Specialized piRNA Pathways Act in Germline and Somatic Tissues of the Drosophila Ovary (original) (raw)
. Author manuscript; available in PMC: 2010 Jun 9.
SUMMARY
In Drosophila gonads, Piwi proteins and associated piRNAs collaborate with additional factors to form a small RNA-based immune system that silences mobile elements. Here, we analyzed nine Drosophila piRNA pathway mutants for their impacts on both small RNA populations and the subcellular localization patterns of Piwi proteins. We find that distinct piRNA pathways with differing components function in ovarian germ and somatic cells. In the soma, Piwi acts singularly with the conserved flamenco piRNA cluster to enforce silencing of retroviral elements that may propagate by infecting neighboring germ cells. In the germline, silencing programs encoded within piRNA clusters are optimized via a slicer-dependent amplification loop to suppress a broad spectrum of elements. The classes of transposons targeted by germline and somatic piRNA clusters, though not the precise elements, are conserved among Drosophilids, demonstrating that the architecture of piRNA clusters has coevolved with the transposons that they are tasked to control.
INTRODUCTION
Eukaryotic genomes harbor a wide variety of transposons, whose maintenance and spread throughout the population requires the colonization of new genomic locations in germ cells. For the host, the deleterious consequences of transposon propagation range from insertional mutagenesis and reductions in the long-term fitness of their progeny to an acute loss of germ cell integrity and sterility.
Transposable elements can be broadly categorized as retrotransposons (class I), which move via an RNA intermediate, or DNA transposons (class II), which mobilize through a “cut-and-paste” mechanism (Slotkin and Martienssen, 2007). Transposons within these classes differ in their structure, evolutionary origins, and both their tissue and developmental expression patterns. Many transposons are expressed in germ cells, where movement can lead to heritable expansions in their number. Examples in Drosophila include TAHRE, TART, HetA, copia, and the I element (Brennecke et al., 2008; Chambeyron et al., 2008; Shpiz et al., 2009; Shpiz et al., 2007; Vagin et al., 2004). Some transposons are exclusively or additionally expressed in somatic cells of the ovary, with gypsy, ZAM, and idefix occupying this category in the Drosophila ovary (Desset et al., 2008; Desset et al., 2003; Mével-Ninio et al., 2007; Pélisson et al., 2007; Prud’homme et al., 1995; Sarot et al., 2004). The diversity of transposition strategies and the overall similarities to host protein-coding genes pose a substantial challenge to their selective silencing (Malone and Hannon, 2009).
In animals, suppression of mobile elements is accomplished by an elegant, small RNA-based immune system, which displays both genetically encoded and adaptive aspects. Its core components are Piwi family proteins, and their associated Piwi-interacting RNAs (piRNAs). Like other members of the Argonaute family, Piwi proteins use bound small RNAs as guides for substrate recognition and target cleavage (Carmell et al., 2002). In Drosophila, the major sources of piRNAs are discrete heterochromatic loci termed piRNA clusters (Supplemental Glossary available online) (Brennecke et al., 2007). These are characterized by an exceptional density of nested, fragmented, and immobilized transposon remnants. Thus, the generation of piRNAs from these loci inherently targets the three Drosophila Piwi proteins, Piwi, Aub, and AGO3, toward mobile elements.
Most piRNA clusters contain transposon fragments in sense and antisense orientations and produce piRNAs from both genomic strands. Nevertheless, piRNAs overall tend to be anti-sense to transposons (Brennecke et al., 2007). Piwi- and Aub-associated piRNAs reflect the antisense bias of the system, whereas AGO3-bound piRNAs are typically sense to transposons. Sense and antisense piRNAs bound by AGO3 and Aub, respectively, show a prevalent relationship with their 5′ ends, overlapping by precisely 10 nt.
These observations coalesced into a model in which Piwi proteins engage in a Slicer-dependent amplification loop (the ping-pong cycle—Supplemental Glossary) between piRNA clusters and active elements (Brennecke et al., 2007; Gunawardane et al., 2007). Cleavage of a transposon transcript by Aub, loaded with an antisense piRNA, triggers production of an AGO3-bound sense piRNA, whose 5′ end is offset by 10 nt. The AGO3-bound piRNA can then catalyze the production of more silencing-competent piRNAs, which associate with Aub, via cleavage of antisense transposon sequences within cluster transcripts. Overall, the ping-pong cycle optimizes the piRNA response against transposons active in a given cell and at a given developmental time point. Signatures of the ping-pong cycle are conserved throughout animals, suggesting that it is a fundamental property of the piRNA pathway (Aravin et al., 2007b; Houwing et al., 2007; Murchison et al., 2008).
Though the majority of Aub- and AGO3-bound piRNAs appear to be generated via the ping-pong cycle, only a small proportion of Piwi-bound piRNAs display ping-pong signatures. Yet, Piwi-bound piRNAs still exhibit a strong antisense bias. This has led to the concept of primary piRNA biogenesis, wherein Piwi acts as a possible recipient of cluster-derived piRNAs that are generated via yet unknown mechanisms.
Primary piRNAs have been proposed as one initiator of the ping-pong cycle. However, a recent study has also highlighted the importance of maternally inherited piRNA populations (Brennecke et al., 2008). For two transposons that were examined, the I and P elements, the lack of a maternal piRNA program prevented silencing in progeny, and this was associated with the lack of a robust ping-pong response. Thus, primary and maternally deposited piRNAs serve as inputs into the pathway, which initiate a cycle of interactions between piRNA clusters and transposon mRNAs.
We sought to determine whether this model applied universally, not only in germ cells but also in somatic support cells wherein a subset of transposons are regulated by Piwi, the sole family member expressed in this compartment. By comparing germline-specific piRNA populations to those derived from whole ovaries, we show that a distinct, ping-pong-independent piRNA pathway operates in somatic cells. Analysis of piRNA profiles from mutant ovaries strongly supports this model and indicates that the somatic pathway depends exclusively upon Piwi and the flamenco piRNA cluster. We also probed the roles of additional factors within the piRNA pathway, examining the impacts of nine such mutants on piRNA populations, on the operation of the pathway, and on the localization of pathway components in germ and somatic cells. We find that Piwi function in the germline depends on the RNA helicase Armitage. The ping-pong cycle acts independently of Piwi and Armitage but requires the function of Aubergine, the RNA helicases Spindle-E and Vasa, and the Tudor-domain protein Krimper. Through these studies, we begin to assemble a scaffold model of the piRNA pathway, which differs substantially in the germline and somatic compartments of the ovary.
RESULTS
Transposons Display Tissue-Specific Control Mechanisms
In the Drosophila ovary, all three Piwi-family members (Piwi, Aub, and AGO3) are expressed in germline cells (Brennecke et al., 2007; Cox et al., 2000; Gunawardane et al., 2007; Harris and Macdonald, 2001; Saito et al., 2006), while Piwi alone is expressed in gonadal somatic cells. This implied possible differences in the architecture of the piRNA pathway, and perhaps the elements that it controls, in germline and somatic tissues. We therefore sought to separately analyze piRNAs present in these two compartments.
Germline cells within the Drosophila egg chamber are syncytial, and nearly all of the cytoplasmic contents of nurse cells are incorporated into late-stage oocytes (Spradling, 1993). In contrast, the follicular epithelium is shed from the laid egg. Thus, we could infer somatic and germline piRNA pools by comparing small RNA libraries derived from wild-type ovaries to those from 0–2 hr old embryos, prior to the activation of the zygotic genome (Brennecke et al., 2008).
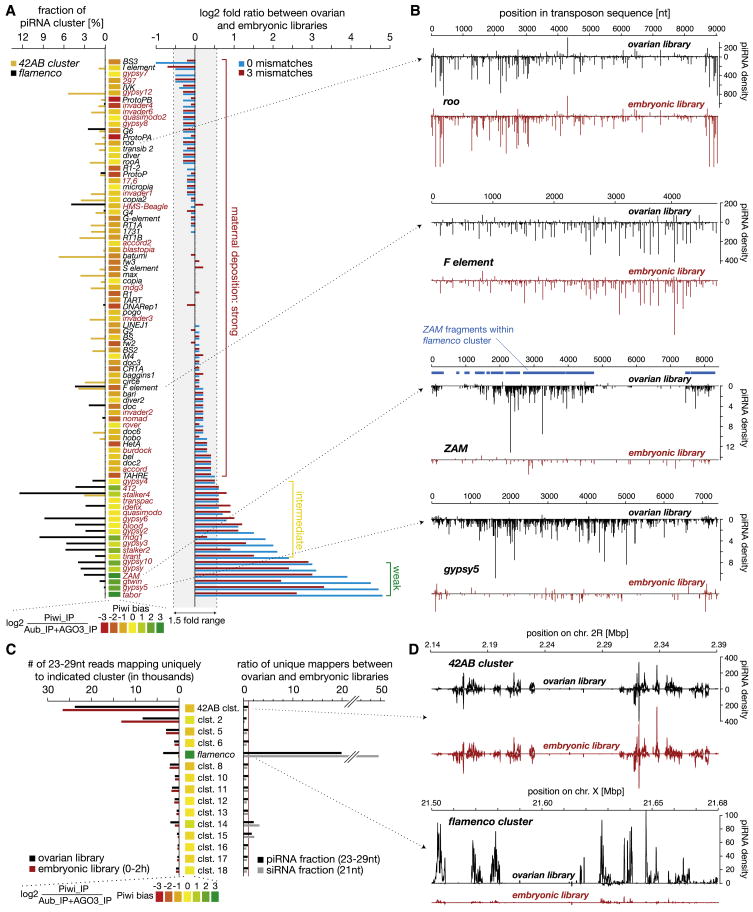
piRNAs (see Figure S1 available online) were mapped to the Repbase collection of known Drosophila melanogaster elements (allowing up to three mismatches) (Supplemental Experimental Procedures). We chose to focus on the 86 elements most heavily targeted by the piRNA pathway. This corresponds to 75% of all elements and includes ~99% of all transposon-derived piRNAs. piRNAs were also assigned to their generative clusters, including only those small RNAs mapping unambiguously to a single site within the Drosophila genome. Our key question was the extent to which piRNAs present in the mixed ovarian sample were maternally deposited.
For the majority of transposons, the piRNA content of early embryos mirrored that of total ovary (Figure 1A). As exemplified by roo and the F element, not only the overall abundance but also the distribution of ovarian piRNAs targeting each transposon was faithfully retained in early embryos (Figure 1A, right). However, piRNAs targeting a number of transposons (e.g., ZAM and gypsy; Figure 1A, right; Figure S2A) were substantially underrepresented in the embryonic piRNA pool. The transposons targeted by these small RNAs are likely subject to selective control by the piRNA system in somatic cells.
Figure 1. Maternal Deposition of piRNAs Defines Somatic and Germline piRNA Pathways.
Ovarian and early embryonic piRNAs were mapped to transposons (independent of mapping number) and piRNA clusters (only piRNAs with unique genome-wide mapping).
(A) The log2 fold ratio between ovarian and embryonic piRNAs over the 86 most targeted transposons is shown (right). The extent of maternal piRNA deposition is defined as strong (red), intermediate (yellow), or weak (green). _Gypsy_-family LTR retrotransposons are shown in red. For each element, the Piwi bias (log2 fold ratio of Piwi-bound piRNAs to Aub/AGO3-bound piRNAs) is shown in heat map form (center; green indicates strong, red weak Piwi bias). To the left, the sequence contribution of each element to the 42AB and flamenco piRNA clusters is shown in orange and black, respectively.
(B) Ovarian (black) and embryonic (red) piRNAs were plotted over elements with strong (roo and F element) or weak (ZAM and gyspy 5) maternal piRNA deposition (identical y axes). For ZAM, a strong correlation between piRNA density and sequence fragments present in the flamenco cluster (blue) was found.
(C) Total number (left) and ratio (right) of ovarian and embryonic piRNAs mapping to the 15 major piRNA clusters is shown. The corresponding Piwi-bias heat map is shown for each cluster as in (A).
(D) piRNA densities over the 42AB and flamenco piRNA clusters (identical y axes) are shown.
Elements with piRNA patterns characteristic of somatic control populate the gypsy family of long terminal repeat (LTR) retrotransposons. Among these are ZAM, gypsy, and idefix, all of which are regulated by the flamenco piRNA cluster. Across the entire spectrum of transposons, a lack of maternally deposited piRNAs correlated strongly with the presence of corresponding transposon fragments in flamenco (Figure 1A, left). In contrast, elements with fragments lying within other piRNA clusters (e.g., the cluster at cytological position 42AB; Figure 1A, left) contribute piRNAs to both ovarian and early embryonic libraries, indicating active control in germline cells.
These observations suggested tissue-specific regulation of certain element classes. This correlated with tissue-specific expression of piRNA clusters. Small RNAs derived from flamenco were highly depleted from early embryonic populations, irrespective of whether piRNA or siRNA pools were analyzed (Figure 1C). All remaining major clusters showed relatively equivalent contribution to ovary and embryo libraries. As with individual elements, the relative pattern of piRNAs mapping to clusters apparently expressed in germ cells was mirrored in embryo libraries (Figure 1D, right).
The overall degree to which transposons display Piwi-biased association (Piwi bias—Supplemental Glossary) strongly correlated with lower representation in maternally deposited small RNA populations (Figures 1A, 1B, and S2A). Also, while the majority of piRNA clusters load small RNAs into all three Piwi family members, _flamenco_-derived piRNAs almost exclusively occupy Piwi complexes (Figures 1C, 1D, and S2B).
These data suggest the existence of two separate piRNA pathways in germline and somatic cells of the gonad. In the soma, Piwi appears to be programmed exclusively or predominantly by the flamenco cluster to target elements from the gypsy family. In the germline, a variety of clusters collaborate with all three Piwi-clade proteins to control a broad range of elements and to contribute a heritable collection of piRNAs that maintains resistance across generations.
The Ping-Pong Cycle Is Germ Cell Specific
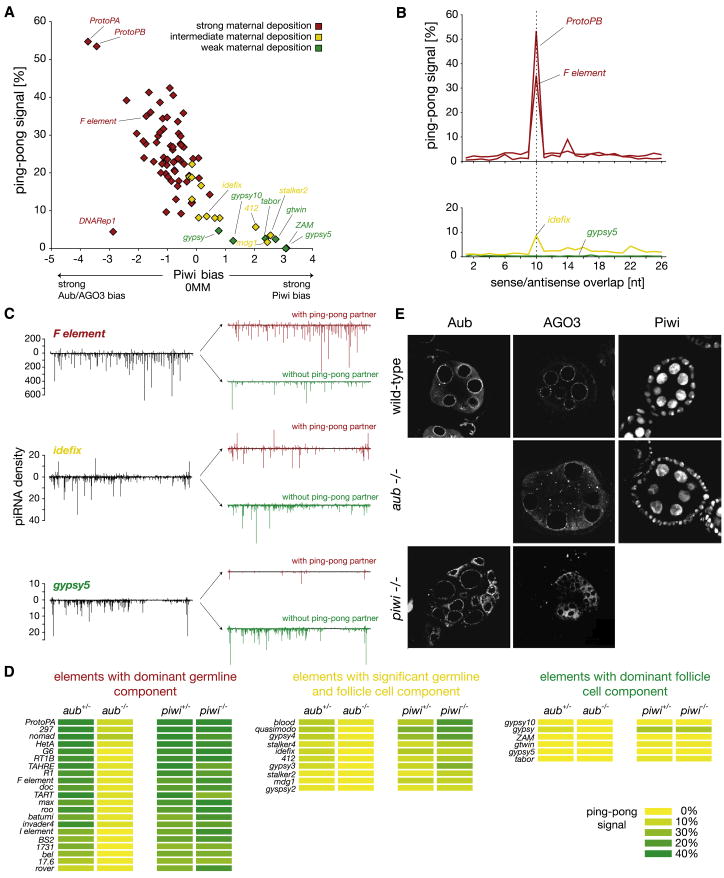
Ping-pong constitutes a feed-forward loop that optimizes the piRNA response against elements active in a given strain and simultaneously creates characteristic relationships between small RNAs that reveal their participation in the cycle (Brennecke et al., 2007; Gunawardane et al., 2007). The strongest ping-pong interactions are found between sense-oriented piRNAs in AGO3 and antisense piRNAs in Aub (Brennecke et al., 2007). In contrast, the participation of Piwi in the ping-pong cycle is less obvious. We therefore probed the degree to which individual transposons participate in the ping-pong cycle and correlated this with their bias toward control by individual Piwi proteins.
For each transposon, we quantified its ping-pong signature (Supplemental Experimental Procedures). In short, this was defined as the likelihood, in percent, for the average piRNA mapping to an element to have a complementary ping-pong partner. If plotted against the degree to which an element has corresponding piRNAs in Piwi versus Aub/AGO3 complexes, we find that Piwi-biased elements show no significant evidence of ping-pong (Figure 2A).
Figure 2. Transposons Segregate into Distinct Regulatory Classes.
Transposons segregated by maternal deposition and Piwi bias display differential levels of ping-pong amplification. (A)–(C) display piRNAs from the wK strain.
(A) Ping-pong signal and Piwi bias of transposons with strong (red), intermediate (yellow), or weak (green) maternal piRNA deposition are displayed as a scatter plot (0 MM, zero mismatches).
(B) Depiction of the ping-pong signature for indicated transposons. Graphs indicate the likelihood (in percent) that a complementary piRNA exists with a 5′ end at the indicated distance (x axis) for the average piRNA mapping to a particular transposon. The ping-pong signal was defined as the value at position 10 nt.
(C) piRNA densities over the indicated transposons are shown in black. Those are split into piRNAs with (red) and piRNAs without (green) a sequenced ping-pong partner.
(D) Ping-pong signals (value at 10 nt in [B]) are displayed as heat maps for aub and piwi heterozygote (+/−) and mutant (−/−) libraries.
(E) Aub, AGO3, and Piwi protein localization in wild-type, aubaubQC42/HN2, and piwi1/2 mutant ovaries. We note some variability in piwi mutants due to their aberrant morphology.
To probe the correlation between ping-pong signatures and maternal inheritance (Supplemental Glossary), we divided transposons into three groups (Figures 1A and 2A). Those with strong maternal deposition (red) are considered to have a dominant germline silencing component. Those with intermediate levels of maternal deposition (yellow) are considered to be expressed and targeted in both germline and follicle cells, while those with weak maternal deposition (green) are defined as having a dominant somatic silencing component (Supplemental Glossary).
Strong ping-pong signatures correlate with germline silencing (Figure 2A). Somatically silenced elements, such as gypsy5, show no enrichment for ping-pong pairs (Figure 2B), while elements such as idefix with mixed germline and somatic silencing show a weak but evident ping-pong signature. Predominantly germline elements such as ProtoP-B and F element show strong ping-pong signals (Figure 2B). For F element, idefix, and gypsy5, we plotted the distribution of piRNAs along each element consensus and then split the total population into piRNAs with an identified ping-pong partner and those without (Figure 2C). While piRNAs that have a ping-pong partner do show an overall antisense bias, this is much more pronounced for piRNAs that appear to arise via primary biogenesis. While an understanding is emerging for how the antisense bias is created for gypsy5 (see below), we still cannot explain how strand information for germline elements is incorporated into the pathway as a whole.
Although somatic Piwi lacks detectable ping-pong activity, we could not rule out roles for Piwi in the ping-pong cycle of germline elements. We therefore compared the impact of mutations in aub and piwi on ping-pong signatures. In a panel of 21 representative transposons with dominant germline expression, loss of Piwi showed no significant impact on ping-pong signals, while loss of Aub essentially ablated the cycle (Figure 2D). Elements targeted in somatic cells fail to enter the cycle (Figure 2D, right). Elements with apparently mixed expression patterns lose their ping-pong signatures in aub mutants but often show elevated signals in piwi mutants. This suggests that piwi and aub mutations impact different piRNA populations and that the germ-line-specific ping-pong cycle operates independently of Piwi.
Piwi resides in the nuclei of both germ and follicle cells (Cox et al., 2000). Aub and AGO3 concentrate in nuage, perinuclear RNP granules characteristic of germ cells (Figures 2E and S3) (Brennecke et al., 2007; Gunawardane et al., 2007; Harris and Macdonald, 2001). This has led to the speculation that that the ping-pong cycle might operate in nuage (Klattenhoff and Theurkauf, 2008). Loss of Aub leads to delocalization of AGO3 from nuage and to its accumulation in discrete cytoplasmic foci, a pattern not seen in piwi mutant germ cells (Figure 2E). These results underscore the link between Aub and AGO3 in germ cells and support a role for nuage in the ping-pong cycle.
Flamenco Programs the Somatic piRNA Pathway
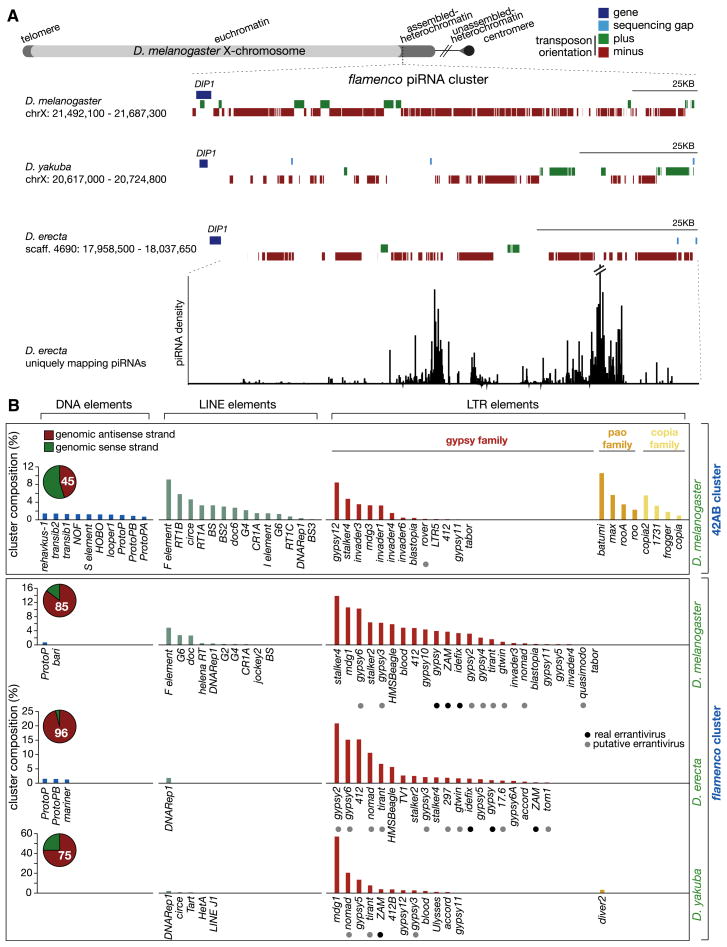
Many features distinguish flamenco from other generative loci. The flamenco locus shows an extreme orientation bias of the elements it harbors (Figure 1B), and unlike clusters expressed in germ cells, this bias seems to have been evolutionarily hardwired. flamenco comprises ~180 KB of pericentromeric heterochromatin on the X chromosome, in which the majority of transposon fragments (~85%) are similarly oriented (Figure 3A). Moreover, _flamenco_-derived piRNAs are produced exclusively from the plus strand of the genome, indicating transcription from the DIP1 gene toward the centromere. Analysis of P element insertions suggests that flamenco generates a long, continuously transcribed precursor, which is converted into a preponderance of antisense piRNAs by primary processing (Figure S4).
Figure 3. Evolutionary Conservation of the flamenco piRNA Cluster.
Transposon composition and chromosomal organization of the somatic flamenco piRNA cluster.
(A) Schematic of the Drosophila melanogaster X chromosome with the flamenco cluster enlarged. Below, the transposon annotation at the flamenco loci in two other Drosophilid species is shown. Uniquely mapping D. erecta piRNAs are plotted over the putative flamenco cluster (bottom).
(B) The transposon makeup of the 42AB, flamenco, and putative flamenco clusters are displayed. Pie charts display transposon orientation percentages. All graphs display the percentage of total annotated Repbase transposons in the cluster. Known and putative errantiviruses are indicated by black and gray dots.
In D. yakuba and D. erecta (Drosophila 12 Genomes Consortium et al., 2007), putative flamenco loci could be identified via their proximity to DIP1, and genomic assemblies were sufficiently complete to allow informative analysis (Figure 3A). In both cases, the _DIP1_-proximal region was enriched in transposon fragments with a consistent genomic orientation, such that transcription from DIP1 across these loci would produce antisense transposon information (Figure 3A). To confirm that these syntenic regions actually represent functional piRNA clusters, we sequenced a small RNA library from D. erecta ovaries. Abundant, uniquely mapping species could be assigned to the putative D. erecta flamenco locus, and as in D. melanogaster, they were derived from only one genomic strand (Figure 3A). Moreover, these RNAs showed no substantial evidence of an active amplification cycle (1U/10A partners with a 10 nt, 5′ overlap) (data not shown). Thus, natural selection seems to have shaped the flamenco clusters of Drosophilids to encode anti-sense piRNAs that can efficiently target homologous elements in the absence of an active ping-pong mechanism.
The transposons that are demonstrably impacted by flamenco mutations in ovary include ZAM, idefix, and gypsy (Desset et al., 2008; Mével-Ninio et al., 2007; Prud’homme et al., 1995). Some of these elements are also impacted in nongonadal somatic cells by flamenco/COM mutations, though the mechanism underlying this regulation is unknown (Desset et al., 2008). D. melanogaster flamenco shows a strong enrichment for sequences derived from gypsy family LTR retrotransposons. In D. yakuba and D. erecta flamenco loci, the enrichment for _gypsy_-family elements is conserved, although the precise elements that colonize each of these species and that populate their respective flamenco orthologs differ (Figure 3B). In contrast, germline clusters, such as that found at 42AB, contain a much broader variety of transposon classes and families, suggesting that evolutionary pressure favored the capture of different elements by that locus (Figure 3B). The conserved nature of the flamenco cluster across 10–12 million years of Drosophilid evolution (Drosophila 12 Genomes Consortium et al., 2007) suggests that specific clusters and transposons coevolve to maintain an effective defense.
Mutational Analysis Defines the Broad Genetic Requirements for the piRNA Pathway
A large number of loci disrupt fertility, ovarian morphology, or proper germ cell development (Klattenhoff and Theurkauf, 2008). Some of these represent strong candidates for piRNA pathway components because of their impacts on transposon silencing or the abundance of subsets of piRNAs. To understand their relationship to the germline and somatic piRNA pathways, we examined the molecular phenotypes of a series of eight mutants, with lesions in Piwi-family proteins, putative helicases, nucleases, and proteins of unknown biochemical function (Klattenhoff and Theurkauf, 2008). For reference, we compared these to a line mutant for the somatic piRNA cluster flamenco.
We isolatedand sequenced small RNAs (18–29 nt in length) from ovaries of age-matched flies, mutant for the genes shown in Figure 4 and Table 1. To avoid confounding our analysis because of interstrain variability in transposon content, we compared each mutant to its heterozygous siblings. Small RNA libraries were normalized using a subset of endogenous, AGO2-bound siRNAs as a reference (Tables S1 and S2, Supplemental Experimental Procedures, and Supplemental Glossary).
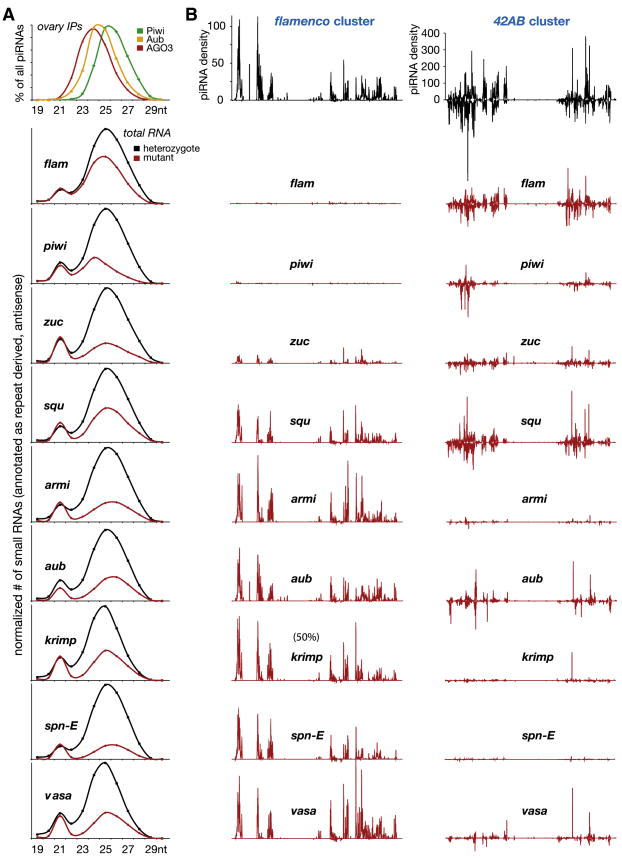
Figure 4. Genetic Dissection of the Germline and Somatic piRNA Pathways.
Analysis of piRNA populations in nine piRNA pathway mutants.
(A) Size profiles of Piwi- (green), Aub- (yellow), and AGO3- (red) bound ovarian piRNAs are plotted as a percentage. Below, siRNA-normalized small RNA size profiles are shown for ovaries mutant (red) or heterozygous (black) for the indicated genes.
(B) Uniquely mapping piRNAs are plotted over the 42AB and flamenco clusters. A typical heterozygote situation (here aub) is shown in black, all mutants in red. Libraries were normalized to allow for a direct comparison of piRNA densities between all libraries (Supplemental Experimental Procedures and Supplemental Glossary). For the krimp mutant, the flamenco density is scaled to 50%, with all other axes identical.
Table 1.
Overview of the Effect of Mutations on the piRNA Pathway
| Mutant | Function | Expression | piRNA Loss | Piwi Protein Localization: | Ping-Pong | piRNA Size | |||
|---|---|---|---|---|---|---|---|---|---|
| S | GL | Aubergine | AGO3 | Piwi | |||||
| flamenco | piRNA cluster | S | +++ | +/− | nuage | nuage | nuclear | functional | ↓↓ |
| piwi | Piwi family protein | S, GL | +++ | ++ | nuage | nuage | n.d. | functional | ↓↓↓ |
| zucchini | putative nuclease | unknown | ++ | ++ | nuage | nuage | nuclear | functional | ↓ |
| squash | putative nuclease | unknown | +/− | + | nuage | nuage | nuclear | functional | → |
| armitage | RNA helicase | GL | +/− | ++ | nuage | nuage | lost in GL | functional | ↑ |
| aubergine | Piwi family protein | GL | +/− | ++ | n.d. | dispersed | nuclear | very weak | ↑↑ |
| krimper | Tudor-domain containing | GL | +/− | +++ | dispersed | dispersed | nuclear | weak | ↑↑ |
| spindle-E | RNA helicase; Tudor domain | GL | +/− | +++ | dispersed | dispersed | low in GL | very weak | ↑↑ |
| vasa | RNA helicase | GL | +/− | ++ | dispersed | dispersed | low in GL | very weak | ↑↑ |
An examination of total piRNA levels and of piRNAs mapping to the major germline and somatic piRNA clusters revealed strong impacts on various aspects of the piRNA pathway for every mutant examined (Figures 4 and S4–S10, Tables 1 and S3–S6, Supplemental Experimental Procedures). Loss of piwi caused a substantial reduction in overall piRNA populations and also resulted in a shift in the overall size of the population to that more characteristic of Aub/AGO3-bound populations (Figure 4A). piwi loss virtually eliminated piRNAs mapping uniquely to the flamenco locus, consistent with the hypothesis that it is the sole family member that acts with this cluster in the somatic pathway. piwi also impacted piRNA production from the germline 42AB cluster, suggesting that it plays an important role in this compartment as well (Figure 4B). Mutation of the flamenco locus itself slightly reduced overall piRNA levels and shifted their average size, consistent with this somatic cluster contributing a substantial fraction of ovarian, Piwi-bound piRNAs (Figure 4A, Table 1). As expected, it had no impact on the production of piRNAs from other piRNA clusters such as 42AB (Figures 4B and S5).
Loss of aubergine strongly impacted overall piRNA populations, this time shifting their average size more toward that characteristic of Piwi-bound species (Figure 4A). flamenco was virtually untouched by the aub mutation. While piRNA production from 42AB was affected to a degree similar to that seen in piwi mutants, the subset of piRNAs that remain were quite different (Figure 4B). 42AB piRNAs remaining in the piwi mutant are Aub-sized and robustly participate in ping-pong, while in the aub mutant, 42AB piRNAs are Piwi-sized and display no ping-pong signatures (data not shown).
A number of mutations shared _aubergine_’s strong impact on the germline 42AB cluster with minimal impact on the somatic piRNA pathway. Loss of spn-E (Gillespie and Berg, 1995), vasa (Liang et al., 1994; Styhler et al., 1998) (Figure S11), krimp (Barbosa et al., 2007; Lim and Kai, 2007), and armi (Cook et al., 2004; Tomari et al., 2004) left the output of piRNAs from flamenco intact while suppressing, to varying degrees, piRNAs uniquely assignable to 42AB (Figure 4B, Table 1). With the exception of _armi_−/−, all of these mutants strongly impacted the ping-pong cycle (Figure S8, Table 1) with a concomitant delocalization of Aub and AGO3 from nuage (Figures S9 and S10). Moreover, the average size of piRNAs in each mutant shifted toward that characteristic of Piwi, consistent with an impact on Aub/AGO3-bound populations (Figure 4A).
zucchini and squash were identified in a screen for female-sterile mutants (Schupbach and Wieschaus, 1991). Both were previously shown to impact the levels of a few abundant piRNAs by northern blotting (Pane et al., 2007). Mutations in zuc reduced somatic and germline piRNA pathways, though both systems still produce some piRNAs in mutant animals. squash shows the least severe impact of all the mutants examined. Substantial production of piRNAs from flamenco and 42AB persist in squash mutants but overall piRNA levels do fall detectably.
The impacts of mutations in each of the genes we examined correlated largely with the expression patterns of the proteins, which they encoded (Table 1). Piwi appears in the nuclei of both germ and somatic cells, while Aubergine, like AGO3, is restricted to germ cells. Vasa is only expressed in germ cells, and similarly restricted expression is seen for Krimper, Spindle-E (M.D. and G.J.H., unpublished data), and Armitage (Cook et al., 2004; Gillespie and Berg, 1995; Lim and Kai, 2007). Expression patterns of Zucchini and Squash are unknown.
spindle-E and flamenco Mutations Define Properties of the Germline and Somatic piRNA Pathways
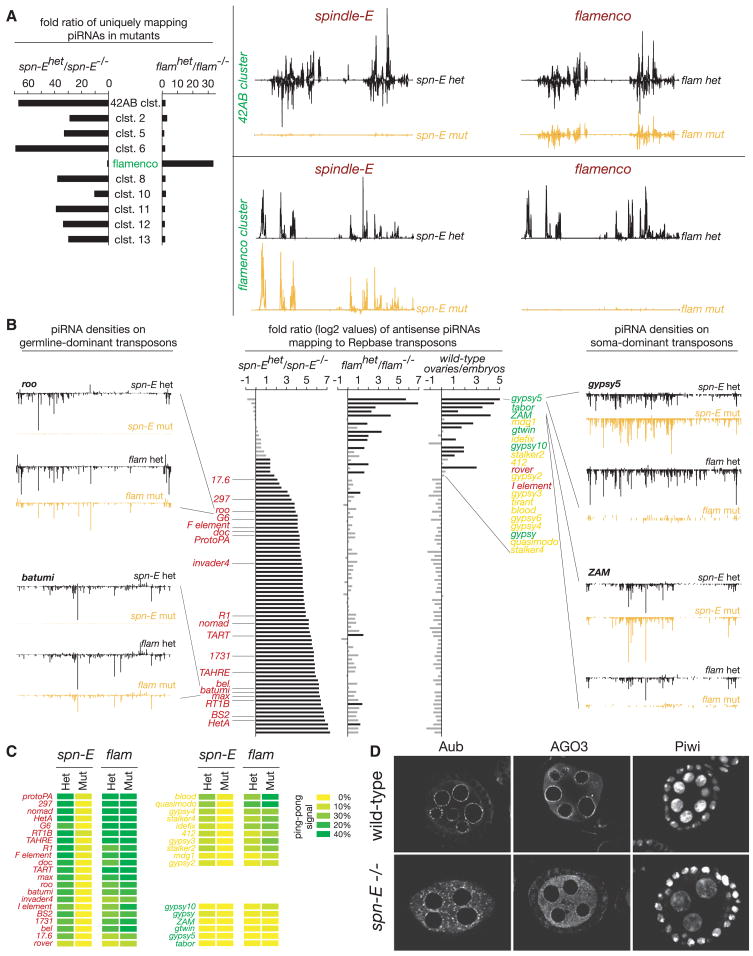
Spindle-E (spn-E) is a putative DExH-box RNA helicase that is critical for silencing of transposons in the Drosophila germline (Aravin et al., 2004; Aravin et al., 2001; Gillespie and Berg, 1995; Kennerdell et al., 2002; Klenov et al., 2007; Vagin et al., 2006). Its strong impact on the 42AB cluster extended to every other prominent piRNA cluster, save flamenco (Figure 5A). Impacts on the germline piRNA pathway were considerably stronger than those observed for aub mutants, suggesting that spn-E also affects piRNA populations bound by AGO3 and Piwi (Figure 4). In fact, in spn-E mutant ovaries (and those lacking other germline components), we consistently detected lower levels of Piwi in germline nuclei as compared to their heterozygote siblings (Figure S9). Thus, spn-E and flamenco mutants can be considered as archetypes for loss of germline and somatic pathways, respectively, allowing us to compare the participation of individual transposons in each system.
Figure 5. Mutation of spindle-E Defines Features of the Germline piRNA Pathway.
Mutations in spn-E and flamenco display reciprocal effects on piRNA profiles and ping-pong signatures.
(A) Shown are the fold changes of cluster-derived piRNAs in spn-E and flamenco mutants compared to their respective heterozygotes (left). To the right, piRNA densities from heterozygote (black) and mutant (yellow) libraries are plotted over the 42AB and flamenco clusters.
(B) A bar diagram shows the log2 fold changes of piRNA levels mapping to all analyzed transposons in spn-E and flamenco mutants compared to their respective heterozygotes (center). The identity of several transposons is given (color coded according to the degree of maternal inheritance [rightmost bar diagram]). Also shown are piRNA densities for selected elements (germline elements to the left, somatic elements to the right).
(C) Heat maps indicating ping-pong signals for typical germline (red), intermediate (yellow), and somatic transposons (green) in flamenco and spn-E mutants.
(D) Immunocytochemical analysis of Aub, AGO3, and Piwi protein localization in wild-type and _spn-E_1/100.37 mutant ovaries.
Elements were sorted according to the fold change in corresponding piRNAs in spn-E mutants, as compared to their heterozygous siblings. Only a few elements responded to loss of flamenco but not spn-E, and these almost perfectly overlapped with those that showed predominantly somatic control and a lack of maternal deposition of corresponding piRNAs (Figure 5B, right). In contrast, the vast majority of elements responded strongly to the spn-E mutation (Figure 5B). These included a wide variety of class I and class II elements, comprising the majority of Drosophila transposons and families (Figure 5B). Examples are roo and batumi. Both are unaffected in flamenco mutants but lose essentially all corresponding piR-NAs in spn-E mutants (Figure 5B). Virtually all of the elements, which respond to spn-E, showed a strong maternal deposition of homologous piRNAs, indicative of germline control. A few elements were impacted by both flamenco and spn-E mutations, consistent with the proposal that some elements are regulated in both somatic and germ cells.
Since the ping-pong cycle operates only in germ cells, it was not surprising that flamenco mutations show little impact on ping-pong signatures for elements that depend heavily on the germline pathway or that are strongly restricted to the soma (Figure 5C, elements designated in red and green, respectively). For elements that appear to be regulated by both germ cell and somatic pathways, flamenco mutations actually increased the proportion of piRNAs with a ping-pong signature. This is expected since the relative contribution from the somatic pathway would have been lost in flamenco mutants. For all elements with a detectable ping-pong signal, spn-E mutations greatly reduced or eliminated detectable partners (Figure 5C).
Mutations in other components of the germ cell pathway impacted both piRNA production and ping-pong signatures similarly to spn-E. For example, loss of the Tudor-domain protein Krimper had very pronounced impacts on piRNA levels and ping-pong signatures. Ovaries mutant for the RNA helicase Vasa resembled aubergine mutants in that both strongly affected the ping-pong cycle, though the impacts on piRNA levels on the germline specific elements was less dramatic than in spn-E and krimp mutants (Figure S6 and S7).
Mutations, which disrupted the ping-pong cycle, shared the impact of aub lesions on the localization of piRNA pathway components to nuage. spn-E mutant cells have altered Aub and AGO3 staining, while maintaining the characteristic localization of Piwi in the nuclei of both somatic and germ cells (Figure 5D). Similar impacts are seen for mutations in the germline pathway components krimp and vasa (Figure S9). Mutations in zuc or squ had no discernable impact on the localization of any Piwi-family protein, despite the impact of the former on the production of piRNAs in both the soma and germline (Figure S10). Notably, these mutations also had no (zuc) or a relatively mild (squ) impact on ping-pong signatures (Figure S8).
Armitage Is Important for Piwi Function in the Germline
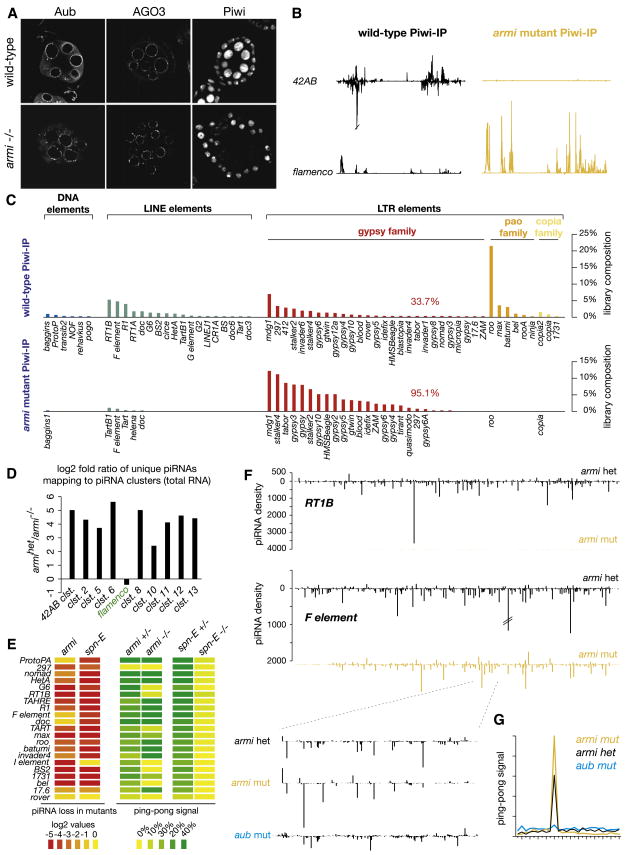
Despite the lack of a role for Piwi in the ping-pong cycle, several germline transposons exhibit a strong loss of piRNAs in piwi mutant animals. To probe the underlying basis of these effects, we wished to examine a situation in which the function of Piwi was specifically impaired in germ cells. We noted that mutations in armitage caused a loss of Piwi from germ cell nuclei (Figure 6A). Armitage encodes a homolog of SDE3, an RNA helicase that is involved in RNAi in Arabidopsis (Tomari et al., 2004) and armi mutations disrupt translational repression and localization of oskar mRNA in Drosophila oocytes (Cook et al., 2004).
Figure 6. Piwi Localization and Loading in Germline Cells Requires Armitage.
(A) Piwi protein localization in wild-type and armi1/72.1 mutant ovaries.
(B) Densities of uniquely mapping, Piwi-bound piRNAs over the 42AB and flamenco clusters from the wild-type (Oregon R, black) and armi mutants (yellow).
(C) Annotation of repeat derived, Piwi-bound piRNAs from wild-type and armi mutant ovaries.
(D) Log2 fold changes of cluster-derived piRNAs in an armi mutant total RNA library compared to heterozygote.
(E) Log2 fold changes of piRNAs mapping antisense to indicated transposons in armi and spn-E mutants compared to respective heterozygotes are shown in heat map form. Corresponding ping-pong signal heat maps are shown (right).
(F) piRNA densities over indicated transposons in heterozygote (black) and armi mutant (yellow) libraries.
(G) F element ping-pong profiles in armi heterozygote (black), armi mutant (yellow), and aub mutant (blue) libraries.
In armi mutant ovaries, loading of _flamenco_-derived piRNAs and piRNAs targeting gypsy-family transposons into Piwi is unaffected, consistent with the maintenance of Piwi expression and localization in somatic cells (Figures 6B and 6C, Table S7). However, we could no longer detect Piwi-associated piRNAs derived from 42AB or piRNAs corresponding to germline-regulated elements in Piwi complexes from mutant animals (Figures 6B and 6C).
Because of its selective effect in the germline, those piRNAs remaining Piwi-associated in armi mutants must represent somatic species. This allowed us to re-evaluate requirements for the somatic pathway among the remaining eight mutants that we characterized (Figure S12). In accord with our prior conclusions, only piwi, flamenco, and zucchini impacted this selected set of RNAs.
An examination of total RNA from armi mutants indicated that piRNAs contributed from all germline clusters were generally depleted (Figure 6D). Of the many germline elements that rely heavily upon the integrity of spn-E for piRNA production, many were also impacted by armi (e.g, RT1B, Figures 6E and 6F). However, a number of elements, including protoP-A, F element, and doc, were relatively insensitive to armi mutations (Figures 6E and 6F). Nearly all germline elements required spn-E for robust ping-pong. In contrast, a minority of elements depended upon armi for their participation in the amplification cycle, indicating that it is not required for the cycle, per se (Figures 6E and 6F). In fact, a detailed examination of piRNA densities across the F element indicated that the complex mixture of small RNAs seen in wild-type ovaries could be split genetically into two pools. armi mutants retain almost exclusively ping-pong pairs, while aubergine mutants retain the antisense-biased pool of small RNAs that lack ping-pong signatures and that likely represent primary piRNAs (Figures 6F and 6G).
Overall, these data were consistent with a model in which Piwi functions analogously in the germline and in the soma, accepting primary piRNAs produced as a result of processing of cluster transcripts. For some elements, these primary piRNAs appear to be essential for sustaining a robust response. However, for others, Aub and AGO3 can sustain a ping-pong response and support substantial piRNA populations in the absence of Piwi complexes loaded with cluster-derived piRNAs (Figures S5–S8). This may be due, in part, to the priming of the pathway against those elements by maternally contributed piRNAs.
DISCUSSION
The piRNA pathway forms an evolutionarily conserved mechanism for recognizing and selectively silencing mobile genetic elements. We have worked to deepen our understanding of the piRNA pathway by comparing piRNA populations from ovaries to early embryos and by examining small RNA populations in nine Drosophila mutants. These studies have revealed the existence of two related but distinct piRNA pathways that operate in the somatic and germline compartments of the Drosophila ovary. Similar conclusions were reached by Zamore and colleagues (Li et al., 2009) though their detailed analysis of piRNA populations AGO3 mutants. Each pathway showed unique features and distinct genetic dependencies.
In germ cells, the integrity of the piRNA pathway was strongly affected by seven different mutations, including piwi, auberinge, spindle-E, vasa, krimper, armitage, and zucchini. For most of these genes, loss of function strongly reduced overall piRNA levels. All but piwi, zucchini, and armitage had a substantial impact on the operation of the ping-pong amplification cycle that forms the adaptive arm of the pathway.
Nuage are a signature feature of germ cells in animals (al-Mukhtar and Webb, 1971; Eddy, 1974; Mahowald, 1968; Snee and Macdonald, 2004; Wilsch-Brauninger et al., 1997), which all share a need to guard their genomes from mobile elements. Previous studies have indicated that Aubergine and AGO3 occupy these structures. We found that this localization was disrupted specifically by mutations that reduce the operation of the ping-pong cycle. In Drosophila germ cells, nuage may concentrate both piRNAs and their targets to facilitate selective amplification of piRNAs targeting active elements (Klattenhoff and Theurkauf, 2008). However, since these structures also contain proteins that have not yet been linked to the piRNA pathway, nuage might also play additional roles in germline RNA metabolism.
Although the piRNA clusters that operate in the germline generally produce small RNAs from both genomic strands and contain element fragments in random orientations, the overall system is strongly biased toward antisense species (Brennecke et al., 2007). This bias is even more evident in small RNAs formed independently of the ping-pong cycle, indicating that primary biogenesis from piRNA clusters somehow perceives strand information. The mechanism by which this occurs remains mysterious, since none of the mutations that we evaluated selectively impacted this feature of the germline pathway.
In somatic follicle cells, a simplified version of the piRNA pathway is driven by the flamenco piRNA cluster. Unlike loci that operate in germ cells, the flamenco cluster generates piRNAs in the absence of ping-pong from only one strand and from a precursor RNA that contains element fragments in a uniform orientation (Brennecke et al., 2007). This arrangement is superficially similar to pachytene piRNA clusters in mammals. However, these do not play a role in transposon control, and their relevant targets remain unknown (Aravin et al., 2007a). Thus, these generative loci share only a propensity to generate piRNAs via a primary biogenesis mechanism.
Unlike the germline system, the antisense bias of the somatic system appears evolutionarily determined by selection for insertion of transposons in a preferred orientation within flamenco. Support for this hypothesis came from our analysis of two related species, D. yakuba and D. erecta. In both of these, flamenco shares a uniformity of transposon orientation and the production of predominantly antisense piRNAs from only one genomic strand.
In addition to its structure, the content of flamenco loci is conserved in all three species examined. This locus specifically targets LTR retrotransposons of the gypsy family. Many elements within the gypsy family are classified as errantiviral transposons (Song et al., 1994), which retain the ability to express envelope proteins. Thus, it has been proposed that _gypsy_-family elements have colonized the somatic gonadal niche and propagate in the population by infecting underlying germ cells with viral particles produced in follicular epithelial cells (Kim et al., 1994; Lécher et al., 1997; Song et al., 1997). In accord with this notion, many of the elements that show a Piwi/flamenco pattern of control can encode envelope proteins (Chalvet et al., 1999). This suggests that retroelements that can potentially form viral particles have long occupied a follicle cell niche and that this strategy has coevolved with a _flamenco_-directed silencing system in the soma.
Overall, the studies presented here have revealed unexpected complexities in the piRNA pathway. Two distinct pathways with different strategies and different genetic compositions are responsible for the silencing of different transposon classes in Drosophila ovaries. This suggests that the pathway adapts specifically to the structure and habits of each element to effectively protect the germline from transposon activity.
EXPERIMENTAL PROCEDURES
Antibodies and Immunocytochemistry
Rabbit polyclonal antisera directed against the N termini of Piwi, Aub, and AGO3 were previously described (Brennecke et al., 2007). Primary antibodies were diluted 1:500 for immunohistochemistry (Findley et al., 2003). In brief, ovaries were fixed in formaldehyde, rinsed, permeabilized in 0.1% Triton and washed. Ovaries were blocked in bovine serum albumin (BSA) and incubated overnight with primary antibodies. Ovaries were then washed and incubated in secondary antibody and washed. Finally, DNA was stained with TO-PRO (Molecular Probes), washed, and mounted in glycerol. Images were acquired with a Carl Zeiss Confocal LSM 510 miscroscope. See the Supplemental Experimental Procedures for further information.
Fly Stocks
The wild-type Drosophila melanogaster strains used in this study are Oregon R and the I element reactive, wK strain (a kind gift of Silke Jensen), (Hazelrigg et al., 1984; Luning, 1981). The following allelic combinations were used for immunolocalization, western, and RNA analyses: armitage, armi1/72.1 (Cook et al., 2004; Tomari et al., 2004), aubergine, _aub_QC42/HN (Schupbach and Wieschaus, 1991; Wilson et al., 1996), flamenco, _flam_KG00476 (Mével-Ninio et al., 2007), krimper, _krimp_f06583 (Barbosa et al., 2007; Lim and Kai, 2007), piwi, _piwi_1/2 (Cox et al., 1998; Lin and Spradling, 1997), spindle-E, _spn-E_1/100.37 (Gillespie and Berg, 1995), squash, _squ_HE47/PP33 (Pane et al., 2007), vasa, _vas_D5/PH165 (Liang et al., 1994; Styhler et al., 1998), zucchini, _zuc_HM27/Df(2I)PRL (Pane et al., 2007). See the Supplemental Experimental Procedures for further information.
Small RNA Cloning and Analysis
Small RNA libraries were generated as previously described (Brennecke et al., 2007). In brief, ovaries of the respective genotype were dissected in 10–50 μl batches into ice cold phosphate-buffered saline (PBS). Total RNA was extracted with Trizol (Invitrogen) and two phenol:chloroform:isoamyl alcohol (ROCHE) extraction steps. For each genotype, 50 μg of total RNA was separated on a 12% denaturing polyacrylamide gel and 18–29nt small RNAs were isolated for cloning. A detailed protocol for the generation of small RNA libraries is available upon request. Corresponding heterozygote libraries were prepared from ovarian RNA of heterozygous siblings (with mutant alleles balanced), which were collected from the same crosses. Only sequences matching the Drosophila release 5 genome (excluding ArmUextra) 100% were retained. Libraries were normalized to a subset of endo-siRNAs (Supplemental Experimental Procedures and Supplemental Glossary) to allow for cross-analysis. We mapped all 23–29 nt small RNAs to known piRNA clusters (Brennecke et al., 2007) and to the complete collection of D. melanogaster transposable elements (Repbase) (Jurka et al., 2005). See the Supplemental Experimental Procedures for further information. Previously published Piwi/AGO3/Aub-ovarian-IP libraries from Oregon R flies (GSE6734) (Brennecke et al., 2007) and total RNA libraries from the wK wild-type strain (GSE13081) (Brennecke et al., 2008) were also analyzed.
Supplementary Material
Supplement
T1
T2
T3
T4
T5
T6
T7
Acknowledgments
We thank members of the Hannon laboratory for helpful discussions and Oliver Tam for computational assistance. We are grateful to Michelle Rooks, Emily Hodges, Danea Rebolini, Laura Cardone, and Mike Regulski (Cold Spring Harbor Laboratory) for help with deep sequencing. We also thank Attilio Pane and Trudi Schüpbach for providing the zuc and squ fly stocks. C.D.M. is a Beckman fellow of the Watson School of Biological Sciences and is supported by a National Science Foundation Graduate Research Fellowship. J.B. is supported by a fellowship from the Ernst Schering foundation. M.D. is an Engelhorn fellow of the Watson School of Biological Sciences. A.S. is supported by a Human Frontier Science Program postdoctoral fellowship. This work was supported in part from grants from the National Institutes of Health to G.J.H. and W.R.M. and a kind gift from Kathryn W. Davis (G.J.H.).
Footnotes
References
- al-Mukhtar KA, Webb AC. An ultrastructural study of primordial germ cells, oogonia and early oocytes in Xenopus laevis. J Embryol Exp Morphol. 1971;26:195–217. [PubMed] [Google Scholar]
- Aravin AA, Naumova NM, Tulin AV, Vagin VV, Rozovsky YM, Gvozdev VA. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr Biol. 2001;11:1017–1027. doi: 10.1016/s0960-9822(01)00299-8. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Klenov MS, Vagin VV, Bantignies F, Cavalli G, Gvozdev VA. Dissection of a natural RNA silencing process in the Drosophila melanogaster germ line. Mol Cell Biol. 2004;24:6742–6750. doi: 10.1128/MCB.24.15.6742-6750.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007a;318:761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007b;316:744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- Barbosa V, Kimm N, Lehmann R. A maternal screen for genes regulating Drosophila oocyte polarity uncovers new steps in meiotic progression. Genetics. 2007;176:1967–1977. doi: 10.1534/genetics.106.069575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Malone CD, Aravin AA, Sachidanandam R, Stark A, Hannon GJ. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science. 2008;322:1387–1392. doi: 10.1126/science.1165171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmell MA, Xuan Z, Zhang MQ, Hannon GJ. The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev. 2002;16:2733–2742. doi: 10.1101/gad.1026102. [DOI] [PubMed] [Google Scholar]
- Chalvet F, Teysset L, Terzian C, Prud’homme N, Santamaria P, Bucheton A, Pelisson A. Proviral amplification of the Gypsy endogenous retrovirus of Drosophila melanogaster involves env-independent invasion of the female germline. EMBO J. 1999;18:2659–2669. doi: 10.1093/emboj/18.9.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambeyron S, Popkova A, Payen-Groschêne G, Brun C, Laouini D, Pelisson A, Bucheton A. piRNA-mediated nuclear accumulation of retrotransposon transcripts in the Drosophila female germline. Proc Natl Acad Sci USA. 2008;105:14964–14969. doi: 10.1073/pnas.0805943105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook HA, Koppetsch BS, Wu J, Theurkauf WE. The Drosophila SDE3 homolog armitage is required for oskar mRNA silencing and embryonic axis specification. Cell. 2004;116:817–829. doi: 10.1016/s0092-8674(04)00250-8. [DOI] [PubMed] [Google Scholar]
- Cox DN, Chao A, Baker J, Chang L, Qiao D, Lin H. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 1998;12:3715–3727. doi: 10.1101/gad.12.23.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DN, Chao A, Lin H. piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development. 2000;127:503–514. doi: 10.1242/dev.127.3.503. [DOI] [PubMed] [Google Scholar]
- Desset S, Meignin C, Dastugue B, Vaury C. COM, a heterochromatic locus governing the control of independent endogenous retroviruses from Drosophila melanogaster. Genetics. 2003;164:501–509. doi: 10.1093/genetics/164.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desset S, Buchon N, Meignin C, Coiffet M, Vaury C. In Drosophila melanogaster the COM locus directs the somatic silencing of two retrotransposons through both Piwi-dependent and -independent pathways. PLoS ONE. 2008;3:e1526. doi: 10.1371/journal.pone.0001526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosophila 12 Genomes Consortium. Clark AG, Eisen MB, Smith DR, Bergman CM, Oliver B, Markow TA, Kaufman TC, Kellis M, Gelbart W, et al. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- Eddy EM. Fine structural observations on the form and distribution of nuage in germ cells of the rat. Anat Rec. 1974;178:731–757. doi: 10.1002/ar.1091780406. [DOI] [PubMed] [Google Scholar]
- Findley SD, Tamanaha M, Clegg NJ, Ruohola-Baker H. Maelstrom, a Drosophila spindle-class gene, encodes a protein that colocalizes with Vasa and RDE1/AGO1 homolog, Aubergine, in nuage. Development. 2003;130:859–871. doi: 10.1242/dev.00310. [DOI] [PubMed] [Google Scholar]
- Gillespie DE, Berg CA. Homeless is required for RNA localization in Drosophila oogenesis and encodes a new member of the DE-H family of RNA-dependent ATPases. Genes Dev. 1995;9:2495–2508. doi: 10.1101/gad.9.20.2495. [DOI] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- Harris AN, Macdonald PM. Aubergine encodes a Drosophila polar granule component required for pole cell formation and related to eIF2C. Development. 2001;128:2823–2832. doi: 10.1242/dev.128.14.2823. [DOI] [PubMed] [Google Scholar]
- Hazelrigg T, Levis R, Rubin GM. Transformation of white locus DNA in drosophila: dosage compensation, zeste interaction, and position effects. Cell. 1984;36:469–481. doi: 10.1016/0092-8674(84)90240-x. [DOI] [PubMed] [Google Scholar]
- Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, van den Elst H, Filippov DV, Blaser H, Raz E, Moens CB, et al. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell. 2007;129:69–82. doi: 10.1016/j.cell.2007.03.026. [DOI] [PubMed] [Google Scholar]
- Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- Kennerdell JR, Yamaguchi S, Carthew RW. RNAi is activated during Drosophila oocyte maturation in a manner dependent on aubergine and spindle-E. Genes Dev. 2002;16:1884–1889. doi: 10.1101/gad.990802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim A, Terzian C, Santamaria P, Pélisson A, Purd’homme N, Bucheton A. Retroviruses in invertebrates: the gypsy retrotransposon is apparently an infectious retrovirus of Drosophila melanogaster. Proc Natl Acad Sci USA. 1994;91:1285–1289. doi: 10.1073/pnas.91.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klattenhoff C, Theurkauf W. Biogenesis and germline functions of piRNAs. Development. 2008;135:3–9. doi: 10.1242/dev.006486. [DOI] [PubMed] [Google Scholar]
- Klenov MS, Lavrov SA, Stolyarenko AD, Ryazansky SS, Aravin AA, Tuschl T, Gvozdev VA. Repeat-associated siRNAs cause chromatin silencing of retrotransposons in the Drosophila melanogaster germline. Nucleic Acids Res. 2007;35:5430–5438. doi: 10.1093/nar/gkm576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lécher P, Bucheton A, Pélisson A. Expression of the Drosophila retrovirus gypsy as ultrastructurally detectable particles in the ovaries of flies carrying a permissive flamenco allele. J Gen Virol. 1997;78:2379–2388. doi: 10.1099/0022-1317-78-9-2379. [DOI] [PubMed] [Google Scholar]
- Li C, Vagin VV, Lee S, Xu J, Ma S, Xi H, Seitz H, Horwich MD, Syrzycka M, Honda BM, et al. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell. 2009;137 doi: 10.1016/j.cell.2009.04.027. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang L, Diehl-Jones W, Lasko P. Localization of vasa protein to the Drosophila pole plasm is independent of its RNA-binding and helicase activities. Development. 1994;120:1201–1211. doi: 10.1242/dev.120.5.1201. [DOI] [PubMed] [Google Scholar]
- Lim AK, Kai T. Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc Natl Acad Sci USA. 2007;104:6714–6719. doi: 10.1073/pnas.0701920104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Spradling AC. A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development. 1997;124:2463–2476. doi: 10.1242/dev.124.12.2463. [DOI] [PubMed] [Google Scholar]
- Luning K. Genetics of inbred Drosophila melanogaster. Hereditas. 1981;95:181–188. doi: 10.1111/j.1601-5223.1983.tb00732.x. [DOI] [PubMed] [Google Scholar]
- Mahowald AP. Polar granules of Drosophila. II. Ultrastructural changes during early embryogenesis. J Exp Zool. 1968;167:237–261. doi: 10.1002/jez.1401670211. [DOI] [PubMed] [Google Scholar]
- Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell. 2009;136:656–668. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mével-Ninio M, Pelisson A, Kinder J, Campos AR, Bucheton A. The flamenco locus controls the gypsy and ZAM retroviruses and is required for Drosophila oogenesis. Genetics. 2007;175:1615–1624. doi: 10.1534/genetics.106.068106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchison EP, Kheradpour P, Sachidanandam R, Smith C, Hodges E, Xuan Z, Kellis M, Grützner F, Stark A, Hannon GJ. Conservation of small RNA pathways in platypus. Genome Res. 2008;18:995–1004. doi: 10.1101/gr.073056.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pane A, Wehr K, Schüpbach T. zucchini and squash encode two putative nucleases required for rasiRNA production in the Drosophila germline. Dev Cell. 2007;12:851–862. doi: 10.1016/j.devcel.2007.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pélisson A, Payen-Groschêne G, Terzian C, Bucheton A. Restrictive flamenco alleles are maintained in Drosophila melanogaster population cages, despite the absence of their endogenous gypsy retroviral targets. Mol Biol Evol. 2007;24:498–504. doi: 10.1093/molbev/msl176. [DOI] [PubMed] [Google Scholar]
- Prud’homme N, Gans M, Masson M, Terzian C. Flamenco, a gene controlling the gypsy retrovirus of Drosophila melanagaster. Genetics. 1995;139:697–711. doi: 10.1093/genetics/139.2.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Nishida KM, Mori T, Kawamura Y, Miyoshi K, Nagami T, Siomi H, Siomi MC. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 2006;20:2214–2222. doi: 10.1101/gad.1454806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarot E, Payen-Groschêne G, Bucheton A, Pélisson A. Evidence for a piwi-dependent RNA silencing of the gypsy endogenous retrovirus by the Drosophila melanogaster flamenco gene. Genetics. 2004;166:1313–1321. doi: 10.1534/genetics.166.3.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schupbach T, Wieschaus E. Female sterile mutations on the second chromosome of Drosophila melanogaster. II. Mutations blocking oogenesis or altering egg morphology. Genetics. 1991;129:1119–1136. doi: 10.1093/genetics/129.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpiz S, Kwon D, Rozovsky Y, Kalmykova A. rasiRNA pathway controls antisense expression of Drosophila telomeric retrotransposons in the nucleus. Nucleic Acids Res. 2009;37:268–278. doi: 10.1093/nar/gkn960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpiz S, Kwon D, Uneva A, Kim M, Klenov M, Rozovsky Y, Georgiev P, Savitsky M, Kalmykova A. Characterization of Drosophila telomeric retroelement TAHRE: transcription, transpositions and RNAi-based regulation of expression. Mol Biol Evol. 2007;24:2535–2545. doi: 10.1093/molbev/msm205. [DOI] [PubMed] [Google Scholar]
- Slotkin RK, Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet. 2007;8:272–285. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- Snee MJ, Macdonald PM. Live imaging of nuage and polar granules: evidence against a precursor-product relationship and a novel role for Oskar in stabilization of polar granule components. J Cell Sci. 2004;117:2109–2120. doi: 10.1242/jcs.01059. [DOI] [PubMed] [Google Scholar]
- Song SU, Gerasimova T, Kurkulos M, Boeke JD, Corces VG. An env-like protein encoded by a Drosophila retroelement: evidence that gypsy is an infectious retrovirus. Genes Dev. 1994;8:2046–2057. doi: 10.1101/gad.8.17.2046. [DOI] [PubMed] [Google Scholar]
- Song SU, Kurkulos M, Boeke JD, Corces VG. Infection of the germ line by retroviral particles produced in the follicle cells: a possible mechanism for the mobilization of the gypsy retroelement of Drosophila. Development. 1997;124:2789–2798. doi: 10.1242/dev.124.14.2789. [DOI] [PubMed] [Google Scholar]
- Spradling AC. Germline cysts: communes that work. Cell. 1993;72:649–651. doi: 10.1016/0092-8674(93)90393-5. [DOI] [PubMed] [Google Scholar]
- Styhler S, Nakamura A, Swan A, Suter B, Lasko P. vasa is required for GURKEN accumulation in the oocyte, and is involved in oocyte differentiation and germline cyst development. Development. 1998;125:1569–1578. doi: 10.1242/dev.125.9.1569. [DOI] [PubMed] [Google Scholar]
- Tomari Y, Du T, Haley B, Schwarz DS, Bennett R, Cook HA, Koppetsch BS, Theurkauf WE, Zamore PD. RISC assembly defects in the Drosophila RNAi mutant armitage. Cell. 2004;116:831–841. doi: 10.1016/s0092-8674(04)00218-1. [DOI] [PubMed] [Google Scholar]
- Vagin VV, Klenov MS, Kalmykova AI, Stolyarenko AD, Kotelnikov RN, Gvozdev VA. The RNA interference proteins and vasa locus are involved in the silencing of retrotransposons in the female germline of Drosophila melanogaster. RNA Biol. 2004;1:54–58. [PubMed] [Google Scholar]
- Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- Wilsch-Brauninger M, Schwarz H, Nusslein-Volhard C. A sponge-like structure involved in the association and transport of maternal products during Drosophila oogenesis. J Cell Biol. 1997;139:817–829. doi: 10.1083/jcb.139.3.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JE, Connell JE, Macdonald PM. aubergine enhances oskar translation in the Drosophila ovary. Development. 1996;122:1631–1639. doi: 10.1242/dev.122.5.1631. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplement
T1
T2
T3
T4
T5
T6
T7