetics and thermodynamics of these processes. The technique we describe here—a robust and facile method to create chimeric bio-molecules and the simple means of monitoring those coupling reactions—should be broadly applicable to a range of problems requiring the tethering of molecules to other molecules or surfaces, and to the site-specific tagging of macromolecules.
Acknowledgments
We thank members of the Marqusee and Bustamante’s labs.
Abbreviations
AFM
Atomic force microscope
RNase H
E. coli ribonuclease HI
DTDP
2,2′-Dithiodipyridine
DTT
Dithiothreitol
RT
Room temperature
GdmCl
Guanidinium chloride
SDS-PAGE
Sodium dodecyl sulphate-polyacrylamide gel electrophoresis
HPLC
High performance liquid chromatography
CD
Circular dichroism
Contributor Information
Susan Marqusee, Email: marqusee@berkeley.edu.
Carlos Bustamante, Email: carlos@alice.berkeley.edu.
References
- Berkemeier F, Schlierf M, Rief M. Mechanically controlled preparation of protein intermediates in single molecule experiments. Phys Status Solidi a-Appl Mater Sci. 2006;203:3492–3495. [Google Scholar]
- Best RB, Li B, Steward A, Daggett V, Clarke J. Can non-mechanical proteins withstand force? Stretching barnase by atomic force microscopy and molecular dynamics simulation. Biophys J. 2001;81:2344–2356. doi: 10.1016/S0006-3495(01)75881-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockwell DJ, Paci E, Zinober RC, Beddard GS, Olmsted PD, Smith DA, Perham RN, Radford SE. Pulling geometry defines the mechanical resistance of a beta-sheet protein. Nat Struct Biol. 2003;10:731–737. doi: 10.1038/nsb968. [DOI] [PubMed] [Google Scholar]
- Bustamante C, Chemla YR, Forde NR, Izhaky D. Mechanical processes in biochemistry. Annu Rev Biochem. 2004;73:705–748. doi: 10.1146/annurev.biochem.72.121801.161542. [DOI] [PubMed] [Google Scholar]
- Bustamante C, Rivetti C, Keller DJ. Scanning force microscopy under aqueous solutions. Curr Opin Struct Biol. 1997;7:709–716. doi: 10.1016/s0959-440x(97)80082-6. [DOI] [PubMed] [Google Scholar]
- Carrion-Vazquez M, Li H, Lu H, Marszalek PE, Oberhauser AF, Fernandez JM. The mechanical stability of ubiquitin is linkage dependent. Nat Struct Biol. 2003;10:738–743. doi: 10.1038/nsb965. [DOI] [PubMed] [Google Scholar]
- Carrion-Vazquez M, Oberhauser AF, Fowler SB, Marszalek PE, Broedel SE, Clarke J, Fernandez JM. Mechanical and chemical unfolding of a single protein: a comparison. Proc Natl Acad Sci USA. 1999;96:3694–3699. doi: 10.1073/pnas.96.7.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecconi C, Shank EA, Bustamante C, Marqusee S. Direct observation of the three-state folding of a single protein molecule. Science. 2005;309:2057–2060. doi: 10.1126/science.1116702. [DOI] [PubMed] [Google Scholar]
- Dabora JM, Marqusee S. Equilibrium unfolding of Escherichia coli ribonuclease H: characterization of a partially folded state. Protein Sci. 1994;3:1401–1408. doi: 10.1002/pro.5560030906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz H, Rief M. Protein structure by mechanical triangulation. Proc Natl Acad Sci USA. 2006;103:1244–1247. doi: 10.1073/pnas.0509217103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz H, Berkemeier F, Bertz M, Rief M. Anisotropic deformation response of single protein molecules. Proc Natl Acad Sci USA. 2006a;103:12724–12728. doi: 10.1073/pnas.0602995103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz H, Bertz M, Schlierf M, Berkemeier F, Bornschlogl T, Junker JP, Rief M. Cysteine engineering of polyproteins for single-molecule force spectroscopy. Nat Protoc. 2006b;1:80–84. doi: 10.1038/nprot.2006.12. [DOI] [PubMed] [Google Scholar]
- Forman JR, Clarke J. Mechanical unfolding of proteins: insights into biology, structure and folding. Curr Opin Struct Biol. 2007;17:58–66. doi: 10.1016/j.sbi.2007.01.006. [DOI] [PubMed] [Google Scholar]
- Garcia-Manyes S, Brujic J, Badilla CL, Fernandez JM. Force-clamp spectroscopy of single-protein monomers reveals the individual unfolding and folding pathways of I27 and ubiquitin. Biophys J. 2007;93:2436–2446. doi: 10.1529/biophysj.107.104422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham GJ, Maio JJ. A rapid and reliable method to create tandem arrays of short DNA sequences. Biotechniques. 1992;13:780–789. [PubMed] [Google Scholar]
- Grassetti DR, Murray JF., Jr Determination of sulfhydryl groups with 2,2′- or 4,4′-dithiodipyridine. Arch Biochem Biophys. 1967;119:41–49. doi: 10.1016/0003-9861(67)90426-2. [DOI] [PubMed] [Google Scholar]
- Ingber DE. Cellular mechanotransduction: putting all the pieces together again. Faseb J. 2006;20:811–827. doi: 10.1096/fj.05-5424rev. [DOI] [PubMed] [Google Scholar]
- Janmey PA, Weitz DA. Dealing with mechanics: mechanisms of force transduction in cells. Trends Biochem Sci. 2004;29:364–370. doi: 10.1016/j.tibs.2004.05.003. [DOI] [PubMed] [Google Scholar]
- Kellermayer MS, Smith S, Bustamante C, Granzier HL. Mechanical manipulation of single titin molecules with laser tweezers. Adv Exp Med Biol. 2000;481:111–126. doi: 10.1007/978-1-4615-4267-4_7. discussion 127–118. [DOI] [PubMed] [Google Scholar]
- Lee G, Abdi K, Jiang Y, Michaely P, Bennett V, Marszalek PE. Nanospring behaviour of ankyrin repeats. Nature. 2006;440:246–249. doi: 10.1038/nature04437. [DOI] [PubMed] [Google Scholar]
- Llinas M, Marqusee S. Subdomain interactions as a determinant in the folding and stability of T4 lysozyme. Protein Sci. 1998;7:96–104. doi: 10.1002/pro.5560070110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manosas M, Wen JD, Li PTX, Smith SB, Bustamante C, Tinoco I, Ritort F. Force unfolding kinetics of RNA using optical tweezers. II. Modeling experiments. Biophys J. 2007;92:3010–3021. doi: 10.1529/biophysj.106.094243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matouschek A. Protein unfolding—an important process in vivo? Curr Opin Struct Biol. 2003;13:98–109. doi: 10.1016/s0959-440x(03)00010-1. [DOI] [PubMed] [Google Scholar]
- Pedersen AO, Jacobsen J. Reactivity of the thiol group in human and bovine albumin at pH 3–9, as measured by exchange with 2,2′-dithiodipyridine. Eur J Biochem. 1980;106:291–295. doi: 10.1111/j.1432-1033.1980.tb06022.x. [DOI] [PubMed] [Google Scholar]
- Rief M, Gautel M, Oesterhelt F, Fernandez JM, Gaub HE. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science. 1997;276:1109–1112. doi: 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]
- Riener CK, Kada G, Gruber HJ. Quick measurement of protein sulfhydryls with Ellman’s reagent and with 4,4′-dithiodipyridine. Anal Bioanal Chem. 2002;373:266–276. doi: 10.1007/s00216-002-1347-2. [DOI] [PubMed] [Google Scholar]
- Robic S, Berger JM, Marqusee S. Contributions of folding cores to the thermostabilities of two ribonucleases H. Protein Sci. 2002;11:381–389. doi: 10.1110/ps.38602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rounsevell R, Forman JR, Clarke J. Atomic force microscopy: mechanical unfolding of proteins. Methods. 2004;34:100–111. doi: 10.1016/j.ymeth.2004.03.007. [DOI] [PubMed] [Google Scholar]
- Seol Y, Li J, Nelson PC, Perkins TT, Betterton MD. Elasticity of short DNA molecules: theory and experiment for contour lengths of 0.6–7 μm. Biophys J. 2007;93:4360–4373. doi: 10.1529/biophysj.107.112995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SB, Cui Y, Bustamante C. Overstretching B-DNA: the elastic response of individual double-stranded and single-stranded DNA molecules. Science. 1996;271:795–799. doi: 10.1126/science.271.5250.795. [DOI] [PubMed] [Google Scholar]
- Smith SB, Cui Y, Bustamante C. Optical-trap force transducer that operates by direct measurement of light momentum. Methods Enzymol. 2003;361:134–162. doi: 10.1016/s0076-6879(03)61009-8. [DOI] [PubMed] [Google Scholar]
- Smith SB, Finzi L, Bustamante C. Direct mechanical measurements of the elasticity of single DNA molecules by using magnetic beads. Science. 1992;258:1122–1126. doi: 10.1126/science.1439819. [DOI] [PubMed] [Google Scholar]
- Steward A, Toca-Herrera JL, Clarke J. Versatile cloning system for construction of multimeric proteins for use in atomic force microscopy. Protein Sci. 2002;11(9):2179–2183. doi: 10.1110/ps.0212702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tskhovrebova L, Trinick J, Sleep JA, Simmons RM. Elasticity and unfolding of single molecules of the giant muscle protein titin. Nature. 1997;387:308–312. doi: 10.1038/387308a0. [DOI] [PubMed] [Google Scholar]
- Vogel V. Mechanotransduction involving multimodular proteins: converting force into biochemical signals. Annu Rev Bio-phys Biomol Struct. 2006;35:459–488. doi: 10.1146/annurev.biophys.35.040405.102013. [DOI] [PubMed] [Google Scholar]
- Walther KA, Grater F, Dougan L, Badilla CL, Berne BJ, Fernandez JM. Signatures of hydrophobic collapse in extended proteins captured with force spectroscopy. Proc Natl Acad Sci USA. 2007;104:7916–7921. doi: 10.1073/pnas.0702179104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams PM, Fowler SB, Best RB, Toca-Herrera JL, Scott KA, Steward A, Clarke J. Hidden complexity in the mechanical properties of titin. Nature. 2003;422:446–449. doi: 10.1038/nature01517. [DOI] [PubMed] [Google Scholar]