Yeast lifespan extension by depletion of 60S ribosomal subunits is mediated by Gcn4 (original) (raw)
. Author manuscript; available in PMC: 2009 Sep 23.
SUMMARY
In nearly every organism studied, reduced caloric intake extends lifespan. In yeast, lifespan extension from dietary restriction is thought to be mediated by the highly-conserved, nutrient-responsive TOR, PKA and Sch9 kinases. These kinases coordinately regulate various cellular processes including stress responses, protein turnover, cell growth and ribosome biogenesis. Here we show that a specific reduction of 60S ribosomal subunit levels slows aging in yeast. Deletion of genes encoding 60S subunit proteins or processing factors or treatment with a small molecule which inhibits 60S subunit biogenesis are each sufficient to significantly increase replicative lifespan. One mechanism by which reduced 60S subunit levels leads to lifespan extension is through induction of Gcn4, a nutrient-responsive transcription factor. Genetic epistasis analyses suggest that dietary restriction, reduced 60S subunit abundance, and Gcn4 activation extend yeast lifespan by similar mechanisms.
INTRODUCTION
Invertebrate model organisms serve as valuable tools for aging research, largely because of their short lifespans and ease of genetic manipulation. The most commonly used model organisms include fruit flies, nematodes, and yeast. In the budding yeast Saccharomyces cerevisiae, two models of cellular aging have been developed: replicative and chronological (Kaeberlein, 2006). Replicative lifespan (RLS) is defined as the number of mitotic cycles completed by a mother cell before senescence and may model the aging of mitotically active cells in multicellular organisms (Mortimer and Johnston, 1959). Chronological lifespan refers to the length of time that a non-dividing yeast cell retains viability during stationary phase and may model the aging of post-mitotic cells (Fabrizio and Longo, 2003).
Dietary restriction (DR) increases lifespan and delays the onset of age-associated diseases in a variety of evolutionarily divergent organisms, including mammals (Masoro, 2005; Weindruch et al., 1988). In yeast, DR by a reduction in either glucose or amino acid concentration in the media results in lifespan extension in both chronological and replicative aging models (Fabrizio and Longo, 2003; Jiang et al., 2000; Lin et al., 2000; Powers et al., 2006). Genetic epistasis experiments support the hypothesis that lifespan extension from DR in yeast is mediated by the coordinated activity of three nutrient responsive kinases: TOR (target of rapamycin), Sch9, and protein kinase A (PKA) (Fabrizio et al., 2004; Fabrizio et al., 2001;Kaeberlein et al., 2005b; Lin et al., 2000; Powers et al., 2006). Decreased activity of these kinases extends both replicative and chronological lifespan (Fabrizio et al., 2004; Fabrizio et al., 2001; Kaeberlein et al., 2005b; Lin et al., 2000; Powers et al., 2006). In response to nutrients, these three kinases regulate multiple important cellular processes, including ribosome biogenesis (Carey, 2003; Jorgensen et al., 2004; Martin et al., 2004; Powers and Walter, 1999), stress response (Beck and Hall, 1999), autophagy (Noda and Ohsumi, 1998), and mitochondrial retrograde metabolism (Dilova et al., 2002). Lifespan extension by deletion of _TOR1_or SCH9 is not additive with DR and is independent of Sir2 protein deacetylase (Kaeberlein et al., 2005b).
TOR was first identified as a regulator of yeast lifespan through a random screen of 564 yeast strains, each lacking a single nonessential gene (Kaeberlein et al., 2005b). Along with TOR1 and SCH9, deletion of several other genes in the TOR signaling pathway were identified to be long-lived, including RPL31A and RPL6B (Kaeberlein et al., 2005b). The observation that rpl31aΔ or rpl6bΔ cells are long-lived suggests that one mechanism by which DR might slow replicative aging is by decreasing ribosomal protein (RP) production through down-regulation of TOR and Sch9 activity. Consistently, several reports have since linked a reduction in RP levels to increased lifespan in both yeast and C. elegans. Deletion of RPL10, RPS6B, or RPS18A, B also increases yeast RLS (Chiocchetti et al., 2007). In C. elegans, individual knockdown of nine different 40S subunit RP genes and seven different 60S subunit RP genes has been reported to extend lifespan (Chen et al., 2007; Curran and Ruvkun, 2007; Hansen et al., 2007). In addition, inhibition of several translation initiation factors including the eIF2β/γ, eIF3A/B/F, eIF4A/E/G, or eIF5A homologues has been shown to increase C. elegans lifespan (Chen et al., 2007; Curran and Ruvkun, 2007; Hamilton et al., 2005; Hansen et al., 2007; Henderson et al., 2006; Pan et al., 2007; Syntichaki et al., 2007). Inhibition of the ribosomal protein S6 kinase has also been linked to lifespan extension in both worms (Chen et al., 2007; Hansen et al., 2007; Pan et al., 2007) and flies (Kapahi et al., 2004), and recent data suggests that Sch9 is the functional ortholog of S6 kinase in yeast (Powers, 2007; Urban et al., 2007).
To better understand the relationship between ribosomal proteins and aging, we measured the RLS for each of 107 RP gene deletion strains present in the yeast deletion collection and determined that multiple different 60S RP gene deletions significantly extend RLS. Consistently, we found that decreasing the abundance of 60S ribosomal subunits by deletion of 60S-specific ribosomal processing factors or by treatment with the small molecule diazaborine also leads to increased RLS. Epistasis analyses allowed us to conclude that depletion of 60S subunits extends lifespan by a mechanism similar to DR and independent of Sir2. Finally, we show that the transcription factor Gcn4 is required for RLS extension in mutants with depleted 60S subunits, demonstrating a novel longevity-promoting function of Gcn4.
RESULTS
Longevity analysis of RP gene deletion strains
The yeast ribosome consists of two subunits, the 40S (small) and the 60S (large), which together contain four discrete rRNA species and 78 ribosomal proteins (RPs). In yeast, about 85% of RP genes are present in duplicate copies, allowing for the viable deletion of either paralog, but generally not both paralogs simultaneously. Of the 137 genes encoding RPs, 107 are present as quality control verified (see Experimental Procedures) deletions in the MATα ORF deletion collection (Winzeler et al., 1999). We measured the RLS for each of these 107 RP single-gene deletion strains, corresponding to 46 RP paralog pairs (e.g., RPL31A and RPL31B) and 15 unpaired RP genes, 3 of which are non-essential single-copy genes and 12 of which have paralogs not represented in the deletion set.
Of the 107 RP gene deletion strains analyzed, 28 were found to be significantly long-lived relative to experiment-matched wild type cells (p < 0.05) in the MATα deletion set. To verify these results, we then measured the RLS of the 28 corresponding deletion strains derived from the MATa ORF deletion collection (Figures 1A–1C, Table S1). In total, 14 RP gene deletion strains were verified to be significantly long-lived in both the MATα and MATa ORF deletion collections. Based on prior studies (Kaeberlein et al., 2005b), we would expect a similar analysis of 107 randomly chosen deletion strains to yield 2–3 (2.5) with significantly increased RLS; therefore the percent of verified long-lived RP gene deletions is enriched approximately 5-fold relative to the entire set of non-essential ORFs. Strikingly, all 14 of the verified long-lived RP gene deletion strains lack protein components of the 60S subunit, whereas none of the deletion strains lacking RP genes encoding 40S subunit proteins were verified to be long-lived. Some long-lived rplΔ mutations, such as rpl22aΔ, and rpp2bΔ, resulted in lifespan extension exceeding 50% (Figures 1A–1C), with longevity comparable to the longest-lived single-gene deletion mutants reported in yeast (Kaeberlein et al., 2005a; Kaeberlein et al., 2005b). Not all rplΔ strains were long-lived, however, and some were short-lived (Table S1), for example rpl20aΔ (Figure 2B). These findings indicate that ribosomal proteins of the large subunit (RPLs) are important determinants of longevity in yeast.
Figure 1. Genome-wide screen of RP gene deletion strains verifies 14 significantly long-lived strains, each lacking an RPL gene.
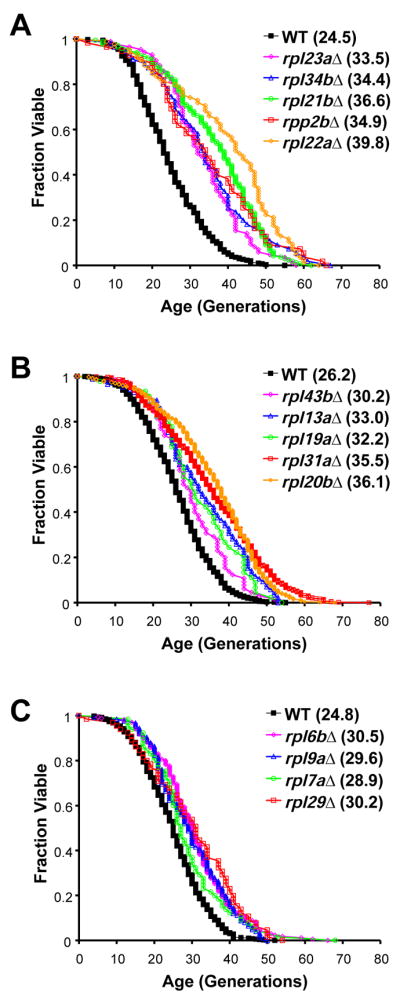
A–C. Survival curves for RP deletion strains that are significantly (p < 0.05) long-lived in both the MATα and MATa ORF deletion collections. Data from MATa and MATα deletion strains are pooled and experiment-matched wild-type cells are shown. Mean lifespans are shown in parentheses. (See also Table S1.)
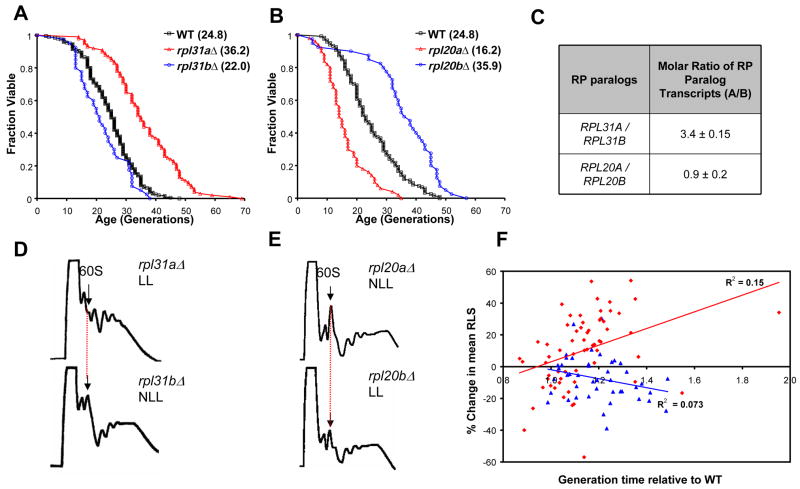
Figure 2. Abundance of 60S ribosomal subunits correlates with RLS.
A–B. Survival curves for RP paralog gene deletions and experiment-matched wild-type cells. Mean lifespans are shown in parentheses. C. Molar ratios of RP paralog transcripts (A/B). (See also Table S2). D–E. Polysome profiles of rplΔ paralogs. Long-lived deletion strains rpl31aΔ or rpl20bΔ show a reduced level of 60S ribosomal subunits relative to its paralog deletion strain (not long-lived). F. Generation time of RPL (red diamonds) and RPS (blue triangles) gene deletion strains relative to wild-type plotted versus the percent change in mean RLS relative to experiment-matched wild-type cells. Linear regressions for RPL (red) and RPS (blue) gene deletions are shown separately.
Differential longevity of RPL paralog deletion strains
Although multiple RPL gene deletions were long-lived, many were not. To characterize this apparent specificity, we further examined paralog pairs for which deletion of one gene significantly increased RLS while deletion of the other did not (Table S1). For example, rpl31aΔ and rpl20bΔ increased RLS, but their corresponding paralog gene deletions did not (Figures 2A–2B). The protein products encoded by the majority of RPL paralog pairs (26 of 33) are greater than 98% identical, and in the case of Rpl31a and Rpl31b, only a single conservative amino acid change differentiates the paralogs. Thus, it seems unlikely that one of the two paralogous RP proteins has evolved a specialized longevity-modulating function in the cases where rplΔ paralog pairs have divergent RLS phenotypes. Furthermore, we found no correlation between RLS and any other functional role reported for ribosomal proteins(Komili et al., 2007).
One possible explanation for divergent RLS phenotypes among paralog pair deletion strains is that one paralog is transcribed at a higher level than the other and, thus, accounts for a disproportionate amount of the total protein produced from both paralogous genes. To test this possibility, we examined expression of two RPL paralog pairs by quantitative RT-PCR analysis of RNA isolated from 3 independent cultures of the wild-type strain, using specific primers (Figure 2C, Table S2). In the case of RPL31A, its mRNA transcript is more abundant than that of RPL31B, consistent with the longevity of rpl31aΔ cells compared to rpl31bΔ cells. This correlation does not extend, however, to the RPL20A/B paralogs, which had nearly equimolar steady-state mRNA levels. Therefore, transcriptional bias among paralog pairs cannot account for divergent RLS phenotypes in every case.
In addition to transcriptional control, yeast cells can use a variety of mechanisms to regulate the level of RPs, including mRNA splicing, translation initiation, and turnover of excess protein (Tsay et al., 1988; Warner et al., 1985). To more directly assay whether deletion of one paralog more robustly affects the total amount of protein produced, we analyzed overall polysome profiles; cells limited for a particular RP should display a reduced abundance of the corresponding subunit. Polysome profiles were generated for several rplΔ paralog pairs using high-salt conditions (to disrupt non-translating 80S monosomes) (Figures 2D–2E, S1, Supplemental Information). In all examples studied, 60S subunit levels and overall polysome profile were more profoundly decreased in the rplΔ paralog with significantly increased RLS.
Depressed polysome profiles (Figures 2D–2E) indicate that translation is reduced in these long-lived mutants. In yeast, a sufficient reduction in translation will slow growth rate. In order to determine whether growth rate among RP deletion strains is a predictor of longevity, we measured the doubling time for each of the 107 rpΔ strains (Table S3) and compared growth rate to the percent change in mean RLS relative to wild-type (Figure 2F). The set of RPL gene deletion strains differed markedly from the set of RPS gene deletions strains with respect to the relationship between growth rate and RLS. For the set of rplΔ strains, growth rate inversely correlated with RLS, while the opposite trend was observed for rpsΔ strains. The growth rate analysis of rpΔ strains is complicated, however, by the strong selection for suppressors of slow growth among RP gene deletion strains in the ORF deletion set. We have observed three different cases where growth rate suppressors are present as spontaneously arising mutations in rplΔ strains from the deletion collection (see Supplemental Information for details), and we suspect that the inverse correlation between growth rate and RLS among rplΔ strains would be more highly significant if it were possible to prevent spontaneous mutations suppressing growth rate defects. Regardless, these data suggest that differential lifespan potential among RPL gene deletion strains is related to the abundance of functional 60S subunits as indicated by polysome profile and, perhaps, overall translation rate.
Loss of non-essential 60S processing factors increases lifespan
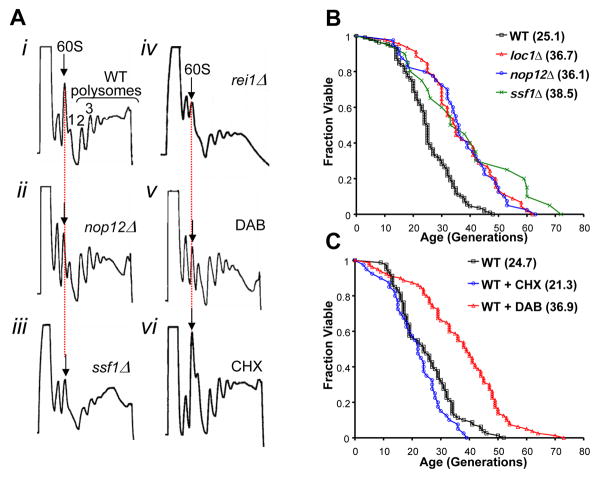
If RLS extension in long-lived rplΔ strains is a result of reduced 60S subunit levels, then mutations in non-ribosomal proteins important for 60S maturation might also extend lifespan. To test this hypothesis, three single-gene deletion strains: nop12Δ, loc1Δ, and ssf1Δ, each of which lacks a factor specifically involved in a different stage of pre-60S subunit maturation, were characterized. Deletion of LOC1 has been previously shown to cause decreased abundance of 60S subunits (Harnpicharnchai et al., 2001), and a similar depletion of 60S subunits was observed in polysome profiles of ssf1Δ or nop12Δ cells relative to wild-type (Figure 3A). Consistent with our prediction, each of these deletion mutants had a RLS significantly longer than wild-type cells (Figure 3B).
Figure 3. Interventions which decrease 60S ribosomal subunits extend RLS.
A. Relative to (i) wild-type, polysome profiles for (ii) nop12Δ, (iii) ssf1Δ, (iv) rei1Δ, and (v) diazaborine-treated (15 μg/ml) cells show significant reduction of 60S subunit levels while (vi) cycloheximide-treated (25 ng/ml) cells do not. B. Deletion of 60S-specific processing factor genes NOP12, SSF1, or LOC1 increases lifespan relative to experiment-matched wild-type cells. C. Treatment of wild-type cells with 15 μg/ml diazaborine extends lifespan relative to experiment-matched wild-type cells while treatment with 25 ng/ml cycloheximide does not. Mean lifespans are shown in parentheses.
In our previously reported screen of 564 random deletion mutants, deletion of either REI1 or YBR266C extended RLS (Kaeberlein et al., 2005b). These two ORFs are encoded on opposite strands and overlap. It has since been determined that the slow-growth phenotype of both of these mutants is due to loss of Rei1 function and unrelated to Ybr266c (Figure S3), which is designated dubious and is unlikely to encode a functional protein. Rei1 has recently been implicated in late-stage pre-60S processing (Hung and Johnson, 2006; Lebreton et al., 2006). Like nop12Δ, ssf1Δ, and loc1Δ cells, rei1Δ cells have reduced 60S subunits (Figure 3A). Thus, we conclude that non-ribosomal mutations which impair 60S maturation can increase RLS in a manner similar to deletion of large subunit RP genes.
Pharmacological inhibition of 60S maturation increases lifespan
We next determined whether RLS extension could also be achieved by a pharmacological intervention that depletes 60S subunit levels. Diazaborine is a synthetic antibiotic effective against Gram-negative bacteria (Baldock et al., 1998) that has been shown to reduce levels of 60S ribosomal subunits in yeast by a mechanism that likely involves pre-rRNA processing (Pertschy et al., 2004). Consistent with the above results, sub-lethal concentrations of diazaborine (15 μg/ml) reduced 60S subunit abundance (Figure 3A) and significantly increased RLS (Figure 3C).
In parallel, we determined the effect of adding sub-lethal concentrations of the general translation inhibitor cycloheximide (10–100 ng/ml) to the media. The lowest concentration of cycloheximide tested resulted in only a modest reduction in growth rate, whereas the highest concentrations substantially slowed growth. In contrast to diazaborine, cycloheximide neither increased RLS (Figures 3C, S4) nor led to reduced levels of 60S subunits (Figure 3A). Thus, pharmacological depletion of 60S subunits with diazaborine, but not general inhibition of translation with cycloheximide, is sufficient to increase yeast RLS.
60S subunit deficiency increases lifespan independently of Sir2
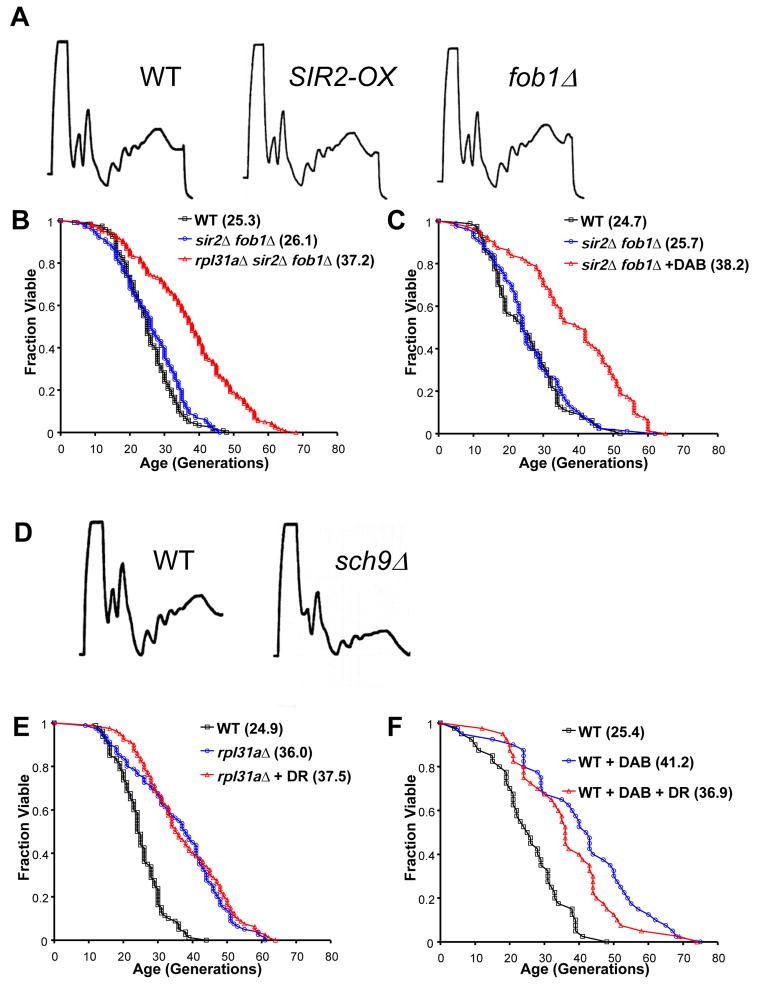
Enhanced Sir2 activity increases yeast RLS, an effect thought to be mediated by repression of extra-chromosomal rDNA circle (ERC) formation (Kaeberlein et al., 1999). Similarly, deletion of FOB1, encoding the rDNA replication fork barrier protein, extends yeast RLS by limiting accumulation of ERCs (Defossez et al., 1999). Because Sir2 and Fob1 regulate rDNA recombination, they might also influence 60S subunit levels by modulating the rate of rDNA transcription. Contrary to this idea, however, neither over-expression of Sir2 nor deletion of FOB1 had a detectable effect on 60S subunit levels or on overall polysome profile relative to wild-type cells (Figure 4A).
Figure 4. Depletion of 60S subunits extends RLS by a mechanism independent of Sir2 and similar to DR.
A. _SIR2_-overexpression and fob1Δ cells show polysome profiles similar to that of wild-type. B–C. Deletion of RPL31A or diazaborine treatment (15 μg/ml) increases the RLS of sir2Δ fob1Δ cells. D. A genetic model of DR (sch9Δ) results in cells with reduced levels of both 40S and 60S ribosomal subunits and polysomes relative to wild-type. E–F. DR does not further extend the RLS of rpl31aΔ cells or cells treated with diazaborine (15 μg/ml). Mean lifespans are shown in parentheses.
As long as ERC levels are kept low by deletion of FOB1, RLS extension by DR (via growth on reduced glucose media or genetic models of DR) is independent of Sir2 (Kaeberlein et al., 2004; Kaeberlein et al., 2006). Similarly, deletion of RPL31A or treatment with diazaborine significantly increased the RLS of sir2Δ fob1Δ cells (Figures 4B–4C). These data indicate that, similar to tor1Δ or sch9Δ (Kaeberlein et al., 2005b), depletion of 60S subunits extends lifespan independently of Sir2.
60S subunit deficiency increases lifespan by a mechanism similar to DR
During DR, the activity of TOR and Sch9 are reduced, resulting in decreased RP transcription (Jorgensen et al., 2004). Therefore, it is possible that TOR and Sch9 mediate lifespan extension in response to DR by depleting 60S subunits. Inhibition of TOR by treatment with rapamycin results in a moderately depressed polysome profile (Powers and Walter, 1999), although unlike diazaborine treatment, inhibition of TOR does not specifically affect 60S subunit abundance. Strains lacking SCH9 display a polysome profile in which both free 40S and 60S subunit levels as well as polysomes are reduced (Figure 4D). The polysome profiles of these genetic models do not show a specific reduction in the abundance of 60S subunits, possibly indicating that the simultaneous decrease of both 40S and 60S subunits is compatible with long lifespan and that the decrease in 60S abundance is dominant. Another possibility is that the young cells used for polysome analysis (log-phase cultures) cannot accurately model aging cells, in which important changes in polysome profile may occur over time.
DR does not further increase the RLS of long-lived tor1Δ or sch9Δ cells (Kaeberlein et al., 2005b), consistent with these genes acting in a genetic pathway with DR. DR also failed to significantly increase the long RLS of rpl31aΔ cells (Figure 4E) or of cells grown on media containing diazaborine (Figure 4F). Together these data support a model in which the depletion of 60S subunits promotes longevity by a mechanism independent of Sir2 and similar to tor1Δ, sch9Δ, and DR.
Gcn4 is required for full lifespan extension by depletion of 60S subunits
Gcn4 is a nutritionally-regulated transcriptional activator important for activating transcription of amino acid biosynthetic genes in response to amino acid starvation (reviewed in (Hinnebusch, 2005) as well as regulating diverse cellular processes including purine biosynthesis, autophagy, biosynthesis of organelles, ER stress response, and induction of mitochondrial transport carrier proteins (Jia et al., 2000; Natarajan et al., 2001; Patil et al., 2004). Gcn4 protein levels are primarily determined by translation and protein degradation rather than by transcription. Translation of GCN4 mRNA is regulated by four small upstream open reading frames (uORFs1-4) in the 5′ leader region of the GCN4 mRNA, and both amino acid starvation (reviewed in (Hinnebusch, 2005) and glucose limitation (similar to DR) (Yang et al., 2000) are known to induce Gcn4 activity in a Gcn2-dependent manner. RPL mutations have also been shown to induce expression of Gcn4 reporters (Foiani et al., 1991; Martin-Marcos et al., 2007), as has inhibition of TOR signaling (Cherkasova and Hinnebusch, 2003; Kubota et al., 2003; Valenzuela et al., 2001).
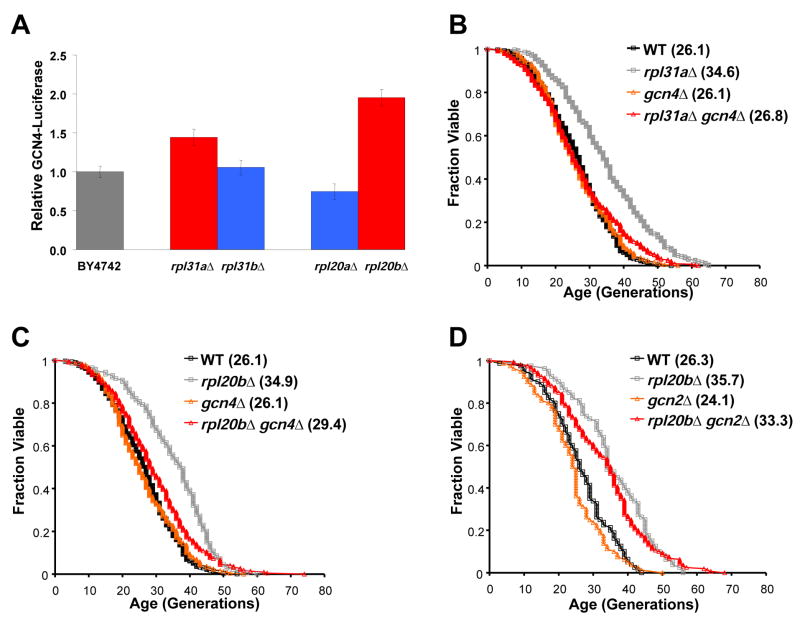
We speculate that in cells limited for 60S subunits, ternary complexes containing initiation factors and a 40S subunit will more frequently scan through the inhibitory uORFs present in the GCN4 5′ leader region before binding a 60S subunit and translating the GCN4 ORF. The resulting increased expression of Gcn4 protein may be related to the increased RLS of these cells. Consistent with this hypothesis, two different long-lived strains, rpl20bΔ and rpl31aΔ, displayed elevated expression of Gcn4-luciferase relative to wild-type in a dual luciferase reporter assay (Figure 5A), while the deletion strains corresponding to their paralogs, which are not long-lived, did not induce Gcn4-luciferase (Figure 5A).
Figure 5. Cells lacking RPL genes require GCN4, but not GCN2, for increased longevity.
A. Gcn4-luciferease levels for rpl31aΔ, rpl31bΔ, rpl20aΔ, and rpl20bΔ relative to wild-type cells show that translation of _GCN4_-luciferase RNA correlates with long lifespan. Red bars represent long-lived strains and blue bars represent strains that are not long-lived. B–C. Long-lived strains rpl20bΔ and rpl31aΔ require _GCN_4 for full lifespan extension. D. GCN2 is not required for lifespan extension by deletion of RPL20B. Mean lifespans are shown in parentheses.
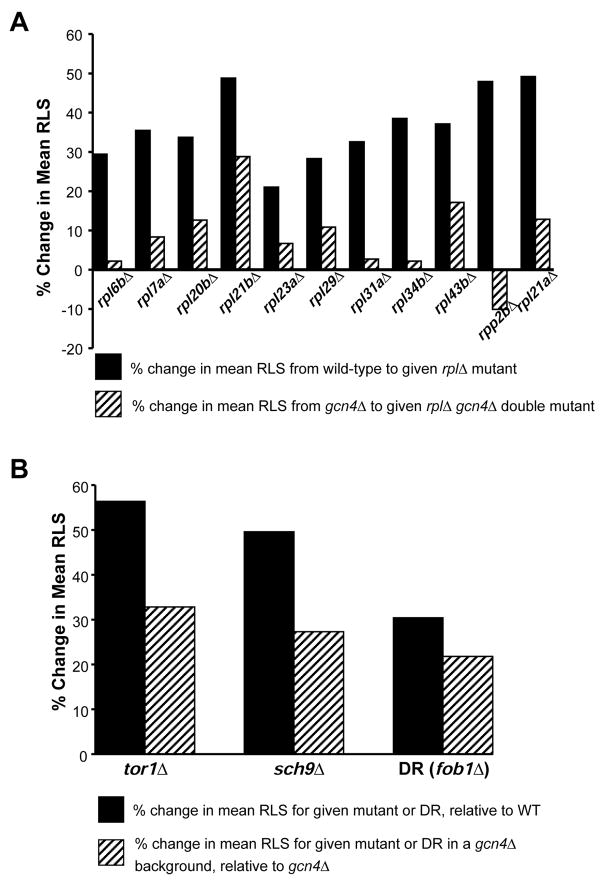
We next tested whether the long lifespan of strains lacking RPL genes is dependent of the presence of Gcn4. In each of the 11 cases examined, the percent increase in mean RLS observed by deletion of an RPL gene was diminished when GCN4 was simultaneously deleted (p < 0.001) (Figures 5B–5C, 6A, S5). In addition, the lifespan extension observed by deletion of RPL20B was independent of the eIF2α kinase Gcn2 (Figure 5D), consistent with a model in which translation of Gcn4 occurs more frequently due to a lack of 60S subunits available to initiate translation at upstream inhibitory uORFs. Together, these data support a model in which cells deficient for 60S subunits induce expression of Gcn4 to achieve maximum lifespan extension, and for the first time identify Gcn4 as a potential longevity factor.
Figure 6. GCN4 is required for full lifespan extension by depletion of 60S subunits or by DR.
A. Mean lifespan extension by deletion of any of 11 different RPL genes is largely dependent on GCN4 (p = <0.001). Solid bars represent the percent change in mean RLS for each rplΔ strain, relative to experiment-matched wild-type cells; hashed bars represent the percent change in mean RLS for each corresponding rplΔ gcn4Δ double mutant, relative to experiment-matched gcn4Δ cells. B. Full lifespan extension by tor1Δ (p = .03), sch9Δ (p = .21), or DR (p = .44) is dependent on GCN4. Solid bars represent the percent change in mean RLS from tor1Δ, sch9Δ or DR, relative to experiment-matched wild-type cells; hashed bars represent the percent change in mean RLS from tor1Δ, sch9Δ or DR in a gcn4Δ background, relative to experiment-matched gcn4Δ cells.
Full lifespan extension by DR is dependent on Gcn4
Since DR or TOR inhibition is known to reduce RP levels and increase Gcn4 translation, we considered the possibility that DR might promote longevity in part via induction of Gcn4. If so, then deletion of Gcn4 should attenuate the RLS extension afforded by DR or TOR inhibition. Consistent with this hypothesis, the lifespan extension from TOR1 deletion is significantly reduced in gcn4Δ cells, relative to wild type cells (p = 0.028) (Figure 6B, S6). A similar trend was observed in sch9Δ cells or in response to DR by growth on 0.05% glucose media; however, statistical significance was not attained in triplicate replicates. Thus, we conclude that Gcn4 is required for full lifespan extension in response to depletion of 60S subunits or reduced TOR signaling, and may also play a role in the response to DR (Figure 7).
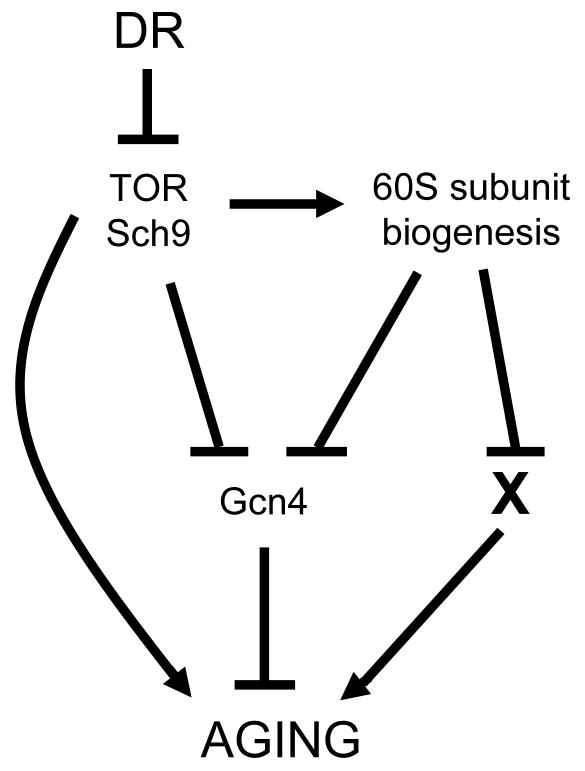
Figure 7. A genetic model for lifespan extension by DR.
Lifespan extension by depletion of 60S subunits, tor1Δ, sch9Δ, or DR, is mediated in part by Gcn4; however, a portion of the lifespan extension in each case can occur via at least one Gcn4-independent mechanism.
DISCUSSION
The 60S ribosomal subunit modulates longevity in yeast
Accumulating evidence suggests that regulation of mRNA translation is an evolutionarily conserved mechanism for modulating longevity (Kaeberlein and Kennedy, 2007). From a comprehensive analysis of 107 different RP gene deletions in yeast, we have determined that at least 14 different RPL gene deletions confer long RLS. In addition, deletion of any one of four different 60S-specific processing factors or treatment of cells with the 60S inhibitor diazaborine is also sufficient to increase RLS. Interestingly, we find no evidence that reduction of 40S subunits has a similar effect on lifespan, even when translation and polysomes are decreased to an extent similar to that of long-lived rplΔ mutants (Figure S7). Thus, we conclude that the RLS extension reported here does not result solely from reduced translation, but is caused by a specific reduction in 60S ribosomal subunit levels.
The relatively greater importance of 60S subunits over 40S subunits for longevity in yeast is interesting given that no such specificity has been observed in C. elegans, where RNAi knockdown of multiple large and small subunit RPs increases adult longevity (Chen et al., 2007; Curran and Ruvkun, 2007; Hansen et al., 2007). In yeast, two 40S proteins have been reported to influence RLS (Chiocchetti et al., 2007). It is possible that a number of RPS gene deletions are long-lived in the set we analyzed, and the change in RLS is not statistically significant at the current level of analysis. Anomalies in the yeast ORF deletion collection could also account for differential results in RLS experiments (see Supplemental Information). One possible explanation for these observations is that a subset of 40S RP mutations can alter the translational machinery in a manner that may induce GCN4. Altered GCN4 translational regulation due to RPS mutations is consistent with previously published data (Mueller et al., 1998). Alternatively, there may be a lifespan benefit derived directly from reduced translation, such as improved protein homeostasis with age (Kaeberlein and Kennedy, 2007). Our data suggest that, at least in yeast, however, the primary contribution to extended RLS is from specific depletion of 60S subunits and enhanced Gcn4 translation.
Gcn4 modulates longevity
We have provided evidence that Gcn4 is required for full lifespan extension by depletion of 60S subunits, DR, or genetic mimics of DR, tor1Δ and sch9Δ. Gcn4 induces the transcription of over 500 target genes, many of which are implicated in processes linked to lifespan regulation (Natarajan et al., 2001). For example, both Gcn4 and TOR are involved in regulating autophagy (Jia et al., 2000; Natarajan et al., 2001), and in C. elegans, lifespan extension by daf-2 requires beclin, the ortholog of yeast ATG6 (Melendez et al., 2003). In future studies, it will be important to determine which of Gcn4’s many target genes are most important for lifespan regulation. The mammalian Gcn4 ortholog (ATF4) is regulated via a similar Gcn2-dependent translational mechanism (Lu et al., 2004), raising the possibility that this pathway could play a similar role in multicellular organisms.
Our data provide evidence that Gcn4 is required for full lifespan extension by deletion of RPL genes in yeast. Although this dependence is significant for all the RPL mutants examined (p = < 0.001), the degree to which different RPL mutants depend on Gcn4 for lifespan extension varied (Figures 5B–5C, 6A, S5). Perhaps this reflects that deletion of single RPL genes can result in varied levels of GCN4 induction. The large number of Gcn4 target genes and the stringent nature of its regulation lead us to suspect that genetic approaches to attain an “optimal level” of Gcn4 for lifespan may be difficult.
A model for translational control of replicative lifespan
Epistasis analyses place reduction of 60S subunits together with DR, tor1Δ, and sch9Δ, characterized by a lack of response to DR and an ability to extend RLS independently of Sir2. However, a single linear path from DR to increased GCN4 synthesis is not entirely supported by the data presented here, because the loss of GCN4 does not completely prevent the lifespan extension observed by either DR (environmental or genetic) or of strains lacking RPL genes (Figures 5B–5C, 6A–6B, S5, S6). This indicates that one or more additional _GCN4_-independent pathways exist for lifespan regulation in response to nutrients (Figure 7).
What might a Gcn4-independent pathway be? A number of other yeast genes encode mRNAs containing 5′ uORFs (Vilela and McCarthy, 2003; Zhang and Dietrich, 2005), including HAP4 and CLN3, both of which are also important for appropriate response to nutrients, and increased expression of Hap4 increases yeast RLS (Lin et al., 2002). The translational regulation of these messages is largely unstudied, leaving open the possibility that reduced 60S subunit levels may be influencing expression of these genes similarly to GCN4. It is also possible that changes in 60S subunit abundance or structural composition (if ribosomes lacking a protein are being composed) affect translation of particular messages independently of uORFs.
Another possible means by which cells depleted for 60S subunits could increase RLS is by modulating the cellular response to ER stress (Miyoshi et al., 2002; Zhao et al., 2003). Yeast carrying the sly1-1 mutation or cells treated with tunicamycin, both of which induce an ER stress response, activate the PKC pathway leading to a dramatic decrease in RP gene transcription. For unknown reasons, this signal is abrogated in cells lacking RPL but not RPS genes (Miyoshi et al., 2002; Zhao et al., 2003). Interestingly, this signal is still abrogated in double mutants lacking both a RPL and a RPS gene (Zhao et al., 2003), perhaps supporting the idea that cells like tor1Δ and sch9Δ, which display polysome profiles with both decreased 40S and 60S subunits, should act similarly to cells in which only 60S subunits are limited. As cells age, they may experience ER stress which results in inhibition of RP gene transcription to a point at which translation can no longer be supported. Reduction of 60S subunits may specifically block this signal, allowing the cells to maintain a level of protein translation sufficient to support additional replicative cycles. Gcn4 is required for activating a majority of unfolded protein response target genes in response to ER stress (Patil et al., 2004), and ER stress has been proposed to play a role in C. elegans lifespan regulation (Viswanathan et al., 2005).
Conclusions
Our findings demonstrate that the abundance of 60S ribosomal subunits is a key determinant of yeast RLS. Genetic evidence places DR, TOR inhibition, SCH9 deletion, and depletion of 60S subunits in a longevity pathway that is partially dependent on the Gcn4 transcription factor. Gcn4 activity is enhanced in long-lived mutants and is required for full lifespan extension by reduction of 60S subunits, tor1Δ, sch9Δ, or DR. Evidence from multicellular eukaryotes is consistent with DR being mediated in part by an altered translational program. Decreased activity of TOR and Sch9 orthologs results in increased lifespan in worms and flies (Hertweck et al., 2004; Jia et al., 2004; Kapahi et al., 2004; Vellai et al., 2003), and inhibition of factors important for translation initiation has been shown to increase the lifespan of C. elegans (Chen et al., 2007; Curran and Ruvkun, 2007; Hamilton et al., 2005; Hansen et al., 2007; Henderson et al., 2006; Pan et al., 2007; Syntichaki et al., 2007). Thus, we propose the existence of an evolutionarily conserved pathway linking DR, protein translation, and longevity. The high level of conservation among Gcn4 orthologs in multicellular eukaryotes merits the future investigation of their potential roles in lifespan regulation.
EXPERIMENTAL PROCEDURES
Strains and media
All yeast strains were derived from the parent strains of the haploid yeast ORF deletion collections (Winzeler et al., 1999), BY4742 (_MAT_α his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0) and BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0). The _MAT_α haploid ORF deletion collection and the MATa haploid ORF deletion collection, along with the parental strains, were obtained from Research Genetics. Of the RP gene deletion strains from the MATα deletion collection, rps12Δ failed to pass quality control during construction and was therefore excluded from our analysis. Figure 1 contains pooled data from both MATa and the corresponding _MAT_α deletion collection strains (and remade strains in the cases of rpl31aΔ and rpl20bΔ, see Supplemental Information). Data represented in all other figures were generated from strains in the MATα deletion collection. The multiple gene deletion strains represented in Figures 4 and 6 were constructed by standard PCR-based gene-disruption as described (Kaeberlein et al., 2004). The SIR2 overexpression strain (Figure 4A) was constructed by genomic integration of an extra copy of SIR2 at the LEU2 locus, as described (Kaeberlein et al., 1999).
Cells were grown in standard YPD containing 1% yeast extract, 2% peptone, and 2% glucose, with the exception of the lifespan assays which were done using YPD containing 0.05% glucose where noted. Diazaborine was a generous gift from Gregor Hogenaur (Graz, Austria).
Replicative lifespan analysis
Lifespan assays were carried out as described previously (Kaeberlein et al., 2005b). All lifespan experiments were carried out on standard YPD plates (2% glucose, unless otherwise noted). DR experiments were carried out on 0.05% glucose; we have previously shown that 0.05% glucose is an optimal concentration for DR in RLS studies using BY4742 (Kaeberlein et al., 2004). For lifespan studies with diazaborine or cycloheximide, the drug was added from frozen stock to melted and cooled YPD at the appropriate concentration. Statistical significance for RP mutants were determined using a Wilcoxon Rank-Sum test (MATLAB ‘ranksum’ function) using a p = 0.05 cutoff. For each of the 107 rpΔ strains analyzed in this study, mean lifespan and p-values can be found in Table S1. For Figure 6A, a T-test was used to determine the statistical significance for the dependence of RPL mutants on GCN4 for long lifespan. Independent T-tests were used to determine the statistical significance for the percent change in mean RLS for DR, tor1Δ and sch9Δ with or without GCN4 (Figure 6B).
Polysome Analysis
Polysome analysis was carried out as described previously (MacKay et al., 2004). Briefly, log phase yeast cultures were quick-chilled with crushed frozen YPD containing 100 μg/ml cycloheximide. Cells were harvested by centrifugation, washed with 10 ml lysis buffer (25 mM Tris-HCl, pH 7.5, 40 mM KCl, 7.5 mM MgCl2, 1 mM DTT, 0.5 mg/ml heparin, 100 μg/ml cycloheximide) and resuspended in 1 ml lysis buffer. Cells were lysed by vortexing with glass beads. Triton X-100 and sodium deoxycholate were added (1% final concentration each) with vortexing and the samples stood on ice for 5 min before the supernatant was clarified by centrifugation. All reagents were ice-cold and all steps were done in a 4°C cold room. For separation on gradients, 1 ml containing 20 (or 25, Figure 2E) A260 units of lysate were loaded onto 11-ml linear 7%–47% sucrose gradients in 50 mM Tris-HCl, pH 7.5, 0.8 M KCl, 15 mM MgCl2, 0.5 mg/ml heparin, 100 μg/ml cycloheximide and sedimented at 39,000 rpm at 4°C in an SW40 Ti swinging bucket rotor (Beckman) for 2 hr (or 1.5 hr, Figure 2E). Gradients were collected from the top and profiles were monitored at 254 nm.
Quantitative real-time PCR analysis of RPL RNAs
Lysates of strain BY4742 were prepared as described above and previously (MacKay et al., 2004), total RNA was purified using Qiagen RNeasy mini-columns, and 2 μg RNA was converted to cDNA with Invitrogen Ss III reverse transcriptase and an oligo(dT)25 primer with a G/C/A 3′ anchor. Lysates, RNA purification, and reverse transcription reactions were performed on different days for three independently-grown cultures. Specific cDNAs were quantitated with an iCycler (Bio-Rad, Hercules, CA) and SYBRGreen detection of products (according to manufacturer’s specifications). See Supplemental Information for primer sequences and details.
Gcn4-Luciferase Assays
Gcn4 expression was assayed using a dual luciferase reporter plasmid pVW31, modified from the _URA3_-2μ plasmid pDB688 (Keeling et al., 2004; Salas-Marco and Bedwell, 2005; kindly provided by David Bedwell). Plasmid pVW31 contains (1) a _GCN4_-firefly luciferase cDNA fusion under transcriptional control of a 772 bp GCN4 5′ fragment containing all of the promoter, upstream open reading frames (uORFs), and other 5′ regulatory elements and the CYC1 transcription terminator, (2) an independent transcriptional unit with a Renilla luciferase cDNA transcribed from the constitutive S. cerevisiae PGK1 promoter and terminated with a 3′ fragment from GCY1. (Plasmid details will be described elsewhere, manuscript in preparation.) Strains transformed with pVW31 were grown in synthetic glucose minimal medium lacking uracil and containing required amino acids as well as isoleucine and valine (Lucchini et al., 1984). A dual luciferase reporter assay system (Promega) and a Perkin Elmer Victor Light Model 1420 luminometer were used to measure luciferase activities. Firefly luciferase activity was normalized to the Renilla luciferase activity.
Growth Rate Analysis
Growth curves for the RP gene deletion strains were generated using a Bioscreen C machine (Growth Curves USA). Overnight cultures of the strains were grown in 250 μl YPD in 96-well plates (inoculated from single colonies). The next day, 8 μl of overnight culture was added to 250 μl fresh YPD medium in Bioscreen C Honeycomb microplates and cultures were grown in the Bioscreen C at 30°C for 24 hours. Optical density was measured every 30 minutes, the plates were shaken every 10 minutes for 20 seconds. The doubling time was calculated between every 30 minute interval. The generation time was defined as the average of the four lowest doubling times (steepest part of the growth curve), after dropping the lowest single number. Independent growth curves were generated for each strain on three different days; the average generation time ± standard deviation for at least three independent assays is given in Table S3. The raw data collected for each of the independent experiments are provided as online supplemental data in Excel file format (Supplemental data downloadable file).
Supplementary Material
01
02
03
04
05
Acknowledgments
We would like to thank Gregor Hogenauer for supplying diazaborine, David Morris for technical comments and advice, and the Ferric Fang lab for assistance generating growth curves. K.K.S. has been supported by NIH training grant P30 AG013280. S.F. is an investigator of the Howard Hughes Medical Institute. This work was funded by an award from the Ellison Medical Foundation to B.K.K. and M.K. and by National Institutes of Health Grant R01 AG024287 to B.K.K.
Footnotes
Supplemental Data
Supplemental Data include Experimental Procedures (including primer nucleotide sequences), three tables, seven figures, and a downloadable Excel data file containing growth curve data.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baldock C, de Boer GJ, Rafferty JB, Stuitje AR, Rice DW. Mechanism of action of diazaborines. Biochem Pharmacol. 1998;55:1541–1549. doi: 10.1016/s0006-2952(97)00684-9. [DOI] [PubMed] [Google Scholar]
- Beck T, Hall MN. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature. 1999;402:689–692. doi: 10.1038/45287. [DOI] [PubMed] [Google Scholar]
- Carey JR. Longevity: The biology and demography of life span. Princeton, NJ: Princeton University Press; 2003. [Google Scholar]
- Chen D, Pan KZ, Palter JE, Kapahi P. Longevity determined by developmental arrest genes in Caenorhabditis elegans. Aging Cell. 2007;6:525–533. doi: 10.1111/j.1474-9726.2007.00305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherkasova VA, Hinnebusch AG. Translational control by TOR and TAP42 through dephosphorylation of eIF2alpha kinase GCN2. Genes Dev. 2003;17:859–872. doi: 10.1101/gad.1069003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiocchetti A, Zhou J, Zhu H, Karl T, Haubenreisser O, Rinnerthaler M, Heeren G, Oender K, Bauer J, Hintner H, et al. Ribosomal proteins Rpl10 and Rps6 are potent regulators of yeast replicative life span. Exp Gerontol. 2007;42:275–286. doi: 10.1016/j.exger.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Curran SP, Ruvkun G. Lifespan regulation by evolutionarily conserved genes essential for viability. PLoS Genet. 2007;3:e56. doi: 10.1371/journal.pgen.0030056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defossez PA, Prusty R, Kaeberlein M, Lin SJ, Ferrigno P, Silver PA, Keil RL, Guarente L. Elimination of replication block protein Fob1 extends the life span of yeast mother cells. Mol Cell. 1999;3:447–455. doi: 10.1016/s1097-2765(00)80472-4. [DOI] [PubMed] [Google Scholar]
- Dilova I, Chen CY, Powers T. Mks1 in concert with TOR signaling negatively regulates RTG target gene expression in S. cerevisiae. Curr Biol. 2002;12:389–395. doi: 10.1016/s0960-9822(02)00677-2. [DOI] [PubMed] [Google Scholar]
- Fabrizio P, Longo VD. The chronological life span of Saccharomyces cerevisiae. Aging Cell. 2003;2:73–81. doi: 10.1046/j.1474-9728.2003.00033.x. [DOI] [PubMed] [Google Scholar]
- Fabrizio P, Pletcher SD, Minois N, Vaupel JW, Longo VD. Chronological aging-independent replicative life span regulation by Msn2/Msn4 and Sod2 in Saccharomyces cerevisiae. FEBS Lett. 2004;557:136–142. doi: 10.1016/s0014-5793(03)01462-5. [DOI] [PubMed] [Google Scholar]
- Fabrizio P, Pozza F, Pletcher SD, Gendron CM, Longo VD. Regulation of longevity and stress resistance by Sch9 in yeast. Science. 2001;292:288–290. doi: 10.1126/science.1059497. [DOI] [PubMed] [Google Scholar]
- Foiani M, Cigan AM, Paddon CJ, Harashima S, Hinnebusch AG. GCD2, a translational repressor of the GCN4 gene, has a general function in the initiation of protein synthesis in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:3203–3216. doi: 10.1128/mcb.11.6.3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton B, Dong Y, Shindo M, Liu W, Odell I, Ruvkun G, Lee SS. A systematic RNAi screen for longevity genes in C. elegans. Genes Dev. 2005;19:1544–1555. doi: 10.1101/gad.1308205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M, Taubert S, Crawford D, Libina N, Lee SJ, Kenyon C. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell. 2007;6:95–110. doi: 10.1111/j.1474-9726.2006.00267.x. [DOI] [PubMed] [Google Scholar]
- Harnpicharnchai P, Jakovljevic J, Horsey E, Miles T, Roman J, Rout M, Meagher D, Imai B, Guo Y, Brame CJ, et al. Composition and functional characterization of yeast 66S ribosome assembly intermediates. Mol Cell. 2001;8:505–515. doi: 10.1016/s1097-2765(01)00344-6. [DOI] [PubMed] [Google Scholar]
- Henderson ST, Bonafe M, Johnson TE. daf-16 protects the nematode Caenorhabditis elegans during food deprivation. J Gerontol A Biol Sci Med Sci. 2006;61:444–460. doi: 10.1093/gerona/61.5.444. [DOI] [PubMed] [Google Scholar]
- Hertweck M, Gobel C, Baumeister R. C. elegans SGK-1 is the critical component in the Akt/PKB kinase complex to control stress response and life span. Dev Cell. 2004;6:577–588. doi: 10.1016/s1534-5807(04)00095-4. [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG. Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol. 2005;59:407–450. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- Hung NJ, Johnson AW. Nuclear recycling of the pre-60S ribosomal subunit-associated factor Arx1 depends on Rei1 in Saccharomyces cerevisiae. Mol Cell Biol. 2006;26:3718–3727. doi: 10.1128/MCB.26.10.3718-3727.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia K, Chen D, Riddle DL. The TOR pathway interacts with the insulin signaling pathway to regulate C. elegans larval development, metabolism and life span. Development. 2004;131:3897–3906. doi: 10.1242/dev.01255. [DOI] [PubMed] [Google Scholar]
- Jia MH, Larossa RA, Lee JM, Rafalski A, Derose E, Gonye G, Xue Z. Global expression profiling of yeast treated with an inhibitor of amino acid biosynthesis, sulfometuron methyl. Physiol Genomics. 2000;3:83–92. doi: 10.1152/physiolgenomics.2000.3.2.83. [DOI] [PubMed] [Google Scholar]
- Jiang JC, Jaruga E, Repnevskaya MV, Jazwinski SM. An intervention resembling caloric restriction prolongs life span and retards aging in yeast. Faseb J. 2000;14:2135–2137. doi: 10.1096/fj.00-0242fje. [DOI] [PubMed] [Google Scholar]
- Jorgensen P, Rupes I, Sharom JR, Schneper L, Broach JR, Tyers M. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 2004;18:2491–2505. doi: 10.1101/gad.1228804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M. In: Longevity and aging in the budding yeast, In Handbook of models for human aging. Conn PM, editor. Boston: Elsevier Press; 2006. pp. 109–120. [Google Scholar]
- Kaeberlein M, Kennedy BK. Protein translation, 2007. Aging Cell. 2007;6:731–734. doi: 10.1111/j.1474-9726.2007.00341.x. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, Kirkland KT, Fields S, Kennedy BK. Sir2-independent life span extension by calorie restriction in yeast. PLoS Biol. 2004;2:E296. doi: 10.1371/journal.pbio.0020296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, Kirkland KT, Fields S, Kennedy BK. Genes determining yeast replicative life span in a long-lived genetic background. Mech Ageing Dev. 2005a;126:491–504. doi: 10.1016/j.mad.2004.10.007. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, Powers RW, 3rd, Steffen KK, Westman EA, Hu D, Dang N, Kerr EO, Kirkland KT, Fields S, Kennedy BK. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005b;310:1193–1196. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, Steffen KK, Hu D, Dang N, Kerr EO, Tsuchiya M, Fields S, Kennedy BK. Comment on “HST2 mediates SIR2-independent life-span extension by calorie restriction”. Science. 2006;312:1312. doi: 10.1126/science.1124608. author reply 1312. [DOI] [PubMed] [Google Scholar]
- Kapahi P, Zid BM, Harper T, Koslover D, Sapin V, Benzer S. Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr Biol. 2004;14:885–890. doi: 10.1016/j.cub.2004.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komili S, Farny NG, Roth FP, Silver PA. Functional specificity among ribosomal proteins regulates gene expression. Cell. 2007;131:557–571. doi: 10.1016/j.cell.2007.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota H, Obata T, Ota K, Sasaki T, Ito T. Rapamycin-induced translational derepression of GCN4 mRNA involves a novel mechanism for activation of the eIF2 alpha kinase GCN2. J Biol Chem. 2003;278:20457–20460. doi: 10.1074/jbc.C300133200. [DOI] [PubMed] [Google Scholar]
- Lebreton A, Saveanu C, Decourty L, Rain JC, Jacquier A, Fromont-Racine M. A functional network involved in the recycling of nucleocytoplasmic pre-60S factors. J Cell Biol. 2006;173:349–360. doi: 10.1083/jcb.200510080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SJ, Defossez PA, Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289:2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- Lin SJ, Kaeberlein M, Andalis AA, Sturtz LA, Defossez PA, Culotta VC, Fink GR, Guarente L. Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature. 2002;418:344–348. doi: 10.1038/nature00829. [DOI] [PubMed] [Google Scholar]
- Lu PD, Harding HP, Ron D. Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J Cell Biol. 2004;167:27–33. doi: 10.1083/jcb.200408003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKay VL, Li X, Flory MR, Turcott E, Law GL, Serikawa KA, Xu XL, Lee H, Goodlett DR, Aebersold R, et al. Gene expression analyzed by high-resolution state array analysis and quantitative proteomics: response of yeast to mating pheromone. Mol Cell Proteomics. 2004;3:478–489. doi: 10.1074/mcp.M300129-MCP200. [DOI] [PubMed] [Google Scholar]
- Martin-Marcos P, Hinnebusch AG, Tamame M. Ribosomal Protein L33 Is Required for Ribosome Biogenesis, Subunit Joining, and Repression of GCN4 Translation. Mol Cell Biol. 2007;27:5968–5985. doi: 10.1128/MCB.00019-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DE, Soulard A, Hall MN. TOR regulates ribosomal protein gene expression via PKA and the Forkhead transcription factor FHL1. Cell. 2004;119:969–979. doi: 10.1016/j.cell.2004.11.047. [DOI] [PubMed] [Google Scholar]
- Masoro EJ. Overview of caloric restriction and ageing. Mech Ageing Dev. 2005;126:913–922. doi: 10.1016/j.mad.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Melendez A, Talloczy Z, Seaman M, Eskelinen EL, Hall DH, Levine B. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 2003;301:1387–1391. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- Miyoshi K, Tsujii R, Yoshida H, Maki Y, Wada A, Matsui Y, Toh EA, Mizuta K. Normal assembly of 60 S ribosomal subunits is required for the signaling in response to a secretory defect in Saccharomyces cerevisiae. J Biol Chem. 2002;277:18334–18339. doi: 10.1074/jbc.M201667200. [DOI] [PubMed] [Google Scholar]
- Mortimer RK, Johnston JR. Life span of individual yeast cells. Nature. 1959;183:1751–1752. doi: 10.1038/1831751a0. [DOI] [PubMed] [Google Scholar]
- Mueller PP, Grueter P, Hinnebusch AG, Trachsel H. A ribosomal protein is required for translational regulation of GCN4 mRNA. Evidence for involvement of the ribosome in eIF2 recycling. J Biol Chem. 1998;273:32870–32877. doi: 10.1074/jbc.273.49.32870. [DOI] [PubMed] [Google Scholar]
- Natarajan K, Meyer MR, Jackson BM, Slade D, Roberts C, Hinnebusch AG, Marton MJ. Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol Cell Biol. 2001;21:4347–4368. doi: 10.1128/MCB.21.13.4347-4368.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T, Ohsumi Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J Biol Chem. 1998;273:3963–3966. doi: 10.1074/jbc.273.7.3963. [DOI] [PubMed] [Google Scholar]
- Pan KZ, Palter JE, Rogers AN, Olsen A, Chen D, Lithgow GJ, Kapahi P. Inhibition of mRNA translation extends lifespan in Caenorhabditis elegans. Aging Cell. 2007;6:111–119. doi: 10.1111/j.1474-9726.2006.00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil CK, Li H, Walter P. Gcn4p and novel upstream activating sequences regulate targets of the unfolded protein response. PLoS Biol. 2004;2:E246. doi: 10.1371/journal.pbio.0020246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertschy B, Zisser G, Schein H, Koffel R, Rauch G, Grillitsch K, Morgenstern C, Durchschlag M, Hogenauer G, Bergler H. Diazaborine treatment of yeast cells inhibits maturation of the 60S ribosomal subunit. Mol Cell Biol. 2004;24:6476–6487. doi: 10.1128/MCB.24.14.6476-6487.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers RW, 3rd, Kaeberlein M, Caldwell SD, Kennedy BK, Fields S. Extension of chronological life span in yeast by decreased TOR pathway signaling. Genes Dev. 2006;20:174–184. doi: 10.1101/gad.1381406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers T. TOR signaling and S6 kinase 1: Yeast catches up. Cell Metab. 2007;6:1–2. doi: 10.1016/j.cmet.2007.06.009. [DOI] [PubMed] [Google Scholar]
- Powers T, Walter P. Regulation of ribosome biogenesis by the rapamycin-sensitive TOR-signaling pathway in Saccharomyces cerevisiae. Mol Biol Cell. 1999;10:987–1000. doi: 10.1091/mbc.10.4.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syntichaki P, Troulinaki K, Tavernarakis N. eIF4E function in somatic cells modulates ageing in Caenorhabditis elegans. Nature. 2007;445:922–926. doi: 10.1038/nature05603. [DOI] [PubMed] [Google Scholar]
- Tsay YF, Thompson JR, Rotenberg MO, Larkin JC, Woolford JL., Jr Ribosomal protein synthesis is not regulated at the translational level in Saccharomyces cerevisiae: balanced accumulation of ribosomal proteins L16 and rp59 is mediated by turnover of excess protein. Genes Dev. 1988;2:664–676. doi: 10.1101/gad.2.6.664. [DOI] [PubMed] [Google Scholar]
- Urban J, Soulard A, Huber A, Lippman S, Mukhopadhyay D, Deloche O, Wanke V, Anrather D, Ammerer G, Riezman H, et al. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol Cell. 2007;26:663–674. doi: 10.1016/j.molcel.2007.04.020. [DOI] [PubMed] [Google Scholar]
- Valenzuela L, Aranda C, Gonzalez A. TOR modulates GCN4-dependent expression of genes turned on by nitrogen limitation. J Bacteriol. 2001;183:2331–2334. doi: 10.1128/JB.183.7.2331-2334.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellai T, Takacs-Vellai K, Zhang Y, Kovacs AL, Orosz L, Muller F. Genetics: influence of TOR kinase on lifespan in C. elegans. Nature. 2003;426:620. doi: 10.1038/426620a. [DOI] [PubMed] [Google Scholar]
- Vilela C, McCarthy JE. Regulation of fungal gene expression via short open reading frames in the mRNA 5′untranslated region. Mol Microbiol. 2003;49:859–867. doi: 10.1046/j.1365-2958.2003.03622.x. [DOI] [PubMed] [Google Scholar]
- Viswanathan M, Kim SK, Berdichevsky A, Guarente L. A role for SIR–2.1 regulation of ER stress response genes in determining C. elegans life span. Dev Cell. 2005;9:605–615. doi: 10.1016/j.devcel.2005.09.017. [DOI] [PubMed] [Google Scholar]
- Warner JR, Mitra G, Schwindinger WF, Studeny M, Fried HM. Saccharomyces cerevisiae coordinates accumulation of yeast ribosomal proteins by modulating mRNA splicing, translational initiation, and protein turnover. Mol Cell Biol. 1985;5:1512–1521. doi: 10.1128/mcb.5.6.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weindruch R, Naylor PH, Goldstein AL, Walford RL. Influences of aging and dietary restriction on serum thymosin alpha 1 levels in mice. J Gerontol. 1988;43:B40–42. doi: 10.1093/geronj/43.2.b40. [DOI] [PubMed] [Google Scholar]
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Yang R, Wek SA, Wek RC. Glucose limitation induces GCN4 translation by activation of Gcn2 protein kinase. Mol Cell Biol. 2000;20:2706–2717. doi: 10.1128/mcb.20.8.2706-2717.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Dietrich FS. Identification and characterization of upstream open reading frames (uORF) in the 5′ untranslated regions (UTR) of genes in Saccharomyces cerevisiae. Curr Genet. 2005;48:77–87. doi: 10.1007/s00294-005-0001-x. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Sohn JH, Warner JR. Autoregulation in the biosynthesis of ribosomes. Mol Cell Biol. 2003;23:699–707. doi: 10.1128/MCB.23.2.699-707.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01
02
03
04
05