A family of Salmonella virulence factors functions as a distinct class of autoregulated E3 ubiquitin ligases (original) (raw)
Abstract
Processes as diverse as receptor binding and signaling, cytoskeletal dynamics, and programmed cell death are manipulated by mimics of host proteins encoded by pathogenic bacteria. We show here that the Salmonella virulence factor SspH2 belongs to a growing class of bacterial effector proteins that harness and subvert the eukaryotic ubiquitination pathway. This virulence protein possesses ubiquitination activity that depends on a conserved cysteine residue. A crystal structure of SspH2 reveals a canonical leucine-rich repeat (LRR) domain that interacts with a unique E3 ligase [which we have termed NEL for _Novel E3 Ligase_] C-terminal fold unrelated to previously observed HECT or RING-finger E3 ligases. Moreover, the LRR domain sequesters the catalytic cysteine residue contained in the NEL domain, and we suggest a mechanism for activation of the ligase requiring a substantial conformational change to release the catalytic domain for function. We also show that the N-terminal domain targets SspH2 to the apical plasma membrane of polarized epithelial cells and propose a model whereby binding of the LRR to proteins at the target site releases the ligase domain for site-specific function.
Keywords: microbial pathogenesis, type III secretion, crystallography, SspH2
The ubiquitination of proteins is a central eukaryotic regulatory mechanism controlling multiple biological processes, such as programmed cell death, cell cycle progression, and signal transduction, which, when defective, can lead to cancer and neurodegenerative disorders (1–4). The ubiquitination system regulates many biological processes through the covalent conjugation of either a single or multiple subunit(s) of ubiquitin to a target protein. The structural form of the ubiquitin modification can result in distinct functional signals. For example, the conjugation of a single ubiquitin molecule can alter protein trafficking, whereas polyubiquitination of proteins can regulate protein function by both proteasome-dependent and independent mechanisms (5–8). Bacterial factors have recently been identified that exploit these pathways, functioning as E3 ubiquitin ligases (9–13), possessing sequences conveying differential half-lives in the host (14), or serving themselves as substrates for monoubiquitination for purposes such as localization within the eukaryotic cell (15).
Protein ubiquitination is a multistep enzymatic process resulting in the formation of an isopeptide bond between the C-terminal glycine of ubiquitin and internal lysine residues of the substrate protein. The process involves a ubiquitin-activating enzyme (E1), which transfers ubiquitin to a family of ubiquitin-conjugating enzymes (E2s). Ubiquitin-loaded E2s are then recruited to their substrates by a family of ubiquitin ligases (E3s), which play a critical role in conferring specificity to the reaction (16). Eukaryotic cells possess 2 known classes of E3 ubiquitin ligases that target substrates for modification, the RING-finger and HECT families, which possess distinct structural and functional properties (9, 17). Whereas some bacterial ubiquitin ligases display structural and functional homology to RING-finger and HECT domains (10, 11), another class of recently discovered bacterial E3 ligases does not show any primary amino acid sequence similarity to any host proteins (12).
To understand how this class of bacterial mimics functions, we undertook a biochemical, structural, and cell biological study of one member of this family from Salmonella enterica serovar Typhimurium (S. typhimurium), the multidomain protein SspH2. Along with its close homologue SspH1, this protein is delivered into host cells by a Type III secretion system (T3SS) machine encoded within the pathogenicity island 2 of the S. typhimurium chromosome (18). Both SspH1 and SspH2 contribute to virulence in animal models of infection by mechanisms that are currently not well understood (18). Whereas SspH2 is widely distributed among Salmonella serotypes, SspH1 has a more restricted distribution. Recent studies have shown that SspH1 belongs to a new class of E3 ubiquitin ligase (12). Like its Shigella spp. homologue IpaH9.8, SspH1 showed E3 ligase activity in vitro and catalyzed the ubiquitination of its proposed host cell target PKN1 (12, 19). Although highly related at the primary amino acid sequence level, SspH1 and SspH2 are likely to play different roles during S. typhimurium infection. For example, whereas SspH1 can be delivered by both S. typhimurium T3SSs encoded in pathogenicity island 1 (SPI-1) and 2 (SPI-2), SspH2 is exclusively delivered by the SPI-2 T3SS, which is only expressed when S. typhimurium resides within cells (18, 19). Furthermore, SspH1 and SspH2 exhibit different localization when transiently expressed in cultured cells (20). Consequently, the differential temporal and spatial regulation of these two highly related effector proteins suggests distinct roles during infection.
Herein, we report the crystal structure of the Salmonella SPI-2 effector SspH2, an E3 ubiquitin ligase with a unique fold. We show that SspH2 localizes to the apical plasma membrane and propose a mechanism for the activation of its enzymatic activity involving a dramatic conformational change between 2 subunits of the protein.
Results
Ubiquitination Activity of SspH2.
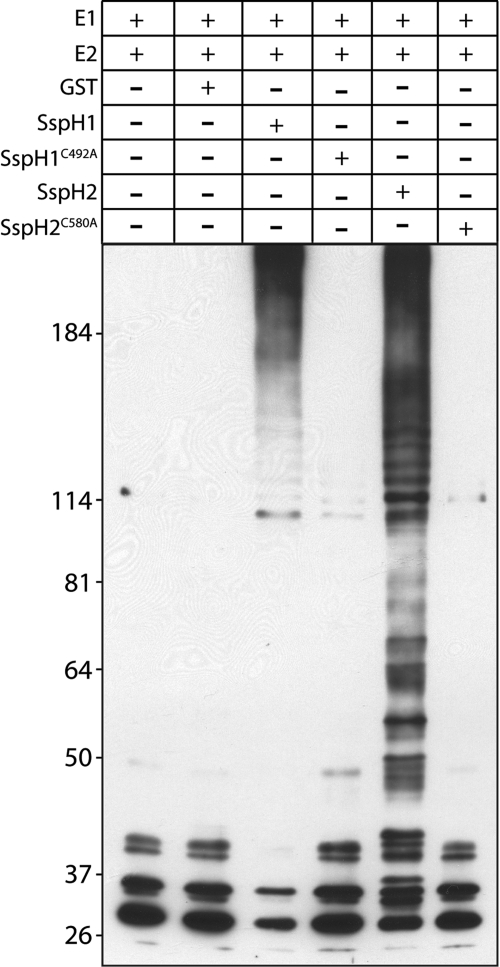
We tested the ubiquitin ligase activity of purified SspH2 in vitro. Like its homologue SspH1 (ref. 12, Fig. 1), SspH2, but not the catalytic mutant SspH2C580A, was able to remove ubiquitin from the ubiquitin-conjugating E2 enzyme UbcH5B, autoubiquitinate, and catalyze the polyubiquitination of FLAG-tagged ubiquitin (Fig. 1), which are properties associated with E3 ubiquitin ligases.
Fig. 1.
SspH2 is an E3 ubiquitin ligase. Ubiquitin ligase activity of SspH2. Purified recombinant GST-SspH2, GST-SspH2C580A, GST-SspH1, or GST-SspH1C492A (as indicated) were incubated with FLAG-tagged ubiquitin in the presence of ATP, E1, and recombinant UbcH5b. Aliquots of the reaction were analyzed by 6–12% SDS-PAGE and immunoblotted with anti-FLAG antibody to detect ubiquitin conjugates. The indicated components of the reaction were omitted as controls.
Crystal Structure of SspH2 Reveals a Unique E3 Ligase Fold.
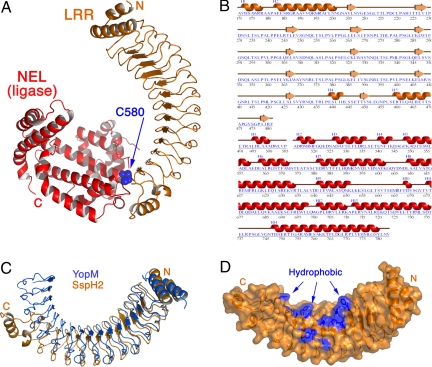
To further characterize this unique class of E3 ligases, we solved the crystal structure of SspH2 (residues 166–783) to a resolution of 1.90 Å(Table S1). The structure reveals a 2-domain architecture (Fig. 2 A and B). Residues 171–481 form a canonical leucine-rich repeat (LRR) domain consisting of 12 repeats of the LRR motif (LxxLPxxLxxLx5VxLxxNPL) that are capped at the N- and C-terminal regions of the domain by α helices. At the C terminus of the LRR domain, a stretch of 10 residues, 5 of them disordered, serves as a connecting loop to the second domain in the protein. The C-terminal domain begins at residue 492 and extends through an all-helical domain of unique fold. We have termed this unique fold “NEL,” for _N_ovel _E_3 _L_igase. The 2 domains possess a small surface of interaction near the end of the LRR domain, burying roughly 578 Å2 of surface area (21).
Fig. 2.
SspH2 consists of an LRR domain and an NEL domain. (A) The overall structure of SspH2 is shown as a ribbon diagram, with the LRR domain in orange and the NEL domain in red. The catalytic cysteine residue is shown in a space-filling rendering (blue). N, NH2 terminus; C, COOH terminus. (B) Sequence and secondary structure of the crystallized region of SspH2 are shown. Secondary structure is numbered separately for the 2 domains. (C) Comparison of YopM (PDB ID 1JL5) and SspH2 LRR domains. Proteins were aligned to maximize the overlap at the N terminus, revealing the divergence between the folds as the chain progresses. (D) Partially transparent molecular surface of the SspH2 LRR domain with the nonpolar residues lining the concave surface (blue) (residues in patch F270, W288, F290, C325, W328, Y365, W368, Y370, I388, M408, and Y430). Orientation similar to panel C.
The LRR domain differs from other known structures and from the most closely related structure of YopM from the pathogen Yersinia pestis (22), both in the number of repeats (12) as well as in the curvature of the fold (Fig. 2C). SspH2 is shorter than YopM by 3 repeats, and the NEL domain occupies the space taken by the final 3 repeats in YopM. The inner, concave surface of SspH2, used by most LRR domains as a protein–protein interaction site, is lined with nonpolar residues, giving it a strongly hydrophobic interaction surface (Fig. 2D). In addition, there are several large charged patches on the protein surface that may be sites of biological interactions in the host cell.
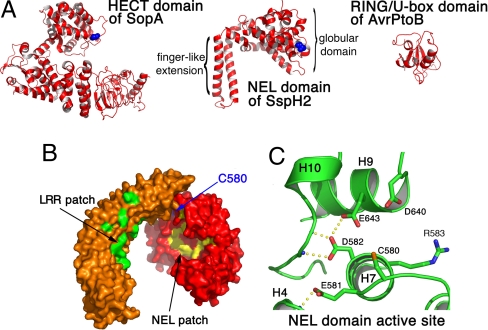
The NEL domain, which harbors the catalytic cysteine, is unrelated in structure to host RING-finger or HECT domain E3 ligases (Fig. 3A) (23). The domain consists of 14 helices roughly divided into 2 subdomains: an N-terminal globular region spanning residues 491–699, followed by 2 long helices that interact with each other, protruding from the globular fold as a long finger-like extension until the end of the ordered structure. The catalytic cysteine residue (Cys 580) is located on the opposite side from the helical extension at the end of a loop spanning residues 574–580 (Figs. 2A and 3B). It is partially buried by a portion of the LRR domain (Figs. 2A and 3B). A large hydrophobic patch is centered on the concave surface of the NEL domain facing toward the LRR domain. This patch is extensive, involving 10 residues, and is proximal to the catalytic cysteine (Fig. 3B).
Fig. 3.
Structure of the NEL domain of SspH2. (A) Comparison of the SspH2 NEL domain with 2 bacterial E3 ligases: SopA, a HECT family of cysteine-dependent E3 ubiquitin ligases from Salmonella (PDB ID 2QYU), and AvrPtoB, a RING finger/U-box protein (PDB ID 2FD4). The catalytic cysteine residues are shown in a space-filling format (blue). (B) Molecular surface representation of the crystallized construct of SspH2 with the LRR and NEL domains in orange and red, respectively. The catalytic cysteine is show in blue. Hydrophobic residues in 2 clusters are show in green (LRR domain) and yellow (NEL domain, residues F570, F586, F587, V594, V597, M619, F620, L622, V633, and W645). (C) Residues near the catalytic cysteine of SspH2. Hydrogen bonds are shown as yellow spheres between atoms, and atoms of nitrogen and oxygen are shown in blue and red, respectively.
The catalytic cysteine is surrounded by several charged residues positioned favorably to participate in the ubiquitination reaction and/or substrate binding, including Arg 583, Asp 640, and Glu 643 (Fig. 3C). Two acidic residues immediately adjacent to Cys 580 (Glu 581 and Asp 582) play a structural role through an extensive hydrogen-bonding network linking several main-chain atoms as well as coordinating Glu 643 through a side chain hydrogen bond (Fig. 3C). The 2 acidic residues are likely important in maintaining the proper conformation (and thereby function) of the loop harboring Cys 580 as well as the surrounding structure. The presence of these residues, which are conserved in SspH2 homologues, in such a clustered region creates a large negative electrostatic patch on the surface of the protein and suggests that these residues may also be involved in the catalytic mechanism.
Mechanism of Activation for the Catalytic Domain.
The buried nature of the catalytic cysteine residue, the partial occlusion of the concave, putative protein-binding surface of the LRR domain by the NEL domain, and the relatively moderate interaction surface between the 2 domains strongly suggest that a conformational change could be involved in the activity of this molecule. Specifically, the relative orientation of the 2 domains of SspH2 would likely be required to change to expose the NEL active site for optimal catalysis as well as the LRR domain for binding to its substrate in the host cell. In such a model, the NEL domain would swing out and away from the LRR, perhaps owing to competition and clash from the binding partner for the LRR binding surface, thereby freeing the hydrophobic binding pocket and catalytic cysteine to function. Furthermore, coupling the binding of the LRR domain and the activation of the NEL domain would allow for SspH2 to be active as a ligase only when properly engaged in the presence of its substrate.
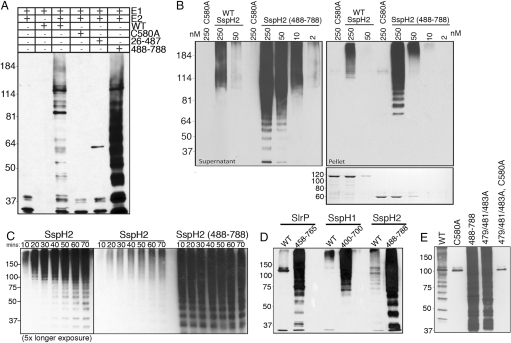
To test this hypothesis, we first examined the ubiquitin ligase activity of different SspH2 domains. As expected, the construct encompassing the SspH2 N terminus plus the LRR domain (residues 26–487) lacked E3 ligase activity (Fig. 4A). In contrast, titration experiments indicated that the NEL domain alone possesses approximately 25-fold higher activity than the full-length protein, both in autoubiquitination and in generating chains of free ubiquitin (Fig. 4 B and C), consistent with the hypothesis that the LRR domain inhibits NEL domain activity. A dramatic increase in the ligase activity of SspH1 and SlrP is also observed upon removal of their respective N-terminal domains (Fig. 4D), suggesting that an autoinhibitory E3 ligase mechanism is common to this multidomain family of proteins. To further explore this premise, we generated 2 types of mutations in the full-length SspH2 protein. First, we replaced the loop connecting the LRR and NEL domains with a glycine linker (482DMAGASAPR490 to 482GGGGSGGGG490), hypothesizing that the additional flexibility might facilitate the proposed hinge-like motion of the C-terminal domain. In addition, a triple mutant of residues involved in interdomain contacts between the LRR and NEL domains was created (I479A, F481A, and M483A of the LRR domain). Both of these mutations increase the E3 ligase activity of full-length SspH2 to nearly that of the NEL domain alone (Fig. 4E and Fig. S1). These experiments further support the proposed mechanism of a required conformation change to free the ligase domain from the LRR domain.
Fig. 4.
The amino terminal domain of SspH2 regulates NEL domain ligase activity. (A) E3 ligase activity of the amino and carboxy terminal domains. Purified recombinant SspH2, SspH2C580A, SspH226–487, and SspH2488–788 were incubated with FLAG-tagged ubiquitin in the presence of ATP, E1, and recombinant UbcH5b. Aliquots of the reaction were analyzed by 6–12% SDS-PAGE and immunoblotted with anti-FLAG antibody to detect ubiquitin conjugates. As controls the indicated components of the reaction were omitted. (B) Comparison of the ubiquitin ligase activities of SspH2 and SspH2488–788. Different amounts (as indicated) of purified recombinant SspH2 or SspH2488–788 were tested for their ubiquitin ligase activity as indicated above. After in vitro ubiquitination reactions, free ubiquitin chains (supernatant) and E3–ubiquitin conjugates (pellet) were separated and analyzed by SDS-PAGE and immunoblotted with anti-FLAG antibody. Bottom: Coomassie-stained gel representing the concentration of GST-tagged E3 used in the above titration. (C) Time course comparison of the ubiquitin conjugation reaction of SspH2 and SspH2488–788. Purified recombinant GST-SspH2 or GST-SspH2488–788 (250 nM) were incubated with FLAG-tagged ubiquitin in the presence of ATP, E1, and recombinant UbcH5b. Aliquots of the reactions were removed at the indicated times and analyzed by 8% SDS-PAGE and immunoblotted with anti-FLAG antibody. (D) Removal of the LRR domain increases the E3 ligase activity of SlrP and SspH1. Purified recombinant GST-SspH1, GST-SspH1400–700, GST-SlrP, GST-SspH1458–765, GST-SspH2, or GST-SspH2488–788 (250 nM) were analyzed as above. (E) Disruption of the interaction of the N-terminal domain with the NEL domain relieves autoinhibition of the E3 ligase activity of full-length SspH2. Recombinant GST-SspH2, GST-SspH2C580A, GST-SspH2488–788, GST-SspH2479/481/483A, and GST-SspH2479/481/483A, C580A were incubated with FLAG-tagged ubiquitin in the presence of ATP, E1, and recombinant UbcH5b. Aliquots of the reactions were removed at the indicated times and analyzed by 8% SDS-PAGE and immunoblotted with anti-FLAG antibody.
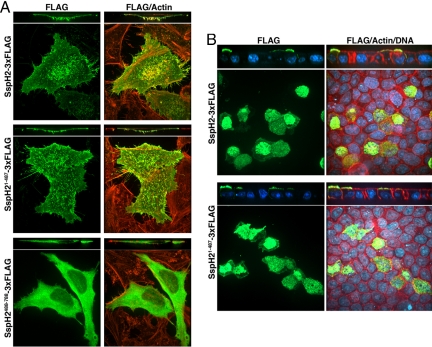
We also examined the localization of the different domains of SspH2 when transiently expressed in cultured cells. We found that full-length SspH2 localized to the plasma membrane, and it was particularly enriched in microvilli (Fig. 5A and Fig. S2). The LRR-containing domain of SspH21–487 exhibits a localization profile virtually indistinguishable from that observed with the full-length protein (Fig. 5A), consistent with the hypothesis that the N-terminal SspH2 region serves to target the enzymatic activity to the appropriate cellular site (20). In contrast, the NEL domain localizes throughout the cell (Fig. 5A). Furthermore, the NEL domain when expressed alone proved to be cytotoxic, although this effect did not depend on its catalytic activity (data not shown).
Fig. 5.
The amino terminal domain of SspH2 mediates localization within host cells. FLAG epitope–tagged SspH2, SspH21–487, or SspH2488–788 were transiently expressed in cultured HeLa (A) or polarized MDCK (B) cells and their localization examined by immunofluorescence confocal microscopy using an antibody directed to the tag (green), rhodamine phallodian to stain polymerized actin (red) and where indicated POPO1 to stain DNA (blue).
The observed enrichment of SspH2 in microvilli prompted us to examine its localization when expressed in polarized epithelial cells. We found that SspH2 localized almost exclusively in the apical membrane of these cells, and more specifically, it was seen in close association to microvilli (Fig. 5B and Movie S1). This unique localization was also dependent on the presence of the N-terminal LRR-containing domain (Fig. 5B). Taken together, these results suggest that the SspH2 N-terminal region containing the LRR domain modulates the host interactions of the NEL domain, directing its activity to the apical plasma membrane to engage its putative substrates and the host ubiquitination machinery.
Discussion
These studies have identified a unique class of cysteine-dependent E3 ubiquitin ligase present in several species of pathogenic bacteria, which differs from all known E3 ligases both in structure and mechanism. However, SspH2 shares some generalized characteristics with the HECT E3 family of ligases, namely the presence of a conserved cysteine residue on a loop that is critical to the ubiquitination reaction, the formation of a stable covalent link with ubiquitin, and the requirement of conformational changes for biochemical activity. The NEL domain of the Salmonella virulence factor, however, does not necessarily mimic the catalytic mechanism of known eukaryotic HECT E3 ligases. A thioester intermediate was not isolated from SspH2 or its homologues SspH1 and IpaH9.8 (12). SspH2, in addition to having a unique fold, may therefore use a different mechanism in the ubiquitination of substrates than a HECT E3.
Putative homologues of SspH2 are present in other pathogenic bacteria, including Shigella and Yersinia spp. (18). In these homologues, as in SspH2, the C-terminal domain is often paired with an LRR domain of variable length. LRR domains provide a versatile structural framework for protein–protein interactions and for coupling enzymatic domains to substrate-binding domains in the LRR (24). As an example, the SspH1 LRR domain was previously shown to bind its substrate, PKN1 (19). Similar modularity can be found in the family of HECT E3 ligases, which are divided into 3 classes according to their N-terminal extensions. Two of these groups contain known protein–protein interaction domains RLD (RCC1-like domain) (25) and WW domains (26, 27), adjacent to the catalytic domain that binds the substrate.
The modular nature of this family of proteins suggests that the NEL domain mediates E2 binding and the ubiquitination reaction, whereas the LRR domain interacts with substrate(s). Sequence comparison reveals that conserved residues among the SspH2 homologues are clustered in 2 regions of the NEL domain. The charged residues surrounding the catalytic cysteine that forms salt bridges or hydrogen bond networks are conserved. Additionally, conserved amino acids line the surface of a groove that includes the hydrophobic patch (depicted in Fig. 3B) in the vicinity of the catalytic cysteine (Fig. S3), suggesting a potential binding site for an E2 enzyme.
Although the catalytic mechanism of SspH2 and its homologues may differ from that of HECT E3 ligases, their mechanism of action may share the similarity of coupling an enzymatic ligase domain with a substrate-binding domain. Thus, a catalytically important cysteine in the C terminus facilitates the transfer of ubiquitin from an activated E2 to a substrate protein bound to the LRR domain.
To control the targeting of the ligase reaction, the pathogen seems to have evolved an autoinhibitory mechanism that sequesters the catalytic site of the NEL domain within the LRR domain of the protein. Data presented in this article demonstrate that the presence of the LRR domain attenuates the ligase activity of SspH2 and its Salmonella homologues SspH1 and SlrP (Fig. 4D). However, when structure-based mutants that disrupt the binding interface between the LRR and NEL domains are made, ligase activity is increased to that observed for the NEL domain alone (Fig. 4E and Fig. S1). Our crystal structure, with its buried active site cysteine and partially occluded access to the inner concave surface of the LRR domain, likely depicts an autoinhibited conformation of the E3 ligase (Fig. 2A). An analogous mode of regulation was observed in the Smurf2 E3 ligase, which exhibits a HECT domain inhibited by intramolecular interactions with a C2 domain (28).
Two structures of Shigella homologues of SspH2, published simultaneously as our manuscript was under review, further support our mechanistic hypothesis (29, 30). A comparison of the IpaH3 and SspH2 crystal structures reveals a striking difference in the positioning of the NEL domain with respect to the LRR (Fig. S4). IpaH3 exhibits a solvent accessible catalytic cysteine and an LRR binding surface that is completely accessible, possibly depicting the activated conformer of these bacterial enzymes. To transition from the buried form in SspH2 to the exposed conformation on IpaH3, the NEL domain must swing out ≈180° to expose its cysteine and free access to the putative LRR substrate-binding site (Fig. S4). This radical hinge motion between these 2 structures is exactly predicted by our hypothesis and consistent with our biochemical data.
The NEL domain by itself is toxic to cells, even with the catalytic cysteine mutated, indicating that it is not the ubiquitination itself but likely the interaction of the NEL domain with host ubiquitination machineries or substrates that induces a toxic effect. Folding of SspH2 into an autoinhibited form likely prevents premature activation resulting in cellular toxicity. The binding of host targets, presumably by the LRR domain, would result in a relative reorientation of the 2 domains, exposing the catalytic site to the host ubiquitination machinery and substrate(s).
The multidomain effector, SspH2, therefore belongs to an exquisitely evolved protein family with interacting subunits, an N-terminal domain that targets the enzymatic activity to the plasma membrane, thereby triggering a conformational change that releases the NEL domain to function in a localized and controlled fashion. The specific localization of SspH2 to the brush border of polarized epithelial cells is intriguing given the prominent role that intestinal epithelial cells play during the interaction of S. typhimurium, an enteropathogen, with its various hosts. Although the actual target(s) of the E3 ligase activity of SspH2 have yet to be identified, its restricted localization should aid this search and help to put its enzymatic activity in a firm physiologic context.
Protein secretion mechanisms like the T3SS used by Salmonella have existed for nearly a billion years (31), bringing bacterial proteins into intimate contact with host cell biology and allowing the selective pressures of coevolutionary forces to shape the biochemical interactions between these very different forms of life. The highly sophisticated nature of bacterial virulence factors underscores this observation, and the targeting of the eukaryotic ubiquitination system represents a remarkable example of biochemical mimicry of host cell biology. In addition, the insights into the structure and function of SspH2 may illuminate the mechanisms of function and activity in other infectious agents sharing homology, opening new directions for the design of novel anti-infectives, as well as providing insights into the host ubiquitination system.
Methods
Protein Expression and Purification.
For crystallization, SspH2166–783 from the Salmonella Typhimurium LT2 genome was expressed from a pCDF-Duet-1 vector (Novagen) that added at the N terminus 2 tandem hexahistidine tags separated by a linker and followed by a downstream rhinovirus 3C protease cleavage site. The protein was expressed in BL21(DE3) at 20 °C and purified by affinity chromatography. After overnight dialysis and cleavage with 3C protease, the protein was further purified by anion exchange and gel filtration chromatography. For biochemical experiments, slrp, sspH1, and sspH2 amplified from the S. typhimurium 14028s genome were cloned in pGEX-KG vector (32), expressed, and purified by affinity chromatography. Catalytically inactive and other site-specific mutants were generated by PCR mutagenesis. For localization studies, a 3xFLAG tag was added to the C terminus of full-length SspH2, SspH21–487, and SspH2488–788 and the resulting constructs cloned into the eukaryotic expression plasmid pcDNA3.1.
Crystallization and Structural Determination.
Crystals were obtained from mixing 2 μL of 14 mg/mL SspH2 (166–783) with 2 μL of the reservoir solution [5% Tacsimate, 0.1 M Hepes (pH 7.2), and 8–10% PEG5K], followed by equilibration against reservoir by the hanging drop method. Crystals were briefly soaked in a cryogenic solution [5% Tacsimate, 0.1 M Hepes (pH 7.2), 12% PEG5K, and 15% ethylene glycol] and then flash-frozen in liquid nitrogen before diffraction data collection. The structure was determined in a 2-step process in which an initial model was obtained from a poorly diffracting crystal form through selenomethionine substituted protein, after which molecular replacement was performed with this partial model into a well-diffracting crystal form with the program PHASER (33). The PHASER solution was used in the flex/ARP Expert System, and a nearly complete model for SspH2 (171–783) was thereby obtained (34). Iterative rounds of refinement using REFMAC5 (35) and manual model building in COOT (36) yielded a final model spanning residues 171–783 with an R/Rfree of 22.0%/26.3% (Table S1).
In Vitro Ubiquitination Assays.
In vitro ubiquitination experiments were carried out in a reaction mixture (40 μL) containing 50 mM Tris-HCl (pH 7.5), 50 mM NaCl, 10 mM MgCl2, 5 mM ATP, 0.1 mM DTT, 2 μg FLAG-ubiquitin (Boston Biochem), 0.5 μg UBE1 (Boston Biochem), and 2 μg UbcH5b (Boston Biochem), and the indicated amount of bacterially expressed SspH1, SspH2, or SlrP were incubated at 37 °C for 90 min. Reactions were quenched with SDS sample buffer containing 100 mM DTT and were resolved on a 6–12% SDS-PAGE gradient gel and subjected to immunoblot analysis with mouse anti-FLAG antibodies (Sigma). For E3 ligase titration experiments, after the in vitro ubiquitination reaction as indicated above, GST-tagged E3 ligases were removed by 2 sequential incubations with glutathione Sepharose. The cleared supernatants and pellets were then separated on SDS-PAGE and subjected to immunoblot analysis as described above. Equivalent amounts of GST-SspH2 and GST-SspH2488–788 and their respective mutants as used in the E3 ligase titration experiment were separated on a 10% SDS-PAGE and detected by Coomassie staining. For time course experiments, 250 nM of GST-SspH2 or GST-SspH2488–788 were incubated as previously described. Aliquots were removed every 10 min, and ubiquitin conjugates were detected as above.
Fluorescence Microscopy.
For localization studies, HeLa cells (70–80% confluent) were transfected with the indicated cDNA using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Cells were fixed in 4% paraformaldehyde for 15 min and permeabilized with 0.2% Triton X-100 in 10 mM glycine in PBS (Gly/PBS) containing 2% BSA for 10 min. For localization in polarized cells, MDCK cells were seeded onto transwells, and once polarized, cells were transfected with the indicated cDNAs using Lipofectamine 2000. Cells were fixed in 4% paraformaldehyde for 15 min and permeabilized with 0.2% Triton X-100 in Gly/PBS buffer containing 1 mg/mL RNase and 2% BSA for 30 min. Coverslips (or transwell membranes) were incubated in mouse anti-FLAG antibody diluted in Gly/PBS plus 2% BSA for 40 min. Coverslips were washed and incubated with antimouse Alexa 488 and rhodamine-phalloidin, and where indicated with POPO-1 (Invitrogen) to stain DNA. Coverslips were then mounted in ProLong Gold (Invitrogen). Confocal images were collected at 0.2-μm intervals for 10 μm (HeLa cells) or 25 μm (MDCK cells) on an Improvision spinning disc confocal microscope equipped with a Hamamatsu EM-CCD digital camera using Volocity software (Improvision).
Cell Fractionation.
HeLa cells were transfected with the indicated cDNA using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Cells were scraped into PBS, centrifuged for 5 min at 400 × g, resuspended in 10 mM Tris (pH 7.6), 15 mM NaCl, 2 mM MgCl2, and protease inhibitor mixture (Complete; Roche) and incubated on ice for 5 min. Sucrose was added to a final concentration of 8.5%, and cells were broken by 30 strokes of a Dounce homogenizer, generating the whole-cell fraction. The lysate was centrifuged at 500 × g for 10 min. The supernatant was centrifuged at 200,000 × g for 1 h. The resulting supernatant was collected (cytosolic fraction), and the pellet (membrane fraction) was resuspended in radio immunoprecipitation assay buffer (1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, 15 mM NaCl, 1 mM sodium phosphate (pH 7.2), 2 mM EDTA, and protease inhibitor mixture). Ten micrograms of each fraction was analyzed by SDS-PAGE and visualized by immunoblot analysis as previously described.
Supplementary Material
Supporting Information
Acknowledgments.
We thank D. Oren at Rockefeller University and Babu Manjesetty and W. Shi of Brookhaven beamlines X3A and X29, respectively, for access to and assistance with crystallographic equipment. S.W.H. was supported by National Research Service Award postdoctoral fellowship AI069704 from the National Institutes of Health (NIH). C.M.Q. was supported by a Rockefeller University Women and Science Fellowship. This work was funded by NIH Grants AI52182 (to C.E.S.) and AI055472 (to J.E.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3G06).
References
- 1.Gutierrez GJ, Ronai Z. Ubiquitin and SUMO systems in the regulation of mitotic checkpoints. Trends Biochem Sci. 2006;31:324–332. doi: 10.1016/j.tibs.2006.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mukhopadhyay D, Riezman H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science. 2007;315:201–205. doi: 10.1126/science.1127085. [DOI] [PubMed] [Google Scholar]
- 3.Newton K, Vucic D. Ubiquitin ligases in cancer: Ushers for degradation. Cancer Invest. 2007;25:502–513. doi: 10.1080/07357900701508041. [DOI] [PubMed] [Google Scholar]
- 4.Reed SI. The ubiquitin-proteasome pathway in cell cycle control. Results Probl Cell Differ. 2006;42:147–181. doi: 10.1007/b136681. [DOI] [PubMed] [Google Scholar]
- 5.d'Azzo A, Bongiovanni A, Nastasi T. E3 ubiquitin ligases as regulators of membrane protein trafficking and degradation. Traffic. 2005;6:429–441. doi: 10.1111/j.1600-0854.2005.00294.x. [DOI] [PubMed] [Google Scholar]
- 6.Sigismund S, Polo S, Di Fiore PP. Signaling through monoubiquitination. Curr Top Microbiol Immunol. 2004;286:149–185. doi: 10.1007/978-3-540-69494-6_6. [DOI] [PubMed] [Google Scholar]
- 7.Herrmann J, Lerman LO, Lerman A. Ubiquitin and ubiquitin-like proteins in protein regulation. Circ Res. 2007;100:1276–1291. doi: 10.1161/01.RES.0000264500.11888.f0. [DOI] [PubMed] [Google Scholar]
- 8.Welchman RL, Gordon C, Mayer RJ. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat Rev Mol Cell Biol. 2005;6:599–609. doi: 10.1038/nrm1700. [DOI] [PubMed] [Google Scholar]
- 9.Angot A, Vergunst A, Genin S, Peeters N. Exploitation of eukaryotic ubiquitin signaling pathways by effectors translocated by bacterial type III and type IV secretion systems. PLoS Pathog. 2007;3:e3. doi: 10.1371/journal.ppat.0030003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diao J, Zhang Y, Huibregtse JM, Zhou D, Chen J. Crystal structure of SopA, a Salmonella effector protein mimicking a eukaryotic ubiquitin ligase. Nat Struct Mol Biol. 2008;15:65–70. doi: 10.1038/nsmb1346. [DOI] [PubMed] [Google Scholar]
- 11.Janjusevic R, Abramovitch RB, Martin GB, Stebbins CE. A bacterial inhibitor of host programmed cell death defenses is an E3 ubiquitin ligase. Science. 2006;311:222–226. doi: 10.1126/science.1120131. [DOI] [PubMed] [Google Scholar]
- 12.Rohde JR, Breitkreutz A, Chenal A, Sansonetti PJ, Parsot C. Type III secretion effectors of the IpaH family are E3 ubiquitin ligases. Cell Host Microbe. 2007;1:77–83. doi: 10.1016/j.chom.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Y, Higashide WM, McCormick BA, Chen J, Zhou D. The inflammation-associated Salmonella SopA is a HECT-like E3 ubiquitin ligase. Mol Microbiol. 2006;62:786–793. doi: 10.1111/j.1365-2958.2006.05407.x. [DOI] [PubMed] [Google Scholar]
- 14.Kubori T, Galan JE. Temporal regulation of salmonella virulence effector function by proteasome-dependent protein degradation. Cell. 2003;115:333–342. doi: 10.1016/s0092-8674(03)00849-3. [DOI] [PubMed] [Google Scholar]
- 15.Patel J, Hueffer K, Lam T, Galán J. Diversification of a Salmonella effector protein function by ubiquitin-dependent differential localization. Cell. 2009 doi: 10.1016/j.cell.2009.01.056. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: Destruction for the sake of construction. Physiol Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 17.Pavletich NP. Structural biology of ubiquitin-protein ligases. Harvey Lect. 2002;98:65–102. [PubMed] [Google Scholar]
- 18.Miao EA, et al. Salmonella typhimurium leucine-rich repeat proteins are targeted to the SPI1 and SPI2 type III secretion systems. Mol Microbiol. 1999;34:850–864. doi: 10.1046/j.1365-2958.1999.01651.x. [DOI] [PubMed] [Google Scholar]
- 19.Haraga A, Miller SI. A Salmonella type III secretion effector interacts with the mammalian serine/threonine protein kinase PKN1. Cell Microbiol. 2006;8:837–846. doi: 10.1111/j.1462-5822.2005.00670.x. [DOI] [PubMed] [Google Scholar]
- 20.Miao E, et al. Salmonella effectors translocated across the vacuolar membrane interact with the actin cytoskeleton. Mol Microbiol. 2003;48:401–415. doi: 10.1046/j.1365-2958.2003.t01-1-03456.x. [DOI] [PubMed] [Google Scholar]
- 21.Laskowski RA, Chistyakov VV, Thornton JM. PDBsum more: New summaries and analyses of the known 3D structures of proteins and nucleic acids. Nucleic Acids Res. 2005;33:D266–268. doi: 10.1093/nar/gki001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Evdokimov AG, Anderson DE, Routzahn KM, Waugh DS. Unusual molecular architecture of the Yersinia pestis cytotoxin YopM: A leucine-rich repeat protein with the shortest repeating unit. J Mol Biol. 2001;312:807–821. doi: 10.1006/jmbi.2001.4973. [DOI] [PubMed] [Google Scholar]
- 23.Holm L, Sander C. Dali: A network tool for protein structure comparison. Trends Biochem Sci. 1995;20:478–480. doi: 10.1016/s0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- 24.Kobe B, Kajava AV. The leucine-rich repeat as a protein recognition motif. Curr Opin Struct Biol. 2001;11:725–732. doi: 10.1016/s0959-440x(01)00266-4. [DOI] [PubMed] [Google Scholar]
- 25.Garcia-Gonzalo FR, Rosa JL. The HERC proteins: Functional and evolutionary insights. Cell Mol Life Sci. 2005;62:1826–1838. doi: 10.1007/s00018-005-5119-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ingham RJ, Gish G, Pawson T. The Nedd4 family of E3 ubiquitin ligases: Functional diversity within a common modular architecture. Oncogene. 2004;23:1972–1984. doi: 10.1038/sj.onc.1207436. [DOI] [PubMed] [Google Scholar]
- 27.Shearwin-Whyatt L, Dalton HE, Foot N, Kumar S. Regulation of functional diversity within the Nedd4 family by accessory and adaptor proteins. Bioessays. 2006;28:617–628. doi: 10.1002/bies.20422. [DOI] [PubMed] [Google Scholar]
- 28.Wiesner S, et al. Autoinhibition of the HECT-type ubiquitin ligase Smurf2 through its C2 domain. Cell. 2007;130:651–662. doi: 10.1016/j.cell.2007.06.050. [DOI] [PubMed] [Google Scholar]
- 29.Zhu Y, et al. Structure of a Shigella effector reveals a new class of ubiquitin ligases. Nat Struct Mol Biol. 2008;15:1302–1308. doi: 10.1038/nsmb.1517. [DOI] [PubMed] [Google Scholar]
- 30.Singer AU, et al. Structure of the Shigella T3SS effector IpaH defines a new class of E3 ubiquitin ligases. Nat Struct Mol Biol. 2008;15:1293–1301. doi: 10.1038/nsmb.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horn M, et al. Illuminating the evolutionary history of chlamydiae. Science. 2004;304:728–730. doi: 10.1126/science.1096330. [DOI] [PubMed] [Google Scholar]
- 32.Guan KL, Dixon JE. Eukaryotic proteins expressed in Escherichia coli: An improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Ann Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- 33.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat Protoc. 2008;3:1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vagin AA, et al. REFMAC5 dictionary: Organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2184–2195. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- 36.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information