PHD fingers in human diseases: disorders arising from misinterpreting epigenetic marks (original) (raw)
. Author manuscript; available in PMC: 2009 Dec 1.
Abstract
Histone covalent modifications regulate many, if not all, DNA-templated processes, including gene expression and DNA damage response. The biological consequences of histone modifications are mediated partially by evolutionarily conserved “reader/effector” modules that bind to histone marks in a modification- and context-specific fashion and subsequently enact chromatin changes or recruit other proteins to do so. Recently, the _P_lant _H_omeo_d_omain (PHD) finger has emerged as a class of specialized “reader” modules that, in some instances, recognize the methylation status of histone lysine residues, such as histone H3 lysine 4 (H3K4). While mutations in catalytic enzymes that mediate the addition or removal of histone modifications (i.e., “writers” and “erasers”) are already known to be involved in various human diseases, mutations in the modification-specific “reader” proteins are only beginning to be recognized as contributing to human diseases. For instance, point mutations, deletions or chromosomal translocations that target PHD fingers encoded by many genes (such as RAG2, ING, NSD1 and ATRX) have been associated with a wide range of human pathologies including immunological disorders, cancers, and neurological diseases. In this review, we will discuss the structural features of PHD fingers as well as the diseases for which direct mutation or dysregulation of the PHD finger has been reported. We propose that misinterpretation of the epigenetic marks may serve as a general mechanism for human diseases of this category. Determining the regulatory roles of histone covalent modifications in the context of human disease will allow for a more thorough understanding of normal and pathological development, and may provide innovative therapeutic strategies wherein “chromatin readers” stand as potential drug targets.
1. Introduction
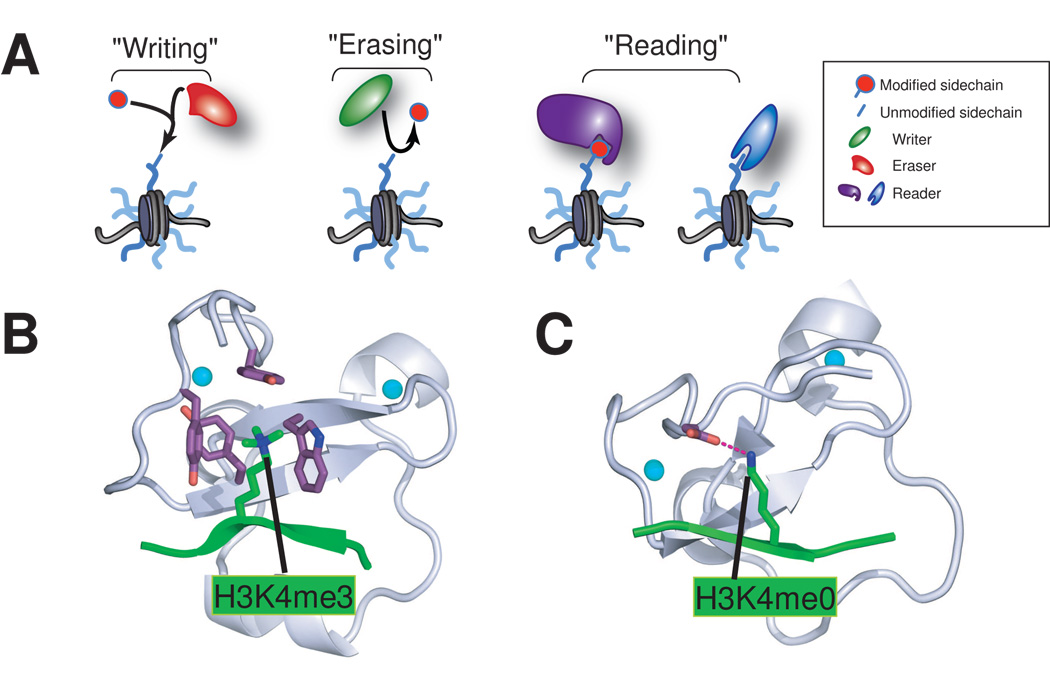
The fundamental repeating unit of chromatin, the nucleosome core particle, consists of ~146 base pairs of DNA wrapped around a histone octamer consisting of two copies each of the core histones – H2A, H2B, H3 and H4 [1]. Covalent modification of histones and DNA methylation may serve as the potential molecular carriers of epigenetic inheritance, ensuring the correct storage, organization, and interpretation of genetic information spatially and temporally during development [2]. Histone post-translational modification is often a dynamic and reversible process mediated by two antagonizing sets of enzymatic complexes: the “writer” and “eraser” proteins and associated factors that site-specifically attach and remove the modifications, respectively (Figure 1A). For example, methylation of histone H3, lysines 4 and 36 (H3K4 and H3K36) is generally associated with “open” euchromatin structure and transcriptional activation, whereas methylation of histone H3, lysines 9 and 27 (H3K9 and H3K27) is generally associated with “closed” heterochromatin structure and gene silencing [2,3]. However, mechanisms by which histone modification marks contribute to their specific functional consequences are not fully understood. While some histone modifications, such as lysine acetylation, alter chromatin structure directly via charge ablation [4], other modifications serve as binding sites, recruiting the so-called “reader/effector” proteins that specifically recognize such marks and translate them into subsequent meaningful biological consequences via either their intrinsic activities or those of their interacting partners (Figure 1A) [5,6]. For example, bromodomains and chromodomains interact with specific histone acetylation and methylation marks respectively [7,8]. Recently, the _P_lant _H_omeo_d_omain (PHD) finger has emerged as a motif that, in some cases, differentially recognizes either methylated [9–13] or unmodified [14,15] lysine residues in histone tails (Figure 1B–C). Notably, many “reader” module-containing factors or complexes also harbor “writer” or “eraser” activities, and these combined activities coordinate “read-write” or “read-erase” processes that might underlie the spreading or erasing of epigenetic marks over a large domain in the genome [16].
Figure 1. PHD fingers are “reader/effector” modules that recognize histone lysine methylation status.
(A) A schematic model that illustrates the “writing”, “erasing” and “reading” of covalent modification marks on histone tails. (B–C) Two specialized subsets of PHD fingers specifically recognize and bind to the highly methylated-H3K4 (B) and unmodified H3K4 (C) respectively. The structures shown are PHD fingers of BPTF [11] and BHC80 [15], with PHD finger depicted in silver, H3K4 peptide in green and Zn2+ ion in cyan sphere. Side chains of the critical engaging residues are shown in purple. For simplicity, only modules engaging single marks are shown, and aspects involving multivalent engaging modules have been described elsewhere [7].
A prediction of the “histone code hypothesis” [17] is that alterations in the “balance” between “on” versus “off” chromatin states lead to inappropriate expression or silencing of gene programs that, in turn, alter states of cellular identity and may lead to human disease. Cancer development has long been recognized as a mingled process of genetic and epigenetic alterations that contribute to its initiation and progression [18], and cancer-associated mutation or dysregulation has been identified in various “writer” and “eraser” enzymes [19,20]. Recently, a variety of diseases including immunodeficiency syndrome, solid and blood cancers, and neurological disorders, have been linked to dysregulation of factors that harbor the chromatin-recognizing “reader/effector” modules, notably PHD fingers in many cases (Table 1). In this review, we discuss the structural features of PHD fingers and elaborate on those diseases associated with PHD finger dysregulation. We propose a category of human diseases that stems from misinterpreting the epigenetic marks, including histone modifications and DNA methylation (exemplified by MeCP2 mutations in Rett’s Syndrome [21]). Understanding the regulatory signals provided by epigenetic marks in the context of human disease will not only broaden the mechanistic appreciation of normal and pathological development, but also pinpoint the significance of “epigenetic codes” in our genome as an additional indexing system that operates beyond the DNA template itself.
Table 1.
Mutations in genes encoding PHD finger proteins are associated with a wide variety of human diseases.
| Protein | Disease Association | PHD finger dysregulation | Ligand for PHD finger | Reference |
|---|---|---|---|---|
| Immune Disorders | ||||
| RAG2 | T-B-NK+ Severe Combined Immunodeficieny (SCID) and Omenn syndrome | Germline mutations | H3K4me3 | [23,38,40,4,6,48–50] |
| AIRE | Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy (APECED), also known as Autoimmune polyglandular syndrome type 1 (APS-1) | Germline mutations (both PHDs) | H3K4me0 (PHD1) Unknown (PHD2) | [30,57,59–62] |
| Cancer | ||||
| ING1 | Breast cancer, melanoma, esophageal squamous cell carcinoma (SSC), head and neck SSC | Somatic mutation | H3K4me3,2 | [13,67,72–75] |
| JARID1A (RBP2) | Myeloid leukemia | Translocation of the third PHD* | Unknown (PHD3) | [82] |
| PHF23 | Myeloid leukemia | Translocation | Unknown | [83] |
| NSD1, NSD3 | Myeloid leukemia | Translocation of PHD fingers | Unknown | [85,86] |
| MMSET (NSD2) | Translocation of PHD fingers | Unknown | [89] | |
| MLL | T-cell lymphoblastic leukaemia | Internal deletion of first PHD | Unknown | [91,92] |
| PHF1 | Endometrial stromal sarcoma | Translocation | Unknown | [93] |
| Neurological Disorders | ||||
| NSD1 | Childhood overgrowth syndromes such as Sotos syndrome and Weaver syndrome | Germline mutations in all 5 PHDs; truncation; microdeletion | Unknown | [88,95,96] |
| ATRX | Various X-linked mental retardation disorders, including Alpha-Thalassemia and Mental Retardation, X-linked (ATRX) Syndrome | Germline mutations (>26 distinct PHD mutations reported) | Unknown | [102] |
| CBP | Rubenstein-Taybi Syndrome (RTS) | Germline mutation | Unknown | [109] |
| PHF6 | Borjeson-Forssman-Lehmann Syndrome (BFLS) | Germline mutations (PHD1 only) | Unknown | [110] |
2. The structure of PHD fingers
Since its initial identification in two plant homeodomain proteins that gave the domain its name, 14 PHD finger-containing proteins have been found in the budding yeast genome, 50 in the fruit fly, and up to several hundred in humans [22]. The typical PHD finger consists of two interleaved atypical zinc fingers, characterized by a Cys4-His-Cys3 architecture that coordinates two Zn2+ ions (Figure 1B–C) [22], although there are noted exceptions, such as the RAG2 PHD finger, containing a Cys3-His2-Cys2-His architecture (Figure 2B) [23,24]. Structurally, the PHD finger resembles the RING finger which functions as an E3 ligase in the ubiquitylation pathway, but the PHD domain generally lacks the E2 ligase-interacting surface that is characteristic of many RING domains [22]. Because many PHD-containing proteins associate with chromatin and regulate its activities, the PHD finger was initially suggested to interact with chromatin [22,25]. Indeed, recent studies have revealed that tri-methylated H3K4 (H3K4me3) and unmodified H3K4 (H3K4me0) serve as ligands for two distinct subclasses of PHD fingers [5,7–11,13].
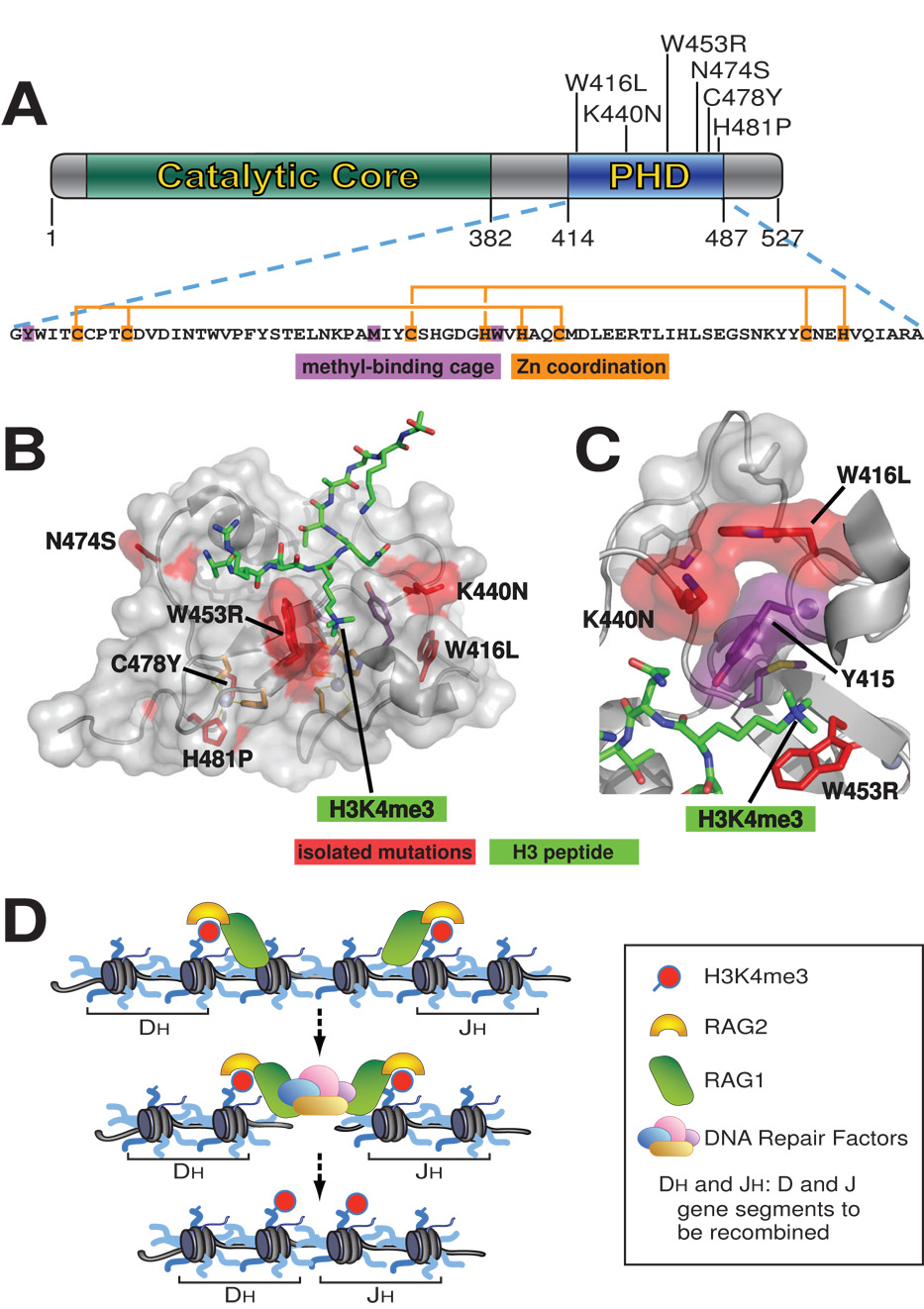
Figure 2. Missense mutations in the RAG2 PHD finger are associated with immunodeficiency syndromes.
(A) Domain structure of RAG2 (NCBI accession number 187423896) showing disease-causing mutations found within the PHD finger. The amino acid sequence of the PHD finger is shown below with H3K4me-caging residues in purple and Zn2+ ion-coordinating residues in orange. (B) Co-crystal structure of the RAG2 PHD finger (in gray) and the H3K4me3 peptide (in green) [24]. Residues mutated in T-B-SCID or Omenn Syndrome are depicted in red, and Y415, which forms part of the aromatic channel, is in purple. (C) Closer view of mutations K440N and W416L as well as the RAG2 PHD aromatic channel with H3K4me3. (D) Schematic model for RAG2’s function in V(D)J recombination. Interactions between the RAG2 PHD and H3K4me3 help to recruit and/or stabilize RAG1/2 recombinases to appropriate V(D)J gene segments marked with this histone modification, where they create double-strand DNA breaks. Then DNA repair factors such as DNA-PKcs, Ku70/80, XRCC4 and DNA Ligase IV assists in ligation of the broken ends of two V(D)J gene segments, creating a functional gene segment. Note here the correlation between the disease-causing mutations and the “reading” function of the RAG2 PHD finger in engaging histone marks.
As founding members of the first subclass, the PHD fingers of BPTF and ING2 engage H3K4me3 and H3R2 simultaneously in two adjacent channels that are separated by a conserved tryptophan in the PHD [9–11,13]. The aromatic or hydrophobic residues that form a channel or cage around H3K4me3 stabilize the interaction between the PHD finger and the H3K4me3 side chain via a composite of cation-π and hydrophobic interactions (Figure 1B). With slight variations, a similar H3K4me3-engaging aromatic cage is also found in other PHD fingers including those of other ING members (ING1, ING3-5, and their yeast homologue Yng1) and RAG2, indicating a common mechanism for H3K4me3 recognition conserved in evolution [5,7,8,23,24,26,27]. On the other hand, the H3R2-engaging channel or pocket differs among these PHD domains. H3R2 methylation inhibits the H3K4me-binding by the ING2, BPTF and Spp1 PHD fingers [24,28,29], whereas H3K4me-binding by the RAG2 PHD finger tolerates H3R2 methylation [24].
A second subclass of PHD fingers, including those of DNMT3L and BHC80/PHF21A interact with unmodified H3K4 (H3K4me0) specifically [14,15]. Instead of utilizing an aromatic cage/channel, the specificity for the H3K4me0-PHD finger association is established through an electrostatic bridge between the unmodified epsilon amino group of H3K4me0 and an acidic residue in PHD finger (Asp90 in DNMT3L or Asp489 in BHC80), and methylation at H3K4 sterically excludes such interaction (Figure 1C). The first PHD finger of the AIRE protein has also been reported as an H3K4me0 binder [30], suggesting that these instances may portend a more generalized mechanism in recognizing unmodified histone tails.
Many other PHD fingers do not seem to fit into the two known subclasses above as they lack those critical engaging residues described. Indeed, emerging evidence shows that some of them associate with different methyl marks, with some PHD fingers in yeast binding to H3K36me and PHD fingers in SMCX and ICBP90 to H3K9me [31–33]. In addition, many PHD fingers may recognize modifications other than methyl-lysines or have unknown functions.
3. PHD finger dysregulation in immunodeficiency syndromes
3.1. Recombination Activating Gene 2 (RAG2)
The immunodeficiency syndromes caused by mutations in the PHD finger of RAG2 provide a paradigmatic example of how PHD mutations contribute to human disease. RAG2 recombinase is the catalytic engine of V(D)J recombination, whereby developing B and T cells fuse different combinations of receptor gene segments to create B and T cell receptor diversity [34,35]. These somatic cell recombination events are the centerpiece of the adaptive immune response. During V(D)J recombination, RAG2 and its associated recombinase RAG1 work together to recognize and create double-strand breaks at recombination signal sequences (RSSs) within specific V(D)J gene segments [36]. Once the breaks are made, repair proteins ligate the broken ends together to generate a functional receptor gene (Figure 2D). Deleting the RAG2 gene in mice results in the disruption of V(D)J recombination, the failure of B/T cell differentiation and a compromised immune system [37]. Loss-of-function point mutations in RAG2 cause similar phenotypes in humans (Figure 2A) [38]. Severe RAG2 mutations completely disrupt V(D)J recombination, causing a condition known as T-B-SCID ( “Severe combined immunodeficiency”) where patients lack functional B or T cells and are susceptible to infections [38]. In a less severe disorder called “Omenn Syndrome”, hypomorphic RAG2 mutations partially impair V(D)J recombination, causing a lack of functional B cells with normal or elevated levels of T cells, which are often activated and only express a limited set of receptors [39,40]. Patients with Omenn Syndrome suffer from chronic infections, alopecia, lymphopenia, diarrhea, and autoimmune problems presumably caused by the inappropriately activated T cells [39,41].
Although the mechanism by which the RAG2-RAG1 complexes are targeted to the correct receptor gene segments remains to be clarified, evidence has linked it to the status of transcription and histone modifications at appropriate recombining loci. Gene segments poised to undergo V(D)J recombination are usually actively transcribed prior to recombination, and are often marked by H3/H4 acetylation and H3K4 methylation [35,42–45]. In addition, recent evidence demonstrated that the RAG2 PHD finger specifically recognizes and binds to the H3K4me3 marks enriched in the V(D)J segments poised to undergo recombination [23,24,46]. Although the RAG2 PHD finger was dispensable for in vitro recombination assays, it was essential for efficient V(D)J recombination in vivo because deletion of the PHD finger resulted in a reduced V(D)J recombination frequency (~20–40% of that for wildtype RAG2) [23,47]. The reduction in H3K4me levels by knocking down WDR5 or over-expressing SMCX in human HT1080 fibroblasts also reduced RAG2 recombination activity [23].
Strikingly, out of the 24 known RAG2 mutations linked to SCID or Omenn Syndrome, six are located within the RAG2 PHD finger domain (Figure 2A), and the severity with which these mutations disrupt the RAG2-H3K4me3 interaction often correlates with the severity of the disease [38,40,48–50]. Mutation W453R (Figure 2B–C), which is found in patients with the less severe form of immunodeficiency, Omenn Syndrome [48,49], targets a highly conserved aromatic residue that participates in the H3K4me3-RAG2 interaction. This mutation destabilizes the H3K4me3 interaction and reduces RAG2 recombination activity in vivo without the perturbation of RAG2 PHD folding [23,24]. Mutations K440N and W416L, which are also linked to Omenn Syndrome [40], have not been tested for recombination activity. These mutations may interfere with either the positioning stability of Y415 in the H3K4me3-binding hydrophobic channel or the hydrogen-bond interaction with the H3 peptide, thus negatively affecting the affinity for H3K4me3-binding (Figure 2C) [24]. Mutations C478Y and H481P, which are linked to the more severe form of immunodeficiency, T-B-SCID [38,49], disrupt residues involving Zn2+ ion coordination, and probably destabilize overall protein folding [24]. Residue N474 forms a hydrogen bond with the N-terminus of histone H3 [24]. Mutation N474S has also been linked to T-B-SCID [50], however it remains unclear whether or not this mutation affects H3K4me3 binding.
Taken together, these results support a general model wherein the interaction between the RAG2 PHD finger and H3K4me3 helps to recruit and/or stabilize the association of the RAG recombinase to gene segments poised to undergo V(D)J recombination, while histone acetylation alters local chromatin accessibility (Figure 2D).
3.2. The autoimmune regulator protein (AIRE)
Mutations in the PHD-containing protein Autoimmune Regulator (AIRE), are associated with autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy (APECED), also known as autoimmune polyglandular syndrome type I [51,52]. APECED patients suffer from various autoimmune problems, due to a failure to negatively select self-recognizing T cells, a process that requires the “promiscuous” expression of tissue restricted antigen (TRA) genes in medullary epithelial cells (MECs) in the thymus [53]. AIRE is crucial to this process because it is required for expression of a subset of the TRA genes in the MECs [54,55]. AIRE was shown to have transcriptional activating properties and to associate with the histone acetyltransferase CBP [56]. In addition, the first PHD finger of AIRE has recently been shown to bind to unmethylated H3K4 (H3K4me0) with a Kd of ~5 µM, and high levels of AIRE binding at the promoters of TRA genes were correlated with low levels of H3K4me3, supporting a model in which PHD-chromatin interactions recruit AIRE/CBP to the TRA promoters to activate those genes [30].
Over 50 APECED-associated mutations in AIRE have been identified, including many point mutations targeting the two PHD domains and truncations lacking one or both PHDs [57–63]. The disease-associated mutation C311Y [57] abolished the interaction between the first PHD finger of AIRE and H3K4me0 [30] by ablating a Zn2+-coordinating cysteine and likely perturbing the fold stability [64]. Mutations P326L and P326Q [57,59] were also reported to perturb the structure of AIRE PHD finger [64]. Interestingly, the disease-associated mutation V301M [60], which is located in a solvent exposed patch on the outside of AIRE PHD1, had no effect on the PHD folding or H3K4me0 interaction, and may perturb some unknown function [30,64]. Mutations R303P (in the first PHD finger) [61] and C446G (in the second PHD finger) [62] are also reported in APECED patients, but how these mutations contribute to APECED remains unclear.
4. PHD finger dysregulation in cancers
Mutations in genes encoding PHD finger-containing factors are not only intimately involved in immune diseases, but also strongly associated with the pathogenesis of various cancers.
4.1. INhibitor of Growth 1 (ING1)
ING1, the ING family founding member, was initially isolated from an elegant screen for tumor suppressor genes [65]. This function was later verified in ING1−/− mice, which exhibit hypersensitivity to gamma-radiation and predisposition to lymphomas [66]. Consistent with their putative tumor suppressive roles, the reduced mRNA expression, allelic loss, or somatic mutation of ING family members (especially ING1, ING3 and ING4), have been reported among many types of human cancers including breast cancer, gastric cancer, melanoma, glioma, esophageal squamous cell carcinoma (SSC), and head and neck SSC [19,67,68]. While the underlying mechanisms are still poorly understood, ING proteins have been linked to many aspects of oncogenesis and cellular growth control, such as cell cycle regulation, senescence, DNA damage repair, apoptosis, and stress signaling [67,69]. In addition to interacting with p53 and PCNA, INGs also recruit and associate with two antagonizing sets of enzymes, histone acetyltransferases (HATs) and histone deacetylases (HDACs) [69,70]. The incorporation of ING proteins into HAT- or HDAC-complexes is conserved in lower organisms such as yeast [27,71].
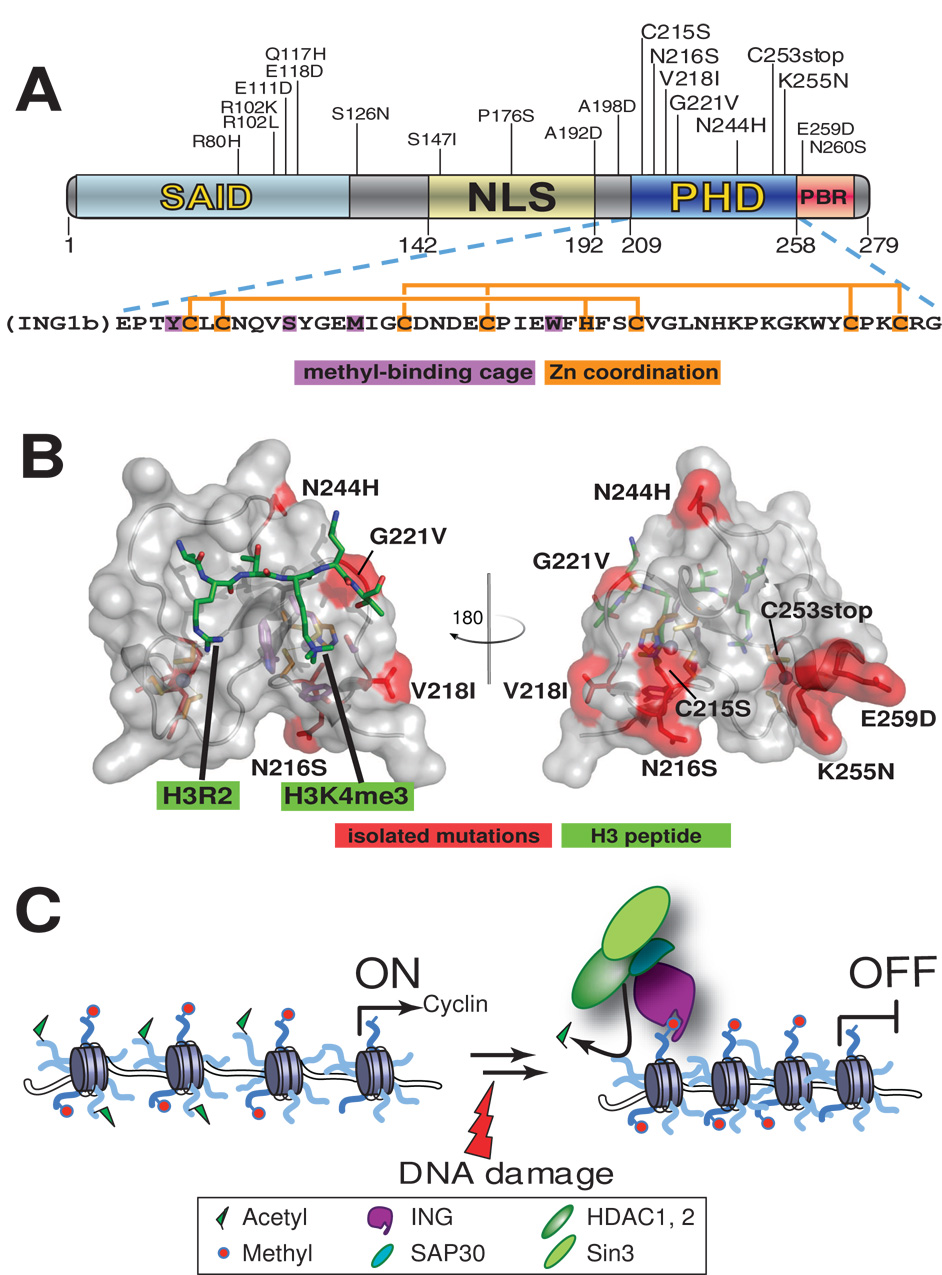
Virtually all ING isoforms contain a conserved C-terminal PHD finger and nearby nuclear localization signals, perhaps suggesting that their functionality relies on a nuclear PHD motif [69]. ING PHD fingers have been shown to recognize H3K4me3/2 specifically [13,27,69]. Mutations within the ING1 PHD finger were identified among various cancers (Figure 3A) [13,67,72–75]. Among them, mutation C253stop [72] results in a truncated PHD domain, and mutation C215S disrupts Zn2+-coordination, presumably disrupting the overall PHD structure and abolishing the H3K4me3 interaction needed for recruitment to target promoters (Figure 3B). Though other PHD mutations are mapped to the outside of the H3K4me3-binding cage, it remains to be seen if any of them interferes with the H3K4me3 binding or other unknown function. Interestingly, missense mutations also cluster in the nuclear localization signals and an N-terminal sequence that overlaps the SAP30-interacting domain [76] (Figure 3A), which may interfere with proper nuclear localization or with SAP30 association. These observations are consistent with a model that upon DNA damage, the Sin3/HDAC repressive complexes are recruited to the promoters of cell cycle regulators such as Cyclin via ING-SAP30 interaction, repressing the transcription of these genes and preventing cell cycle progression (Figure 3C) [10,77,78].
Figure 3. Mutations in the ING1b PHD finger isolated from cancers.
(A) Domain structure of ING1b (p33ING1b, NCBI accession number 38201667) showing missense mutations reported in various solid cancers. The amino acid sequence of PHD finger is shown below, with the same color depiction as in Figure 2. SAID, SAP30-interacting domain [76]; NLS, nuclear localization signal; PBR, poly-basic region that mediates the binding to phosphoinositide [69].
(B) A structural model of f the ING1b PHD finger (in gray) associated with an H3K4me3 peptide (in green). Cancer-associated missense mutations as shown in red, and aromatic cage residues are in purple. This model is based on an co-crystal structure of the ING2 PHD finger and H3K4me3 peptide [13].
(C) A schematic model for ING-mediated cell cycle arrest. Upon DNA damage, the PHD finger of INGs that recognizes H3K4me3 marks enriched in promoters helps to stabilize the targeting of INGs to proliferative genes such as Cyclin wherein INGs subsequently recruit the SAP30/Sin3/HDAC complexes that remove acetyl marks from histone tails, repressing the transcription of proliferative genes and decelerating cell cycle progression.
4.2. PHD fingers fused to NUP98 in blood cancers
Translocation of the Nucleoporin 98 (NUP98) gene represents one of the most promiscuous chromosomal abnormalities in human hematopoietic malignancies such as acute myeloid leukemia (AML) [79]. NUP98, a nuclear pore complex (NPC) component, has been reported to shuttle between the NPC and a specialized nuclear body that associates with active transcription [80]. All leukemic NUP98 fusion proteins retain the N-terminal NUP98 FG-repeats, which recruit p300/CBP and harbor transcriptional activation activities [81].
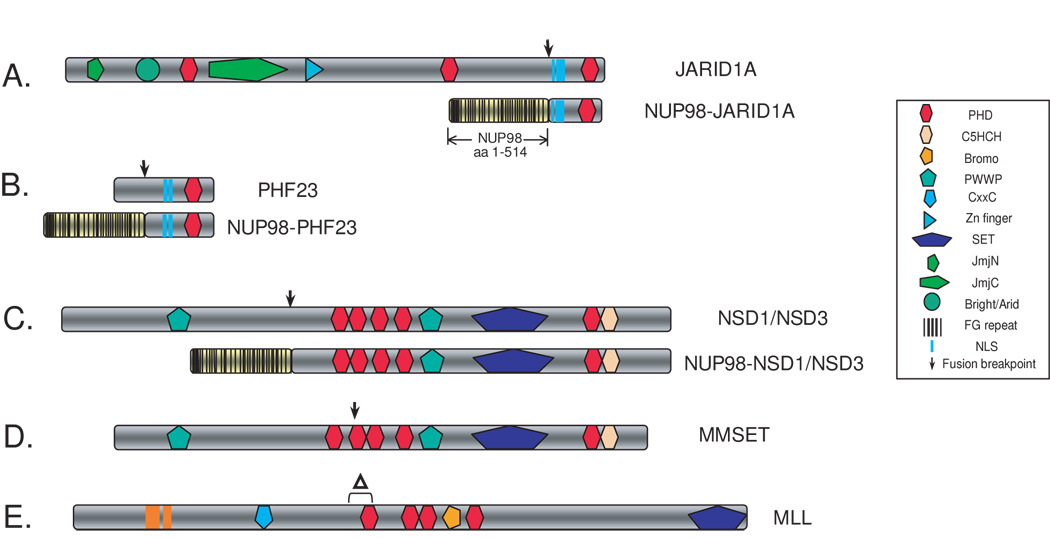
In rare AML cases, cryptic translocations fuse the NUP98 FG-repeats to the C-terminus of PHD finger-containing factors such as the H3K4 demethylase JARID1A (Jumonji, AT-rich interactive domain 1A) (Figure 4A) or PHF23 (PHD finger-protein 23) (Figure 4B) [82–84]. The two fusion products share a high similarity, and the only functional motifs incorporated from JARID1A or PHF23 are the PHD finger and nuclear localization signals, indicating a common leukemogenic mechanism. Little is known about the chromatin-associating properties of these PHD fingers, except that the PHD finger of PHF23 shares similarity to H3K4me-binding PHD fingers [5]. Furthermore, the mechanistic contribution of these PHDs to malignancies is poorly understood.
Figure 4. Chromosomal aberrations that target PHD finger-containing proteins in hematopoietic malignancies.
The NUP98 gene was found translocated to genes encoding the PHD finger-containing proteins JARID1A (panel A), PHF23 (panel B), NSD1 and NSD3 (panel C) in as subset of AML cases, and as a result, the leukemic fusion proteins harbor one or more PHD fingers. (D) MMSET, another PHD finger-containing protein, is commonly translocated in multiple myeloma. (E) Internal deletion (Δ) in MLL seen in T-cell leukaemia cases causes the disruption of the first PHD finger. Motif identities are specified in the boxed panel, and chromosomal translocation breakpoints are designated with arrows. The positions of nuclear localization signals (NLS) are only shown for panels A–B.
In ~5% of AML cases, Nuclear receptor-binding SET domain-containing 1 (NSD1) and the related gene NSD3 were found fused to NUP98 [85,86], and the fusion products retain a large part of NSD that contains the five PHD fingers, the proline-tryptophan-proline-tryptophan (PWWP) motif and the SET (SU[VAR]3–9,E[Z],trithorax) domain (Figure 4C). NSD1 was isolated as a versatile interacting partner of nuclear hormone receptors, acting as co-repressor or co-activator depending on the cellular context [87]. NSD1 and its related genes (MMSET/NSD2/WHSC1, NSD3/WHSC1L1) have been linked to various human diseases including myeloid leukemia, multiple myeloma and childhood overgrowth syndrome [88,89]. NUP98-NSD1 has been shown to be a potent oncoprotein, efficiently transforming myeloid progenitors and inducing leukemia in murine models where NUP98-NSD1 directly binds to and activates various HOX genes [90]. Interestingly, while the NUP98 portion and the SET domain are required to maintain HOX gene activation, the fifth NSD1 PHD finger and a nearby cysteine/histidine-rich C5HCH motif were essential for recruiting the fusion protein to target promoters [90]. How the NSD1 PHD fingers contribute to promoter recruitment is unclear, but the sequences of these domains lack the critical H3K4-engaging residues seen in the known H3K4-binding PHDs, suggesting that they may interact with chromatin using a novel mechanism.
4.3. Mixed lineage leukaemia gene (MLL)
MLL and its Drosophila homologue Trithorax are required for the maintenance of HOX gene expression [91]. In addition to a C-terminal SET domain that specifically methylates H3K4, MLL harbors multiple chromatin-associated motifs, including four PHD fingers, AT-hooks, CxxC motifs, and an atypical bromodomain. While MLL translocation is among the most common causes of leukemias, internal deletion of exon 8 that excludes critical cysteine residues of the first PHD finger without changing the reading frame was found in rare cases of acute lymphoblastic T-cell leukemia (Figure 4E) [91,92]. How this internal deletion alters the function of MLL and whether it is sufficient to promote leukemogenesis have yet to be determined.
4.4 Other PHD proteins implicated in cancer
While we have discussed cases where PHD finger dysregulation is clearly associated with malignancies, the literature also documents many cases where tumorigenesis is associated with dysregulation of genes encoding PHD finger proteins, without an apparent link to the PHD fingers per se. For example, _M_ultiple _M_yeloma _SET_-domain protein (MMSET, aka, NSD2) was found translocated to the immunoglobin locus in >15% of multiple myeloma cases, which leads to over-expression of truncated forms of MMSET containing PHD fingers and a PWWP domain [89]. PHD finger protein 1 (PHF1), which harbors two PHD fingers and a tudor domain, was found rearranged in endometrial stromal sarcoma, a malignant tumor of endometrial stromal cells [93]. As a result, full-length PHF1, fused to the promoter of JAZF1 and its zinc finger, is ectopically expressed in the endometrium [93]. PHF1 shares a similarity to the Drosophila Polycomb-like proteins, and has recently been shown to be a novel component of EZH2 Polycomb complexes and required for global H3K27me [94]. Interestingly, Ezh2 itself has been reported to be overexpressed in various cancers [19].
5. PHD finger dysregulation in neurological disorders
5.1. NSD1
NSD1 mutation causes not only leukemias but also Sotos syndrome, a childhood overgrowth syndrome characterized by pre- and postnatal somatic overgrowth with facial dysmorphism, advanced bone age, seizures, and mental retardation [88,95,96]. NSD1 mutations are also detected in Weaver syndrome, a disorder phenotypically overlapping with Sotos syndrome [96]. NSD1−/− mice die early with defects in gastrulation and embryonic development, suggesting this protein plays a key role in development [97]. In Sotos syndrome, NSD1 missense mutations occur only in the SET, PWWP, PHD and cysteine/histidine-rich C5HCH domain, indicating that these domains are crucial to the proper function of NSD1 [96]. Strikingly, mutations in the PHD domains exhibit a very strong bias toward the Zn2+-coordinating cysteines/histidines, with mutations targeting 15 different cysteines/histidines, which are likely to disrupt PHD folding [88]. In Weaver syndrome, NSD1 mutations cluster within the fifth PHD and the adjacent C5HCH motif [96], the same motifs required for tethering NUP98-NSD1 to target promoters in myeloid leukemic cells [90], indicating a mechanistic commonality underlying these diseases.
5.2. Alpha Thalassaemia and Mental Retardation Syndrome, X-linked (ATRX)
Mutations in ATRX are associated with the ATR-X syndrome, a disorder characterized by severe mental retardation, genital abnormalities, alpha thalassaemia, microcephaly, seizures, and growth retardation [98–100]. ATRX is crucial to neuronal survival in the developing mouse brain, and this function may explain why ATRX mutations associate with mental retardation [101]. ATRX interacts with the heterochromatin-associated proteins HP1alpha, EZH2 and MeCP2, and has been implicated in chromatin remodeling and gene regulation [102–107]. However, its exact function and contribution to ATR-X syndrome remain unclear.
ATRX features an N-terminal ADD (_A_TRX-_D_NMT3-_D_NMT3L) domain, containing a GATA-like zinc finger, an atypical “PHD-like” domain and an alpha helix [102,103]. The ATRX PHD lacks the aromatic caging residues found in the PHD fingers of BPTF or RAG2, indicating that it might interact with chromatin by a different mechanism [102]. Over 40 disease-causing ATRX mutations have been identified within the ADD domain, including 26 in the PHD finger itself [102]. Some mutations may disrupt the folding of the PHD domain and probably destabilize the entire ATRX protein, while other mutations that lie on the surface of the fold probably do not affect PHD structure, and may cause disease by interfering with other unknown interactions or functions [102]. Future research focusing on the function of the ATRX PHD finger will provide valuable insights of the molecular pathogenesis of ATR-X syndrome.
5.3. CREB Binding Protein (CBP/CREBBP) and PHD Finger Protein 6 (PHF6)
Haploinsufficiency of CBP leads to Rubenstein-Taybi Syndrome, a disorder characterized by mental retardation, facial abnormalities, and growth retardation [108]. Many disease-associated mutations have been reported in CBP, including point mutations or internal deletions in the PHD domain [109]. Mutations in the PHF finger protein 6 (PHF6) are associated with Börjeson-Forssman-Lehmann Syndrome, a recessive X-linked disorder characterized by mental retardation, hypogonadism, hypometabolism and obesity [110,111], and one disease-associated mutation (C99F) targets the first PHD finger of PHF6 [110]. While both CBP and PHF6 are implicated in chromatin regulation and neural development, the exact mechanisms underlying their respective neurological disorders are still poorly understood [111,112].
6. Conclusions
In summary, PHD finger mutations have been associated with a wide variety of human diseases. It should be noted that, while evidence supporting a causal role of PHD finger mutation in the pathogenesis of some diseases is convincing (such as RAG2-PHD finger mutation in immunodeficiency syndrome [23]), direct cause-effect relationships for some other diseases still remain to be established and beg for further investigation. In this review, we have chosen to focus on diseases linked to aberrations of the PHD domains themselves. However, there are many other PHD-containing proteins for which mutations in other regions of the protein or loss of the entire protein associate with diseases, and the PHD domains of many of these proteins are likely to contribute to appropriate chromatin targeting. For instance, mutations in SMCX are associated with X-linked mental retardation, and the protein contains several PHD fingers thought to be important for regulating neural development genes [31]. In addition, PHD fingers often occur in conjunction with other chromatin-reading motifs, and such combined action is proposed to be required for precisely interpreting complicated chromatin modification patterns, a phenomenon known as “multivalency” [7]. The contribution of PHD fingers and other chromatin-“reading” motifs to the functional consequences of the epigenetic marks and ultimately, to human diseases is a topic that has only recently received scrutiny. Understanding the regulatory roles of chromatin modifications in the context of human disease will greatly broaden our mechanistic appreciation of normal and pathological development.
Acknowledgements
We thank Drs Alexander Ruthenberg, Ping Chi and Chengran Xu for critical reading and advice on this manuscript. Special thanks to Dr. Alex Ruthenberg for analyzing disease-associated missense mutations in RAG2 and ING structure, and thanks to Dr. Haotai Li for helping on illustration. We apologize to all those whose work we could not cite due to space constraints. C.D.A. is supported by the NIH Merit Grant GM 53512, and L.A.B. by NIH training grant CA09673. G.W. is supported by a C.H.Li Memorial Scholar Fund Award and a Leukemia & Lymphoma Society Postdoctoral Fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 2.Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 3.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 4.Shahbazian MD, Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- 5.Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 6.Kouzarides T. SnapShot: Histone-Modifying Enzymes. Cell. 2007;131:822. doi: 10.1016/j.cell.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 7.Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Biol. 2007;8:983–994. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol. 2007;14:1025–1040. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, Wu C, Allis CD. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 10.Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, Carney D, Pena P, Lan F, Kaadige MR, Lacoste N, Cayrou C, Davrazou F, Saha A, Cairns BR, Ayer DE, Kutateladze TG, Shi Y, Cote J, Chua KF, Gozani O. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD, Patel DJ. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442:91–95. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li H, Fischle W, Wang W, Duncan EM, Liang L, Murakami-Ishibe S, Allis CD, Patel DJ. Structural basis for lower lysine methylation state-specific readout by MBT repeats of L3MBTL1 and an engineered PHD finger. Mol Cell. 2007;28:677–691. doi: 10.1016/j.molcel.2007.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pena PV, Davrazou F, Shi X, Walter KL, Verkhusha VV, Gozani O, Zhao R, Kutateladze TG. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature. 2006;442:100–103. doi: 10.1038/nature04814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, Cheng X, Bestor TH. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lan F, Collins RE, De Cegli R, Alpatov R, Horton JR, Shi X, Gozani O, Cheng X, Shi Y. Recognition of unmethylated histone H3 lysine 4 links BHC80 to LSD1-mediated gene repression. Nature. 2007;448:718–722. doi: 10.1038/nature06034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang K, Mosch K, Fischle W, Grewal SI. Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat Struct Mol Biol. 2008;15:381–388. doi: 10.1038/nsmb.1406. [DOI] [PubMed] [Google Scholar]
- 17.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 18.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang GG, Allis CD, Chi P. Chromatin remodeling and cancer, Part I: Covalent histone modifications. Trends Mol Med. 2007;13:363–372. doi: 10.1016/j.molmed.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 20.Wang GG, Allis CD, Chi P. Chromatin remodeling and cancer, Part II: ATP-dependent chromatin remodeling. Trends Mol Med. 2007;13:373–380. doi: 10.1016/j.molmed.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bienvenu T, Chelly J. Molecular genetics of Rett syndrome: when DNA methylation goes unrecognized. Nat Rev Genet. 2006;7:415–426. doi: 10.1038/nrg1878. [DOI] [PubMed] [Google Scholar]
- 22.Bienz M. The PHD finger, a nuclear protein-interaction domain. Trends Biochem Sci. 2006;31:35–40. doi: 10.1016/j.tibs.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 23.Matthews AG, Kuo AJ, Ramon-Maiques S, Han S, Champagne KS, Ivanov D, Gallardo M, Carney D, Cheung P, Ciccone DN, Walter KL, Utz PJ, Shi Y, Kutateladze TG, Yang W, Gozani O, Oettinger MA. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 2007;450:1106–1110. doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramon-Maiques S, Kuo AJ, Carney D, Matthews AG, Oettinger MA, Gozani O, Yang W. The plant homeodomain finger of RAG2 recognizes histone H3 methylated at both lysine-4 and arginine-2. Proc Natl Acad Sci U S A. 2007;104:18993–18998. doi: 10.1073/pnas.0709170104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schindler U, Beckmann H, Cashmore AR. HAT3.1, a novel Arabidopsis homeodomain protein containing a conserved cysteine-rich region. Plant J. 1993;4:137–150. doi: 10.1046/j.1365-313x.1993.04010137.x. [DOI] [PubMed] [Google Scholar]
- 26.Palacios A, Munoz IG, Pantoja-Uceda D, Marcaida MJ, Torres D, Martin-Garcia JM, Luque I, Montoya G, Blanco FJ. Molecular basis of histone H3K4ME3 recognition by ING4. J Biol Chem. 2008 doi: 10.1074/jbc.M710020200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taverna SD, Ilin S, Rogers RS, Tanny JC, Lavender H, Li H, Baker L, Boyle J, Blair LP, Chait BT, Patel DJ, Aitchison JD, Tackett AJ, Allis CD. Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs. Mol Cell. 2006;24:785–796. doi: 10.1016/j.molcel.2006.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iberg AN, Espejo A, Cheng D, Kim D, Michaud-Levesque J, Richard S, Bedford MT. Arginine methylation of the histone H3 tail impedes effector binding. J Biol Chem. 2008;283:3006–3010. doi: 10.1074/jbc.C700192200. [DOI] [PubMed] [Google Scholar]
- 29.Kirmizis A, Santos-Rosa H, Penkett CJ, Singer MA, Vermeulen M, Mann M, Bahler J, Green RD, Kouzarides T. Arginine methylation at histone H3R2 controls deposition of H3K4 trimethylation. Nature. 2007 doi: 10.1038/nature06160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Org T, Chignola F, Hetenyi C, Gaetani M, Rebane A, Liiv I, Maran U, Mollica L, Bottomley MJ, Musco G, Peterson P. The autoimmune regulator PHD finger binds to non-methylated histone H3K4 to activate gene expression. EMBO Rep. 2008;9:370–376. doi: 10.1038/embor.2008.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iwase S, Lan F, Bayliss P, de la Torre-Ubieta L, Huarte M, Qi HH, Whetstine JR, Bonni A, Roberts TM, Shi Y. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell. 2007;128:1077–1088. doi: 10.1016/j.cell.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 32.Karagianni P, Amazit L, Qin J, Wong J. ICBP90, a novel methyl K9 H3 binding protein linking protein ubiquitination with heterochromatin formation. Mol Cell Biol. 2008;28:705–717. doi: 10.1128/MCB.01598-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi X, Kachirskaia I, Walter KL, Kuo JH, Lake A, Davrazou F, Chan SM, Martin DG, Fingerman IM, Briggs SD, Howe L, Utz PJ, Kutateladze TG, Lugovskoy AA, Bedford MT, Gozani O. Proteome-wide analysis in Saccharomyces cerevisiae identifies several PHD fingers as novel direct and selective binding modules of histone H3 methylated at either lysine 4 or lysine 36. J Biol Chem. 2007;282:2450–2455. doi: 10.1074/jbc.C600286200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jung D, Alt FW. Unraveling V(D)J recombination; insights into gene regulation. Cell. 2004;116:299–311. doi: 10.1016/s0092-8674(04)00039-x. [DOI] [PubMed] [Google Scholar]
- 35.Oettinger MA. How to keep V(D)J recombination under control. Immunol Rev. 2004;200:165–181. doi: 10.1111/j.0105-2896.2004.00172.x. [DOI] [PubMed] [Google Scholar]
- 36.Fugmann SD, Lee AI, Shockett PE, Villey IJ, Schatz DG. The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annu Rev Immunol. 2000;18:495–527. doi: 10.1146/annurev.immunol.18.1.495. [DOI] [PubMed] [Google Scholar]
- 37.Shinkai Y, Rathbun G, Lam KP, Oltz EM, Stewart V, Mendelsohn M, Charron J, Datta M, Young F, Stall AM, et al. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell. 1992;68:855–867. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]
- 38.Schwarz K, Gauss GH, Ludwig L, Pannicke U, Li Z, Lindner D, Friedrich W, Seger RA, Hansen-Hagge TE, Desiderio S, Lieber MR, Bartram CR. RAG mutations in human B cell-negative SCID. Science. 1996;274:97–99. doi: 10.1126/science.274.5284.97. [DOI] [PubMed] [Google Scholar]
- 39.Marrella V, Poliani PL, Sobacchi C, Grassi F, Villa A. Of Omenn and mice. Trends Immunol. 2008;29:133–140. doi: 10.1016/j.it.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 40.Sobacchi C, Marrella V, Rucci F, Vezzoni P, Villa A. RAG-dependent primary immunodeficiencies. Hum Mutat. 2006;27:1174–1184. doi: 10.1002/humu.20408. [DOI] [PubMed] [Google Scholar]
- 41.Omenn GS. Familial Reticuloendotheliosis with Eosinophilia. N Engl J Med. 1965;273:427–432. doi: 10.1056/NEJM196508192730806. [DOI] [PubMed] [Google Scholar]
- 42.Goldmit M, Ji Y, Skok J, Roldan E, Jung S, Cedar H, Bergman Y. Epigenetic ontogeny of the Igk locus during B cell development. Nat Immunol. 2005;6:198–203. doi: 10.1038/ni1154. [DOI] [PubMed] [Google Scholar]
- 43.Morshead KB, Ciccone DN, Taverna SD, Allis CD, Oettinger MA. Antigen receptor loci poised for V(D)J rearrangement are broadly associated with BRG1 and flanked by peaks of histone H3 dimethylated at lysine 4. Proc Natl Acad Sci U S A. 2003;100:11577–11582. doi: 10.1073/pnas.1932643100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sleckman BP, Gorman JR, Alt FW. Accessibility control of antigen-receptor variable-region gene assembly: role of cis-acting elements. Annu Rev Immunol. 1996;14:459–481. doi: 10.1146/annurev.immunol.14.1.459. [DOI] [PubMed] [Google Scholar]
- 45.Krangel MS. Gene segment selection in V(D)J recombination: accessibility and beyond. Nat Immunol. 2003;4:624–630. doi: 10.1038/ni0703-624. [DOI] [PubMed] [Google Scholar]
- 46.Liu Y, Subrahmanyam R, Chakraborty T, Sen R, Desiderio S. A plant homeodomain in RAG-2 that binds Hypermethylated lysine 4 of histone H3 is necessary for efficient antigen-receptor-gene rearrangement. Immunity. 2007;27:561–571. doi: 10.1016/j.immuni.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Elkin SK, Ivanov D, Ewalt M, Ferguson CG, Hyberts SG, Sun ZY, Prestwich GD, Yuan J, Wagner G, Oettinger MA, Gozani OP. A PHD finger motif in the C terminus of RAG2 modulates recombination activity. J Biol Chem. 2005;280:28701–28710. doi: 10.1074/jbc.M504731200. [DOI] [PubMed] [Google Scholar]
- 48.Gomez CA, Ptaszek LM, Villa A, Bozzi F, Sobacchi C, Brooks EG, Notarangelo LD, Spanopoulou E, Pan ZQ, Vezzoni P, Cortes P, Santagata S. Mutations in conserved regions of the predicted RAG2 kelch repeats block initiation of V(D)J recombination and result in primary immunodeficiencies. Mol Cell Biol. 2000;20:5653–5664. doi: 10.1128/mcb.20.15.5653-5664.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Noordzij JG, de Bruin-Versteeg S, Verkaik NS, Vossen JM, de Groot R, Bernatowska E, Langerak AW, van Gent DC, van Dongen JJ. The immunophenotypic and immunogenotypic B-cell differentiation arrest in bone marrow of RAG-deficient SCID patients corresponds to residual recombination activities of mutated RAG proteins. Blood. 2002;100:2145–2152. [PubMed] [Google Scholar]
- 50.Villa A, Sobacchi C, Notarangelo LD, Bozzi F, Abinun M, Abrahamsen TG, Arkwright PD, Baniyash M, Brooks EG, Conley ME, Cortes P, Duse M, Fasth A, Filipovich AM, Infante AJ, Jones A, Mazzolari E, Muller SM, Pasic S, Rechavi G, Sacco MG, Santagata S, Schroeder ML, Seger R, Strina D, Ugazio A, Valiaho J, Vihinen M, Vogler LB, Ochs H, Vezzoni P, Friedrich W, Schwarz K. V(D)J recombination defects in lymphocytes due to RAG mutations: severe immunodeficiency with a spectrum of clinical presentations. Blood. 2001;97:81–88. doi: 10.1182/blood.v97.1.81. [DOI] [PubMed] [Google Scholar]
- 51.An autoimmune disease, APECED, caused by mutations in a novel gene featuring two PHD-type zinc-finger domains. Nat Genet. 1997;17:399–403. [Google Scholar]
- 52.Nagamine K, Peterson P, Scott HS, Kudoh J, Minoshima S, Heino M, Krohn KJ, Lalioti MD, Mullis PE, Antonarakis SE, Kawasaki K, Asakawa S, Ito F, Shimizu N. Positional cloning of the APECED gene. Nat Genet. 1997;17:393–398. [Google Scholar]
- 53.Cheng MH, Shum AK, Anderson MS. What's new in the Aire? Trends Immunol. 2007;28:321–327. doi: 10.1016/j.it.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 54.Anderson MS, Venanzi ES, Klein L, Chen Z, Berzins SP, Turley SJ, von Boehmer H, Bronson R, Dierich A, Benoist C, Mathis D. Projection of an immunological self shadow within the thymus by the aire protein. Science. 2002;298:1395–1401. doi: 10.1126/science.1075958. [DOI] [PubMed] [Google Scholar]
- 55.Derbinski J, Gabler J, Brors B, Tierling S, Jonnakuty S, Hergenhahn M, Peltonen L, Walter J, Kyewski B. Promiscuous gene expression in thymic epithelial cells is regulated at multiple levels. J Exp Med. 2005;202:33–45. doi: 10.1084/jem.20050471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pitkanen J, Doucas V, Sternsdorf T, Nakajima T, Aratani S, Jensen K, Will H, Vahamurto P, Ollila J, Vihinen M, Scott HS, Antonarakis SE, Kudoh J, Shimizu N, Krohn K, Peterson P. The autoimmune regulator protein has transcriptional transactivating properties and interacts with the common coactivator CREB-binding protein. J Biol Chem. 2000;275:16802–16809. doi: 10.1074/jbc.M908944199. [DOI] [PubMed] [Google Scholar]
- 57.Bjorses P, Halonen M, Palvimo JJ, Kolmer M, Aaltonen J, Ellonen P, Perheentupa J, Ulmanen I, Peltonen L. Mutations in the AIRE gene: effects on subcellular location and transactivation function of the autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy protein. Am J Hum Genet. 2000;66:378–392. doi: 10.1086/302765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ruan QG, She JX. Autoimmune polyglandular syndrome type 1 and the autoimmune regulator. Clin Lab Med. 2004;24:305–317. doi: 10.1016/j.cll.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 59.Saugier-Veber P, Drouot N, Wolf LM, Kuhn JM, Frebourg T, Lefebvre H. Identification of a novel mutation in the autoimmune regulator (AIRE-1) gene in a French family with autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy. Eur J Endocrinol. 2001;144:347–351. doi: 10.1530/eje.0.1440347. [DOI] [PubMed] [Google Scholar]
- 60.Soderbergh A, Rorsman F, Halonen M, Ekwall O, Bjorses P, Kampe O, Husebye ES. Autoantibodies against aromatic L-amino acid decarboxylase identifies a subgroup of patients with Addison's disease. J Clin Endocrinol Metab. 2000;85:460–463. doi: 10.1210/jcem.85.1.6266. [DOI] [PubMed] [Google Scholar]
- 61.Stolarski B, Pronicka E, Korniszewski L, Pollak A, Kostrzewa G, Rowinska E, Wlodarski P, Skorka A, Gremida M, Krajewski P, Ploski R. Molecular background of polyendocrinopathy-candidiasis-ectodermal dystrophy syndrome in a Polish population: novel AIRE mutations and an estimate of disease prevalence. Clin Genet. 2006;70:348–354. doi: 10.1111/j.1399-0004.2006.00690.x. [DOI] [PubMed] [Google Scholar]
- 62.Wolff AS, Erichsen MM, Meager A, Magitta NF, Myhre AG, Bollerslev J, Fougner KJ, Lima K, Knappskog PM, Husebye ES. Autoimmune polyendocrine syndrome type 1 in Norway: phenotypic variation, autoantibodies, and novel mutations in the autoimmune regulator gene. J Clin Endocrinol Metab. 2007;92:595–603. doi: 10.1210/jc.2006-1873. [DOI] [PubMed] [Google Scholar]
- 63.Peterson P, Pitkanen J, Sillanpaa N, Krohn K. Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy (APECED): a model disease to study molecular aspects of endocrine autoimmunity. Clin Exp Immunol. 2004;135:348–357. doi: 10.1111/j.1365-2249.2004.02384.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bottomley MJ, Stier G, Pennacchini D, Legube G, Simon B, Akhtar A, Sattler M, Musco G. NMR structure of the first PHD finger of autoimmune regulator protein (AIRE1). Insights into autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy (APECED) disease. J Biol Chem. 2005;280:11505–11512. doi: 10.1074/jbc.M413959200. [DOI] [PubMed] [Google Scholar]
- 65.Garkavtsev I, Kazarov A, Gudkov A, Riabowol K. Suppression of the novel growth inhibitor p33ING1 promotes neoplastic transformation. Nat Genet. 1996;14:415–420. doi: 10.1038/ng1296-415. [DOI] [PubMed] [Google Scholar]
- 66.Kichina JV, Zeremski M, Aris L, Gurova KV, Walker E, Franks R, Nikitin AY, Kiyokawa H, Gudkov AV. Targeted disruption of the mouse ing1 locus results in reduced body size, hypersensitivity to radiation and elevated incidence of lymphomas. Oncogene. 2006;25:857–866. doi: 10.1038/sj.onc.1209118. [DOI] [PubMed] [Google Scholar]
- 67.Campos EI, Chin MY, Kuo WH, Li G. Biological functions of the ING family tumor suppressors. Cell Mol Life Sci. 2004;61:2597–2613. doi: 10.1007/s00018-004-4199-4. [DOI] [PubMed] [Google Scholar]
- 68.Shi X, Gozani O. The fellowships of the INGs. J Cell Biochem. 2005;96:1127–1136. doi: 10.1002/jcb.20625. [DOI] [PubMed] [Google Scholar]
- 69.Soliman MA, Riabowol K. After a decade of study-ING, a PHD for a versatile family of proteins. Trends Biochem Sci. 2007;32:509–519. doi: 10.1016/j.tibs.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 70.Doyon Y, Cayrou C, Ullah M, Landry AJ, Cote V, Selleck W, Lane WS, Tan S, Yang XJ, Cote J. ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol Cell. 2006;21:51–64. doi: 10.1016/j.molcel.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 71.Doyon Y, Selleck W, Lane WS, Tan S, Cote J. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol Cell Biol. 2004;24:1884–1896. doi: 10.1128/MCB.24.5.1884-1896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Campos EI, Martinka M, Mitchell DL, Dai DL, Li G. Mutations of the ING1 tumor suppressor gene detected in human melanoma abrogate nucleotide excision repair. Int J Oncol. 2004;25:73–80. [PubMed] [Google Scholar]
- 73.Chen L, Matsubara N, Yoshino T, Nagasaka T, Hoshizima N, Shirakawa Y, Naomoto Y, Isozaki H, Riabowol K, Tanaka N. Genetic alterations of candidate tumor suppressor ING1 in human esophageal squamous cell cancer. Cancer Res. 2001;61:4345–4349. [PubMed] [Google Scholar]
- 74.Chen B, Campos EI, Crawford R, Martinka M, Li G. Analyses of the tumour suppressor ING1 expression and gene mutation in human basal cell carcinoma. Int J Oncol. 2003;22:927–931. [PubMed] [Google Scholar]
- 75.Gong W, Suzuki K, Russell M, Riabowol K. Function of the ING family of PHD proteins in cancer. Int J Biochem Cell Biol. 2005;37:1054–1065. doi: 10.1016/j.biocel.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 76.Kuzmichev A, Zhang Y, Erdjument-Bromage H, Tempst P, Reinberg D. Role of the Sin3-histone deacetylase complex in growth regulation by the candidate tumor suppressor p33(ING1) Mol Cell Biol. 2002;22:835–848. doi: 10.1128/MCB.22.3.835-848.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Garate M, Campos EI, Bush JA, Xiao H, Li G. Phosphorylation of the tumor suppressor p33(ING1b) at Ser-126 influences its protein stability and proliferation of melanoma cells. Faseb J. 2007;21:3705–3716. doi: 10.1096/fj.07-8069com. [DOI] [PubMed] [Google Scholar]
- 78.Takahashi M, Seki N, Ozaki T, Kato M, Kuno T, Nakagawa T, Watanabe K, Miyazaki K, Ohira M, Hayashi S, Hosoda M, Tokita H, Mizuguchi H, Hayakawa T, Todo S, Nakagawara A. Identification of the p33(ING1)-regulated genes that include cyclin B1 and proto-oncogene DEK by using cDNA microarray in a mouse mammary epithelial cell line NMuMG. Cancer Res. 2002;62:2203–2209. [PubMed] [Google Scholar]
- 79.Moore MA, Chung KY, Plasilova M, Schuringa JJ, Shieh JH, Zhou P, Morrone G. NUP98 dysregulation in myeloid leukemogenesis. Ann N Y Acad Sci. 2007;1106:114–142. doi: 10.1196/annals.1392.019. [DOI] [PubMed] [Google Scholar]
- 80.Griffis ER, Altan N, Lippincott-Schwartz J, Powers MA. Nup98 is a mobile nucleoporin with transcription-dependent dynamics. Mol Biol Cell. 2002;13:1282–1297. doi: 10.1091/mbc.01-11-0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kasper LH, Brindle PK, Schnabel CA, Pritchard CE, Cleary ML, van Deursen JM. CREB binding protein interacts with nucleoporin-specific FG repeats that activate transcription and mediate NUP98-HOXA9 oncogenicity. Mol Cell Biol. 1999;19:764–776. doi: 10.1128/mcb.19.1.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.van Zutven LJ, Onen E, Velthuizen SC, van Drunen E, von Bergh AR, van den Heuvel-Eibrink MM, Veronese A, Mecucci C, Negrini M, de Greef GE, Beverloo HB. Identification of NUP98 abnormalities in acute leukemia: JARID1A (12p13) as a new partner gene. Genes Chromosomes Cancer. 2006;45:437–446. doi: 10.1002/gcc.20308. [DOI] [PubMed] [Google Scholar]
- 83.Reader JC, Meekins JS, Gojo I, Ning Y. A novel NUP98-PHF23 fusion resulting from a cryptic translocation t(11;17)(p15;p13) in acute myeloid leukemia. Leukemia. 2007;21:842–844. doi: 10.1038/sj.leu.2404579. [DOI] [PubMed] [Google Scholar]
- 84.Klose RJ, Yan Q, Tothova Z, Yamane K, Erdjument-Bromage H, Tempst P, Gilliland DG, Zhang Y, Kaelin WG., Jr The retinoblastoma binding protein RBP2 is an H3K4 demethylase. Cell. 2007;128:889–900. doi: 10.1016/j.cell.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 85.Cerveira N, Correia C, Doria S, Bizarro S, Rocha P, Gomes P, Torres L, Norton L, Borges BS, Castedo S, Teixeira MR. Frequency of NUP98-NSD1 fusion transcript in childhood acute myeloid leukaemia. Leukemia. 2003;17:2244–2247. doi: 10.1038/sj.leu.2403104. [DOI] [PubMed] [Google Scholar]
- 86.Rosati R, La Starza R, Veronese A, Aventin A, Schwienbacher C, Vallespi T, Negrini M, Martelli MF, Mecucci C. NUP98 is fused to the NSD3 gene in acute myeloid leukemia associated with t(8;11)(p11.2;p15) Blood. 2002;99:3857–3860. doi: 10.1182/blood.v99.10.3857. [DOI] [PubMed] [Google Scholar]
- 87.Huang N, vom Baur E, Garnier JM, Lerouge T, Vonesch JL, Lutz Y, Chambon P, Losson R. Two distinct nuclear receptor interaction domains in NSD1, a novel SET protein that exhibits characteristics of both corepressors and coactivators. Embo J. 1998;17:3398–3412. doi: 10.1093/emboj/17.12.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tatton-Brown K, Douglas J, Coleman K, Baujat G, Cole TR, Das S, Horn D, Hughes HE, Temple IK, Faravelli F, Waggoner D, Turkmen S, Cormier-Daire V, Irrthum A, Rahman N. Genotype-phenotype associations in Sotos syndrome: an analysis of 266 individuals with NSD1 aberrations. Am J Hum Genet. 2005;77:193–204. doi: 10.1086/432082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Keats JJ, Maxwell CA, Taylor BJ, Hendzel MJ, Chesi M, Bergsagel PL, Larratt LM, Mant MJ, Reiman T, Belch AR, Pilarski LM. Overexpression of transcripts originating from the MMSET locus characterizes all t(4;14)(p16;q32)-positive multiple myeloma patients. Blood. 2005;105:4060–4069. doi: 10.1182/blood-2004-09-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang GG, Cai L, Pasillas MP, Kamps MP. NUP98-NSD1 links H3K36 methylation to Hox-A gene activation and leukaemogenesis. Nat Cell Biol. 2007;9:804–812. doi: 10.1038/ncb1608. [DOI] [PubMed] [Google Scholar]
- 91.Ayton PM, Cleary ML. Molecular mechanisms of leukemogenesis mediated by MLL fusion proteins. Oncogene. 2001;20:5695–5707. doi: 10.1038/sj.onc.1204639. [DOI] [PubMed] [Google Scholar]
- 92.Lochner K, Siegler G, Fuhrer M, Greil J, Beck JD, Fey GH, Marschalek R. A specific deletion in the breakpoint cluster region of the ALL-1 gene is associated with acute lymphoblastic T-cell leukemias. Cancer Res. 1996;56:2171–2177. [PubMed] [Google Scholar]
- 93.Micci F, Panagopoulos I, Bjerkehagen B, Heim S. Consistent rearrangement of chromosomal band 6p21 with generation of fusion genes JAZF1/PHF1 and EPC1/PHF1 in endometrial stromal sarcoma. Cancer Res. 2006;66:107–112. doi: 10.1158/0008-5472.CAN-05-2485. [DOI] [PubMed] [Google Scholar]
- 94.Sarma K, Margueron R, Ivanov A, Pirrotta V, Reinberg D. Ezh2 requires PHF1 to efficiently catalyze H3 lysine 27 trimethylation in vivo. Mol Cell Biol. 2008;28:2718–2731. doi: 10.1128/MCB.02017-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kurotaki N, Imaizumi K, Harada N, Masuno M, Kondoh T, Nagai T, Ohashi H, Naritomi K, Tsukahara M, Makita Y, Sugimoto T, Sonoda T, Hasegawa T, Chinen Y, Tomita Ha HA, Kinoshita A, Mizuguchi T, Yoshiura Ki K, Ohta T, Kishino T, Fukushima Y, Niikawa N, Matsumoto N. Haploinsufficiency of NSD1 causes Sotos syndrome. Nat Genet. 2002;30:365–366. doi: 10.1038/ng863. [DOI] [PubMed] [Google Scholar]
- 96.Douglas J, Hanks S, Temple IK, Davies S, Murray A, Upadhyaya M, Tomkins S, Hughes HE, Cole TR, Rahman N. NSD1 mutations are the major cause of Sotos syndrome and occur in some cases of Weaver syndrome but are rare in other overgrowth phenotypes. Am J Hum Genet. 2003;72:132–143. doi: 10.1086/345647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rayasam GV, Wendling O, Angrand PO, Mark M, Niederreither K, Song L, Lerouge T, Hager GL, Chambon P, Losson R. NSD1 is essential for early post-implantation development and has a catalytically active SET domain. Embo J. 2003;22:3153–3163. doi: 10.1093/emboj/cdg288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gibbons R. Alpha thalassaemia-mental retardation, X linked. Orphanet J Rare Dis. 2006;1:15. doi: 10.1186/1750-1172-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Raymond FL. X linked mental retardation: a clinical guide. J Med Genet. 2006;43:193–200. doi: 10.1136/jmg.2005.033043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gibbons RJ, Picketts DJ, Villard L, Higgs DR. Mutations in a putative global transcriptional regulator cause X-linked mental retardation with alpha-thalassemia (ATR-X syndrome) Cell. 1995;80:837–845. doi: 10.1016/0092-8674(95)90287-2. [DOI] [PubMed] [Google Scholar]
- 101.Berube NG, Mangelsdorf M, Jagla M, Vanderluit J, Garrick D, Gibbons RJ, Higgs DR, Slack RS, Picketts DJ. The chromatin-remodeling protein ATRX is critical for neuronal survival during corticogenesis. J Clin Invest. 2005;115:258–267. doi: 10.1172/JCI22329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Argentaro A, Yang JC, Chapman L, Kowalczyk MS, Gibbons RJ, Higgs DR, Neuhaus D, Rhodes D. Structural consequences of disease-causing mutations in the ATRX-DNMT3-DNMT3L (ADD) domain of the chromatin-associated protein ATRX. Proc Natl Acad Sci U S A. 2007;104:11939–11944. doi: 10.1073/pnas.0704057104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Stayton CL, Dabovic B, Gulisano M, Gecz J, Broccoli V, Giovanazzi S, Bossolasco M, Monaco L, Rastan S, Boncinelli E, et al. Cloning and characterization of a new human Xq13 gene, encoding a putative helicase. Hum Mol Genet. 1994;3:1957–1964. doi: 10.1093/hmg/3.11.1957. [DOI] [PubMed] [Google Scholar]
- 104.Cardoso C, Timsit S, Villard L, Khrestchatisky M, Fontes M, Colleaux L. Specific interaction between the XNP/ATR-X gene product and the SET domain of the human EZH2 protein. Hum Mol Genet. 1998;7:679–684. doi: 10.1093/hmg/7.4.679. [DOI] [PubMed] [Google Scholar]
- 105.Le Douarin B, Nielsen AL, Garnier JM, Ichinose H, Jeanmougin F, Losson R, Chambon P. A possible involvement of TIF1 alpha and TIF1 beta in the epigenetic control of transcription by nuclear receptors. EMBO J. 1996;15:6701–6715. [PMC free article] [PubMed] [Google Scholar]
- 106.Berube NG, Smeenk CA, Picketts DJ. Cell cycle-dependent phosphorylation of the ATRX protein correlates with changes in nuclear matrix and chromatin association. Hum Mol Genet. 2000;9:539–547. doi: 10.1093/hmg/9.4.539. [DOI] [PubMed] [Google Scholar]
- 107.Nan X, Hou J, Maclean A, Nasir J, Lafuente MJ, Shu X, Kriaucionis S, Bird A. Interaction between chromatin proteins MECP2 and ATRX is disrupted by mutations that cause inherited mental retardation. Proc Natl Acad Sci U S A. 2007;104:2709–2714. doi: 10.1073/pnas.0608056104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Petrij F, Giles RH, Dauwerse HG, Saris JJ, Hennekam RC, Masuno M, Tommerup N, van Ommen GJ, Goodman RH, Peters DJ, et al. Rubinstein-Taybi syndrome caused by mutations in the transcriptional co-activator CBP. Nature. 1995;376:348–351. doi: 10.1038/376348a0. [DOI] [PubMed] [Google Scholar]
- 109.Kalkhoven E, Roelfsema JH, Teunissen H, den Boer A, Ariyurek Y, Zantema A, Breuning MH, Hennekam RC, Peters DJ. Loss of CBP acetyltransferase activity by PHD finger mutations in Rubinstein-Taybi syndrome. Hum Mol Genet. 2003;12:441–450. doi: 10.1093/hmg/ddg039. [DOI] [PubMed] [Google Scholar]
- 110.Lower KM, Turner G, Kerr BA, Mathews KD, Shaw MA, Gedeon AK, Schelley S, Hoyme HE, White SM, Delatycki MB, Lampe AK, Clayton-Smith J, Stewart H, van Ravenswaay CM, de Vries BB, Cox B, Grompe M, Ross S, Thomas P, Mulley JC, Gecz J. Mutations in PHF6 are associated with Borjeson-Forssman-Lehmann syndrome. Nat Genet. 2002;32:661–665. doi: 10.1038/ng1040. [DOI] [PubMed] [Google Scholar]
- 111.Voss AK, Gamble R, Collin C, Shoubridge C, Corbett M, Gecz J, Thomas T. Protein and gene expression analysis of Phf6, the gene mutated in the Borjeson-Forssman-Lehmann Syndrome of intellectual disability and obesity. Gene Expr Patterns. 2007;7:858–871. doi: 10.1016/j.modgep.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 112.Lee J, Hagerty S, Cormier KA, Ferrante RJ, Kung AL, Ryu H. Monoallele Deletion of CBP Leads to Pericentromeric Heterochromatin Condensation through ESET Expression and Histone H3 (K9) Methylation. Hum Mol Genet. 2008 doi: 10.1093/hmg/ddn067. [DOI] [PMC free article] [PubMed] [Google Scholar]