Ars2 regulates both miRNA- and siRNA-dependent silencing and suppresses RNA virus infection in Drosophila (original) (raw)
. Author manuscript; available in PMC: 2010 Jul 23.
Summary
Intrinsic immune responses autonomously inhibit viral replication and spread. One pathway that restricts viral infection in plants and insects is RNA interference (RNAi), which targets and degrades viral RNA to limit infection. To identify additional genes involved in intrinsic antiviral immunity, we screened Drosophila cells for modulators of viral infection using an RNAi library. We identified Ars2 (CG7843) as a key component of Drosophila antiviral immunity. Loss of Ars2 in cells, or in flies, increases susceptibility to RNA viruses. Consistent with its antiviral properties, we found that Ars2 physically interacts with Dcr-2, modulates its activity in vitro and is required for siRNA-mediated silencing. Furthermore, we show that Ars2 plays an essential role in miRNA-mediated silencing, interacting with the Microprocessor and stabilizing pri-miRNAs. The identification of Ars2 as a player in these small RNA pathways provides new insight into the biogenesis of small RNAs that may be extended to other systems.
Introduction
Innate immunity is the most ancient line of defense against pathogens. In mammals, the innate immune system provides the initial response to infection and primes the adaptive immune response. In contrast, invertebrates and plants lack adaptive immunity and therefore rely solely on innate mechanisms to combat infections. Recent studies have identified RNA interference (RNAi) as an ancient, cell-intrinsic immune mechanism that controls RNA viruses in plants and insects (Ding and Voinnet, 2007). RNAi is one of several small RNA-dependent silencing pathways that control gene expression in a sequence-specific manner in plants and animals. Small RNA-driven silencing is initiated by an RNase III enzyme Dicer, which produces RNA duplexes of approximately 21 nucleotides through the cleavage of longer precursor molecules (Bernstein et al., 2001; Tomari and Zamore, 2005; Zamore et al., 2000). Once generated, the duplex is incorporated into the multi-protein RNA-induced silencing complex (RISC) where one strand of the duplex is preferentially retained (Hammond et al., 2000; Khvorova et al., 2003; Schwarz et al., 2003). This guide strand directs RISC to a homologous target, where an Argonaute protein (Ago) mediates post-transcriptional gene silencing (Hutvagner and Simard, 2008).
In Drosophila, small interfering RNAs (siRNAs), which are derived from exogenous or endogenous sources of double stranded RNA (dsRNA), are generated by Dicer-2 (Dcr-2) and incorporated into an Ago2-dependent RISC, leading to the degradation of complementary mRNAs or viral RNAs (Kim et al., 2006). In contrast, endogenous primary miRNA (pri-miRNA) transcripts are processed into pre-miRNAs in the nucleus by the Microprocessor, which includes the RNase III enzyme Drosha and its binding partner Pasha (Denli et al., 2004). Next, the pre-miRNAs are exported and further processed by cytoplasmic Dicer-1 (Dcr-1) into mature miRNAs (Lee et al., 2004). Mature miRNAs are incorporated into an Ago1-dependent RISC and mediate translational inhibition of target transcripts (Okamura et al., 2004).
Studies using mutants in the known components of the classical siRNA pathway, including Dcr-2, r2d2 and AGO2, revealed the essential antiviral role that RNAi plays against positive-stranded RNA viruses in Drosophila (Galiana-Arnoux et al., 2006; van Rij et al., 2006; Wang et al., 2006). We set out to identify additional cellular components of the Drosophila intrinsic antiviral arsenal. An unbiased screen for factors that when lost, led to increased viral replication, identified Ars2 (CG7843). Ars2 is a poorly characterized gene that is highly conserved and required for development in Arabidopsis, zebrafish, and mice (Amsterdam et al., 2004; Prigge and Wagner, 2001; Wilson et al., 2008). The best-characterized Ars2 homolog is Arabidopsis SERRATE, which recently emerged as a component of the miRNA biogenesis pathway (Lobbes et al., 2006; Yang et al., 2006). In plants, Ars2 genetically interacts with the nuclear cap binding complex (CBC) components ABH1/CBP80 and CBP20 (Laubinger et al., 2008). Both our study and the work in the accompanying manuscript now reveal a physical interaction between Ars2 and the CBC in Drosophila and mammals (Gruber et al., 2009). We further demonstrate that Ars2, along with the CBC, plays a role in antiviral immunity against a battery of RNA viruses, and is required for both siRNA- and miRNA-mediated silencing, controlling the biogenesis of small regulatory RNAs.
Results
Ars2 is a restriction factor for VSV replication
To identify novel genes that control viral replication, we performed a small-scale screen for cellular factors that allow increased viral replication in cells when depleted by RNAi. We used a Drosophila cell culture system for these studies due to the high potency of RNAi, low genomic redundancy, and the lack of a complicating interferon response. To screen for such host factors, we treated Drosophila cells with dsRNA, incubated the cells for three days to allow for loss-of-function phenotypes, and challenged the cells with the mammalian virus Vesicular Stomatitis Virus (VSV). VSV is an enveloped, negative-stranded RNA virus that is naturally transmitted to mammals from insects (Letchworth et al., 1999; Lyles and Rupprecht, 2007). To monitor infection, we used a recombinant virus that expresses the reporter gene GFP upon replication (Ramsburg et al., 2005). Treatment with dsRNA against GFP is used as a positive control for silencing, as it is expressed by the virus upon replication. Luciferase dsRNA is used as a non-targeting control, as it is not expressed in this system. Using a fluorescence assay we found that VSV readily infects Drosophila cells, and that RNAi-mediated depletion of GFP can be monitored by microscopy (Figure 1A). Using this strategy we screened approximately 100 genes and identified CG7843, the Drosophila homolog of mammalian Ars2, which led to an increase in the percentage of VSV infected cells when depleted by RNAi (Figure 1A). To validate that the increase in infection was due to a depletion of Ars2 rather than an off-target effect, we generated an independent dsRNA to Ars2 and observed a similar increase in VSV infection (Figure 1A). Effective knockdown of Ars2 was verified by Northern blot analysis (Figure 1C).
Figure 1. Ars2 controls VSV infection in cell culture.
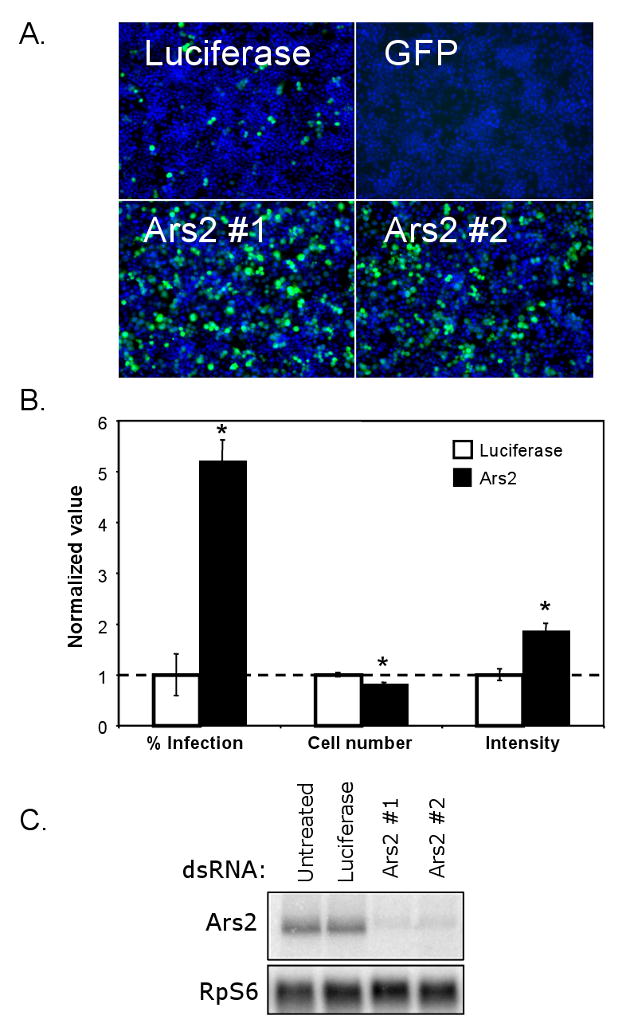
(A) Cells were treated with the indicated dsRNAs; Ars2 #1 and #2 denote independent dsRNA amplicons. Infected cells expressing a VSV-encoded GFP reporter are shown in green, nuclei in blue. (B) Infected Ars2-deficient cells were normalized to infected control cells and percent infection, cell number and average intensity were quantified. * denotes p<1×0-5; error bars indicate +/- SD from a representative experiment. Experiment was repeated 5 times. (C) Northern analysis of cells treated with the indicated dsRNAs.
We next measured several characteristics of control and Ars2-deficient cells during VSV infection (Figure 1B). The percentage of infected cells was quantified using automated image analysis; an average 5-fold increase in the percentage of infected cells was observed in Ars2 knockdown cells compared to control cells. In addition, we found that depletion of Ars2 resulted in a small but consistent 10-15% decrease in cell number, suggesting that Ars2 may play a role in cell proliferation or viability.
Since the expression of the GFP reporter can be measured directly by fluorescence, the average intensity of the GFP signal in each infected cell serves as surrogate for the amount of viral replication per cell. Using automated image analysis we found that the average GFP intensity in Ars2-deficient cells was ∼1.5-fold higher than control cells (Figure 1B). Because Ars2-deficient cells display enhanced permissivity to VSV infection and support increased viral replication upon infection, our data suggest that Ars2 may normally act in an antiviral manner to limit viral replication in Drosophila cells.
Ars2 restricts the replication of a panel of RNA viruses
To determine whether this cellular restriction factor is a specific inhibitor of VSV or whether it plays a more general antiviral role, we tested whether depletion of Ars2 influenced the infectivity of a panel of disparate RNA viruses (Figure 2A). These viruses included Drosophila C Virus (DCV), Flock House Virus (FHV) and Sindbis virus (SIN), which were chosen because they have all been previously shown to infect Drosophila and because they represent diverse families of RNA viruses that are arthropod-borne. DCV, a dicistrovirus, is related to mammalian picornaviruses and is a natural Drosophila pathogen (Cherry and Perrimon, 2004; Johnson and Christian, 1998). DCV and the nodavirus FHV, another insect pathogen, represent non-enveloped, positive sense RNA viruses (Friesen, 2007; Venter and Schneemann, 2008). SIN is an alphavirus and serves as a model for enveloped, positive sense RNA viruses (Griffin, 2007). In addition, we chose to test Vaccinia virus, a large dsDNA virus, which is the prototypical member of the poxvirus family of viruses (Damon, 2007).
Figure 2. Ars2 controls infection of a panel of RNA viruses in cell culture.
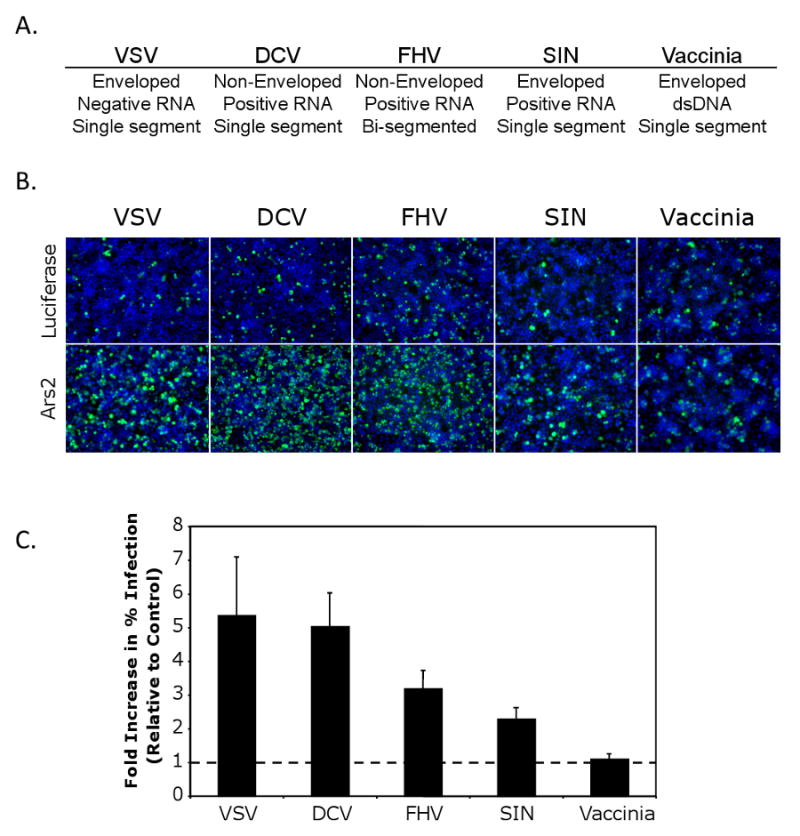
(A) Table of the name and the physical properties of each virus. (B) Immunofluorescence of DL1 cells infected with a diverse panel of viruses. Cells were treated with dsRNAs against Ars2 or control. Virus antigen is shown in green, nuclei in blue. (C) Quantification of images as in (B). All values were significant at p<0.0001; error bars indicate +/- SD of three independent experiments.
To investigate the role of Ars2 during infection with this diverse panel of viruses, control and Ars2-deficient cells were challenged with each virus, visualized by immunofluorescence microscopy and quantified using automated image analysis (Figure 2B). Knockdown of Ars2 increased infection of DCV, FHV, and SIN by at least 2-fold (Figure 2C). In contrast, Vaccinia virus infection was unaffected by Ars2 depletion. Vaccinia virus has a DNA genome, while the genomes of Ars2-sensitive viruses are encoded by RNA. The differential effects of Ars2 depletion on RNA and DNA viruses suggest that Ars2 may act in a RNA virus-specific manner.
Ars2 restricts viral RNA production in cultured cells
Viral infection involves a sequential series of steps from entry to RNA transcription, RNA replication, translation, and finally assembly and release. To characterize the mechanism of Ars2 antiviral action, we investigated the step in the RNA virus life cycle that is inhibited by Ars2. To independently verify that Ars2 knockdown allows enhanced production of viral protein products, immunoblot analysis of VSV-infected cells was performed (Figure 3A). We treated cells with dsRNA against VSV M as a positive control. Using this assay, we found that cells depleted of Ars2 expressed higher levels of viral M protein than control cells, consistent with the immunofluorescence assay. Likewise, we tested whether loss of Ars2 also led to an increase in DCV antigen production by immunoblot. While treatment with dsRNA against the viral genome inhibited viral antigen production compared to a non-targeting control, depletion of Ars2 led to a significant increase in the levels of DCV capsid protein production (Figure 3B).
Figure 3. Ars2 controls viral RNA levels during RNA virus infection.
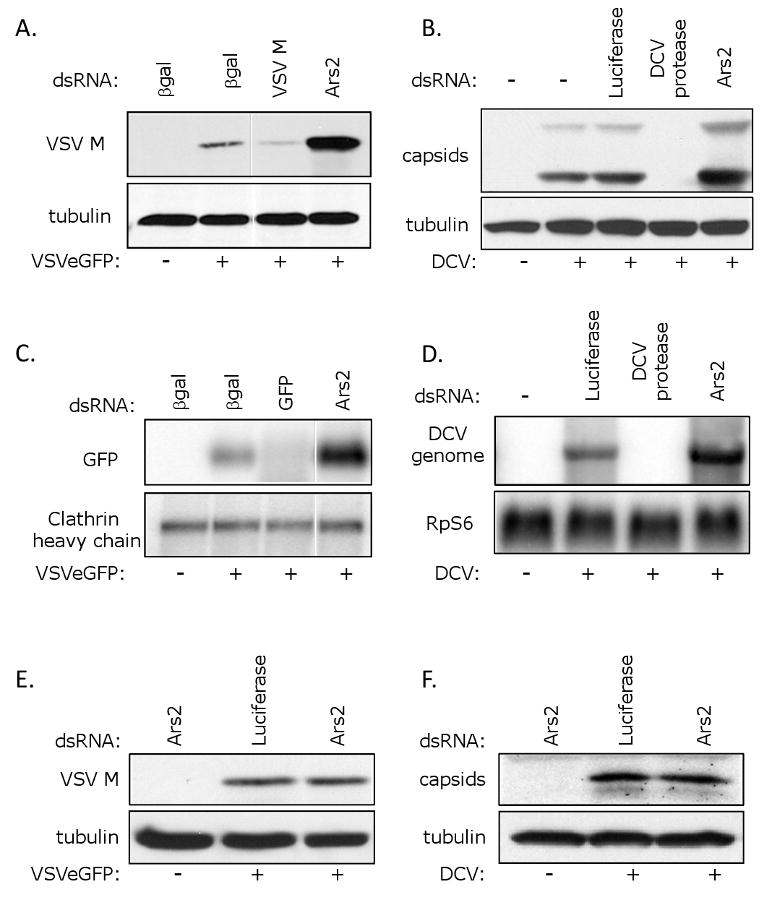
(A) Immunoblot analysis of cells pre-treated with the indicated dsRNAs either uninfected, or infected with VSV as indicated. (B) Immunoblot analysis of cells pre-treated with the indicated dsRNAs either uninfected, or infected with DCV as indicated. (C) Northern blot analysis of cells pre-treated with the indicated dsRNAs either uninfected, or infected with VSV as indicated. (D) Northern blot analysis of cells pre-treated with the indicated dsRNAs either uninfected, or infected with DCV as indicated. (E) Entry assay by immunoblot analysis. Cells pre-treated with the indicated dsRNAs were either uninfected or were infected with VSV, extracellular virus was removed, and immunoblot analysis was performed against internalized VSV (VSV M) or control (tubulin). (F) Entry assay by immunoblot analysis. Cells pre-treated with the indicated dsRNAs were either uninfected or were infected with DCV, extracellular virus was removed, and immunoblot analysis was performed against internalized DCV (capsids) or the control (tubulin).
Increased viral protein production in Ars2-deficient cells may be due to enhanced translation of viral mRNA or an increase in any step prior to translation. To determine whether there is a difference in viral RNA production in Ars2-deficient cells, we measured viral RNA levels by Northern blot. We monitored VSV infection by measuring viral GFP mRNA, and found that VSV mRNA levels are increased in Ars2-depleted cells compared to negative control cells (Figure 3C). Similarly, we found that DCV RNA levels were increased in Ars2-deficient cells compared to control cells (Figure 3D). Together, this suggests that the restriction of viral replication is not due to reduced translation of viral transcripts, but rather that Ars2 is acting at the level of RNA replication or stability or at an earlier step in the viral life cycle such as entry.
To determine whether Ars2 depletion affects viral entry, we performed an entry assay similar to that used by Otsuka et al. (Otsuka et al., 2007). Drosophila cells were treated with dsRNA against Ars2 or a control, infected with VSV, and levels of internalized viral M protein were measured by immunoblot. Ars2-deficient cells did not display an increase in internalized VSV antigen when compared to control cells (Figure 3E). We next tested whether loss of Ars2 affected DCV entry, and did not observe a difference in the amount of internalized DCV capsid protein when comparing Ars2-deficient cells to the negative control (Figure 3F). These findings suggest that Ars2 restricts RNA virus infection at a common step in the viral life cycle that is downstream of entry, and is likely at the level of RNA replication or stability.
Ars2 plays an antiviral role in adult flies
The above findings indicate a critical role for Ars2 in cultured cells. To extend the requirements for Ars2 to the organismal level during virus infection, we obtained two independent transposon insertion fly lines in Ars2 (c00735, e01128) which are lethal alleles that are unable to complement one another (Figure 4A). Therefore, Ars2 is an essential gene in Drosophila, prohibiting studies during adult stages. To generate hypomorphic alleles, we used the Gal4/UAS system to drive expression of a UAS-Ars2 inverted repeat transgene (UAS-Ars2 IR), which bears a long hairpin dsRNA construct targeting endogenous Ars2. Expression of UAS-Ars2 IR is controlled by the UAS-binding protein Gal4. UAS-Ars2 IR serves as a substrate for RNAi, leading to loss-of-function phenotypes in vivo. To characterize the developmental requirements of Ars2, we expressed the Ars2 IR dsRNA ubiquitously, at high levels using an Actin promoter or at low levels using a daughterless (da) promoter driving Gal4. In both cases, ubiquitous expression of the dsRNA was lethal (Figure 4A). To bypass the developmental requirement of Ars2, we drove the expression of UAS-Ars2 IR using a heat shock (hs) promoter that can be induced at 37°C. Raising the flies at 25°C allowed the generation of wild type numbers of flies that were induced to express Ars2 dsRNA as adults (Figure 4A).
Figure 4. Ars2 is required for antiviral immunity in adult flies.
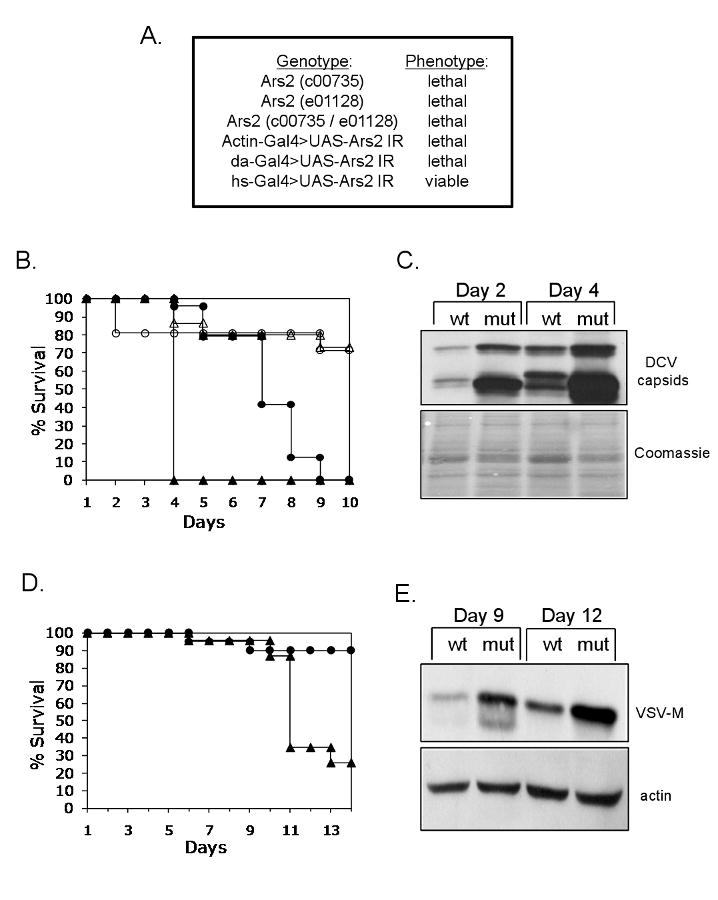
(A) Ars2 is developmentally required in Drosophila. Table of Ars2 mutants and their phenotypes. (B) Adult flies carrying heat shock inducible Gal4 were crossed to flies that can be induced to express dsRNA against Ars2 (hs-Gal4>UAS-Ars2 IR) or controls (hs-Gal4>+). Adult progeny controls (open circles) or Ars2-deficient flies (open triangles) were challenged with vehicle (PBS) and mortality was monitored as a function of time post-injection. In addition, control (closed circles) or Ars2-deficient flies (closed triangles) were challenged with DCV and mortality was monitored as a function of time post-infection. Percent survival is graphed as a function of time (p<0.001 log rank test). A representative of at least three experiments is shown. (C) Immunoblot analysis of flies that are either depleted for Ars2 (mut) or controls (wt) as described in (B), infected with DCV and collected at the indicated time points post infection. (D) Ars2-deficient flies (triangles) or controls (circles) as described in (B) were challenged with VSV and monitored daily for mortality; percent survival is graphed as a function of time (p<0.001 log rank test). A representative of at least three experiments is shown. (E) Immunoblot analysis of flies that are either depleted for Ars2 (mut) or controls (wt) as described in (B), infected with VSV and collected at the indicated time points post infection.
We expressed the Ars2 hairpin in transgenic flies by pulsing hs-Gal4> UAS-Ars2 IR or controls at 37°C. We challenged the Ars2-depleted or control flies with either carrier (PBS) or DCV and monitored mortality as a function of time post-injection. Depletion of Ars2 does not affect viability post-PBS challenge (Figure 4B). In contrast, there was a significant shift in the survival curve upon loss of Ars2 when challenged with the natural pathogen DCV. The Ars2-depleted flies succumb to the infection earlier than the control flies (Figure 4B). To determine if lethality is due to increased viral replication, we challenged control or Ars2-depleted flies with DCV and monitored viral protein production. Under these conditions we observed increased production of viral capsid proteins upon depletion of Ars2 (Figure 4C).
Next, we determined whether flies challenged with VSV were also sensitive to loss of Ars2. Again, we infected either the Ars2-depleted flies or control flies with VSV and found that while control flies or wild type flies do not succumb to infection by VSV, there was a significant increase in mortality upon loss of Ars2 (Figure 4D, data not shown). Again, we observed increased viral protein production upon depletion of Ars2 (Figure 4E). This demonstrates that Ars2 plays an antiviral role against disparate RNA viruses both in cultured cells and in vivo in adult flies.
Ars2 is required for siRNA- and esiRNA-mediated silencing
Since Ars2 is antiviral against a panel of RNA viruses that have been shown to be susceptible to antiviral RNAi (Galiana-Arnoux et al., 2006; van Rij et al., 2006; Wang et al., 2006), we tested whether loss of Ars2 impacts siRNA-mediated silencing. For these studies we used a dual-reporter system that allows us to simultaneously monitor transfection efficiency along with siRNA-mediated silencing. Three inducible constructs were transfected into Drosophila cells: a Firefly Luciferase control reporter, a Renilla Luciferase reporter, and a vector that produces a long Renilla-specific dsRNA. Transfected cells were depleted of RNA silencing components, and Renilla levels were normalized to Firefly control levels. Depletion of Dcr-2 or Ago2 led to a significant derepression of Renilla, whereas treatment with a non-targeting control or Ago1 had no effect on Renilla silencing (Figure 5A). In this assay, loss of Ars2 resulted in a twofold induction of Renilla, suggesting that Ars2 plays a role in siRNA-mediated gene silencing.
Figure 5. Ars2 is required for siRNA-mediated silencing.
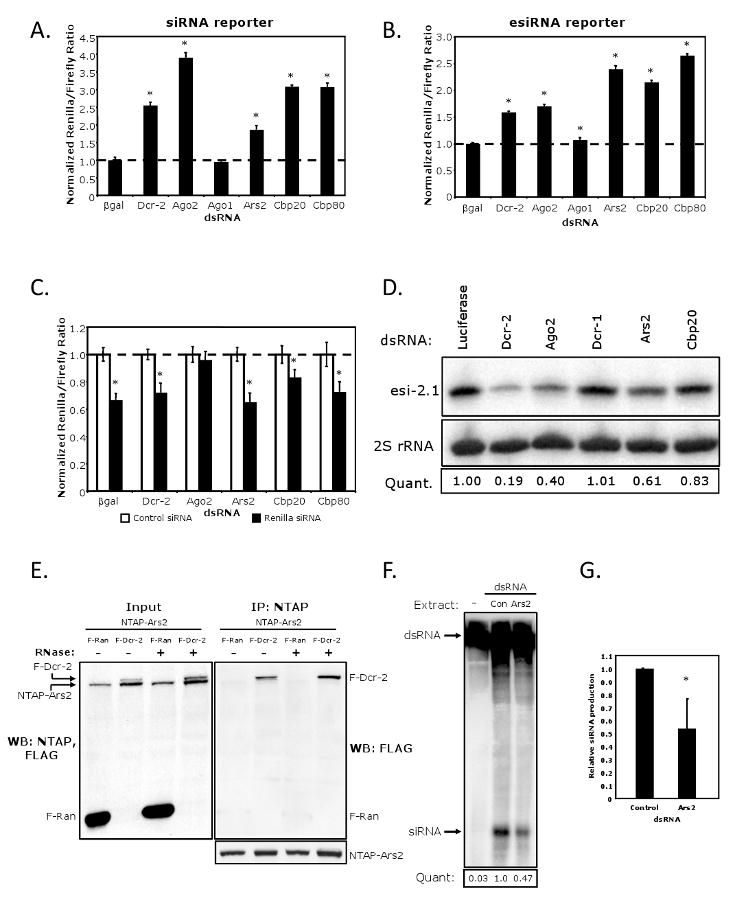
(A) Cells were transfected with a control reporter (Firefly), a Renilla reporter, and a vector expressing a Renilla hairpin. The indicated proteins were depleted by RNAi and cells were monitored for siRNA-mediated silencing. * denotes p<1×0-5. (B) Cells were transfected with a control vector (Firefly) and Renilla-mus308. The indicated proteins were depleted by RNAi and the cells were monitored for esiRNA-mediated silencing. * denotes p<1×0-5. (C) Cells were transfected as in (A) however siRNA duplexes against Renilla were transfected in lieu of the long hairpin trigger vector. * denotes p<0.001. (D) Northern blot analysis of esi-2.1 in cells depleted as indicated. esi-2.1 expression was quantified relative to the 2S rRNA, and values were normalized to the Luciferase negative control. (E) Cells expressing NTAP-Ars2 and either FLAG-Ran or FLAG-Dcr-2 were immunoprecipitated, treated with RNase as indicated, and immuoblotted for FLAG. Left panel shows 1/10 input and the right panel shows the co-immunoprecipitate. (F) Cytoplasmic extracts from cells expressing FLAG-Dcr-2 were depleted for Ars2 or Luciferase control (Con), and incubated with a uniformly radiolabeled dsRNA substrate to measure siRNA-generating activity. (G) Quantification of siRNA production normalized to the siRNA-generating activity of the control extract. * denotes p<0.05.
Error bars represent +/- SD of three independent experiments.
Due to the finding that Ars2 is involved in siRNA silencing, we hypothesized that Ars2 may also play a role in silencing mediated by endogenous siRNAs (esiRNAs) that are normally produced via a Dcr-2/Ago2-dependent pathway (Czech et al., 2008; Ghildiyal et al., 2008; Kawamura et al., 2008; Okamura et al., 2008). To test this possibility, a Renilla reporter fused to mus308 was employed. The mus308 transcript is a natural cellular target of esi-2.1 (Okamura et al., 2008), thus the reporter is silenced by the endogenous pools of esi-2.1. Knockdown of negative controls βgal and Ago1 had no effect on reporter silencing, while Dcr-2 and Ago2 depletion alleviated the repression of the Renilla-mus308 reporter (Figure 5B). Loss of Ars2 led to more than a twofold induction of Renilla; similar results were also observed using a Renilla reporter bearing two esi-2.1 target sites (Figure S1). Taken together, these data suggest that Ars2 is required for esiRNA-mediated silencing.
Ars2 is required upstream of RISC
To determine the step in the siRNA pathway for which Ars2 function is required, we tested whether introduction of siRNAs rather than long dsRNAs could bypass the requirement for Ars2. Cells were cotransfected with Firefly and Renilla reporters and bathed in dsRNAs to deplete RNA silencing components. Cells were then transfected with non-targeting or Renilla-specific siRNAs. We found that the Renilla-specific siRNAs reduced Renilla expression by 40% in control cells treated with βgal dsRNA (Figure 5C). Renilla expression in Ago2-deficient cells was unaffected by the introduction of Renilla siRNAs, since Ago2 is the catalytic core of RISC and mediates the downstream effector steps of the pathway (Okamura et al., 2004). When Dcr-2 or Ars2-deficient cells were transfected with Renilla-specific siRNAs, the reporter was silenced as efficiently as in βgal control cells. These data suggest that Ars2 is important for siRNA biogenesis but is dispensable for the effector functions of the pathway.
Ars2 controls the production of endogenous siRNAs
Since our Luciferase reporter assays suggested a role for Ars2 in an upstream biogenesis step of RNA silencing, and given that the repression of the esiRNA reporter was controlled by esi-2.1 (Figure 5B), we examined the steady state levels of esi-2.1 upon depletion of either controls, or Ars2 by Northern blot (Figure 5D). As expected, loss of Dcr-2 or Ago2 but not Dcr-1 led to a decrease in the accumulation of esi-2.1 compared to a non-targeting control (Luciferase). Furthermore, we found that depletion of Ars2 reduced the levels of esi-2.1, demonstrating that Ars2 controls the bona fide endogenous siRNA pools.
Ars2 physically interacts with Dcr-2
Given that Ars2 is required upstream of RISC, we tested whether Ars2 can physically interact with the most upstream siRNA pathway component known, Dcr-2. To this end, we expressed NTAP-Ars2 in Drosophila cells along with either a control protein (FLAG-Ran) or FLAG-Dcr-2. Immunoprecipitation of NTAP-Ars2 followed by immunoblot analysis showed that Ars2 co-precipitates Dcr-2 but not the control protein (Figure 5E). Moreover, the interaction with Dcr-2 is RNA-independent. Therefore, our data suggest that Ars2 mediates its effects on the siRNA pathway through an interaction with cytoplasmic Dcr-2.
Cytoplasmic dsRNA processing requires Ars2
Our findings implicate Ars2 in the early steps of siRNA silencing, both functionally and biochemically. To test the relevance of this interaction on Dcr-2 activity directly, we determined whether the dsRNA processing activity of Dcr-2 was dependent upon Ars2 in vitro. Drosophila cells expressing FLAG-Dcr-2 were depleted of endogenous Ars2 or a negative control (Luciferase) by RNAi, and cytoplasmic extracts were incubated with a uniformly radiolabeled dsRNA. siRNA production was significantly reduced in Ars2-depleted lysates (Figure 5F). Replicate experiments indicate that loss of Ars2 results in an approximately two-fold attenuation of dsRNA processing (Figure 5G). These data suggest that Ars2 is required for the efficient processing of siRNA pathway substrates by Dcr-2, and provide a mechanism for the Ars2 requirement in cytoplasmic siRNA silencing.
Ars2 is required for miRNA-mediated silencing
Studies in plants have found that SERRATE, the plant homolog of Ars2, plays a role in miRNA biogenesis (Lobbes et al., 2006; Yang et al., 2006). Moreover in Drosophila, loss of Ars2 is lethal, as are mutants in the miRNA pathway but not mutants in the siRNA pathway (Figure 4A) (Lee et al., 2004; Okamura et al., 2004). Therefore, we tested whether Ars2 is also involved in miRNA silencing using another Luciferase-based reporter assay. Cells were transfected with a Firefly Luciferase control, a construct expressing pri-miR-277, and a Renilla reporter bearing four bulged miR-277 target sites. miR-277 is a Drosophila miRNA that was shown to silence bulged targets through the canonical miRNA silencing pathway (Forstemann et al., 2007). Knockdown of negative controls βgal, Dcr-2 and Ago2 had little effect on reporter silencing, while depletion of Ago1, a core component of miRNA RISC, resulted in a significant loss of repression of the Renilla reporter (Figure 6A). Under these conditions, loss of Ars2 resulted in a significant increase in Renilla expression, revealing that Ars2 is required for miRNA-mediated silencing in addition to siRNA-mediated silencing.
Figure 6. Ars2 is required for miRNA-mediated silencing.
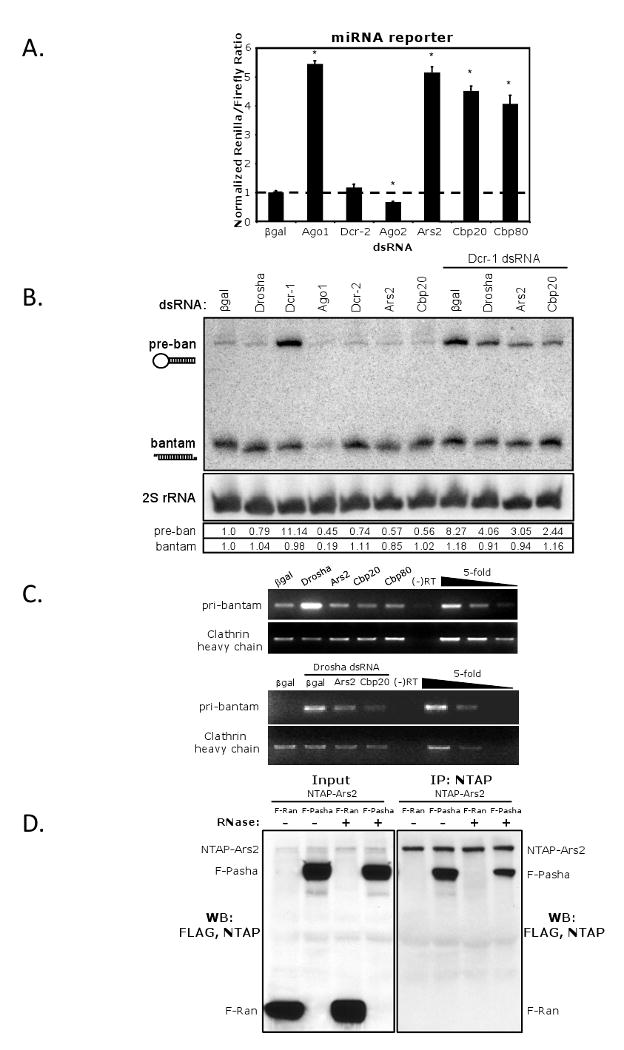
(A) Cells were transfected with a control reporter (Firefly), a vector expressing pri-miR-277, and a Renilla reporter bearing bulged miR-277 target sites. The indicated proteins were depleted by RNAi and the cells were monitored for miRNA-mediated silencing. * denotes p<0.0001; error bars represent +/- SD of 3 independent experiments. (B) Northern blot analysis of pre- and mature bantam miRNA. Expression of pre- and mature bantam was quantified relative to the 2S rRNA, and values were normalized to βgal. (C) RT-PCR analysis of pri-bantam and cellular control transcript (clathrin heavy chain) levels in cells depleted as indicated. Serial five-fold dilutions indicate amplification in the linear range. (D) Cells expressing NTAP-Ars2 and either FLAG-Ran or FLAG-Pasha were immunoprecipitated, treated with RNase as indicated, and immuoblotted for FLAG. Left panel shows the 1/10 input and the right panel shows the co-immunoprecipitate.
Ars2 is required upstream of pre-miRNA production
To further characterize the role of Ars2 in the miRNA pathway, we monitored the steady state levels of endogenous bantam miRNA. First, we examined the levels of mature bantam upon depletion of controls or Ars2 by Northern blot. As expected, loss of Ago1 had a significant effect on the levels of the mature miRNA (Figure 6B). In contrast, while Dcr-1 is required to produce mature miRNAs, it has been previously shown that transient Dcr-1 knockdown has little effect on the steady state levels of mature bantam, and instead has a much stronger effect on the levels of pre-bantam (Figure 6B) (Forstemann et al., 2005). Depletion of Ars2 had little effect on the accumulation of mature bantam, but resulted in an approximately twofold reduction in pre-bantam. We reasoned that if Ars2 was required upstream of Dcr-1, then loss of Ars2 would suppress the accumulation of pre-bantam observed upon loss of Dcr-1. As a positive control we depleted cells of both Dcr-1 and Drosha, the core component of the upstream Microprocessor, and indeed found that loss of Drosha suppressed the accumulation of pre-bantam. When Ars2 was depleted in combination with Dcr-1, there was a reduction in the accumulated levels of pre-bantam (Figure 6B). This observation places Ars2 upstream of Dcr-1 activity, likely at the level of the Microprocessor.
Primary miRNA transcripts are depleted in the absence of Ars2
Given that the steady-state levels of pre-bantam are reduced upon loss of Ars2, we next examined the levels of primary bantam (pri-bantam) miRNA transcripts by RT-PCR. Loss of Drosha resulted in a strong accumulation of pri-bantam, as the enzyme is required for the liberation of pre-bantam from the primary transcript (Figure 6C, top panel) (Lee et al., 2003). In contrast, depletion of Ars2 did not result in an accumulation of pri-bantam; rather, the levels were slightly less than the βgal negative control cells. A decrease in pri-miRNA levels upon Ars2 knockdown was also observed in mammalian cells (Gruber et al., 2009). One possible explanation is that Ars2 is important for the stability of primary miRNA transcripts. To test this hypothesis, we co-depleted both Drosha and Ars2 from cells, reasoning that if Ars2 stabilizes pri-miRNAs, then pri-bantam will not accumulate in cells lacking both Drosha and Ars2, since unprocessed transcripts will be degraded. Indeed, we found that, compared to βgal/Drosha-depleted cells, Ars2/Drosha-depleted cells had decreased levels of pri-bantam (Figure 6C, bottom panel). These data suggest that Ars2 is required upstream or at the level of the Microprocessor during miRNA biogenesis, and support the idea that Ars2 is necessary to stabilize primary transcripts.
Ars2 physically interacts with the Microprocessor
We next tested whether Ars2 could physically interact with the Microprocessor. To this end, we expressed NTAP-Ars2 in Drosophila cells along with either a control protein (FLAG-Ran) or FLAG-Pasha. Immunoprecipitation of NTAP-Ars2 followed by immunoblot analysis showed that Ars2 co-precipitates Pasha but not the control protein in an RNA-independent manner (Figure 6D). Therefore, our data suggest that Ars2 mediates its effects on the miRNA pathway through an interaction with the Microprocessor.
The CBC controls small RNA silencing pathways
In mammalian systems, Ars2 physically interacts with and shuttles between the nucleus and cytoplasm with the nuclear cap binding complex (CBC) proteins CBP20 and CBP80 (Gruber et al., 2009). Therefore, we tested whether the corresponding biochemical complex exists in Drosophila. We generated a polyclonal antibody against Ars2 and used it to detect overexpressed Ars2 since endogenous Ars2 levels were undetectable in input lysates (Figure 7A). We found that NTAP-Cbp20 specifically immunoprecipitates Ars2 and not a control protein (FLAG-Ran), suggesting that the Ars2 interaction with the CBC is conserved in Drosophila (Figure 7A).
Figure 7. The CBC interacts with Ars2 and restricts RNA virus infection.
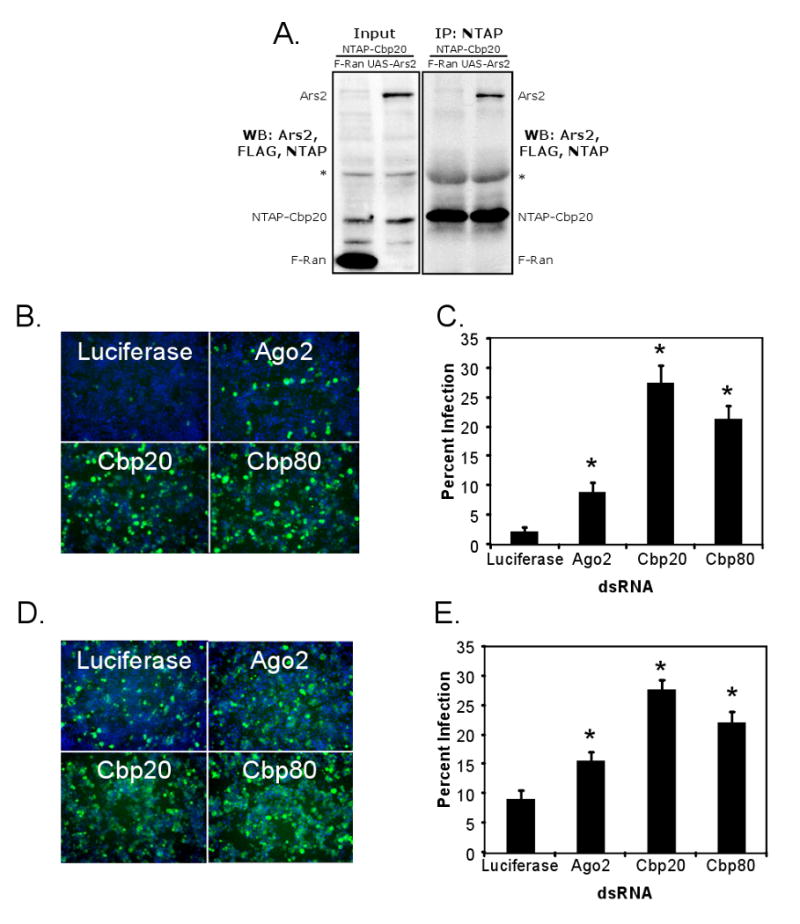
(A) Cells expressing NTAP-Cbp20 and either control (FLAG-Ran) or Ars2 (pMT-Gal4>UAS-Ars2) were immunoprecipitated and immunoblotted for FLAG and Ars2. Left panel shows the 1/10 input and the right panel shows the co-immunoprecipitate. * denotes nonspecific cross-reacting proteins. (B) Fluorescence of cells pre-treated with dsRNAs against the indicated genes and infected with VSV. Infected cells expressing GFP are shown in green, nuclei in blue. (C) Percent infection was calculated by dividing the number of GFP-positive cells by the total number of cells. * denotes p<1×0-8. (D) Immunofluorescence of cells pre-treated with dsRNAs against the indicated genes and infected with DCV. Infected cells expressing capsids are shown in green, nuclei in blue. (E) Percent infection was calculated by dividing the number of DCV-positive cells by the total number of cells. * denotes p<1×0-8.
Error bars represent +/- SD of three independent experiments.
Since Ars2 is important for small RNA silencing, we hypothesized that Ars2 binding partners may also be required for siRNA, esiRNA and miRNA-mediated silencing. Indeed, cells depleted of Cbp20 or Cbp80 are defective in all three modes of silencing (Figure 5A, 5B, 6A). Furthermore, we found that the CBC phenocopies Ars2 in each of the assays used to place Ars2 in the RNA silencing pathways. Like Ars2, the CBC components function in the early steps of the siRNA pathway; mature Renilla-specific siRNAs are able to effectively silence the Renilla reporter in CBC-depleted cells (Figure 5C), and steady-state levels of esi-2.1 are reduced in the absence of Cbp20 (Figure 5D). Moreover, the CBC acts at the level of the Microprocessor in the miRNA pathway; depletion of Cbp20 leads to a decrease in pre- and pri-bantam accumulation (Figures 6B and 6C), implicating the CBC in pri-miRNA stability.
The CBC controls viral replication
Given that the CBC binds Ars2 and is necessary for siRNA silencing, we tested whether the CBC also plays a role in restricting viral infection. To this end, cells were depleted by RNAi for Luciferase as a negative control, Ago2 as a positive control for the antiviral RNAi pathway, or our candidates Cbp20 and Cbp80. The cells were then challenged with VSV or DCV and imaged by fluorescence microscopy (Figures 7B and 7D). Automated image analysis quantified the percentage of infected cells (Figure 7C and 7E). Depletion of Ago2, Cbp20 or Cbp80 resulted in enhanced infection of two disparate RNA viruses VSV (Figure 7C) and DCV, which replicates using an uncapped viral RNA (Figure 7E).
Discussion
Our findings demonstrate that Ars2 and the CBC are required for miRNA- and siRNA-mediated silencing as well as antiviral defense in Drosophila. In the accompanying manuscript, Gruber et al. report a similar role for Ars2 in mammalian miRNA-mediated silencing (Gruber et al., 2009). Ars2 and the CBC are required at upstream steps in both Drosophila and mammalian RNA silencing pathways, which is consistent with recent data from plants; the Ars2 homolog SERRATE and homologs of the CBC (ABH1 and CBP20) control pri-miRNA processing in Arabidopsis (Gregory et al., 2008; Laubinger et al., 2008; Lobbes et al., 2006; Yang et al., 2006). Ars2 functionally and biochemically interacts with the nuclear Microprocessor, as does SERRATE, which physically interacts with HYL1, a component of the Arabidopsis miRNA biogenesis pathway similar in function to Drosophila Pasha and mammalian DGCR8 (Lobbes et al., 2006; Yang et al., 2006).
The results of our studies, combined with the evidence from existing literature, have led us to propose two potential models for Ars2 function. The first is a bridging model in which Ars2 serves as a recruitment factor to guide the RNA processing machinery to the proper substrates. Under this model, Ars2 and the CBC bind pri-miRNA transcripts through recognition of their 5′ cap, and Ars2 actively recruits the Microprocessor to the transcript, promoting its cleavage into a pre-miRNA. This bridging model suggests a mechanism by which Ars2 and the CBC increase the efficiency of pri-miRNA processing by acting as chaperones to stabilize and deliver the primary transcripts directly to the Microprocessor. It then follows that in the absence of Ars2 or the CBC, Drosha-directed pri-miRNA processing is impaired, since the recruitment of the Microprocessor to primary transcripts is less efficient, and the unprocessed transcripts are destabilized. Our finding that pri-bantam levels are reduced in Ars2 or CBC-depleted cells (Figure 6C) along with similar findings in mammalian cells (Gruber et al., 2009) supports this model. While the bridging model provides a compelling mechanism for the Ars2 and CBC requirement in the miRNA pathway, it is less likely to be relevant for the siRNA pathway, as the substrates of the pathway (dsRNA, viral RNAs) are not necessarily 5′ capped. Although it is possible that Ars2 recruits Dcr-2 to uncapped substrates through the targeting of secondary RNA structure, our data favor an RNA recognition-independent model.
This second model proposes that Ars2 serves as a cofactor for the enzymatic activity of RNase III enzymes. Under this model, the presence of Ars2 in Drosha or Dcr-2-containing complexes promotes robust enzymatic cleavage of RNA substrates and increases the fidelity of processing. Consistent with this model, recent work by Dong et al. has shown that the addition of recombinant SERRATE to an in vitro pri-miRNA processing assay enhances both the activity and the accuracy of DCL1 substrate cleavage (Dong et al., 2008). Moreover, Gruber et al. demonstrate in the accompanying manuscript that mammalian pri-miRNA processing is altered in the absence of Ars2 (Gruber et al., 2009). Our functional dicing assay demonstrates that Dcr-2-mediated processing of long dsRNA is impaired in the absence of Ars2 (Figures 5F, 5G), lending support to the idea that Ars2 is an essential accessory factor for Dcr-2 activity on uncapped RNAs. Since the substrates of the cytoplasmic siRNA pathway are not necessarily capped, this further argues that the CBC may be required for the cofactor activity of Ars2 rather than for substrate recognition.
The underlying difference between the two models is the precise step of substrate recognition and processing for which Ars2 is required; the bridging model poses that Ars2 and the CBC physically bind the RNA substrate, recruiting the proper processing activity for cleavage. Conversely, the cofactor model suggests that the binding of Ars2 and the CBC to the processing machinery allows the enzymes to execute robust and accurate substrate cleavage. Of course, these models are not mutually exclusive, and the true function of Ars2 may combine aspects of both models. Ars2 may also play distinct roles depending on its particular binding partners or intracellular localization. Ultimately, this study implicates Ars2 as a fundamental component of several modes of RNA silencing and contributes to the growing body of evidence that RNA silencing pathways are more interconnected than previously appreciated (Zhou et al., 2008). The RNA content of a cell is influenced by the contributions of many transcriptional and post-transcriptional regulatory pathways that have evolved to respond quickly and sensitively to the needs of the cell. The identification of novel components of these pathways, such as Ars2, aids in our efforts to uncover the mechanisms by which cellular processes such as proliferation or antiviral defense are exquisitely regulated.
Experimental Procedures
Cells, Viruses, Antibodies and Reagents
Drosophila cells were grown and maintained as previously described (Cherry and Perrimon, 2004). VSV-eGFP (gift from J. Rose) was grown in BHK cells as described (Ramsburg et al., 2005). FHV (gift from P. Ahlquist) was grown in DL1 cells. Sindbis/GFP (gift from R. Hardy) was grown in C636 cells (Burnham et al., 2007). DCV was grown and purified as described (Cherry and Perrimon, 2004). Vaccinia strain vPRA13 (gift from R. Doms) was grown in HeLa S3 suspension cells (Alexander et al., 1992). Antibodies were obtained from the following sources: anti-GFP (Invitrogen), anti-tubulin (Sigma), anti-VSVM (gift from D. Lyles), anti-FHVB2 (gift from P. Ahlquist), anti-DCV (Cherry and Perrimon, 2004), anti-β gal (Cappel). Rabbit polyclonal anti-Ars2 was generated against purified GST-mouse Ars2 (Prosci). Fluorescently labeled secondary antibodies were obtained from Jackson Immunochemicals, and HRP-conjugated antibodies from Amersham. Additional chemicals were obtained from Sigma.
RNAi
dsRNAs were generated as described (Boutros et al., 2004). For RNAi, cells were passaged into serum free media and plated into wells containing dsRNA. One hour later, complete media was added and cells were incubated for three days.
Viral Infections
Three days post-dsRNA addition, DL1 cells were infected with the indicated viral inoculum. VSV (MOI=5), FHV (MOI=5), and DCV (MOI=1.5) were added to cells and fixed at 24 hours post infection. SIN (MOI=15) was spinoculated at 1200 rpm for 2 hours, and fixed at 36 hours post-infection. Vaccinia (MOI=1.5) was added to cells in 2% serum and the cells were fixed at 48 hours post-infection.
Immunofluorescence
Cells were fixed in 4% formaldehyde/PBS, washed in PBS/0.1% TritonX-100 (PBST) twice, and blocked in 2% BSA/PBST. Primary antibody was diluted in block and incubated overnight at 4°C. Cells were washed, and incubated in secondary antibody for one hour at 25°C. Cells were counterstained with Hoescht33342 (Sigma) and imaged using automated microscopy (ImageXpress Micro). Images of three sites per well were collected, with a minimum of three wells per treatment, and percent infection was measured using MetaXpress software.
Immunoblotting and Immunoprecipitations
For immunoblotting, cells or flies were collected and were lysed in radioimmunoprecipitation (RIPA) buffer supplemented with a protease inhibitor cocktail (Boehringer). Samples were separated and blotted as previously described (Cherry and Perrimon, 2004). For immunoprecipitations, S2 cells were processed as previously described (Saito et al., 2005).
RNA analysis
Total RNA was extracted from DL1 cells using Trizol (Invitrogen). To visualize RNA species greater than 300bp in length, total RNA was separated on a 1% agarose/formaldehyde gel and blotted as previously described (Cherry et al., 2005). For small RNA blots, total RNA was separated on a denaturing polyacrylamide/urea gel and transferred to Hybond N+ (Amersham). Samples were normalized against controls using ImageQuant. For RT-PCR, cDNA was prepared from total RNA using M-MLV reverse transcriptase (Invitrogen) random primers, and the indicated transcripts were amplified by PCR.
Entry Assay
DL1 cells were infected with either VSV (MOI=25) or DCV (MOI=15) for two hours in the presence of cycloheximide (10μg/mL). Cells were washed, treated with 0.25% trypsin/0.025% EDTA, then pelleted and washed again. Cells were lysed in RIPA buffer supplemented with a protease inhibitor cocktail (Boehringer) and samples were immunoblotted as described above.
Adult Infections
Ars2 (c00735) and Ars2 (e01128) were obtained from the Exelixis collection at Harvard Medical School. Flies carrying UAS-Ars2 IR (transformant 22574) were obtained from the Vienna Drosophila RNAi Center. All other flies were obtained from the Bloomington stock center. Flies carrying UAS-Ars2 IR or control (wild type or UAS-lacZ IR) were crossed to actin-Gal4, da-Gal4 or hs-Gal4 at room temperature. On the day of injection the progeny from the hs-Gal4 crosses were heat shocked at 37°C for one hour and shocked every two days throughout the experiment. 4-7 day old adults of the stated genotypes were inoculated with virus as previously described (Cherry and Perrimon, 2004). Flies were monitored daily for mortality or were processed at the indicated time point post infection. For survival curves, statistical significance was determined with a log rank test.
Luciferase Reporter Assays
To measure siRNA, esiRNA or miRNA mediated silencing, S2 cells were transiently transfected with 1.2 μg pMT-Renilla, 40 ng pMT-Firefly, and 0.8 μg pMT-Renilla-hairpin or with 2μg pMT-Renilla-mus308 and 40 ng pMT-Firefly, or with 100 ng pMT-Renilla 4B, 40 ng pMT-Firefly, and 2 μg pMT-miR277. Two days post- transfection, cells were bathed in the indicated dsRNAs for 3 days. For siRNA transfection experiments, three days post-bathing, Effectene was used to transfect cells with Renilla-specific or non-targeting control siRNAs (Ambion) at a final concentration of 20nM. Reporters were induced with 200 μM CuSO4 and luminescence assays were performed 24 hours later using DualGlo following the manufacturer's protocol (Promega).
Dicing Assay
S2 cells were transfected with FLAG-Dcr-2 and bathed in dsRNA to deplete the indicated protein. Cytoplasmic extracts were prepared as described (Saito et al., 2005) and the dicing substrate was a 240bp dsRNA that was uniformly labeled with [α-32P]UTP during in vitro synthesis. Cleavage reactions were carried out essentially as described (Liu et al., 2003) for 60 minutes at 29°C after which the RNA was extracted, and products were separated on a denaturing polyacrylamide/urea gel, transferred to Hybond N+ (Amersham) and exposed to a phosphoimager cassette.
Supplementary Material
01
Acknowledgments
We thank N. Bonini, M. Tudor for critically reading the manuscript; members of the Cherry laboratory for technical advice and helpful discussions; S. Poethig and M.Y. Park for technical advice; J. Rose for VSV-eGFP; D. Lyles for anti-VSV M; P. Ahlquist for FHV anti-B2; R. Hardy for SlN/GFP; R. Doms for Vaccinia; Harvard and VDRC for Ars2 transgenic flies; The Bloomington Stock center for other fly stocks. R.Z. is supported by the Leukemia and Lymphoma Society. L.R.S. was supported by NIH training grant T32 GM-07229 and NIH grant RO1AI07451 to S.C.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alexander WA, Moss B, Fuerst TR. Regulated expression of foreign genes in vaccinia virus under the control of bacteriophage T7 RNA polymerase and the Escherichia coli lac repressor. J Virol. 1992;66:2934–2942. doi: 10.1128/jvi.66.5.2934-2942.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amsterdam A, Nissen RM, Sun Z, Swindell EC, Farrington S, Hopkins N. Identification of 315 genes essential for early zebrafish development. Proc Natl Acad Sci U S A. 2004;101:12792–12797. doi: 10.1073/pnas.0403929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Boutros M, Kiger AA, Armknecht S, Kerr K, Hild M, Koch B, Haas SA, Paro R, Perrimon N. Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science. 2004;303:832–835. doi: 10.1126/science.1091266. [DOI] [PubMed] [Google Scholar]
- Burnham AJ, Gong L, Hardy RW. Heterogeneous nuclear ribonuclear protein K interacts with Sindbis virus nonstructural proteins and viral subgenomic mRNA. Virology. 2007;367:212–221. doi: 10.1016/j.virol.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Cherry S, Doukas T, Armknecht S, Whelan S, Wang H, Sarnow P, Perrimon N. Genome-wide RNAi screen reveals a specific sensitivity of IRES-containing RNA viruses to host translation inhibition. Genes Dev. 2005;19:445–452. doi: 10.1101/gad.1267905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry S, Perrimon N. Entry is a rate-limiting step for viral infection in a Drosophila melanogaster model of pathogenesis. Nat Immunol. 2004;5:81–87. doi: 10.1038/ni1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, Dus M, Perrimon N, Kellis M, Wohlschlegel JA, Sachidanandam R, et al. An endogenous small interfering RNA pathway in Drosophila. Nature. 2008 doi: 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damon IK. Poxviruses. In: Knipe DM, Howley PM, editors. Fields Virology. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 2948–2975. [Google Scholar]
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- Ding SW, Voinnet O. Antiviral immunity directed by small RNAs. Cell. 2007;130:413–426. doi: 10.1016/j.cell.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Z, Han MH, Fedoroff N. The RNA-binding proteins HYL1 and SE promote accurate in vitro processing of pri-miRNA by DCL1. Proc Natl Acad Sci U S A. 2008;105:9970–9975. doi: 10.1073/pnas.0803356105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forstemann K, Horwich MD, Wee L, Tomari Y, Zamore PD. Drosophila microRNAs are sorted into functionally distinct argonaute complexes after production by dicer-1. Cell. 2007;130:287–297. doi: 10.1016/j.cell.2007.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forstemann K, Tomari Y, Du T, Vagin VV, Denli AM, Bratu DP, Klattenhoff C, Theurkauf WE, Zamore PD. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 2005;3:e236. doi: 10.1371/journal.pbio.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesen PD. Insect Viruses. In: Knipe DM, Howley PM, editors. Fields Virology. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 707–736. [Google Scholar]
- Galiana-Arnoux D, Dostert C, Schneemann A, Hoffmann JA, Imler JL. Essential function in vivo for Dicer-2 in host defense against RNA viruses in drosophila. Nat Immunol. 2006;7:590–597. doi: 10.1038/ni1335. [DOI] [PubMed] [Google Scholar]
- Ghildiyal M, Seitz H, Horwich MD, Li C, Du T, Lee S, Xu J, Kittler EL, Zapp ML, Weng Z, et al. Endogenous siRNAs Derived from Transposons and mRNAs in Drosophila Somatic Cells. Science. 2008 doi: 10.1126/science.1157396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory BD, O'Malley RC, Lister R, Urich MA, Tonti-Filippini J, Chen H, Millar AH, Ecker JR. A link between RNA metabolism and silencing affecting Arabidopsis development. Dev Cell. 2008;14:854–866. doi: 10.1016/j.devcel.2008.04.005. [DOI] [PubMed] [Google Scholar]
- Griffin DE. Alphaviruses. In: Knipe DM, Howley PM, editors. Fields Virology. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 1023–1067. [Google Scholar]
- Gruber JJ, Sabin LR, Zatechka SD, Yong J, Lum JJ, Kong M, Zong WX, Zhang Z, Lau CK, Cherry S, et al. Ars2 links the nuclear cap binding complex to RNA interference and cell proliferation. 2009 doi: 10.1016/j.cell.2009.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- Johnson KN, Christian PD. The novel genome organization of the insect picorna-like virus Drosophila C virus suggests this virus belongs to a previously undescribed virus family. J Gen Virol. 1998;79(Pt 1):191–203. doi: 10.1099/0022-1317-79-1-191. [DOI] [PubMed] [Google Scholar]
- Kawamura Y, Saito K, Kin T, Ono Y, Asai K, Sunohara T, Okada TN, Siomi MC, Siomi H. Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature. 2008 doi: 10.1038/nature06938. [DOI] [PubMed] [Google Scholar]
- Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- Kim K, Lee YS, Harris D, Nakahara K, Carthew RW. The RNAi pathway initiated by Dicer-2 in Drosophila. Cold Spring Harb Symp Quant Biol. 2006;71:39–44. doi: 10.1101/sqb.2006.71.008. [DOI] [PubMed] [Google Scholar]
- Laubinger S, Sachsenberg T, Zeller G, Busch W, Lohmann JU, Ratsch G, Weigel D. Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2008 doi: 10.1073/pnas.0802493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, Carthew RW. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- Letchworth GJ, Rodriguez LL, Del cbarrera J. Vesicular stomatitis. Vet J. 1999;157:239–260. doi: 10.1053/tvjl.1998.0303. [DOI] [PubMed] [Google Scholar]
- Liu Q, Rand TA, Kalidas S, Du F, Kim HE, Smith DP, Wang X. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science. 2003;301:1921–1925. doi: 10.1126/science.1088710. [DOI] [PubMed] [Google Scholar]
- Lobbes D, Rallapalli G, Schmidt DD, Martin C, Clarke J. SERRATE: a new player on the plant microRNA scene. EMBO Rep. 2006;7:1052–1058. doi: 10.1038/sj.embor.7400806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyles DS, Rupprecht CE. Rhabdoviridae. In: Knipe DM, Howley PM, editors. Fields Virology. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 1363–1408. [Google Scholar]
- Okamura K, Chung WJ, Ruby JG, Guo H, Bartel DP, Lai EC. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature. 2008 doi: 10.1038/nature07015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Ishizuka A, Siomi H, Siomi MC. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev. 2004;18:1655–1666. doi: 10.1101/gad.1210204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka M, Jing Q, Georgel P, New L, Chen J, Mols J, Kang YJ, Jiang Z, Du X, Cook R, et al. Hypersusceptibility to vesicular stomatitis virus infection in Dicer1-deficient mice is due to impaired miR24 and miR93 expression. Immunity. 2007;27:123–134. doi: 10.1016/j.immuni.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Prigge MJ, Wagner DR. The arabidopsis serrate gene encodes a zinc-finger protein required for normal shoot development. Plant Cell. 2001;13:1263–1279. doi: 10.1105/tpc.13.6.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsburg E, Publicover J, Buonocore L, Poholek A, Robek M, Palin A, Rose JK. A vesicular stomatitis virus recombinant expressing granulocyte-macrophage colony-stimulating factor induces enhanced T-cell responses and is highly attenuated for replication in animals. J Virol. 2005;79:15043–15053. doi: 10.1128/JVI.79.24.15043-15053.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Ishizuka A, Siomi H, Siomi MC. Processing of pre-microRNAs by the Dicer-1-Loquacious complex in Drosophila cells. PLoS Biol. 2005;3:e235. doi: 10.1371/journal.pbio.0030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- Tomari Y, Zamore PD. Perspective: machines for RNAi. Genes Dev. 2005;19:517–529. doi: 10.1101/gad.1284105. [DOI] [PubMed] [Google Scholar]
- van Rij RP, Saleh MC, Berry B, Foo C, Houk A, Antoniewski C, Andino R. The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster. Genes Dev. 2006;20:2985–2995. doi: 10.1101/gad.1482006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter PA, Schneemann A. Recent insights into the biology and biomedical applications of Flock House virus. Cell Mol Life Sci. 2008 doi: 10.1007/s00018-008-8037-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XH, Aliyari R, Li WX, Li HW, Kim K, Carthew R, Atkinson P, Ding SW. RNA interference directs innate immunity against viruses in adult Drosophila. Science. 2006;312:452–454. doi: 10.1126/science.1125694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MD, Wang D, Wagner R, Breyssens H, Gertsenstein M, Lobe C, Lu X, Nagy A, Burke RD, Koop BF, et al. ARS2 is a conserved eukaryotic gene essential for early mammalian development. Mol Cell Biol. 2008;28:1503–1514. doi: 10.1128/MCB.01565-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Liu Z, Lu F, Dong A, Huang H. SERRATE is a novel nuclear regulator in primary microRNA processing in Arabidopsis. Plant J. 2006;47:841–850. doi: 10.1111/j.1365-313X.2006.02835.x. [DOI] [PubMed] [Google Scholar]
- Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- Zhou R, Hotta I, Denli AM, Hong P, Perrimon N, Hannon GJ. Comparative analysis of argonaute-dependent small RNA pathways in Drosophila. Mol Cell. 2008;32:592–599. doi: 10.1016/j.molcel.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01