Modularity and interactions in the genetics of gene expression (original) (raw)
Abstract
Understanding the effect of genetic sequence variation on phenotype is a major challenge that lies at the heart of genetics. We developed GOLPH (GenOmic Linkage to PHenotype), a statistical method to identify genetic interactions, and used it to characterize the landscape of genetic interactions between gene expression quantitative trait loci. Our results reveal that allele-specific interactions, in which a gene only exerts an influence on the phenotype in the presence of a particular allele at the primary locus, are widespread and that genetic interactions are predominantly nonadditive. The data portray a complex picture in which interacting loci influence the expression of modules of coexpressed genes involved in coherent biological processes and pathways. We show that genetic variation at a single gene can have a major impact on the global transcriptional response, altering interactions between genes through shutdown or activation of pathways. Thus, different cellular states occur not only in response to the external environment but also result from intrinsic genetic variation.
Keywords: computational biology, gene regulation, molecular networks, systems biology
Understanding the effect of genetic sequence variation on phenotype is a major challenge that lies at the heart of genetics. Recent technological advances in genotyping have now made it possible to obtain a comprehensive view of genomewide variation in a large number of individuals. However, association studies involving tens of thousands of individuals (1) have, for the most part, only been able to detect loci that collectively account for 3% of the heritable phenotype. This finding suggests that the connection between genotype and phenotype is more complex than previously assumed and that more sophisticated approaches are needed to interpret the data.
Quantitative trait mapping of gene expression abundances [expression quantitative trait locus (eQTL)] has proved a powerful model system for studying genetic traits in a number of organisms (2–5). To study gene–gene interactions between QTL, we use gene expression and genotype data on segregants generated in a cross between a laboratory strain (BY) and a wild strain (RM) of Saccharomyces cerevisiae (6, 7). We developed GOLPH (GenOmic Linkage to PHenotype), a statistical algorithm to identify multiple genetic factors influencing gene expression abundance. Our premise is that the modular organization of gene regulation can be used to enhance the statistical power of linkage to eQTLs.
GOLPH identifies an unprecedented number of linked regions for each gene. We used GENATOMY, our custom-built analysis tool, to visualize and analyze the resulting genetic interactions between QTL. Our results portray a complex picture in which multiple interacting loci influence the expression of modules of coexpressed genes that define coherent biological processes. The data show that genetic polymorphism can give rise to distinct cellular states in which entire metabolic pathways and biological processes are activated to different extents between individuals. In this regard, genotypic differences are similar to environmental perturbations in their effect on the internal state of the cell. Most interacting loci demonstrate allele-specific genetic interactions, in which the secondary locus exerts an influence on phenotype only when the primary locus has a particular allele.
A possible explanation is that the primary locus switches the cell among states or predisposes it toward adopting a cellular state. The secondary locus only has an effect in one of these states. For example, we observe differences in the cellular state mediated by variation at the IRA2 locus. Genetic variation in IRA2, an inhibitor of RAS/PKA signaling, predisposes strains with the IRA2-RM allele toward aerobic respiration (7). We identify several loci containing genes with critical functions involved with mitochondria and respiration that exhibit _IRA2-RM_-specific influences on entire transcriptional programs. Our data depict a complex relationship between genotype and phenotype resulting from the dynamic nature of genetic interaction networks that are responsive to both the environment and genetic variation.
Results
We developed GOLPH, a statistical approach to find multilocus linkage or association to gene expression traits. It is based on the detection of interacting QTL (iQTL) that involve 2 or 3 loci. Each iQTL consists of a primary locus and up to 2 secondary interacting loci, which significantly link to the trait when the primary locus has a specific allele, represented as a decision tree. GOLPH constructs iQTL modules consisting of the iQTL decision tree and all of the genes that link to that combination of interacting loci. These iQTL modules are further partitioned into subsets of coexpressed genes, referred to as expression patterns.
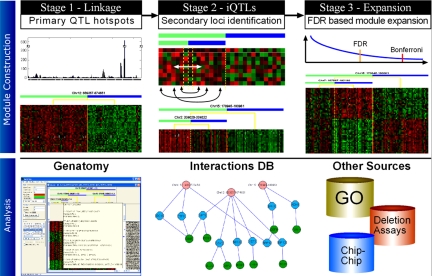
We applied GOLPH to genotype and gene expression data obtained from 108 segregants and their parents (2, 6, 7). GOLPH works in 3 stages, each increasing the number of detected linkages. Similar to previous studies (8, 9), GOLPH begins with a stepwise search. In the first stage, primary QTLs are detected for each trait, and in the second stage, secondary interacting loci are detected. In contrast to previous studies (8), a secondary QTL is identified independently for every allele at the primary locus. In the final phase we exploit the modular organization of gene regulation to link genes that are not significant alone, but which share a pattern with significantly linked genes (see Fig. 1 and Materials and Methods). We analyzed the resulting linkages by using GENATOMY, a purpose-built visualization tool, to gain insight into the architecture of interacting loci.
Fig. 1.
Overview of the GOLPH algorithm. GOLPH takes as input gene expression and genotype data for a set of individuals. (Upper) The computation occurring at each stage. In stage 1 genes are linked to a primary locus; in stage 2 iQTL are constructed by partitioning the samples based on the primary locus and linkage to the secondary locus; and in stage 3, FDR is used to expand significant linkages. See Figs. S1–S3 for more detail. (Lower) Once all iQTL modules have been constructed they are analyzed by using GENATOMY, our interactive visualization and data analysis tool. GENATOMY uses additional resources such as sequence, GO annotations, protein–DNA interactions, and genetic interactions to help interpret the data.
The GOLPH Algorithm Significantly Increases the Number of Linkages.
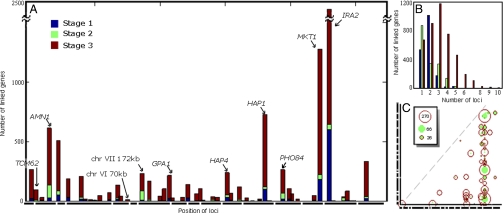
Stage 1 of our analysis (Fig. S1 and Table S1) identified 44 hotspots, including many previously reported regions (AMN1, GPA1, HAP1, IRA2, MKT1, PHO84) (2, 5, 7, 10, 11). Using these hotspots, stage 2 identifies secondary loci that interact with each of the primary loci. We found 81 pairs of iQTL that link to 5 or more genes, resulting in an increase in the number of multilocus genes (Fig. 2 A and B).
Fig. 2.
Stage 3 results in a marked increase in linked genes. The number of linked genes increases at each of the 3 stages, with the greatest expansion in module size occurring at stage 3. (A) Number of linkages at each locus color-coded by stage 1 (blue), 2 (green), and 3 (red). The x axis represents the location of the locus, each of the bold lines below the axis represents yeast chromosomes I–XVI. The y axis represents the number of genes linked to that locus. (B) Histogram representing the number of loci linking to each gene at each of the 3 stages. The color code is the same as in A. (C) Plot showing the size of each iQTL at stages 2 (green) and 3 (red). The size of the circle is proportional to the number of genes linked to the iQTL. Both axes relate to chromosomal location with the position of the chromosome marked in bold.
In stage 3, GOLPH uses the modularity of gene expression to gain additional power. Our premise is that gene regulatory networks are organized into modules of coregulated genes (12, 13), deletion studies have shown that when a regulator is deleted, the expression of hundreds of genes are influenced (14) and therefore weaker linkage of additional genes to the iQTL identified in stage 2 are more likely to be real. Stage 3 leads to a dramatic increase in both the number of genes linked to each marker and the number of markers linked to each gene (Fig. 2). After stage 3, >2,500 genes linked to 2 or more loci and >800 genes linked to 5 or more loci, matching previous analysis estimating that the expression of more than half the genes is likely influenced by at least 5 different loci (6). To ensure our method does not report spurious linkage, we performed careful randomization testing for each step, and indeed no signal was detected for the randomized data (see Fig. S3). We conclude that GOLPH detects an unprecedented number of loci for each gene expression trait and demonstrates that genetic interactions between loci are more common than previously estimated (10).
iQTLs Generate Coordinated Biological Programs of Gene Expression.
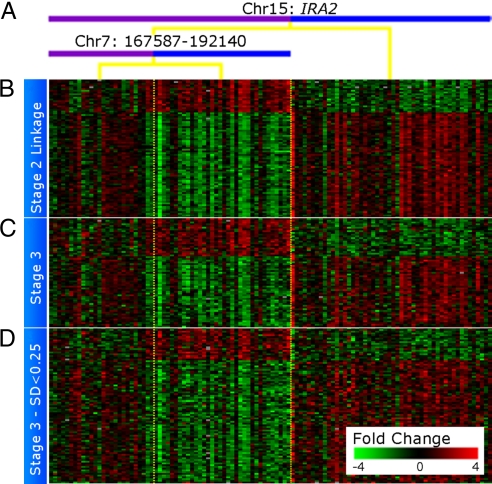
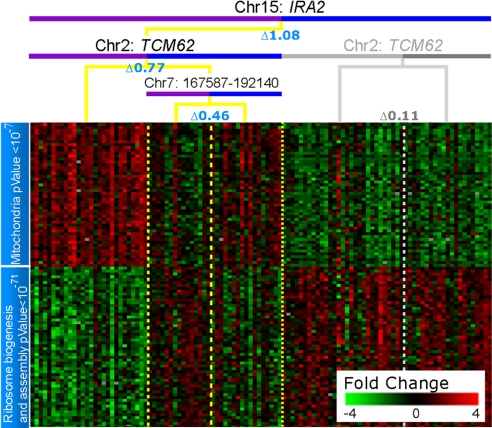
Although genes were added to each iQTL module based on their linkage alone, the resulting sets of genes form tightly coexpressed clusters. Fig. 3 shows the set of genes that are added to an iQTL module involving IRA2 (chromosome XV:170945–180961) and the chromosome VII locus (chromosome VII:167587–192140) at each stage. The genes added in stage 3 have the same pattern of expression as the genes added during the more rigorous stage 2. In addition, the genes added in stage 3 share Gene Ontology (GO) annotations and binding sites with those chosen in stages 1 and 2, significantly improving the functional enrichment of the modules, and further supporting their linkage (Fig. S4_d_). Examples of improved enrichment include ribosome biogenesis and assembly: 10−47 in stage 2 to 10−102 after stage 3, mitochondrion from 10−18 to 10−76, iron ion transport 10−4 to 10−10, and aerobic respiration 10−3 to 10−12. We conclude that iQTL do not influence a single gene but rather entire biological processes and pathways.
Fig. 3.
Gene expression in iQTL modules resemble environmental response. A heat map showing the _IRA2_–chrVII iQTL module and the expression of the genes linked at stages 2 and 3. Each row represents a gene, and each column represents a strain. The module is organized as a decision tree based on the strain's genotype and whether it inherited the BY (blue) or RM (purple) genotype for each of the interacting loci. (A) Top split based on the primary locus, chromosome XV:IRA2. The lower split is based on the secondary locus chromosome VII:167587–192140. (B) Eighty genes linked in stage 2. The columns represent strains and are arranged according to the tree; the vertical dotted yellow lines show the split point in the genotype. (C) Sixty-two genes linked in stage 3. The variance in expression of these genes is >0.25 SD. These genes were considered in stages 1 and 2, but did not pass the higher threshold for significance. (D) An additional 88 genes are linked in stage 3. These genes are not considered in stage 2 because their variance in expression is <0.25 SD and are hence noisier. The names of the genes represented in B–D are provided in Table S3.
Fig. 3 shows 2 distinct patterns of coexpressed genes that are inverted, i.e., down-regulation on one side of the heat map is accompanied by up-regulation of equivalent magnitude on the other side and vice versa. This is a widespread phenomenon involving 122 modules and 3,638 linked genes, resembling the response to environmental perturbation, in which entire processes are coordinately up- or down-regulated (see Fig. S5 for an additional example). The existence of inverse expression patterns suggests that many iQTL not only regulate single pathways, but rather orchestrate entire cellular responses involving multiple biological processes.
Our results provide a view of genetic variation as an internal cue that predisposes the organism toward or away from a cellular state. The presence of a single allele can tip the balance between one state and another. The most striking example is provided by the IRA2 locus that links to >2,000 genes. Ira2 is a GTPase-activating protein that negatively regulates RAS. The RAS/PKA pathway plays a central role in coordinating processes such as growth and stress tolerance in response to nutrient availability. The _IRA2_-RM sequence differs from BY by 87 nonsynonymous coding SNPs and 3 gaps and segregants with the RM allele of IRA2 correspondingly inhibit Ras/PKA signaling better than segregants with the BY allele (7). Although all of the segregants were grown in glucose (and might be expected to undergo fermentative growth), the presence of the RM allele correlates with the up-regulation of genes annotated for mitochondria (10−14), aerobic respiration (10−9), response to stress (10−8), and the down-regulation of genes annotated for ribosome biogenesis and assembly (10−95), rRNA processing (10−57) and the nucleolus (10−56), suggesting a transcriptional response consistent with respiration.
In contrast to IRA2, the phenotypic differences that link to the HAP4 (chromosome XI: 247944-247956) locus are likely to be driven by allelic differences in the promoter. Hap4 is part of a transcriptional activator complex that regulates the transcription of genes in response to heme/oxygen and/or growth on nonfermentable substrates (15) and the locus is linked to >200 genes. HAP4 is a _cis_-eQTL, i.e., a gene that links to its own locus, and the RM strain has 14 promoter SNPs. Moreover, the presence of the HAP4-RM allele correspondingly correlates with the up-regulation of HAP4 along with genes it activates: Hap4 bound genes (10−19), those annotated for mitochondria (10−90), and aerobic respiration (10−13).
The Landscape of Genetic Interactions.
GOLPH detected 83 pairs of interacting loci in 205 modules with 542 expression patterns. We used the multilocus phenotypes to characterize the genetic interactions between QTLs. Most methods for multilocus traits assume an additive model, y ∼aX + bY. For example, the iQTL module involving the IRA2 and HAP4 loci influence different aspects of mitochondrial function. The IRA2-RM allele represses the PKA pathway, predisposing the strain toward respiratory growth. Hap4, an activator of aerobic respiration is up-regulated in segregants with the HAP4-RM locus. Therefore, the presence of IRA2-RM and HAP4-RM each push the cell toward respiration through independent mechanisms and their joint influence is an additive combination of their individual influences (Fig. S4_a_).
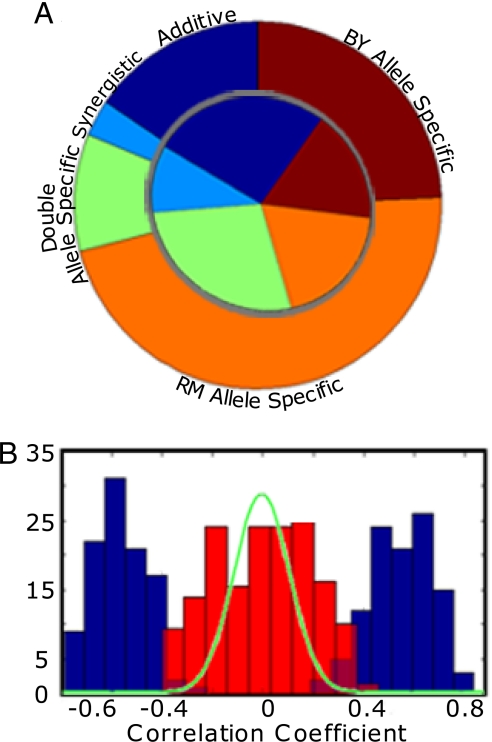
One of the most striking aspects of the data is the dominance of allele-specific interactions, i.e., situations in which the secondary locus exerts an influence on the phenotype only when the primary locus has a particular allele (and has little or no influence when the primary locus has another allele). GOLPH is able to detect allele-specific interactions because each primary allele is tested for linkage independently and the secondary locus need not link to both. We have already encountered such an effect in the iQTL module in Fig. 3, the chromosome VII locus interacts with IRA2-RM only and has no influence on IRA2-BY. We note that whereas 196 genes link to the chromosome VII locus in with the presence of IRA2-RM, none of these linkage signals were significant in stage 1. To confirm that these interacting loci are indeed allele-specific and do not reflect borderline effects, we compared the regression coefficient of the linked versus nonlinked alleles and found that coefficients for the nonlinked alleles resemble a random distribution (Fig. 4B). GOLPH detected a remarkable number of allele-specific genetic interactions. These involve 78 interacting loci, organized into 94 iQTL modules that contain 1,856 unique genes and 2,891 allele-specific interactions, 81% of the total interactions identified (Fig. 4A). We conclude that allele-specific genetic interactions are prevalent in our data.
Fig. 4.
Landscape of genetic interactions between loci. (A) A pie chart representing the types of interactions between loci in our analysis. The outer circle represents genes, and the inner circle represents modules. RM allele-specific interactions are orange, BY allele-specific interactions are brown. Blue represents situations in which the secondary allele links to both sides, additive interactions are dark blue, and synergistic interactions are light blue. Green represents modules with 2 different allele-specific interactions, one for each side. The dominance of allele-specific interactions is evident. (B) Histogram of correlation coefficients in allele-specific modules. The data show that the effect of the secondary locus on the noninteracting allele is negligible. The x axis is the correlation coefficient between the secondary locus and the mean expression level for genes in the module. The y axis shows the number of modules. The blue bars represent data from the interacting primary allele and the red bars represent the other noninteracting allele. The green line shows that the distribution for randomly chosen pairs of loci is similar to the histogram in red demonstrating that the interactions are indeed with only one allele and not the other.
The same secondary locus was found to influence both primary alleles in 50 iQTL modules containing a total of 562 genes. We tested each of these for epistasis (see Materials and Methods), and in cases where the secondary locus links to both primary alleles, the majority of interactions (423/562) do not show a significant interaction term. Because most other methods do not detect allele-specific interactions, these found that genetic interactions are typically additive. Although there are only a few epistatic modules, these can exhibit dramatic effects; a number of iQTL modules had secondary locus effects in opposing directions between the 2 primary alleles. For instance, in the iQTL involving IRA2 and chromosome VI:70818-75460, the effect of the chromosome VI locus depends on the IRA2 allele. The RM allele of chromosome VI:70818-75460 up-regulates the genes in the module in the presence of IRA2-BY and down-regulates the genes in the presence of IRA2-RM (see Fig. S4_b_ and Table S2).
The Prevalence of Allele-Specific Genetic Interactions.
To understand how allele specificity might arise, we analyzed an iQTL module linked to IRA2-RM and a locus on chromosome II: 334020-334022 (Fig. 5). The causal gene on chromosome II is likely to be TCM62, which encodes a protein that supports biogenesis of the mitochondrial succinate dehydrogenase complex by acting as a molecular chaperone (16). Strains deleted for TCM62 grow slowly on rich glycerol medium and are respiration deficient. TCM62-RM has 3 coding SNPs and 57 promoter SNPs compared with the BY sequence, including SNPs in 2 Pho2 binding sites. One of these SNPs is predicted to increase the binding affinity of Pho2 (17); indeed, TCM62 is a strong _cis_-eQTL and is up-regulated in segregants bearing the TCM62-RM allele.
Fig. 5.
Allele-specific IRA2 module (see also Fig. S6). The IRA2-TCM62 iQTL module is graphically represented as described in Fig. 3. For compactness, representative genes were chosen for each pattern. The full list of genes for each pattern is provided in Table S4. We manually added an additional partition by using the chromosome VII locus from Fig. 3 to TCM62-BY to demonstrate that the chromosome VII locus represents an alternative pathway to that affected by TCM62-RM.
When segregants have both IRA2-RM and TCM62-RM, mitochondrial genes (10−7) and Skn7 targets (10−5) are up-regulated, whereas ribosome biogenesis and assembly (10−63), nucleolar (10−37), and rRNA processing genes (10−32) are down-regulated. Fig. 5 shows the expression pattern of genes in the module and that the TCM62 locus has a strong influence in segregants with the IRA2-RM allele and negligible influence in segregants with the IRA2-BY allele (see also Fig. S6. The PKA pathway is inhibited in segregants bearing the IRA2-RM allele and the balance is tipped toward the expression of genes associated with respiratory growth and the up-regulated TCM62-RM allele likely further tips the cell toward respiratory growth.
Previous work reports that linkage to a particular locus often depends on environment, the locus exerting an influence in one environment, but not in another (7). Both external environmental signals and genetically-driven internal cues can drive cells to switch between states, as reflected by different metabolic fluxes and stresses acting on the cell. We postulate that the allele-specific linkages we detect are largely caused by such events. These switches can sometimes be subtle, such as a release of inhibition or a shift in bottlenecks, making certain genes more critical in some conditions than others. Thus, genetic variation leads to internal change, altering interactions between genes through shutdown or activation of pathways, release of inhibition, or shifting of bottlenecks.
The IRA2-RM allele plays a dominant role among the allele-specific iQTL modules accounting for 41% of the genes influenced by allele-specific interactions with RM; however, other loci also exhibit this phenomenon. Each of HAP1-RM and MKT1-RM has allele-specific effects on the expression of >200 genes. Although there are fewer allele-specific interactions with BY, 886 genes are affected by allele-specific interactions on the BY side. The loci that dominate BY allele-specific interactions include HAP1-BY and a locus on chromosome I:41483-42639, likely to be caused by polymorphism in OAF1, an oleate-activated transcription factor involved in the β-oxidation of fatty acids and peroxisome organization and biogenesis. Together there are >10 different hotspots that exert allele-specific influences over a large number of genes.
Discussion
The emergence of new technological advances in high-throughput genotyping and sequencing has enabled large-scale characterization of genetic variation at high resolution. However, novel computational approaches are needed to detect causal sequence variants and model how genotype influences phenotype. A first step is to characterize the landscape of genetic interactions between naturally-occurring variants and elucidate how multiple loci combine to affect phenotype.
Applying GOLPH to yeast detected between 2 and 10 linkages for each of 2,745 genes, providing an expansive view on the architecture of multilocus traits and the genetic interactions between them. A remarkable finding is a large-scale occurrence of allele-specific interactions, indicating that the landscape of multilocus traits is predominantly nonadditive. A likely mechanism for allele-specific interactions stems from the observation that genetic variation can mimic the response to environmental change. Thus, different biological states occur not only in response to the external environment but also as a result of intrinsic genetic variation.
Genetic variation in both coding and regulatory regions of transcription factors can lead to responses that alter cellular state (e.g., HAP1, HAP4). More intriguing, such large-scale transcriptional responses are not only caused by variation in classical transcriptional regulators, but also caused by polymorphism in metabolic enzymes, regulators of translation, and molecular chaperones (e.g., LEU2, MKT1, TCM62). These demonstrate that genetic variation in a single gene may trigger a cascade of events, leading to an alternative cellular state, by predisposing the cell toward shutdown or activation of pathways. In this way the molecular network can be considered an intricate web of interacting factors in which dynamic entities may rewire their connectivity in response to perturbations in the environment and as a result of intrinsic genetic variation.
The prevalence of complex, nonadditive gene–gene interactions is likely to play a large role in human and disease-related genetics and offers clues as to why recent association studies involving over tens of thousands of individuals have accounted only for a very small fraction of the heritable variation observed (1). We believe that state changes driven by intrinsic genetic variation and the resulting allele specific interactions are likely common in human and disease-associated genetics. In multicellular organisms, genetic variation can lead not only to an altered cellular state, but can propagate to changes at the level of the entire organism. Detecting such allele-specific association in human is significantly more challenging as the genome is 2 orders of magnitude larger than yeast and the population structure is more complex.
Nevertheless, there are multiple lines of evidence in support of such allele specificity. The influence of genetic variation on phenotype is condition-dependent and is influenced both by external (18) and internal factors such as tissue type. For example, alleles of IRGM, that confer risk and protection for Crohn's disease, show different patterns of tissue-specific expression (19) and allelic imbalances that confer tissue specificity are likely to be common (20). Moreover, there are a number of “hotspot” genes associated to a broad range of diseases including MHC and ApoE. The MHC is associated with autoimmune, infectious, and inflammatory diseases including multiple sclerosis, type 1 diabetes, systemic lupus erythematosus, ulcerative colitis, Crohn's disease, and rheumatoid arthritis (21). ApoE is associated with lipid level and Alzheimer's disease (18) spanning the range from metabolic to neurodegenerative disorders. We hypothesize that variation in genes such as HLA and ApoE perturbs the molecular network, altering multiple biological processes and pathways relevant to many of the associated diseases.
Although much of the challenge in genetic association lies in detecting the factors involved in disease, understanding the complex nature of molecular networks that give rise to phenotype can be leveraged to gain insight into the transmission of information from genotype to phenotype. A number of approaches that take the molecular network into account have proved successful. Some of these involve integrating different data sources including gene annotations, transcription factor–protein and protein–protein interactions to identify paths between the locus and the linked gene, thus helping to pinpoint the causal gene within the locus (22, 23). The use of Bayesian networks (24) to reconstruct the regulatory network and the perturbations to it arising from genetic variation has also proved to be particularly powerful (25–27). For example, Chen et al. (27) used this approach to identify molecular networks conserved between human and mouse that are perturbed by susceptibility loci in metabolic syndrome. One of the factors leading to the success of this approach is that DNA sequence polymorphisms are effective perturb-agens that provide a rich source of variation, which helps to uncover regulatory relations in the molecular network and direct their causality. Thus, there is complementary duality between 2 long-standing computational challenges: Genetic variation of gene expression helps reveal the regulatory network, which subsequently aids in identifying the genetic factors underlying complex traits.
Materials and Methods
Data.
The strains, genotypes, and gene expression measurements were those of ref. 7. We merged adjacent, highly-correlated markers, to obtain a total of 526 markers (25). For our analysis we normalized expression data mean of 0 and variance 1. For stages 1 and 2 of our algorithm we only used data from the 1,733 genes that showed significant variation (SD >0.25) in their expression level. GO categories from www.yeastgenome.org with >5 genes were used for the evaluation of biological function. Putative transcription factor binding sites were obtained from http://fraenkel.mit.edu/yeast_map_2006.
GOLPH Algorithm.
GOLPH is a multistep procedure for identifying multilocus linkage and pairs of interacting loci. We briefly describe the algorithm, deferring detailed explanation to SI Text. Two key features in GOLPH enable the ability to identify multiple locus linkages. First, GOLPH permits the identification of allele-specific interactions in which secondary QTL are specific for the allele at the primary locus. This is in contrast to a secondary QTL that contributes irrespective of the allele at the primary locus. Our model can be written as expression y ∼ baseline + aX + α_bY_ + (1 − α)cZ, α = 1 for X = BY and α = 0 for X = RM, where X is the primary locus, and Y and Z are 2 secondary loci.
Second is the use of modularity: as opposed to searching for interacting QTLs at each gene independently, we group genes into modules based on the hotspots identified for each one. This step greatly increases the number of linkages detected and reduces measurement artifacts and noise.
Stage 1.
The first stage of our analysis applies classic genetic analysis (2, 28) to look for linkage of gene expression traits to a primary locus. For each gene and marker, we use a Welch's t test statistic (29) and permutation testing with a stringent cutoff to evaluate the significance of the linkage, with cutoffs of 0.05 for the t test's P value and 10−5 for the permutation testing. Because genes linked to one marker are also likely to have linkage signals in neighboring markers, we merge small peaks with proximal larger peaks into chromosomal hotspots. After the merging of peaks, we identified 44 locus hotspots that link to at least 5 genes for stage 2.
Stage 2.
For each of the 44 modules identified in stage 1 and each gene that links to these, we partitioned segregants on the basis of inheritance (either BY or RM) at the primary locus and similarly tested each subgroup for further secondary loci. This process was carried out independently for the BY or RM allele at the primary locus. Secondary loci are considered significant if Welch's t test P <0.05 and P <10−4. Each detected secondary linkage defines an iQTL represented as a decision tree. The resulting tree can have secondary splits on the BY (right) side, the RM (left) side, or both. Because close loci link to overlapping sets of genes, we merged similar iQTL modules (see SI Text). After removing modules that have <5 genes, we obtained 91 iQTL modules.
Stage 3.
As discussed above, GOLPH uses the modularity of gene expression to gain additional power. We seed our search with the iQTL detected by using highly-stringent criteria in stage 2, ensuring that the loci selected are likely to exert causal regulatory influence on gene transcripts. We go over the regulation trees one by one and evaluate all 4,338 genes in our set for that module. Each tree involves 2 independent tests, depending on the structure of the tree. For each module, we generate a distribution of P values over all 4,338 genes independently for each of the 2 tests above. A gene is assigned to the module by using a genomewide false discovery rate (FDR) of 1% (30) for both tests. Hence our threshold is adaptive to the number of genes and the strength of linkage signal for each locus, so a large number of weak signals that point to the same locus increase the significance.
Module Annotation.
To biologically annotate the resulting modules, we calculated the hypergeometric enrichment for all modules against all annotations and carried out an FDR correction for multiple independent hypotheses. We considered values of _P_corrected < 0.005 to be significant.
Additional Information.
For interactive viewing and analysis of all of the constructed iQTL modules we have generated a file formatted for visualization with our interactive GENATOMY analysis tool.*
Supplementary Material
Supporting Information
Acknowledgments.
We thank Ran Reshef, Itsik Pe'er, and Lyle Ungar for valuable comments. This work was supported by the National Institutes of Health Roadmap Initiative, National Institutes of Health Director's New Innovator Award Program Grant 1-DP2-OD002414-01, and National Centers for Biomedical Computing Grant 1U54CA121852-01A1. D.P. holds a Career Award at the Scientific Interface from the Burroughs Wellcome Fund.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Consortium WTCC. Genomewide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brem RB, Yvert G, Clinton R, Kruglyak L. Genetic dissection of transcriptional regulation in budding yeast. Science. 2002;296:752–755. doi: 10.1126/science.1069516. [DOI] [PubMed] [Google Scholar]
- 3.Cheung VG, et al. Natural variation in human gene expression assessed in lymphoblastoid cells. Nat Genet. 2003;33:422–425. doi: 10.1038/ng1094. [DOI] [PubMed] [Google Scholar]
- 4.Schadt EE, et al. Genetics of gene expression surveyed in maize, mouse, and man. Nature. 2003;422:297–302. doi: 10.1038/nature01434. [DOI] [PubMed] [Google Scholar]
- 5.Yvert G, et al. Trans-acting regulatory variation in Saccharomyces cerevisiae and the role of transcription factors. Nat Genet. 2003;35:57–64. doi: 10.1038/ng1222. [DOI] [PubMed] [Google Scholar]
- 6.Brem RB, Kruglyak L. The landscape of genetic complexity across 5,700 gene expression traits in yeast. Proc Natl Acad Sci USA. 2005;102:1572–1577. doi: 10.1073/pnas.0408709102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith EN, Kruglyak L. Gene–environment interaction in yeast gene expression. PLoS Biol. 2008;6:e83. doi: 10.1371/journal.pbio.0060083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Storey JD, Akey JM, Kruglyak L. Multiple locus linkage analysis of genomewide expression in yeast. PLoS Biol. 2005;3:e267. doi: 10.1371/journal.pbio.0030267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brem RB, Storey JD, Whittle J, Kruglyak L. Genetic interactions between polymorphisms that affect gene expression in yeast. Nature. 2005;436:701–703. doi: 10.1038/nature03865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu J, et al. Integrating large-scale functional genomic dadta to dissect the complexity of yeast regulatory networks. Nat Genet. 2008;40:854–861. doi: 10.1038/ng.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perlstein EO, Ruderfer DM, Roberts DC, Schreiber SL, Kruglyak L. Genetic basis of individual differences in the response to small-molecule drugs in yeast. Nat Genet. 2007;39:496–502. doi: 10.1038/ng1991. [DOI] [PubMed] [Google Scholar]
- 12.Hartwell LH, Hopfield JJ, Leibler S, Murray AW. From molecular to modular cell biology. Nature. 1999;402(Suppl):C47–C52. doi: 10.1038/35011540. [DOI] [PubMed] [Google Scholar]
- 13.Segal E, et al. Module networks: Identifying regulatory modules and their condition-specific regulators from gene expression data. Nat Genet. 2003;34:166–176. doi: 10.1038/ng1165. [DOI] [PubMed] [Google Scholar]
- 14.Hughes TR, et al. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- 15.Kwast KE, Burke PV, Poyton RO. Oxygen sensing and the transcriptional regulation of oxygen-responsive genes in yeast. J Exp Biol. 1998;201:1177–1195. doi: 10.1242/jeb.201.8.1177. [DOI] [PubMed] [Google Scholar]
- 16.Dibrov E, Fu S, Lemire BD. The Saccharomyces cerevisiae TCM62 gene encodes a chaperone necessary for the assembly of the mitochondrial succinate dehydrogenase (complex II) J Biol Chem. 1998;273:32042–32048. doi: 10.1074/jbc.273.48.32042. [DOI] [PubMed] [Google Scholar]
- 17.MacIsaac KD, et al. An improved map of conserved regulatory sites for Saccharomyces cerevisiae. BMC Bioinformatics. 2006;7:113. doi: 10.1186/1471-2105-7-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Corder EH, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late-onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 19.McCarroll SA, et al. Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn's disease. Nat Genet. 2008;40:1107–1112. doi: 10.1038/ng.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Campbell CD, Kirby A, Nemesh J, Daly MJ, Hirschhorn JN. A survey of allelic imbalance in F1 mice. Genome Res. 2008;18:555–563. doi: 10.1101/gr.068692.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fernando MM, et al. Defining the role of the MHC in autoimmunity: A review and pooled analysis. PLoS Genet. 2008;4:e1000024. doi: 10.1371/journal.pgen.1000024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iossifov I, Zheng T, Baron M, Gilliam TC, Rzhetsky A. Genetic-linkage mapping of complex hereditary disorders to a whole-genome molecular-interaction network. Genome Res. 2008;18:1150–1162. doi: 10.1101/gr.075622.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suthram S, Beyer A, Karp RM, Eldar Y, Ideker T. eQED: An efficient method for interpreting eQTL associations using protein networks. Mol Syst Biol. 2008;4:162. doi: 10.1038/msb.2008.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Friedman N, Linial M, Nachman I, Pe'er D. Using Bayesian networks to analyze expression data. J Comput Biol. 2000;7:601–620. doi: 10.1089/106652700750050961. [DOI] [PubMed] [Google Scholar]
- 25.Lee SI, Pe'er D, Dudley AM, Church GM, Koller D. Identifying regulatory mechanisms using individual variation reveals key role for chromatin modification. Proc Natl Acad Sci USA. 2006;103:14062–14067. doi: 10.1073/pnas.0601852103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu J, et al. Increasing the power to detect causal associations by combining genotypic and expression data in segregating populations. PLoS Comput Biol. 2007;3:e69. doi: 10.1371/journal.pcbi.0030069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Y, et al. Variations in DNA elucidate molecular networks that cause disease. Nature. 2008;452:429–435. doi: 10.1038/nature06757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lander E, Kruglyak L. Genetic dissection of complex traits: Guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- 29.Welch BL. The significance of the difference between two means when the population variances are unequal. Biometrika. 1938;29:350–362. [Google Scholar]
- 30.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information