Ribosomal protein L3 functions as a ‘rocker switch’ to aid in coordinating of large subunit-associated functions in eukaryotes and Archaea (original) (raw)
Abstract
Although ribosomal RNAs (rRNAs) comprise the bulk of the ribosome and carry out its main functions, ribosomal proteins also appear to play important structural and functional roles. Many ribosomal proteins contain long, nonglobular domains that extend deep into the rRNA cores. In eukaryotes and Archaea, ribosomal protein L3 contains two such extended domains tethered to a common globular hub, thus providing an excellent model to address basic questions relating to ribosomal protein structure/function relationships. Previous work in our laboratory identified the central ‘W-finger’ extension of yeast L3 in helping to coordinate ribosomal functions. New studies on the ‘N-terminal’ extension in yeast suggest that it works with the W-finger to coordinate opening and closing of the corridor through which the 3′ end of aa-tRNA moves during the process of accommodation. Additionally, the effect of one of the L3 N-terminal extension mutants on the interaction between C75 of the aa-tRNA and G2921 (Escherichia coli G2553) of 25S rRNA provides the first evidence of the effect of a ribosomal protein on aa-tRNA positioning and peptidyltransfer, possibly through the induced fit model. A model is presented describing how all three domains of L3 may function together as a ‘rocker switch’ to coordinate the stepwise processes of translation elongation.
INTRODUCTION
One of the primary goals of nanotechnology is to create extremely small machines. Two central concerns of machine design center around how the structure of each component contributes to overall function, and how all of the different parts dynamically interact to ensure that the machine operates in an orderly manner. The ribosome can be viewed as a model nanomachine that has been refined for optimal performance over the course of ∼2.5 to 3 billion years of evolutionary selective pressure. It is complex: depending on the kingdom, ribosomes contain three or four distinct ribosomal RNAs (rRNAs) and up to 80 proteins (1). Its function is to convert a set of input data into functional output, in this case translating genetic information encoded by mRNAs into proteins. This process involves numerous steps, all of which must be coordinated with one another. Within the past few years, technical breakthroughs in X-ray crystallography and cryo-EM have significantly enhanced our understanding of ribosome fine structure [(2), and reviewed in references (3) and (4)]. These have proved to be invaluable guides, providing new views and insights toward the ultimate goal of understanding how ribosome structure determines function. Complementary approaches using combinations of biochemical, biophysical and molecular genetics methods are also being used to shed light on the dynamic processes involved in coordinating the various aspects of ribosome function (5–14).
Historically, it was first thought that ribosomal proteins were the central players in ribosome function while rRNA was relegated to a minor, scaffolding function. However, as understanding of the ribosome has progressed, these roles have been almost completely reversed. More recently, ribosomal proteins have become to be appreciated as being more than mere structural glue (15–20). Examination of the atomic scale data reveals that many of the core ribosomal proteins contain long, nonglobular extensions (21), giving rise to questions regarding the functional significance of these structures. Ribosomal protein L3 provides an excellent model to address basic questions relating to ribosomal protein structure/function relationships. The high extent of sequence and structure conservation in L3 proteins among all three domains of life attest to its biological significance (22). L3 is only one of two proteins capable of initiating assembly of Escherichia coli large ribosomal subunits in vitro (23), and it is one of the few ribosomal proteins required for peptidyltransferase activity (24). Molecular genetics and biochemical studies in many organisms have shown that L3 plays an important role in aa-tRNA binding, peptidyltransferase activity, drug resistance, translational frame maintenance, virus replication and as a binding site for a ribosome inhibitory protein (25–34). Saturation mutagenesis suggested that L3 may function to transmit information between the Sarcin/Ricin loop (SRL) and the peptidyltransferase center (PTC) (35). Like many ribosomal proteins, L3 has a globular domain at the cytoplasmic face of the large subunit, and a long extended domain that projects deep into the mostly rRNA core. The eukaryotic and archael L3 proteins are particularly interesting because they contain two such extensions. The universally conserved central extension, also called the tryptophan- or W-finger, projects to the A site side of the PTC, where the tryptophan located at its tip closely approaches the peptidyltransferase center active site (35,36). In the yeast Saccharomyces cerevisiae, mutagenesis of this and nearby amino acids revealed that the W-finger helps synchronize the processes of aa-tRNA accommodation, peptidyltransfer and translocation by functioning as sensor of the tRNA occupancy status of the A site region of the PTC (20). The focus of the current study is the second extension of L3, which is composed of sequences at the N-terminus of the protein. Complementary molecular genetics, biochemical and rRNA structure probing experiments suggest that this domain functions as a piston, which we hypothesize helps to open and close the first of two ‘gates’ located along the corridor through which aa-tRNA moves during the process of accommodation, thus controlling aa-tRNA access to the PTC. In combination with our earlier studies on the W-finger, which interacts with the second ‘gate’, we propose a model in which ribosomal protein L3 may function as a ‘rocker switch’, coordinating the sequential steps of ternary complex binding, aa-tRNA accommodation, peptidyltransfer and elongation factor 2 binding/translocation.
MATERIALS AND METHODS
Strains, plasmids, genetic manipulation and media
Escherichia coli DH5α was used to amplify plasmid DNA. Transformation of E. coli and yeast, and preparation of yeast growth media (YPAD, synthetic drop out medium, and 4.7 MB plates for testing the Killer phenotype), were as previously reported (37). Restriction enzymes were obtained from MBI Fermentas (Vilnius, Lithuania). The QuikChange XL II Site-Directed Specific Mutagenesis Kit was obtained from Stratagene (La Jolla, CA). Genewiz (South Plainfield, NJ) performed DNA sequence analysis. Oligonucleotide primers were purchased from IDT (Coralville, IA). The yeast strains used in this study were all derived from the rpl3_-gene disruption (rpl3Δ) strain JD1090 [MATα ura3-52 lys2-801 trp1δ leu2− his3 RPL3::HIS3 p_RPL3-URA3-CEN6 (L-A HN M1)] (31). Mutants of rpl3 were generated using the wild-type RPL3 gene in pJD225 (31), synthetic oligonucleotides and the QuikChange XL II Kit, and standard ‘plasmid shuffle’ techniques using 5-flouroorotic acid (5-FOA) (38) were used to generate cells expressing only mutant alleles of rpl3. Ten-fold dilution spot assays were performed on rich media alone and on media containing 50 μg/ml of anisomycin, and growth of cells in the presence of drug relative to no drug was used to score for anisomycin resistance.
Characterization of peptidyltransferase activity, tRNA and eEF2 binding studies
Yeast phenylalanyl-tRNAs were aminoacylated with unlabeled phenylalanine or with [14C]Phe to make Phe-tRNA and [14C]Phe-tRNA, respectively. [14C]Phe-tRNA was used to monitor enzymatic binding to the A site of poly(U) primed nonsalt washed ribosomes, and acetylated-[14C]Phe-tRNA (Ac-[14C]Phe-tRNA) was generated to monitor nonenzymatic P site binding using poly(U) primed salt washed ribosomes. Phe-tRNA and poly(U) were used in chemical protection experiments. The charged tRNAs were purified by HPLC, and equilibrium binding studies were performed as previously described (20). The site-specificity of charged tRNA binding was confirmed using the puromycin reaction (39). Peptidyltransfer assays were performed essentially as previously described (35,40), 6 × His-tagged eEF2 was purified from TKY675 yeast cells (kindly provided by Dr. T. Kinzy) (41), and eEF2 binding experiments were performed using salt-washed ribosomes as previously described (20). _K_d values were determined assuming single binding sites using GraphPad Prism software.
Isolation and chemical probing of mutant ribosomes
Ribosomes were isolated as described (35) and were synchronized by puromycin treatment in buffer B10 [20 mM Tris–HCl (pH 7.5), 10 mM magnesium acetate, 1 mM PMSF, 2 mM DTE, 0.5 M KCl, 1 mg/ml heparin and 10% glycerol] containing 1 mM GTP, 1 mM puromycin and 500 pmol of ribosomes. After incubation for 30 min at 30°C, ribosomes were sedimented through a cushion of buffer B25 [20 mM Tris–HCl (pH 7.5), 10 mM magnesium acetate, 0.1 mM PMSF, 2 mM DTE, 0.5 M KCl, 1 mg/ml heparin and 25% glycerol] and suspended in buffer C [50 mM Tris–HCl (pH 7.5), 5 mM magnesium acetate, 50 mM ammonium chloride, 0.1 mM PMSF, 2 mM DTE and 25% glycerol] at 10 pmol/μl. For structure probing of ribosomes with occupied A sites, ribosomal P sites were blocked with 4 × excess of deacylated tRNA, and A sites were loaded by subsequent incubation with 10-fold excess of Phe-tRNA. Chemical probing with DMS, kethoxal and CMCT, followed by RT primer extension analysis of modified RNAs, was performed as described (42). Primers (numbered from the first base of yeast 25S rRNA) employed for these analyses were 2957 (5′-AACCTGTCTCACGACGG-3′), 3043 (5′-CCTGATCAGACAGCCGC-3′), 2877 (5′-GGTATGATAGGAAGAGC-3′), 2435 (5′-CCTCTATGTCTCTTCAC-3′), and 2675 (5′-GTTCTACTGGAGATTTCTG-3′).
Computational analysis of ribosome structure
The X-ray crystal structure of the Haloarcula marismortui 50S ribosomal subunit (1VQ6) (21), the cryo-electron microscopy (cryo-EM) reconstruction of S. cerevisiae ribosomal proteins threaded onto the X-ray crystal structure of the H. marismortui 50S ribosomal subunit (PDB IS1I) (43), the Thermus thermophilus 70S ribosome complexed with two tRNAs at 2.8 Å resolution (PDB 2J01) (44), the E. coli ribosome complexed with three tRNAs at 3.5 Å (2AW4) (45), a cryo-EM reconstruction of the D. radiodurans ribosome complexed with thiostrepton at 3.3 Å–3.7 Å (2ZJR) (46), and the T. thermophilus 70S ribosome with a model mRNA and tRNAs at 5.5 Å (2HGU) were visualized using PyMOL (DeLano Scientific LLC).
RESULTS
Viability of L3 N-terminal mutants
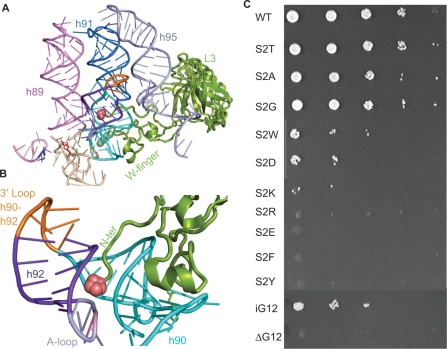
As shown in Figure 1A, ribosomal protein L3 has a solvent accessible globular domain, the inner face of which interacts with the base of helix 95 (atop which is located the SRL). It also contains two separate domains that extend deep into the rRNA core of the large subunit. The tryptophan at the tip of the central extension approaches the A site side of the peptidyltransferase center and studies of this extension suggested that it functions as an allosteric switch to coordinate the functions of the ribosomal elongation factor binding site (20). The N-terminal domain of L3 also extends into the core of the large subunit. A close-up view shows that the L3 N-terminus is nestled in the center of a complex structure formed by Helices 90–92 (Figure 1B). Supplementary Figure 1 shows a multiple sequence alignment of the N-terminal regions of L3 proteins from representative bacterial, archael and eukaryotic species. Examination of this alignment shows that the N-termini of eukaryotic and archael L3 proteins contain an additional ∼50 amino acids as compared to their bacterial counterparts. Mapping of the L3 N-termini to representative high-resolution ribosome structures reveals that these additional amino acids extend the N-terminus out from the globular domain deep into the core of the large subunit where it is nestled in a space between Helices 90 and 92. Interestingly, no analogous mass is present in bacterial ribosomes, but bacterial L27 appears to interact with the 3′ end of the A-site tRNA (44). The potential significance of this is discussed below. In yeast, although methionine is the first amino acid encoded by the RPL3 gene, mass spectroscopic analysis of intact ribosomes indicates that this is absent, suggesting that it is post-translationally removed (47). Thus, in ribosomes the N-terminal amino acid of L3 is a serine. Since this is the second amino acid encoded by the RPL3 gene, this serine has been designated S2. Similarly, in H. marismortu, although methionine is the first amino acid encoded by its Rpl3 gene, the atomic resolution structure indicates that the N-terminal moiety is a proline (21). Analysis of the atomic resolution structure shows that the N-terminal amino acid of L3 is within H-bonding distance of the phosphate backbone of a universally conserved large subunit rRNA base that has been implicated in formation of the first of the two ‘gates’ through which the 3′ end of aa-tRNA passes during the process of accommodation (48). In E. coli this is C2556 (C2591 in H. marismortui, and C2924 in yeast). In yeast, the S2T mutant was previously identified in a screen for anisomycin resistance, a competitive inhibitor for aa-tRNA binding to the A site (35). To follow up on that observation, oligonucleotide site-specific mutagenesis and classic 5-FOA-mediated ‘plasmid shuffle’ (38) methods were used to generate a set of S2 substitution mutants in yeast cells covering a broad range of biochemical properties. Specifically, S2 was changed to residues with the following chemical properties of sidechains: aliphatic (G, A); acetic (D, E); basic (K, R); and aromatic (W, F, Y). Three of the mutants (S2T, S2A and S2G) were able to support robust cell growth as the sole forms of L3 (Figure 1C, Table 1). Three additional mutants supported weak levels of cell growth (S2W, S2D and S2K), and the remainder were lethal. As discussed below, these findings suggest a strong correlation between viable substitutions of S2 and amino acids preferred for N-terminal acetylation (49).
Figure 1.
The N-terminal extension of ribosomal protein L3. (A) The globular domain of ribosomal protein L3 (green) abuts the base of helix 95. The elongation factor binding site is at the top of helices 89, 91 and 95, and the aa-tRNA accommodation corridor is between Helices 91 and 89. The peptidyltransferase center (E. coli A2451, H. marismortui A2484, yeast A2819), lies at the bottom of this corridor and is colored red. The W-finger of L3 extends to under the base of Helix 90, on the A site side of the peptidyltransferase center. The _N_-terminal extension of L3 (denoted as pink spheres) extends into a complex structure formed by Helices 90 and 92. (B) Close-up view of the _N_-terminus of L3 nestled between Helices 90 and 92. (C) Ten-fold dilution spot assays of mutants in the N-terminal extension of L3. The serine at the _N_-terminus of L3 (S2) was mutated to the indicated amino acids. In iG12, an alanine was inserted between histidine at position 11 and the glycine at position 12. ΔG12 indicates deletion of glycine at position 12.
Table 1.
Summary of the L3 N-terminal mutants
| Anisoa | Growthb | Killerc | PTased | A-site _K_de | P-site _K_df | eEF2 _K_dg | |
|---|---|---|---|---|---|---|---|
| WT | s | + | + | 0.26 ± 0.02 | 78.86 ± 8.6 | 56.14 ± 6.12 | 18.73 ± 2.68 |
| S2T | R | + | w | 0.14 ± >0.01 | 14.91 ± 1.6 | 45.36 ± 4.58 | 20.66 ± 2.67 |
| S2A | s | + | w | 0.15 ± >0.01 | 85.14 ± 7.7 | 66.32 ± 6.97 | 19.91 ± 3.15 |
| S2G | s | + | + | ||||
| S2W | s | ↓ | + | ||||
| S2D | s | ↓↓ | + | ||||
| S2K | r | ↓↓↓ | – | 0.03 ± >0.01 | 23.82 ± 3.3 | 53.44 ± 8.86 | 20.43 ± 1.74 |
| S2R | Inviable | ||||||
| S2E | Inviable | ||||||
| S2F | Inviable | ||||||
| S2Y | Inviable | ||||||
| iG12 | s | ↓ | – | 0.19 ± 0.02 | 434.8 ± 33.1 | ND | 10.27 ± 1.59 |
| ΔG12 | Inviable |
Examination of Figure 1A and B suggested that the N-terminal extension of L3 might also have the ability to be ‘pushed into’ and ‘pulled out of’ the accommodation corridor, i.e. it may function analogously to a piston. To genetically mimic this, an amino acid could be inserted or deleted from this structure. This strategy was complicated by the fact that the nuclear localization signal of L3 is encoded in its N-terminal 21 amino acids (50). However, in mapping this region, we had observed that only the glycine at position 12 could be altered without affecting nuclear localization (Dinman lab, unpublished data). Thus, two additional mutants were constructed: one containing an alanine inserted N-terminally to G12 (iG12), and second in which this glycine was removed (ΔG12). iG12 should mimic the piston being pushed into the accommodation corridor, while ΔG12 was expected to mimic retraction of the piston away from this structure. As shown in Figure 1C, the iG12 mutant was viable as the sole form of L3, while ΔG12 was lethal.
Genetic and biochemical studies: the N-terminus of L3 is important for virus propagation, drug resistance, peptidyltransfer and elongation factor binding
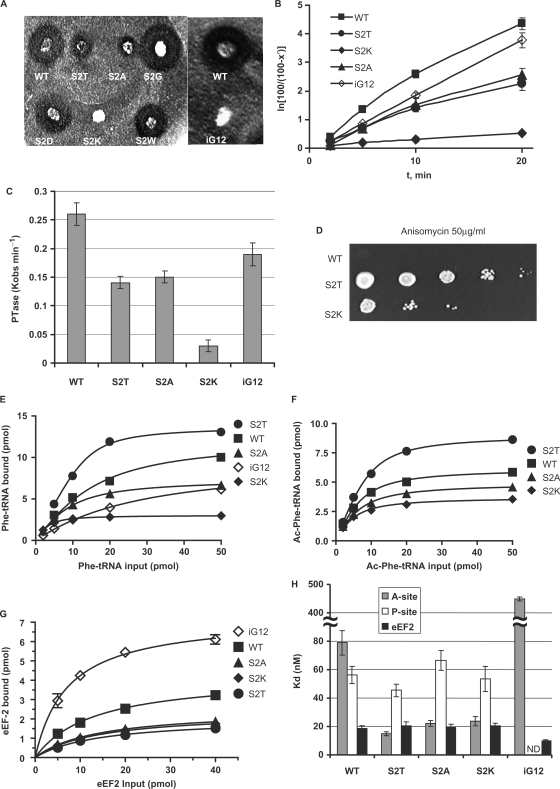
L3 mutants have long been associated with defects in the ability of cells to propagate the endogenous ‘killer’ virus of S. cerevisiae (28,31). To assay this, colonies of cells expressing the viable S2 mutants were replica plated onto a lawn of killer toxin-sensitive cells, and their Killer phenotypes were assayed by scoring for changes in the radii of growth inhibition surrounding the mutants. This experiment revealed that the S2K and iG12 mutants completely abrogated the ability of cells to maintain the killer virus, and that the S2T and S2A mutants produced significantly decreased zones of killer activity (Figure 2A). Peptidyltransferase defects have also been linked to defects in virus propagation (31). Figure 2B shows a first-order time plot derived from time course data of single cycle peptidyltransferase reactions, from which the observed rates of peptidyltransfer (_K_obs) were determined (Figure 2C). All of the killer-defective mutants also promoted decreased peptidyltransferase activity, supporting the relationship between killer virus maintenance and peptidyltransferase activity (summarized in Table 1). The very low level of PTase activity by the S2K mutant may also reflect its strong effect on cell viability (Figure 1C).
Figure 2.
Genetic and biochemical characterization of ribosomes and cells expressing the L3 N-terminal extension mutants. (A) Killer virus maintenance profiles of L3 N-terminal mutants. Presence of, and relative amount of intracellular killer virus is indicated by the diameter of the zone of growth inhibition around the indicated colonies. (B and C) First-order time plots of Ac-[14C]Phe-puromycin formation were used to calculate observed rates of peptidyltransferase activity (_K_obs). (D) Anisomycin resistance phenotypes of S2T and S2K mutants. Ten-fold dilutions of cells harboring the indicated rpl3 alleles were spotted onto complete synthetic medium lacking tryptophan containing anisomycin (50 μg/ml), and were incubated at 30°C for 3 days. (E) Single site isothems of eEF-1A stimulated binding of [14C]Phe-tRNA to A-sites of poly(U) primed ribosomes. (F) Binding isotherms of Ac-[14C]Phe-tRNA to P-sites of poly(U) primed ribosomes. (G) eEF2 binding isotherms for wild-type and mutant ribosomes. (H) Dissociation constants were calculated from data shown in panels E, F and G. Error bars denote standard deviations. N.D. denotes not determined.
Anisomycin is a competitive inhibitor of aa-tRNA binding (51), and we previously demonstrated that the S2T mutant was resistant to this drug (35). Ten-fold dilution spot assays using all of the viable mutants demonstrated that the S2K mutant also conferred anisomycin resistance (Figure 2D). Previous studies have shown a correlation between anisomycin resistance and increased ribosomal affinity for aa-tRNA in the ribosomal A-site but not of acetylated aa-tRNA (Ac-aa-tRNA) in the P-site (20,35). Equilibrium binding studies using purified ribosomes and tRNAs were performed, and dissociation constants were generated from curves fitted to single site binding models (Figure 2E). Since the A-site binding was enzymatic, i.e. using poly(U) primed nonsalt-washed ribosome preparations containing eEF1A and GTP, the major substrates that were bound in these experiments were a combination composed mostly of the eEF1A•[14C]Phe-tRNA•GTP ternary complex (TC) and [14C]Phe-tRNA enzymatically accommodated into the ribosomal A-site (to simplify, this is referred to as aa-tRNA binding for the remainder of the text). As summarized Figure 2H and Table 1, the correlation between anisomycin resistance and increased affinity for aa-tRNA in the A-site held true in the current study. In particular, the extent of anisomycin resistance correlated well with the level to which affinity for aa-tRNA was increased in the S2T and S2K mutants (compare Figure 2D and H). In contrast, the iG12 mutant promoted an almost 8-fold increase in the dissociation constant for aa-tRNA (discussed below). None of the mutants conferred appreciable changes in nonenzymatic binding of Ac-aa-tRNA to the P-sites of poly(U) primed salt-washed ribosomes (Figure 2F and H). We also previously showed an inverse correlation between affinities for aa-tRNA and eEF2, which gave rise to an allosteric model of elongation factor recognition by the ribosome (20). Interestingly, while this pattern was repeated for the iG12 mutant, it did not hold true for the S2T and S2K mutants (Figure 2G and H, Table 1).
rRNA structure analysis: effects of the L3 N-terminal mutants on the intrinsic conformational changes in the aa-tRNAaccommodation corridor
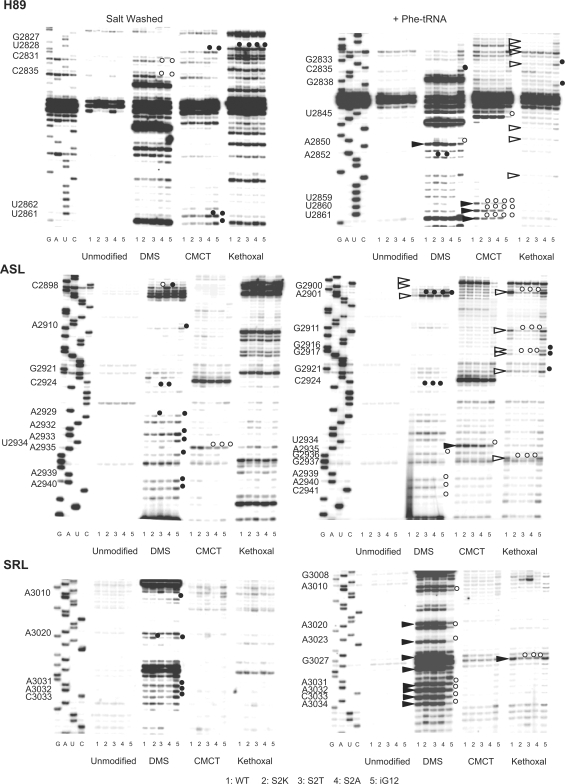
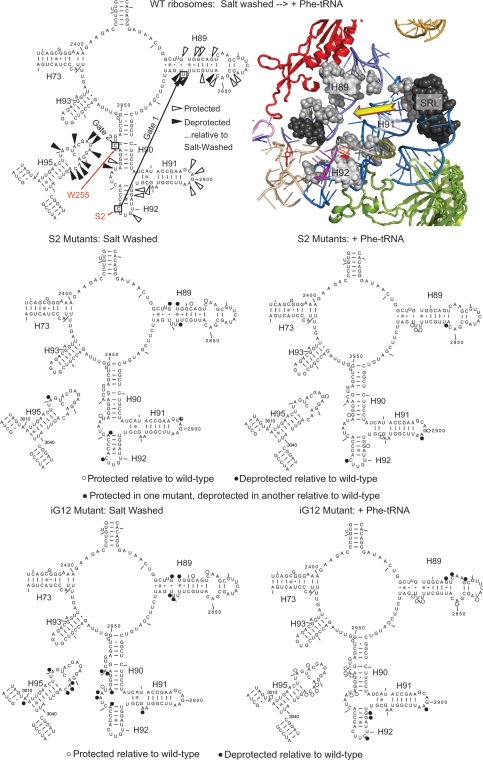
Previously, chemical protection methods demonstrated that mutants of tryptophan 255 at the tip of the W-finger promoted an ‘open’ conformation of the aa-tRNA accommodation corridor, thus implying that this structure is intrinsically conformationally dynamic (20). To address this, purified ribosomes that were either salt-washed, or nonsalt washed and incubated with aa-tRNA, were treated with the base modifying reagents DMS, CMCT or kethoxal, and the resulting patterns of rRNA base modification were determined by reverse transcriptase primer extension as described in the Materials and methods section. The results of these experiments are shown in Figure 3 and summarized in Figure 4. Focusing on analysis of the wild-type ribosomes (lane 1 in Figure 3, and the top two panels in Figure 4), addition of aa-tRNA resulted in significant changes in rRNA conformation along the aa-tRNA accommodation corridor and in the adjacent SRL. As summarized in the top panel of Figure 4, addition of aa-tRNA resulted in increased protection of a large number of bases lying along the accommodation corridor route (indicated by open arrows on the left and gray spheres on the right) with simultaneous deprotection of almost the entire SRL (black arrows and spheres). Analysis of these data suggests that accommodation of aa-tRNA into the ribosome results in movement of the Helix 90–92 structure toward Helix 89 and away from the SRL. The accessibility of rRNA bases in the accommodation corridor in salt-washed ribosomes relative to those loaded with aa-tRNA suggests that prior to addition of aa-tRNA, the accommodation corridor is in ‘open’ conformation, and that addition of aa-tRNA causes it to close, leaving the SRL more exposed to the exterior solvent. Importantly, G2921 (E. coli G2553) became protected from chemical attack after aa-tRNA is loaded into the ribosome. The significance of these changes is discussed below.
Figure 3.
rRNA structure probing of wild-type and the L3 N-terminal extension mutant ribosomes. Autoradiograms at left are of salt-washed wild-type, S2K, S2T, S2A and iG12 mutant ribosomes, while those at right are of nonsalt-washed ribosomes loaded with aa-tRNA. Ribosomes were unmodified or treated with DMS, CMCT or kethoxal as indicated. Reverse transcriptase primer extension reactions spanned sequences in Helix 89 (top panels), Helices 90–92 (ASL, middle panels) and Helix 95 (SRL, lower panels). Sequencing reactions (left sides of panels) are labeled corresponding to the rRNA sense strand. Bases in wild-type ribosomes loaded with aa-tRNAs that are deprotected from chemical attack relative to salt-washed ribosomes are indicated by black arrowheads. Similarly, those that become protected upon addition of aa-tRNA are indicated by white arrowheads. Bases in mutant ribosomes that are deprotected relative to wild-type are indicated by black circles, and those that are hyperprotected are indicated by white circles.
Figure 4.
rRNA protection of the S2 mutants mapped onto yeast 25S rRNA and the _H. marismortui/_yeast large subunit structure. (Top) Left panel shows yeast 25S rRNA bases whose chemical protection patterns change in the presence of aa-tRNA in wild-type ribosomes. Base numbering follows the S. cerevisiae sequence. Black arrows indicate deprotection, and white arrows show hyperprotection in Phe-tRNA containing ribosomes as compared to salt-washed ribosomes. The positions of bases involved in formation of the two aa-tRNA accommodation corridor ‘gates’ (48) are indicated, as well as bases that interact with S2 and W255. In the right panel, these bases have been mapped onto the H. marismortui/yeast structure. Ribosomal protein L3 is colored green, and L10 is in red. Black and gray spheres indicate deprotected and hyperprotected bases, respectively. Yellow arrow connotes movement of Helix 91–92 structure toward Helix 89 to close the accommodation corridor. (Center) Bases of 25S rRNA in the S2 series of mutants whose modification patterns differ from wild-type ribosomes. Left and right panels map changes in salt-washed and aa-tRNA loaded ribosomes. Black and white circles indicate deprotected and hyperprotected bases, respectively. (Bottom) Bases of 25S rRNA in the iG12 mutant whose modification patterns differ from wild-type ribosomes. Left and right panels map changes in salt-washed and aa-tRNA loaded ribosomes. Black and white circles indicate deprotected and hyperprotected bases, respectively.
Extension of this analysis to the S2K, S2T, S2A and iG12 mutants revealed some intriguing differences. The data generated from these experiments are shown in Figure 3 (lanes 2–5), and are mapped onto the 2D rRNA structures in the central panels of Figure 4. Similar to their effects on aa-tRNA and eEF2 binding, the S2K and S2T mutants conferred similar changes on rRNA structure, while the S2A mutant promoted relatively few affects. The iG12 mutant was in a class by itself. Examination of the S2 mutants reveals two interesting trends. First, C2924 (E. coli C2556) was deprotected from chemical attack in all three mutants relative to wild-type ribosomes, irrespective of the presence or absence of aa-tRNA. This base interacts with the N-terminus of L3 in the H. marismortui atomic resolution structure (43), and also forms one of the partners of the first gate in the accommodation corridor (48). Its deprotection in the S2 mutants suggests that the interaction between these two molecules has been diminished by the mutants. The second salient observation is that regions that tended to change their protection patterns upon addition of aa-tRNA in wild-type ribosomes showed opposing protection patterns in the S2 mutants. For example, the 5′ side of H89 remained relatively deprotected in the salt-washed mutants relative to wild type, and the 3′ side of this structure was protected in the presence of aa-tRNA in the mutants relative to wild-type ribosomes. Interestingly, this is where H89 interacts with the N-terminal ‘hook’ extension of ribosomal protein L10 (see accompanying manuscript). Similarly, A3021 (E. coli A2654) in the SRL was deprotected in the salt-washed ribosomes but did not become even more so upon addition of aa-tRNA. As discussed below, these data suggest that the first gate of the aa-tRNA accommodation corridor is broken in these mutants, resulting in decreased amplitude of movement, and promoting a ‘more open than closed’ conformation of this structure. Note that the strong modification in the kethoxal lanes between G3008 and A3010 are not G's. Additionally, although some unevenness in loading of the CMCT and kethoxal is shown in the upper region of SRL panel of the salt-washed ribosomes, no differences were observed in this region of other gels for these reactions. To err on the conservative side, none of these apparent differences are indicated or modeled here.
Chemical protection analysis of the iG12 mutant revealed a significantly different pattern. In the absence of aa-tRNA, the protection patterns in H89 at its site of interaction with the L10 hook, in the loop between H90 and H92, in the 3′ half of H90 and in the SRL more resemble that of aa-tRNA loaded wild-type ribosomes (see lane 5 of Figure 3, and the bottom left panel of Figure 4). This suggests that the accommodation corridor is intrinsically more ‘closed’ in the absence of aa-tRNA in this mutant. Conversely however, many of these same bases remained relatively protected upon addition of aa-tRNA, suggesting that this mutant was not able to attain a completely closed conformation (Figure 4, bottom right panel). Two additional base-specific changes in the chemical protection patterns were of note. First, G2921 (E. coli G2553) did not become protected from chemical attack after aa-tRNA loading. Second, bases in the vicinity of A2940 (E. coli A2572) were relatively deprotected in salt-washed, and hyperprotected in aa-tRNA loaded iG12 relative to wild-type ribosomes. The significance of the former observation with regard to peptidyltransferase center activation, and of the latter with respect to allosteric coordination of ribosome function are discussed below. We also suggest that the inability to close the accommodation corridor may be the underlying reason for the lethality of the ΔG12 mutant.
DISCUSSION
With atomic resolution ribosomes in hand, the major challenge is linking structure to function. Three significant questions in the field are: ‘how does the ribosome coordinate the sequential processes of translation elongation’; ‘what are the allosteric signals that activate the peptidyltransferase center’; and ‘what are the functions of the extended domains of ribosomal proteins?’ Recent work from our laboratory demonstrated that the central extended domain, or ‘W-finger’ of L3 acts as a molecular switch to help coordinate binding of the two elongation factors, leading us to describe L3 as the ‘gatekeeper’ to the ribosomal A site (20). The focus of the current study is the second extended domain located at the N-terminus of L3. Here, a combination of molecular genetics, biochemistry, rRNA structure probing and molecular modeling approaches were used to address these questions.
As noted above, the N-terminus of L3 appears to form a hydrogen bond with C2924 (E. coli C2556), which in turn is one of the two partners of the first accommodation gate. We suggest the N-terminal extension of L3 may act as a piston to either directly induce or indirectly support conformational changes in this accommodation gate. We further suggest that this interaction is broken in the S2 series of mutants, interfering with its ability to fully open or close, while lengthening it by insertion of an additional amino acid forces the accommodation corridor into more closed conformation. The changes in the vicinity of the other side of the first gate [around U2860 (E. coli U2492)] and at the second gate [in the vicinity of C2941 (E. coli C2573)] also suggest that the coordination between the two gates is inhibited by these mutants. Importantly, one of these bases, A2940 (E. coli A2572), appears to interact with the W-finger of L3. The deprotection of C2924 (E. coli C2556), A2940 and C2941 by the W255C mutation (20) suggests that both the N-terminal and central extensions of L3 work in concert to coordinate opening and closing of the aa-tRNA accommodation corridor.
Together with the studies on the W-finger domain, we propose a more detailed, mechanical model of L3 functioning as a ‘rocker switch’ to help coordinate an allosteric signaling pathway between the elongation factor binding site and the peptidyltransferase center. This model is cartooned in Figure 5. Starting with the open conformation (Figure 5, left side), positioning of the N-terminal extension away from the accommodation corridor pulls C2924 (E. coli C2556, the Helix 92 gate 1 base) along with it, away from U2861 (E. coli U2493, the Helix 89 gate 1 base). This favors closure of the proximal loop in Helix 89 (protecting these bases), and pulls bases along the H90–92 structure away from the corridor, exposing them to solvent (deprotection). The fully open conformation positions Helices 89, 90–92, and 95 to form the binding site for the aa-tRNA•eEF1A•GTP ternary complex, promoting increased affinity for aa-tRNA. We suggest that the ‘more open than closed’ conformation of the S2T and S2K mutants accounts for their increased affinities for aa-tRNA and anisomycin resistance. Notably, the fewer number of changes in rRNA structure promoted by the S2A mutant suggests a reason for its lack of effect on aa-tRNA binding and anisomycin sensitivity. In the fully open conformation, the W-finger is in the ‘extended’ conformation, where its tip occupies the A site of the PTC. In this conformation, H256/H259 interact with A2940 of 25S rRNA (E. coli A2572, H. marismortui U2607). This is mimicked by mutations to W255, which also increase ribosomal affinity for aa-tRNA presumably by inducing the open conformation of accommodation corridor (20).
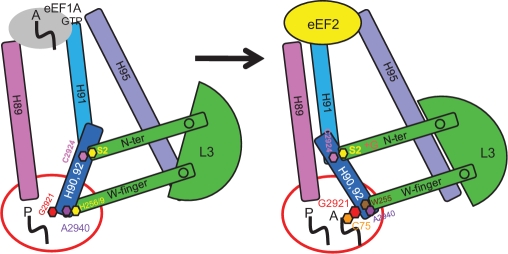
Figure 5.
L3 functions as a ‘rocker switch’ to coordinate elongation factor binding, aa-tRNA accommodation and PTC activation. (Left) Ribosome in ground state with P site occupied by peptidyl-tRNA. L3 W-finger is in the ‘extended’ conformation, maintaining the A site in the closed conformation. Interaction between H259 of L3 and A2940 (E. coli A2572) of 25S rRNA stabilize this state. The L3 N-terminal extension is in the ‘retracted’ conformation, opening the accommodation corridor. (Right) accommodation of aa-tRNA into the A site leads to the following events. (A) Opening of A site and retraction of the W-finger, stabilized by interaction between W255 of L3 and A2940 of 25S rRNA. (B) Rotation of L3 globular domain. (C) Extension of the N-terminal domain toward Helix 89. (D) Displacement of Helix 91, and closure of the accommodation corridor. (E) Repositioning of the H90/H92 structure and of G2921 (E. coli G2553) in the A site, where it can interact with C75 of the aa-tRNA to activate peptidyltransfer (55). (F) Movement of H91 away from H95 results in formation of the eEF2-binding site.
Accommodation of aa-tRNA into the A site displaces the W-finger, repositioning W255 to interact with A2940 (E. coli A2572), a state which is favored by mutagenesis of either H256 or H259 to alanine (20). We propose that this movement is transduced through the globular domain of L3, which in turn pushes the N-terminal extension toward the accommodation corridor. This in turn causes the H90–92 structure to move to close the accommodation corridor (with the resulting changes in protection patterns along this structure) and away from the SRL (resulting in its deprotection). We suggest that this state is somewhat mimicked by the iG12 mutant, although as noted above, this mutant is not able to fully close due to its having broken the coordination between the two gates. The closed state provides the structural basis for eEF2 binding (perhaps by making the SRL available for binding) as evidenced by the increased affinity of iG12 (this study), H259A and H256A (20) mutants for this elongation factor. In addition, closing of the accommodation corridor conferred protection from chemical attack on the bases in Helix 89 that interact with the N-terminal ‘hook’ extension of L10. In light of the data presented in the accompanying manuscript (see accompanying paper by Petrov et al.), we suggest that this provides a point of information transfer between L3 and L10 in helping to coordinate the different functions performed by the large subunit.
The reciprocal relationship between aa-tRNA and eEF2 binding appeared to be partially decoupled by the S2T and S2K mutants. We suggest that the explanation for this lies in the decreased strength of the interaction between the N-terminal extension and C2924 (E. coli C2556). We hypothesize that the resulting ‘more open than closed’ conformation allows increased affinity for aa-tRNA, but does not stabilize interaction between the loop of H91 and the SRL to the extent that significant inhibition of eEF2 binding was observed. However, this conformation of the accommodation corridor should enhance entry of aa-tRNA into the A site, consistent with the decreased _K_D values for the S2T and S2K mutants for aa-tRNA. As noted previously (20), the ability of anisomycin to access the A site should not be as sensitive to the conformational status of the accommodation due to its much smaller molecular radius. Thus, in these mutants, the kinetic partitioning ratio between anisomycin and aa-tRNA is lowered, providing an explanation for their drug resistance.
Examination of Supplementary Figures 1 and 2 reveals that only the eukaryotic and archael L3 proteins contain the N-terminal extension, and that no comparable structure exists in bacteria. Why might this be? Although the nuclear location signal is situated in this extension in eukaryotes, the absence of a nucleus in the Archaea suggests that the N-terminal extension originated for another reason. One possible explanation may come from the observation that N-terminus of bacterial ribosomal protein L27 extends into the PTC, where it is predicted to interact with the A76 phosphate of the A-site tRNA (44). Deletion of the N-terminus of L27 resulted in reduced rates of peptidyltransfer and decreased affinity for aa-tRNA (52). Eukaryotes and Archaea do not have a homologous protein. Thus, it is possible that they evolved the N-terminal extension of L3 to fulfill a similar function. A second explanation is suggested by the fact that the therapeutic basis for many antibiotics derives from their specific inhibition of bacterial as opposed to eukaryotic ribosomes (53). Examples specific to the large subunit include chloramphenicol, many of the macrolides and the streptogramin group of antibiotics (54). Thus, it is possible that the N-terminal extension of L3 may affect the structure of the A-site to favor aa-tRNA binding over that of antibiotics, and/or better coordinate peptidyltransfer, thus providing Archaea and eukaryotes with a selective advantage against small molecule inhibitors of protein synthesis. Experimentally, if addition of an N-terminal extension of L3 could be tolerated by E. coli, it would be interesting to note whether this would confer resistance to a class(s) of large-subunit-specific antibiotics. Alternatively, this could also be computationally modeled and subjected to molecular dynamics free energy simulations.
The effects of the iG12 mutant on G2921 (E. coli G2553, H. marismortui G2588) is of particular interest. It has been proposed that Watson–Crick pairing between this base and C75 of the aa-tRNA is a critical step in positioning of A-site substrate in the induced fit model of peptidyltransfer (55). Consistent with this model, G2921 became relatively protected when the A-site of wild-type ribosomes was occupied by aa-tRNA (Figures 3 and 4). Importantly, this protection failed to occur in the iG12 mutant. This could account for the strong increase in the _K_D of this mutant for aa-tRNA. Why then did the iG12 mutant have but a small effect on peptidyltransferase activity, even though its growth rate was significantly affected? We suggest that this can be explained by the fact that this parameter was monitored using puromycin instead of aa-tRNA, and that absence of a moiety like C75 on this drug served to minimize the observed effect by iG12 (we predict, that peptidyltransferase activity would be significantly reduced in this mutant if C-puromycin or aa-tRNA was used as substrate). In contrast, G2921 was strongly protected in salt-washed W255C mutant ribosomes, which did promote a very strong peptidyltransferase defect (20). These structural data suggest that both the N-terminal extension and W-finger L3 mutants may affect the C75-G2921 interaction and thus be indirectly involved in the induced fit mechanism of peptidyltransfer (55). Alternatively/additionally, changes in local rRNA structure affecting G2921 could cause a shift in the 3′ end of the aa-tRNA, thus misaligning it and promoting the observed defects in peptidyltransfer activity and aa-tRNA binding. Lastly, the defects of these mutants in their ability to maintain the yeast killer virus is consistent with the link between decreased peptidyltransferase activity and virus propagation (31).
The pattern of viability of the S2 mutants could not be classified by any of the biophysical properties of their amino acid sidechains. Although lethality could be due to defects in the interaction between the N-terminus of L3 and C2924, the observed pattern followed the ‘N-end rule’ of post-translational modification (56), suggesting that L3 stability may be the cause. Specifically, there was a strong correlation between amino acid identity at the second residue of the protein (Ser, Ala, Gly and Thr) and amino acids preferred for removal of the N-terminal methionine followed by acetylation in eukaryotes (49,57). Curiously, this is inconsistent with mass spectroscopic analyses of yeast ribosomes, where there is strong evidence showing that while N-terminal amino acid of L3 is serine, it is not acetylated (47,58,59) Such processed but unacetylated N-termini were also observed for many other yeast ribosomal proteins. Do these proteins simply bypass the stability requirements for co-translational N-terminal processing and acetylation, or are they initially processed and acetylated during translation, but later deacetylated for ribosome biogenesis? The latter idea is supported by the notion that ribosome assembly may be facilitated by interactions between highly basic, nonstructured extensions on ribosomal proteins and negatively charged rRNA phosphate groups. Acetylation of N-termini could interfere with assembly by reducing positive charges, potentially inhibiting RP–rRNA interactions, and promoting structural motifs in regions of RPs that need to be unstructured in order to assemble with rRNA. Thus, we suggest that although acetylation is required to stabilize RPs in the cytoplasmic compartment of the cell, this chemical modification must be removed for the process of ribosome biogenesis in the nucleolus. This in turn raises the question of the identity of the deacetylase. A prior study showed that mutation or deletion of RPD3, better known as a histone deacetylase, and of proteins that target Rpd3p to heterochromatin but not to euchromatin, resulted in phenotypic defects similar to those observed with many L3 mutants (60). The hypothesis that Rpd3p may also play a critical role in deacetylating ribosomal proteins prior to their incorporation into nascent ribosomes is currently being tested in our laboratory.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (GM058859 to J.D.D.); and the American Heart Association (AHA 0630163N to A.M.). Funding for open access charge: NIH GM058859.
Conflict of interest statement. None declared.
Supplementary Material
[Supplementary Data]
ACKNOWLEDGEMENTS
We wish to thank the members of our laboratory, with special thanks to Rasa Rakauskaitė, Karen Jack, Ashton Trey Belew and Michael Rhodin. Special thanks to Marat Yusupov and Joachim Frank for providing deep insights into ribosome structure.
REFERENCES
- 1.Spirin AS. The ribosome as an RNA-based molecular machine. RNA Biol. 2004;1:3–9. [PubMed] [Google Scholar]
- 2.Yusupova G, Jenner L, Rees B, Moras D, Yusupov M. Structural basis for messenger RNA movement on the ribosome. Nature. 2006;444:391–394. doi: 10.1038/nature05281. [DOI] [PubMed] [Google Scholar]
- 3.Noller HF. Structure of the bacterial ribosome and some implications for translational regulation. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational Control in Biology and Medicine. Cold Spring Harbor, NY: Cold Spring Harbor Press; 2007. pp. 41–58. [Google Scholar]
- 4.Mitra K, Frank J. Ribosome dynamics: insights from atomic structure modeling into cryo-electron microscopy maps. Annu. Rev. Biophys. Biomol. Struct. 2006;35:299–317. doi: 10.1146/annurev.biophys.35.040405.101950. [DOI] [PubMed] [Google Scholar]
- 5.Vanzi F, Vladimirov S, Knudsen CR, Goldman YE, Cooperman BS. Protein synthesis by single ribosomes. RNA. 2003;9:1174–1179. doi: 10.1261/rna.5800303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Unbehaun A, Marintchev A, Lomakin IB, Didenko T, Wagner G, Hellen CU, Pestova TV. Position of eukaryotic initiation factor eIF5B on the 80S ribosome mapped by directed hydroxyl radical probing. EMBO J. 2007;26:3109–3123. doi: 10.1038/sj.emboj.7601751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daviter T, Gromadski KB, Rodnina MV. The ribosome's response to codon-anticodon mismatches. Biochimie. 2006;88:1001–1011. doi: 10.1016/j.biochi.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 8.Cochella L, Green R. An active role for tRNA in decoding beyond codon:anticodon pairing. Science. 2005;308:1178–1180. doi: 10.1126/science.1111408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sergiev PV, Lesnyak DV, Burakovsky DE, Kiparisov SV, Leonov AA, Bogdanov AA, Brimacombe R, Dontsova OA. Alteration in location of a conserved GTPase-associated center of the ribosome induced by mutagenesis influences the structure of peptidyltransferase center and activity of elongation factor G. J. Biol. Chem. 2005;280:31882–31889. doi: 10.1074/jbc.M505670200. [DOI] [PubMed] [Google Scholar]
- 10.Rakauskaite R, Dinman JD. An arc of unpaired “hinge bases” facilitates information exchange among functional centers of the ribosome. Mol. Cell. Biol. 2006;26:8992–9002. doi: 10.1128/MCB.01311-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hennelly SP, Antoun A, Ehrenberg M, Gualerzi CO, Knight W, Lodmell JS, Hill WE. A time-resolved investigation of ribosomal subunit association. J. Mol. Biol. 2005;346:1243–1258. doi: 10.1016/j.jmb.2004.12.054. [DOI] [PubMed] [Google Scholar]
- 12.Amort M, Wotzel B, Bakowska-Zywicka K, Erlacher MD, Micura R, Polacek N. An intact ribose moiety at A2602 of 23S rRNA is key to trigger peptidyl-tRNA hydrolysis during translation termination. Nucleic Acids Res. 2007;35:5130–5740. doi: 10.1093/nar/gkm539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Munro JB, Vaiana A, Sanbonmatsu KY, Blanchard SC. A new view of protein synthesis: mapping the free energy landscape of the ribosome using single-molecule FRET. Biopolymers. 2008;89:565–577. doi: 10.1002/bip.20961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cornish PV, Ermolenko DN, Noller HF, Ha T. Spontaneous intersubunit rotation in single ribosomes. Mol. Cell. 2008;30:578–588. doi: 10.1016/j.molcel.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilson DN, Nierhaus KH. Ribosomal proteins in the spotlight. Crit. Rev. Biochem. Mol. Biol. 2005;40:243–267. doi: 10.1080/10409230500256523. [DOI] [PubMed] [Google Scholar]
- 16.Stelzl U, Zengel JM, Tovbina M, Walker M, Nierhaus KH, Lindahl L, Patel DJ. RNA-structural mimicry in Escherichia coli ribosomal protein L4-dependent regulation of the S10 operon. J. Biol. Chem. 2003;278:28237–28245. doi: 10.1074/jbc.M302651200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Agrawal RK, Linde J, Sengupta J, Nierhaus KH, Frank J. Localization of L11 protein on the ribosome and elucidation of its involvement in EF-G-dependent translocation. J. Mol. Biol. 2001;311:777–787. doi: 10.1006/jmbi.2001.4907. [DOI] [PubMed] [Google Scholar]
- 18.Maguire BA, Beniaminov AD, Ramu H, Mankin AS, Zimmermann RA. A protein component at the heart of an RNA machine: the importance of protein l27 for the function of the bacterial ribosome. Mol. Cell. 2005;20:427–435. doi: 10.1016/j.molcel.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 19.Takyar S, Hickerson RP, Noller HF. mRNA helicase activity of the ribosome. Cell. 2005;120:49–58. doi: 10.1016/j.cell.2004.11.042. [DOI] [PubMed] [Google Scholar]
- 20.Meskauskas A, Dinman JD. Ribosomal protein L3: gatekeeper to the A-site. Mol. Cell. 2007;25:877–888. doi: 10.1016/j.molcel.2007.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 22.Brodersen DE, Nissen P. The social life of ribosomal proteins. FEBS J. 2005;272:2098–2108. doi: 10.1111/j.1742-4658.2005.04651.x. [DOI] [PubMed] [Google Scholar]
- 23.Nowotny V, Nierhaus KH. Initiator proteins for the assembly of the 50S subunit from Escherichia coli ribosomes. Proc. Natl Acad. Sci. USA. 1982;79:7238–7242. doi: 10.1073/pnas.79.23.7238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schulze H, Nierhaus KH. Minimal set of ribosomal components for reconstitution of the peptidyltransferase activity. EMBO J. 1982;1:609–613. doi: 10.1002/j.1460-2075.1982.tb01216.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schindler D, Grant P, Davies J. Trichodermin resistance–mutation affecting eukaryotic ribosomes. Nature. 1974;248:535–536. doi: 10.1038/248535a0. [DOI] [PubMed] [Google Scholar]
- 26.Jimenez A, Sanchez L, Vazquez D. Simultaneous ribosomal resistance to trichodermin and anisomycin in Saccharomyces cerevisiae mutants. Biochim. Biophys. Acta. 1975;383:427–434. doi: 10.1016/0005-2787(75)90312-3. [DOI] [PubMed] [Google Scholar]
- 27.Fried HM, Warner JR. Cloning of yeast gene for trichodermin resistance and ribosomal protein L3. Proc. Natl Acad. Sci. USA. 1981;78:238–242. doi: 10.1073/pnas.78.1.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wickner RB, Porter-Ridley S, Fried HM, Ball SG. Ribosomal protein L3 is involved in replication or maintenance of the killer double-stranded RNA genome of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 1982;79:4706–4708. doi: 10.1073/pnas.79.15.4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peltz SW, Hammell AB, Cui Y, Yasenchak J, Puljanowski L, Dinman JD. Ribosomal protein L3 mutants alter translational fidelity and promote rapid loss of the yeast killer virus. Mol. Cell. Biol. 1999;19:384–391. doi: 10.1128/mcb.19.1.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hudak KA, Dinman JD, Tumer NE. Pokeweed antiviral protein accesses ribosomes by binding to L3. J. Biol. Chem. 1999;274:3859–3864. doi: 10.1074/jbc.274.6.3859. [DOI] [PubMed] [Google Scholar]
- 31.Meskauskas A, Harger JW, Jacobs KLM, Dinman JD. Decreased peptidyltransferase activity correlates with increased programmed -1 ribosomal frameshifting and viral maintenance defects in the yeast Saccharomyces cerevisiae. RNA. 2003;9:982–992. doi: 10.1261/rna.2165803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bosling J, Poulsen SM, Vester B, Long KS. Resistance to the peptidyl transferase inhibitor tiamulin caused by mutation of ribosomal protein L3. Antimicrob. Agents Chemother. 2003;47:2892–2896. doi: 10.1128/AAC.47.9.2892-2896.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Petrov A, Meskauskas A, Dinman JD. Ribosomal protein L3: influence on ribosome structure and function. RNA Biol. 2004;1:59–65. doi: 10.4161/rna.1.1.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pringle M, Poehlsgaard J, Vester B, Long KS. Mutations in ribosomal protein L3 and 23S ribosomal RNA at the peptidyl transferase centre are associated with reduced susceptibility to tiamulin in Brachyspira spp. isolates. Mol. Microbiol. 2004;54:1295–1306. doi: 10.1111/j.1365-2958.2004.04373.x. [DOI] [PubMed] [Google Scholar]
- 35.Meskauskas A, Petrov AN, Dinman JD. Identification of functionally important amino acids of ribosomal protein L3 by saturation mutagenesis. Mol. Cell. Biol. 2005;25:10863–10874. doi: 10.1128/MCB.25.24.10863-10874.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klein DJ, Moore PB, Steitz TA. The roles of ribosomal proteins in the structure assembly, and evolution of the large ribosomal subunit. J. Mol. Biol. 2004;340:141–177. doi: 10.1016/j.jmb.2004.03.076. [DOI] [PubMed] [Google Scholar]
- 37.Dinman JD, Wickner RB. Ribosomal frameshifting efficiency and Gag/Gag-pol ratio are critical for yeast M1 double-stranded RNA virus propagation. J. Virol. 1992;66:3669–3676. doi: 10.1128/jvi.66.6.3669-3676.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boeke JD, LaCroute F, Fink GR. A positive selection for mutants lacking orotidine-5'-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 39.Rheinberger HJ, Geigenmuller U, Wedde M, Nierhaus KH. Parameters for the preparation of Escherichia coli ribosomes and ribosomal subunits active in tRNA binding. Methods Enzymol. 1988;164:658–670. doi: 10.1016/s0076-6879(88)64076-6. [DOI] [PubMed] [Google Scholar]
- 40.Dresios J, Panopoulos P, Frantziou CP, Synetos D. Yeast ribosomal protein deletion mutants possess altered peptidyltransferase activity and different sensitivity to cycloheximide. Biochemistry. 2001;40:8101–8108. doi: 10.1021/bi0025722. [DOI] [PubMed] [Google Scholar]
- 41.Ortiz PA, Ulloque R, Kihara GK, Zheng H, Kinzy TG. Translation elongation factor 2 anticodon mimicry domain mutants affect fidelity and diphtheria toxin resistance. J. Biol. Chem. 2006;281:32639–32648. doi: 10.1074/jbc.M607076200. [DOI] [PubMed] [Google Scholar]
- 42.Stern S, Moazed D, Noller HF. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol. 1988;164:481–489. doi: 10.1016/s0076-6879(88)64064-x. [DOI] [PubMed] [Google Scholar]
- 43.Spahn CM, Gomez-Lorenzo MG, Grassucci RA, Jorgensen R, Andersen GR, Beckmann R, Penczek PA, Ballesta JP, Frank J. Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation. EMBO J. 2004;23:1008–1019. doi: 10.1038/sj.emboj.7600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Selmer M, Dunham CM, Murphy FV, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 45.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 46.Harms JM, Wilson DN, Schluenzen F, Connell SR, Stachelhaus T, Zaborowska Z, Spahn CM, Fucini P. Translational regulation via L11: molecular switches on the ribosome turned on and off by thiostrepton and micrococcin. Mol. Cell. 2008;30:26–38. doi: 10.1016/j.molcel.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 47.Lee SW, Berger SJ, Martinovic S, Pasa-Tolic L, Anderson GA, Shen Y, Zhao R, Smith RD. Direct mass spectrometric analysis of intact proteins of the yeast large ribosomal subunit using capillary LC/FTICR. Proc. Natl Acad. Sci. USA. 2002;99:5942–5947. doi: 10.1073/pnas.082119899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sanbonmatsu KY, Joseph S, Tung CS. Simulating movement of tRNA into the ribosome during decoding. Proc. Natl Acad. Sci. USA. 2005;102:15854–15859. doi: 10.1073/pnas.0503456102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Polevoda B, Sherman F. N-terminal acetyltransferases and sequence requirements for N-terminal acetylation of eukaryotic proteins. J. Mol. Biol. 2003;325:595–622. doi: 10.1016/s0022-2836(02)01269-x. [DOI] [PubMed] [Google Scholar]
- 50.Moreland RB, Nam HG, Hereford LM, Fried HM. Identification of a nuclear localization signal of a yeast ribosomal protein. Proc. Natl Acad. Sci. USA. 1985;82:6561–6565. doi: 10.1073/pnas.82.19.6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grollman AP. Inhibitors of protein biosynthesis. II. Mode of action of anisomycin. J. Biol. Chem. 1967;242:3226–3233. [PubMed] [Google Scholar]
- 52.Wower IK, Wower J, Zimmermann RA. Ribosomal protein L27 participates in both 50 S subunit assembly and the peptidyl transferase reaction. J. Biol. Chem. 1998;273:19847–19852. doi: 10.1074/jbc.273.31.19847. [DOI] [PubMed] [Google Scholar]
- 53.Pestka S. Inhibitors of ribosome functions. Annu. Rev. Microbiol. 1971;25:487–562. doi: 10.1146/annurev.mi.25.100171.002415. [DOI] [PubMed] [Google Scholar]
- 54.Pestka S. Inhibitors of protein synthesis. In: Weissbach H, Pestka S, editors. Molecular Mechanismns of Protein Biosynthesis. New York: Academic Press; 1977. pp. 467–553. [Google Scholar]
- 55.Schmeing TM, Huang KS, Strobel SA, Steitz TA. An induced-fit mechanism to promote peptide bond formation and exclude hydrolysis of peptidyl-tRNA. Nature. 2005;438:520–524. doi: 10.1038/nature04152. [DOI] [PubMed] [Google Scholar]
- 56.Varshavsky A. The N-end rule. Cell. 1992;69:725–735. doi: 10.1016/0092-8674(92)90285-k. [DOI] [PubMed] [Google Scholar]
- 57.Bradshaw RA, Brickey WW, Walker KW. N-terminal processing: the methionine aminopeptidase and N alpha-acetyl transferase families. Trends Biochem. Sci. 1998;23:263–267. doi: 10.1016/s0968-0004(98)01227-4. [DOI] [PubMed] [Google Scholar]
- 58.Takakura H, Tsunasawa S, Miyagi M, Warner JR. NH2-terminal acetylation of ribosomal proteins of Saccharomyces cerevisiae. J. Biol. Chem. 1992;267:5442–5445. [PubMed] [Google Scholar]
- 59.Arnold RJ, Polevoda B, Reilly JP, Sherman F. The action of N-terminal acetyltransferases on yeast ribosomal proteins. J. Biol. Chem. 1999;274:37035–37040. doi: 10.1074/jbc.274.52.37035. [DOI] [PubMed] [Google Scholar]
- 60.Meskauskas A, Baxter JL, Carr EA, Yasenchak J, Gallagher JEG, Baserga SJ, Dinman JD. Delayed rRNA processing results in significant ribosome biogenesis and functional defects. Mol. Cell Biol. 2003;23:1602–1613. doi: 10.1128/MCB.23.5.1602-1613.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplementary Data]