The ARF tumor suppressor can promote the progression of some tumors (original) (raw)
. Author manuscript; available in PMC: 2009 Dec 1.
Abstract
p14/p19ARF (ARF) is a tumor suppressor gene that is frequently mutated in human cancer. ARF has multiple tumor suppressor functions, some of which are mediated by signalling to p53. Surprisingly, a significant fraction of human tumors retain persistently high levels of ARF, suggesting that ARF may possess a pro-survival function. We show that ARF protein is markedly upregulated in cells exposed to nutrient starvation. Cells with silenced ARF demonstrate reduced autophagy and reduced viability when placed under conditions of starvation. We show for the first time that ARF silencing can limit the progression of some tumors, such as lymphoma, but not others, such as E1A/ras-induced tumors. Specifically, myc-driven lymphomas with mutant p53 tend to overexpress ARF; we show that silencing ARF in these tumors greatly impedes their progression. These data are the first to show that ARF can act in a p53-independent manner to promote the progression of some tumors.
Keywords: ARF, mitochondria, autophagy
Introduction
The ARF tumor suppressor is best known for its ability to stabilize p53, mediated by inhibition of the p53 ubiquitin ligase MDM2. Additionally, ARF has multiple tumor suppressor functions that are independent of p53: these involve interaction with a number of proteins with roles in cell proliferation, including nucleophosmin, NIAM, c-myc, E2F-1, DP1, and others (see 1 for review). Despite its multiple growth suppression functions, ARF is inexplicably overexpressed in a significant fraction of human tumors, including up to 50% of Burkitt’s lymphomas (2, 3), as well as the majority of tumors with mutant p53 (4). To date, the overwhelming majority of studies on ARF have focused on its tumor suppressor roles, and no groups have addressed the possibility that ARF might promote the survival of a subset of tumors.
A role for ARF in autophagy has recently been discovered. Specifically, a small molecular weight variant of this protein generated by translation from an internal methionine has been shown to localize to mitochondria and induce autophagy (5, 6). More recently, another group has shown that full-length ARF can likewise induce autophagy (7). Autophagy is a process of lysosome-mediated self-digestion that occurs during periods of nutrient deprivation (see 8 for review). This process is necessary for cell survival; following nutrient starvation proteins and organelles are degraded, and the released amino acids are utilized for the synthesis of essential proteins. Whereas recent studies have shown that ARF can induce autophagy, the relevance of ARF to starvation-induced autophagy, and the significance of this finding to cancer development, have not been addressed. In this study we show that nutrient deprivation leads to significant increases in ARF protein. We show that ARF-mediated autophagy can protect cells from periods of nutrient deprivation. Further, we show that lymphomas with ARF silenced demonstrate impaired progression in immunocompromised mice. Our data support the premise that ARF can enhance tumor development under certain circumstances, and that some tumors with mutant p53 may retain ARF in order to promote survival under metabolic stress.
Materials and Methods
Cell culture, retroviral infections, cell viability
Wild type, p53-null, and p53/ARF double knockout MEFs were cultured as described (9). B cell lymphomas B2998 and E330 were cultured in RPMI with 20% heat-inactivated fetal calf serum, 55 uM β-mercaptoethanol and 10 ng/mL IL-7 (R&D Systems). Retroviral infection, p53 short hairpins and short hairpin control vector are described (10). shRNA vectors for ARF were generated by PCR amplification of 97mer DNA oligonucleotides as described (11) and cloned into the LMP vector (12). Oligonucleotide sequences for shARF 157:
5′TGCTGTTGACAGTGAGCGACGCTCTGGCTTTCGTGAACATTAGTGAAGCCACAGATGTAA
TGTTCACGAAAGCCAGAGCGCTGCCTACTGCCTCGGA3′.
shARF 56:
5′TGCTGTTGACAGTGAGCGATTGGTCACTGTGAGGATTCAGTAGTGAAGCCACAGATGTAC
TGAATCCTCACAGTGACCAAGTGCCTACTGCCTCGGA3′.
Unless otherwise indicated, shARF56 was used for all experiments to silence ARF. Cell viability assays were performed on using the ViaCount assay and the Guava PCA (Guava Technologies); statistical significance was calculated using the two-sided student’s t test.
Mitochondria isolation, western analysis, Immuno-electron microscopy
Mitochondria were purified as described (13). Western blotting was performed on 100 ug whole cell lysate or 20 ug mitochondrial lysate; antisera used were anti-p19ARF (GeneTex), anti-actin (AC15, Sigma), anti-LC3 (Novus), and anti-Beclin-1 (Santa Cruz Biotechnology), anti-p62SQSTM1 (Santa Cruz Biotechnology). For electron microscopy, cells were fixed with 2% glutaraldehyde/2% formaldehyde in cacodylate buffer, fixed in 1% osmium tetroxide, embedded in epoxy resin and stained with uranyl acetate and lead citrate. The area of autophagosomes was calculated using NIH Image. Immuno-electron microscopy was performed as described (14) and visualized by secondary labeling with Protein A-colloidal gold. All samples were viewed in a FEI Tecnai 12 TEM operated at 80 kV.
Long-lived Protein Degradation Assay
Long-lived protein degradation was assayed as described (15). The percentage of long-lived protein degraded was obtained by the formula % degradation = (14C counts at timepoint/sum of 14C counts at each time point + total cell-associated radioactivity) × 100.
Bioluminescent imaging
For xenograft analyses, MEFs were co-infected with retrovirus encoding luciferase along with retroviruses encoding vector, shBec1, shp53, or E1a/Ras before being injected (1×106 cells/injection) into SCID mice. E1A, Ras and luciferase levels were monitored by western analysis. Prior to injection, luciferase activity in infected cells was tested and silencing of all genes was confirmed by western analysis. For biophotonic imaging on the IVIS® Spectrum (Xenogen Biosciences), 1% isofluorane/oxygen-anesthetized mice were injected with luciferin (4 mg/animal) into the intraperitoneal cavity. In all cases the averaged data from each of two independent experiments using 3 animals for each sample were calculated; caliper measurements of tumors corroborated these analyses. For the lymphoma studies, cells were infected with shControl or shARF four times and selected in 1.5 ug/mL puromycin for 48 hours. After one week cells were resuspended in PBS and 1×106 cells were injected into the tail vein of SCID mice. For these studies the averaged data from each of two independent experiments (3 animals per sample) plus standard error of the mean were calculated and analyzed by the Wilcoxin two sample test, and p values were combined according to the method of Fisher.
Results
ARF plays a role in autophagy induced by nutrient deprivation
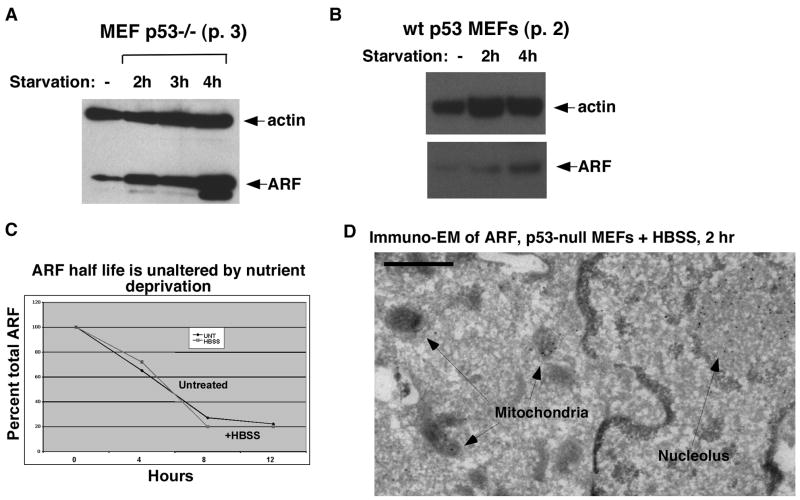
ARF expression is negligible in normal non-transformed cells. This gene is also potently repressed by p53; consequently p53-null mouse embryo fibroblasts (MEFs) express high levels of ARF (4, 16). To determine the relevance of ARF to starvation-induced autophagy, we exposed p53-null MEFs to nutrient deprivation by incubating them in Hanks Buffered Salt Solution (HBSS), which is amino acid and growth factor-free. HBSS incubation led to a rapid (2 h) and significant up-regulation of ARF protein (Fig. 1_A_); this occurred independent of p53 status (Fig. 1_B_). Data quantitated from several experiments indicate that ARF is up-regulated approximately 1.6-fold in wt MEFs, and 6.5-fold in p53 −/− MEFs after 4 hours incubation in HBSS (data not shown). Induction of ARF mRNA by serum starvation has been noted previously (17). However, in response to HBSS, the up-regulation of ARF was not accompanied by an increase in ARF mRNA (data not shown). Further, there were no differences in ARF half-life under these conditions (Fig. 1_C_). The increase in ARF may be due to enhanced translation of ARF mRNA during nutrient deprivation.
Figure 1. Nutrient deprivation causes significant upregulation of ARF, along with mitochondrial localization of ARF.
(A) Western analysis of whole cell lysate from p53 −/− MEFs (passage 3) untreated or incubated in Hank’s Buffered Salt Solution (HBSS) for the indicated timepoints (starvation) using antisera to actin (loading control) and ARF.
(B) Western analysis of MEFs with wt p53 (passage 2), untreated or incubated in Hank’s Buffered Salt Solution (HBSS) for the indicated timepoints (starvation) using antisera to actin (loading control) and ARF.
(C) Analysis of the half life of ARF in untreated and nutrient-deprived cells. p53-null MEFs were untreated (UNT) or incubated for 2 h in HBSS, followed by supplementation with complete media containing 40 ug/uL cycloheximide to halt new protein synthesis. Cells were harvested and analyzed by western blotting for ARF at the indicated timepoints; the data depicted are representative of two independent experiments.
(D) Immuno-electron microscopy using ARF antisera followed by Protein G-Gold in mouse embryo fibroblasts from the p53-knockout mouse following two hours treatment with HBSS (starvation). Arrows point to gold particles in mitochondria and nucleoli. Scale bar is 500 nm.
ARF has been shown to play a role in autophagy via its localization to mitochondria (5). We could find no evidence for ARF localization at mitochondria in unstressed p53-null MEFs (Supplemental Fig. 1_A_); however, immuno-electron microscopy analyses revealed considerable localization of ARF to mitochondria after 2 hours of nutrient deprivation (Fig. 1_D_). These data were consistent with western analyses of purified mitochondria, which showed demonstrable ARF co-purifying with mitochondria after 2 hours of HBSS (Supplemental Fig. 1_B_). Quantitative western analysis and counting of grains from immuno-EM indicated that between five to ten percent of ARF localized to mitochondria following HBSS treatment.
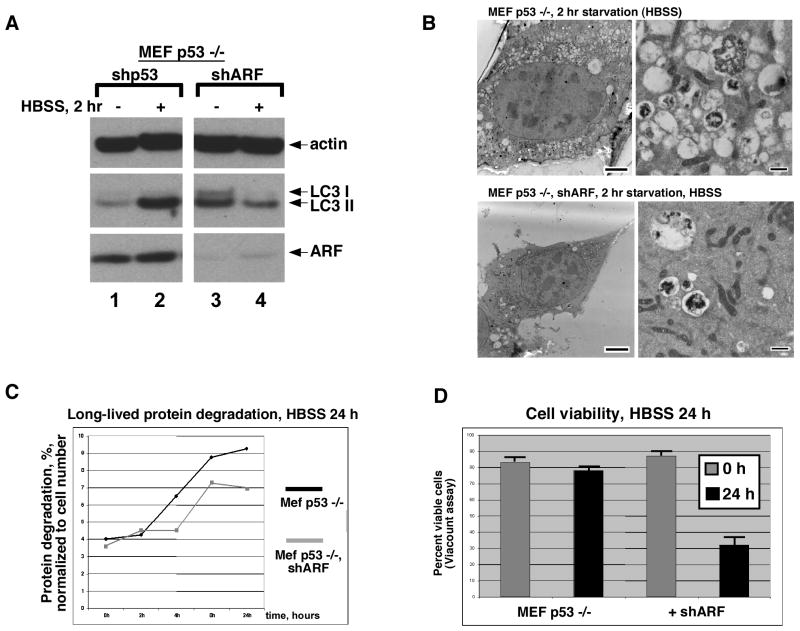
The relevance of ARF upregulation to starvation-induced autophagy was assessed in p53-null MEFs using a short hairpin to silence this gene (shARF); this hairpin reduces over 90% of ARF protein, but does not silence p16ink4a expression (for example see Fig. 4A). In these experiments, the extent of autophagy was assessed by western analysis for the protein LC3, which becomes processed to a faster mobility species during autophagy (LC3 II), as well as by autophagosome formation and the analysis of long-lived protein degradation. Infection of p53-null MEFs with a parental retrovirus, or a virus encoding a short hairpin for p53 (which has no target in p53-null cells) did not influence the accumulation of LC3 II following HBSS treatment (Fig. 2_A_). Notably, however, ARF-silencing in p53-null MEFs led to markedly decreased accumulation of LC3 II following HBSS treatment (Fig. 2_A_). Moreover, electron microscopy indicated that following HBSS treatment, cells with silenced ARF had fewer, and often smaller, autophagosomes than control cells (Fig. 2_B_). The area of autophagosome coverage in HBSS-treated cells decreased from 48 +/− 5% to 7+/− 2% when ARF was silenced. Consistent with this, silencing ARF in p53-null MEFs reduced the degradation of long-lived proteins in response to HBSS treatment (Fig. 2_C_). Not surprisingly, this decrease in autophagy was accompanied by a decrease in viability; after 24 hours of nutrient deprivation in HBSS the majority of p53-null MEFs remained viable, while only 30% of cells with silenced ARF were viable at this timepoint (Fig. 2_D_). The combined data point to a previously unseen role for ARF in promoting cell survival during metabolic stress.
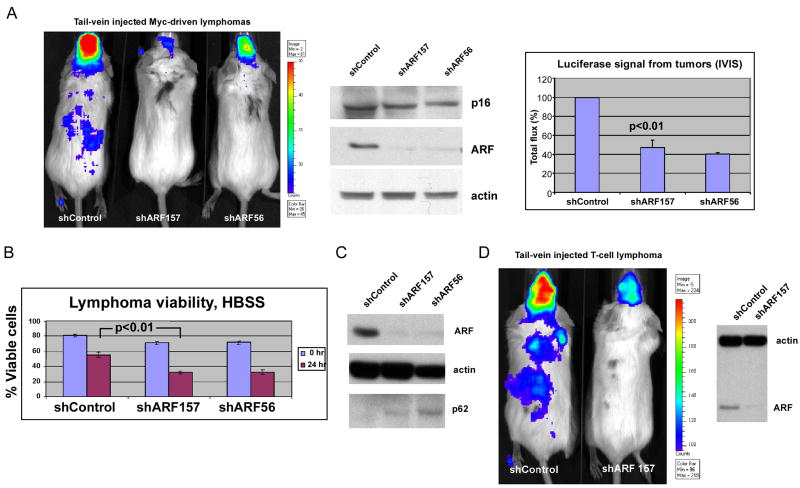
Figure 4. Silencing of ARF inhibits lymphoma development.
(A) Bioluminescent imaging of tail-vein injected, luciferase-expressing B2998 B-cell lymphoma cells from the Eμ-myc mouse, infected with vector control or two different short hairpins for ARF, shARF157 or shARF56. Most tumor cells track to the lymph nodes and meninges. Results are at day 7 following tail vein injection; equivalent results were obtained at day 14. Data depicted are representative of two independent experiments, using 3 mice for each retrovirus (shControl, shARF157, shARF56). The middle panel depicts western analysis for p16, ARF and actin control in infected lymphoma cells from the B2998 lymphoma, 48 hours after retroviral infection. The right panel depicts the combined data from 2 independent experiments using 3 mice per virus, along with standard error of the mean. Silencing of ARF, but not infection with short hairpin control vector, reduces tumor volume by 60% (p<0.01).
(B) The viability of B2998 cells with silenced ARF is decreased following HBSS treatment for 24 hours. Cell viability was determined using the ViaCount assay and the Guava Personal Cell Analysis machine. Data depicted are the averaged results from three independent experiments plus standard error. The p value reflects comparison between the second and fourth column.
(C) Silencing ARF in B2998 lymphoma cells reduces the steady state level of autophagy. Western analysis for ARF, actin and p62SQSTM1 (which is degraded by autophagy) in cells infected with shControl or each of two ARF short hairpins (shARF157 and shARF56).
(D) Silencing ARF in a primary T cell lymphoma from the p53 −/− mouse impedes tumor progression. Results are at day 2 following tail vein injection; equivalent results were obtained at day 4. Results are representative of two independent experiments. The right panel depicts western analysis for ARF and loading control (actin).
Figure 2. ARF-silencing reduces starvation-induced autophagy and survival.
(A) Western analysis for LC3 II in p53 −/− MEFs following nutrient starvation (HBSS). Cells were infected for 48 hours with a short hairpin for p53 (shp53, negative control) or short hairpin for ARF (shARF), and incubated for 2 hrs in HBSS. Data depicted are representative of three independent experiments; the depicted data were obtained from a single blot that was cropped for optimal data presentation. The difference in LC3 II in unstressed cells in lanes 1 and 3 was not consistent or reproducible from experiment to experiment.
(B) Electron microscopy of p53-null MEFs infected with shp53 (negative control, top panels) or shARF (bottom panels) following 2 hour incubation in HBSS (starvation). Scale bars: Left panels, 5 um. Right panels 500 nm.
(C) The degradation of long-lived proteins in p53-null MEFs infected with short hairpin to p53 (negative control, black line) is increased compared to those infected with short hairpin to ARF (shARF, gray line) following starvation in HBSS for 24 hours; results are normalized to cell number. The x axis indicates timepoints of sampling. Data are representative of two independent experiments.
(D) The viability of p53-null cells infected with short hairpin to p53 (MEF p53 −/−) or shARF following 24 hour treatment with HBSS is depicted; equal numbers of viable cells were plated at the 0 timepoint (1 × 106), and cell viability was determined using the ViaCount assay and the Guava Personal Cell Analysis machine. Data depicted are the averaged results from three independent experiments plus standard error.
Tumors without ARF are more sensitive to autophagy inhibition
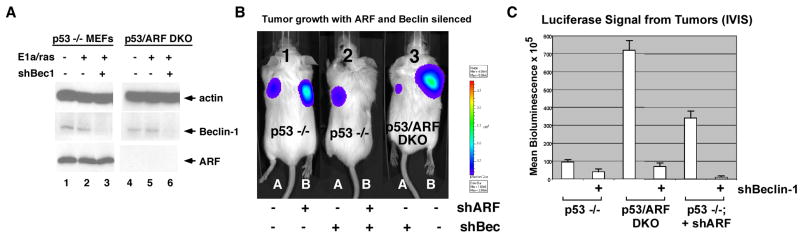
We next sought to determine whether ARF-mediated autophagy might promote tumor growth. For this analysis we chose to induce metabolic stress in tumors by silencing Beclin-1, a critical mediator of autophagy. Other groups have found that autophagy inhibition causes tumor regression (18, 19); similarly, we found that silencing Beclin-1 inhibited xenograft tumor development (Supplemental Fig. 3_A_). Interestingly, we found that the effect of Beclin-1 silencing on tumor growth was influenced by ARF status. For these studies we created luciferase-expressing transformed MEF cell lines that differed in ARF status (p53-null versus p53/ARF double knockout MEFs infected with E1A and Ras); these were subjected to Beclin-1 silencing and monitored for tumor development in a xenograft assay. We were able to achieve significant silencing of Beclin-1 in these lines (Fig. 3_A_). Bioluminescent imaging indicated that tumors with high ARF (p53-null) were consistently resistant to the growth suppressive effects of silencing Beclin-1, compared to tumors without ARF (p53/ARF double knockout, Fig. 3_B_, compare mouse 2A to 3A). Nearly identical results were obtained when we silenced ARF with a short hairpin (shARF, Fig. 3_B_, compare mouse 2A to 2B). The combined results from two independent experiments using three mice per virus are plotted in Figure 3_C_; these data indicate that tumors without ARF are markedly more sensitive to growth inhibition induced by Beclin-1 silencing.
Figure 3. Tumors without ARF are more sensitive to metabolic stress (silencing of Beclin-1).
(A) Western analysis of p53-null MEFS (high ARF) and MEFs from the p53/ARF double knockout mouse (p53/ARF DKO) following infection with short hairpin to silence Beclin-1 (shBec1) for the levels of actin (loading control), ARF and Beclin-1.
(B) Bioluminescent imaging of xenograft tumors derived from early passage MEFs from the p53 −/− mouse, or p53/ARF double knockout (DKO) stably-infected with luciferase virus, and then infected with the retroviruses indicated (shARF or shBec) for 48 hours. Tumors were imaged after 21 days; equivalent results were seen after 30 days, and all measurements were consistent with caliper measurements.
(C) Analysis of the combined imaging data from two independent experiments depicted in B, using three mice for each sample per experiment (6 samples total per retrovirus combination); all values represent the raw mean value of bioluminescence (mean BLI) along with standard error of the mean (SEM).
Silencing of ARF inhibits the progression of lymphomas with mutant or no p53
To assess the physiological relevance of ARF-mediated autophagy to tumor development, we silenced ARF in tumors that overexpress this protein and assessed the impact on tumorigenesis. Toward this end we utilized a primary murine B cell lymphoma cell line (B2998) from the Eμ-myc transgenic mouse that contains mutant p53 (C.M. Eischen, unpublished results) and high levels of ARF. We infected this lymphoma cell line with one of two different short hairpins for ARF (shARF157 or shARF56) or a short hairpin control virus (shControl), followed by a luciferase retrovirus. Following confirmation that luciferase activity was equivalent and that ARF was silenced, cells were injected into the tail vein of immunocompromised mice and imaged after 7 days. As depicted in Figure 4_A_, infection with either of two different ARF short hairpins, but not with a short hairpin control, led to markedly impaired lymphoma development. This result was consistent in another lymphoma cell line containing mutant p53 (E330, Supplemental Fig. 3_B_), and was consistent when tumors were imaged after 7 or 14 days (data not shown). The combined data from two independent experiments using three mice per virus are plotted in Figure 4_A_; these data indicate that the inhibition of tumor progression by ARF silencing was reproducible and statistically significant (p< 0.01). Notably, ARF silencing also led to decreased viability of B2998 cells following HBSS treatment (Fig. 4_B_) and hypoxia (Supplemental Fig. 3_C_) and to decreased levels of steady state autophagy, as evidenced by accumulation of p62SQSTM1, which is degraded by autophagy (Fig. 4_C_). In contrast, ARF silencing did not affect the proliferation of lymphoma cells cultured under nutrient-rich conditions (Supplemental Fig. 3D). To confirm these findings in another cell line, we silenced ARF in a primary T cell lymphoma from the p53-null mouse; here again we found that ARF silencing reduced tumor development (Fig. 4_D_), suggesting that ARF can confer a survival advantage to certain tumors.
Discussion
In this manuscript we provide data indicating that ARF has a previously undiscovered tumor-promoting activity, and that this activity may be mediated by its role in starvation-induced autophagy. These data do not discount the ample data indicating that ARF is a tumor suppressor, as all of our studies are performed in cells with mutant or null p53, exposed to metabolic stress. While some groups have reported that ARF-mediated autophagy is cytotoxic (5–7), we find that ARF is cytoprotective to tumors. This discrepancy may be due to differences in cell types analyzed, or the use of transfection and supra-physiologic overexpression of ARF by these other groups. We find that ARF-silencing in myc-driven lymphoma cells containing mutant p53 impedes their survival under nutrient-starved conditions and their development as xenografts. These data are the first to indicate that the ARF tumor suppressor has a survival function, and further that this survival function may be selected for in certain tumors. It should be noted, however, that this survival function is likely to be tumor-type specific; consistent with the data of others (9), we find that ARF silencing enhances the development of E1A/Ras-driven tumors (Fig. 3_C_). Ras can inhibit autophagy (20), and this may explain why ARF-mediated autophagy has no selective benefit to Ras-driven tumors. The findings described here-in have important clinical relevance: autophagy modulators like chloroquine and rapamycin are currently used in clinical trials for human cancer. Our data suggest that tumors without ARF may be more susceptible to such treatment.
Supplementary Material
Supp Figure 1
Supp Figure 2
Acknowledgments
The authors thank Gerry Zambetti, Scott Lowe and Shengkan Jin for reagents. We thank Eileen White and Carol Prives for critical reading of the manuscript, and Cory Abate-Shen for communicating results prior to publication. This work was supported by NIH R01 CA080854 and CA150002 (M.M.), and by the Philippe Foundation (O.H).
References
- 1.Sherr CJ, Bertwistle D, Den Besten W, et al. p53-Dependent and -independent functions of the Arf tumor suppressor. Cold Spring Harb Symp Quant Biol. 2005;70:129–37. doi: 10.1101/sqb.2005.70.004. [DOI] [PubMed] [Google Scholar]
- 2.Basso K, Margolin AA, Stolovitzky G, Klein U, Dalla-Favera R, Califano A. Reverse engineering of regulatory networks in human B cells. Nat Genet. 2005;37:382–390. doi: 10.1038/ng1532. [DOI] [PubMed] [Google Scholar]
- 3.Eischen CM, Weber JD, Roussel MF, Sherr CJ, Cleveland JL. Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev. 1999;13:2658–69. doi: 10.1101/gad.13.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kamijo T, Weber JD, Zambetti G, Zindy F, Roussel MF, Sherr CJ. Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2. Proc Natl Acad Sci USA. 1998;95:8292–7. doi: 10.1073/pnas.95.14.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reef S, Zalckvar E, Shifman O, et al. A short mitochondrial form of p19ARF induces autophagy and caspase-independent cell death. Mol Cell. 2006;22:463–75. doi: 10.1016/j.molcel.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 6.Ueda Y, Koya T, Yoneda-Kato N, Kato JY. Small mitochondrial ARF (smARF) is located in both the nucleus and cytoplasm, induces cell death, and activates p53 in mouse fibroblasts. FEBS Lett. 2008;582:1459–64. doi: 10.1016/j.febslet.2008.03.032. [DOI] [PubMed] [Google Scholar]
- 7.Abida WM, Gu W. p53-dependent and independent activation of autophagy by ARF. Cancer Res. 2008;68:352–357. doi: 10.1158/0008-5472.CAN-07-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jin S, White E. Role of autophagy in cancer: management of metabolic stress. Autophagy. 2007;3:28–31. doi: 10.4161/auto.3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weber JD, Jeffers JR, Rehg JE, et al. p53-independent functions of the p19(ARF) tumor suppressor. Genes Dev. 2000;14:2358–65. doi: 10.1101/gad.827300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hemann MT, Fridman JS, Zilfou JT, et al. An epi-allelic series of p53 hypomorphs created by stable RNAi produces distinct tumor phenotypes in vivo. Nat Genet. 2003;33:396–400. doi: 10.1038/ng1091. [DOI] [PubMed] [Google Scholar]
- 11.Paddison PJ, Cleary M, Silva JM, et al. Cloning of short hairpin RNAs for gene knockdown in mammalian cells. Nat Methods. 2004;1:163–7. doi: 10.1038/nmeth1104-163. [DOI] [PubMed] [Google Scholar]
- 12.Dickins RA, Hemann MT, Zilfou JT, et al. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nat Genet. 2005;37:1289–95. doi: 10.1038/ng1651. [DOI] [PubMed] [Google Scholar]
- 13.Pietsch EC, Leu JI, Frank A, Dumont P, George DL, Murphy ME. The Tetramerization Domain of p53 is Required for Efficient BAK Oligomerization. Cancer Biol Ther. 2007;6:1576–83. doi: 10.4161/cbt.6.10.4719. [DOI] [PubMed] [Google Scholar]
- 14.Tokuyasu KT. Immunochemistry on ultrathin frozen sections. Histochem J. 1980;12:381–403. doi: 10.1007/BF01011956. [DOI] [PubMed] [Google Scholar]
- 15.Pattingre S, Petiot A, Codogno P. Analyses of G-alpha-interacting protein and activator of G-protein-signaling-3 functions in macroautophagy. Methods Enzymol. 2004;390:17–31. doi: 10.1016/S0076-6879(04)90002-X. [DOI] [PubMed] [Google Scholar]
- 16.Robertson KD, Jones PA. The human ARF cell cycle regulatory gene promoter is a CpG island which can be silenced by DNA methylation and down-regulated by wild-type p53. Mol Cell Biol. 1998;18:6457–73. doi: 10.1128/mcb.18.11.6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Inoue R, Asker C, Klangby U, Pisa P, Wiman KG. Induction of the human ARF protein by serum starvation. Anticancer Res. 1999;19:2939–43. [PubMed] [Google Scholar]
- 18.Amaravadi RK, Yu D, Lum JJ, et al. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Invest. 2007;117:326–36. doi: 10.1172/JCI28833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carew JS, Nawrocki ST, Kahue CN, et al. Targeting autophagy augments the anticancer activity of the histone deacetylase inhibitor SAHA to overcome Bcr-Abl-mediated drug resistance. Blood. 2007;110:313–22. doi: 10.1182/blood-2006-10-050260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Furuta S, Hidaka E, Ogata A, Yokota S, Kamata T. Ras is involved in the negative control of autophagy through the class I PI3-kinase. Oncogene. 2004;23:3898–904. doi: 10.1038/sj.onc.1207539. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supp Figure 1
Supp Figure 2