Telomere shortening relaxes X chromosome inactivation and forces global transcriptome alterations (original) (raw)
Abstract
Telomeres are heterochromatic structures at chromosome ends essential for chromosomal stability. Telomere shortening and the accumulation of dysfunctional telomeres are associated with organismal aging. Using telomerase-deficient TRF2-overexpressing mice (K5TRF2/_Terc_−/−) as a model for accelerated aging, we show that telomere shortening is paralleled by a gradual deregulation of the mammalian transcriptome leading to cumulative changes in a defined set of genes, including up-regulation of the mTOR and Akt survival pathways and down-regulation of cell cycle and DNA repair pathways. Increased DNA damage from dysfunctional telomeres leads to reduced deposition of H3K27me3 onto the inactive X chromosome (Xi), impaired association of the Xi with telomeric transcript accumulations (Tacs), and reactivation of an X chromosome-linked K5TRF2 transgene that is subjected to X-chromosome inactivation in female mice with sufficiently long telomeres. Exogenously induced DNA damage also disrupts Xi-Tacs, suggesting DNA damage at the origin of these alterations. Collectively, these findings suggest that critically short telomeres activate a persistent DNA damage response that alters gene expression programs in a nonstochastic manner toward cell cycle arrest and activation of survival pathways, as well as impacts the maintenance of epigenetic memory and nuclear organization, thereby contributing to organismal aging.
Keywords: aging, chromosome X inactivation, DNA damage, epigenetics, telomeres
Dysfunctional, critically short telomeres elicit a DNA damage response (DDR) that triggers senescence or apoptosis in mammalian cells, two processes that are associated with organismal aging (1–9). Mice with a targeted deletion of the RNA component of telomerase (_Terc_−/−) display accelerated telomere shortening, premature loss of tissue renewal, and decreased longevity (3, 7–9). DNA damage signals originating from critically short telomeres in these mice is in line with current models proposing a causative role for DNA damage in organismal aging (10–13). Interestingly, epigenetic alterations at heterochromatic regions are proposed to lead to changes in gene expression associated with aging (14–16). In S. cerevisiae, induction of DNA double-strand breaks (DSBs) or cellular stress causes a dramatic redistribution of telomeric silent information regulator (Sir) proteins and yKU proteins (17–19), thus linking changes in telomere chromatin to global epigenetic alterations. Sir complex relocalization is known to alter the expression of stress response genes, survival factors, and ribosomal biogenesis (20, 21). In functional analogy to yeast, mammalian SIRT1 is redistributed upon induction of DNA damage, causing broad alterations in global gene expression (22). Collectively, these findings suggest that aging-related DNA damage drives gene expression alterations that could promote the development of aging pathologies.
An important question to determine is how the various types of DNA damage impact gene expression changes associated with organismal aging. In this study, we focused on the isolated effect of dysfunctional telomeres on global genome regulation. Using a mouse model system, we provide evidence that progressive telomere shortening in stratified epithelia, such as the skin, is linked to global deregulation of the mammalian transcriptome and loss of maintenance of epigenetic silencing mechanisms, exemplified by the re-expression of an Xi-linked transgene. Indicative of the induction of a stress response, we find a down-regulation of genes promoting cell cycle progression and up-regulation of the mTOR and Akt survival pathways. In addition, cells with critically short telomeres show down-regulation of various DNA repair pathways. These findings suggest that progressive telomere shortening and the accumulation of dysfunctional telomeres with age may constitute a unique source of DNA damage, sufficient to induce global alterations in genome regulation.
Results
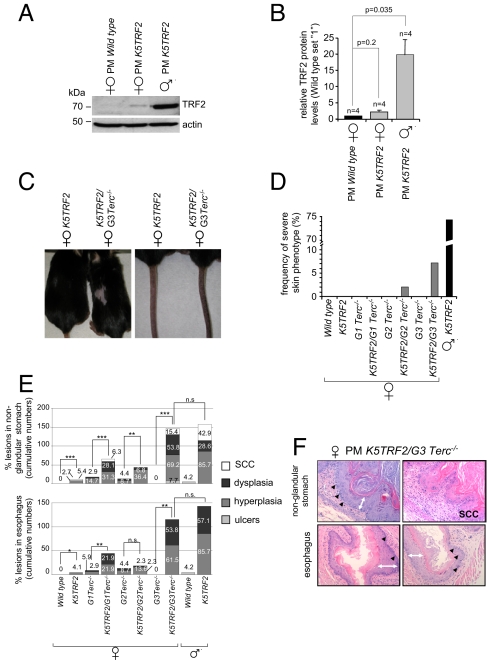
We previously generated mice overexpressing the telomere-binding protein TRF2 under the control of the 5′ regulatory region of the keratin 5 gene (PM K5TRF2 trangenic line) (23). TRF2 is a key player in the regulation of telomere length and telomere protection (23–25). In accordance with this, K5TRF2 mice showed severe telomere shortening, increased sensitivity to UV radiation, premature skin aging (hair loss, skin hyperpigmentation, skin dryness), and increased skin cancer (23, 26). In this transgenic line, skin phenotypes and embryonic lethality were restricted to male mice, whereas female littermates remained phenotypically normal (23). We show here that PM K5TRF2 females display TRF2 protein levels only slightly above wild-type levels, compared with robust TRF2 overexpression in littermate transgenic males (Fig. 1 A and B). These findings suggest that the K5TRF2 transgene is located at the X chromosome and specifically silenced in females. To address this, we performed DNA FISH on male PM K5TRF2 keratinocytes and mapped the integration site for the PM K5TRF2 transgene to the X chromosome (supporting information (SI) Fig. S1_A_). These findings suggest that the K5TRF2 transgene is silenced by a nonrandom X inactivation event in female K5TRF2 mice, thereby preventing TRF2 overexpression and the onset of severe skin pathologies.
Fig. 1.
An X-linked transgene is re-expressed upon telomere shortening. (A) Male PM K5TRF2 mice display elevated TRF2 protein levels compared with female littermates. Actin, loading control. (B) Quantification of Western blots; n, number of keratinocyte preparations; standard error is indicated. A Student's t test was used to calculate statistical significance. (C) Skin phenotypes in PM K5TRF2/_Terc_−/− females. (D) Quantification of skin disorders. (E) Quantification of abnormalities in stratified epithelia. Female mice: wild type, n = 68; K5TRF2, n = 74; _G1 Terc_−/−, n = 34; K5TRF2/_G1 Terc_−/−, n = 32; _G2 Terc_−/−, n = 23; K5TRF2/_G2 Terc_−/−, n = 44; _G3 Terc_−/−, n = 4; K5TRF2/_G3 Terc_−/−, n = 13. Male mice: wild type, n = 24; K5TRF2, n = 7. A Fisher's exact test was used to calculate statistical significance. (F) Histopathological findings in stratified epithelia. Black arrowheads, displastic nuclei; white double-headed arrows, hyperplastic areas.
Next, we crossed PM K5TRF2 mice into a telomerase-deficient (_Terc_−/−) background to address the impact of telomere shortening on global epigenetic alterations, including chromosome X inactivation. To this end, we generated increasing generations (G1–G3) of female PM K5TRF2 transgenic mice in a telomerase-deficient background (K5TRF2/_G1–G3 Terc_−/−; see Methods). Progressive telomere shortening in K5TRF2/_G1–G3 Terc_−/− females resulted in gradual appearance of K5TRF2-associated skin phenotypes. K5TRF2/_G2–G3 Terc_−/− females display hair loss, skin hyperpigmentation, and increased skin lesions, reaching a severity in K5TRF2/_G3 Terc_−/− females that is similar to that in K5TRF2 males (Fig. 1 C and D and Fig. S1_B_). Histopathological analyses further showed that K5TRF2/_G2–G3 Terc_−/− females also develop preneoplastic (dysplasia, hyperplasia) and neoplastic (squamous cell carcinoma, SCC) lesions in other stratified epithelia with reported K5 promoter activity, such as nonglandular stomach and esophagus (Fig. 1 E and F). Again, the penetrance of these epithelial lesions in PM K5TRF2/_G3 Terc_−/− females was comparable with that of male PM K5TRF2 mice (Fig. 1 E and F). Littermate _G3 Terc_−/− females did not develop any of these pathologies (Fig. 1 C–F).
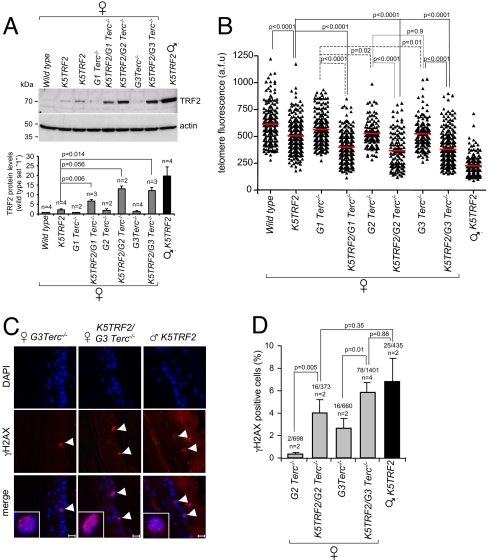
Interestingly, telomere shortening in K5TRF2/_G1–G3 Terc_−/− females coincided with a gradual increase in TRF2 protein levels, reaching the highest levels in K5TRF2/_G3 Terc_−/− females and in PM K5TRF2 males (Fig. 2A). These findings suggest that progressive telomere shortening in K5TRF2/_G1–G3 Terc_−/− females drives a gradual loss of silencing of the Xi-linked K5TRF2 transgene and increased expression of TRF2, which in turn triggers epithelial pathologies in PM K5TRF2/_Terc_−/− females that recapitulate those of PM K5TRF2 transgenic males.
Fig. 2.
Loss of silencing of the X-linked PM K5TRF2 transgene leads to telomere dysfunction in K5TRF2/_Terc_−/− females. (A) (Top) TRF2 protein levels in back skin keratinocytes; (Bottom) quantification of TRF2 levels after normalizing against β-actin. n, 2–4 experiments. (B) Telomere Q-FISH in tail skin. Red bars, average telomere fluorescence intensity. A Student's t test was used to calculate statistical significance. a.f.u, arbitrary fluorescence units. (C) γH2AX-positive cells (white arrowheads) in the indicated skin sections. (Scale bar: 10 μm.) (D) Quantification of γH2AX immunostainings. n, mice analyzed; γH2AX-positive nuclei and total number of cells analyzed are indicated. A Fisher's exact test was used to calculate statistical significance.
TRF2 overexpression leads to dramatic telomere shortening in PM K5TRF2 males (23). To investigate whether reexpression of the silenced PM K5TRF2 transgene induces further telomere shortening in K5TRF2/_G1–G3 Terc_−/− females compared with their _G1–G3 Terc_−/− controls, we performed quantitative telomere Q-FISH on skin sections (see Methods). Telomerase deficiency in _G1–G3 Terc_−/− mice results in slow but continuous telomere shortening with increasing mouse generations (Fig. 2B). Consistent with our previous findings (23), the skin of PM K5TRF2 males displays a dramatic telomere shortening (Fig. 2B). Slightly elevated TRF2 levels in PM K5TRF2 females cause only mild telomere shortening (Fig. 2 A and B). Importantly, loss of silencing of the K5TRF2 transgene in K5TRF2/_G1 Terc_−/− females induces accelerated telomere shortening compared with their _G1 Terc_−/− controls (Fig. 2B), and this effect is exacerbated in K5TRF2/_G2–G3 Terc_−/− females (Fig. 2B). We confirmed these results by Southern blot-based telomere restriction fragment (TRF) analysis (Fig. S1_C_).
Critically short telomeres elicit a DDR provoking entry into cell cycle arrest/senescence or apoptosis (1–9). To test whether reactivation of the K5TRF2 transgene in K5TRF2/_Terc_−/− females resulted in increased DNA damage signaling at dysfunctional telomeres, we quantified the abundance of γH2AX foci in the skin of experimental mice. Consistent with their shorter telomeres (Fig. 2B), we observed a significant increase of γH2AX-positive nuclei in K5TRF2/_G2 Terc_−/− females compared with their G2 _Terc_−/− female littermates (Fig. 2 C and D). This effect was further increased in females lacking telomerase activity for 3 generations, K5TRF2/_G3 Terc_−/− mice, reaching similar DNA damage levels to those of K5TRF2 males (Fig. 2 C and D).
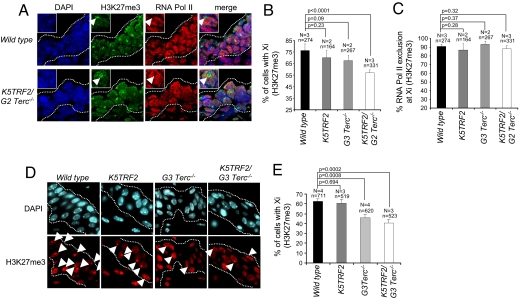
Mammalian X chromosome inactivation (XCI) ensures dosage-compensated expression of X-linked genes in male and female cells. XCI is initiated during female embryonic development by the up-regulation and spreading of the noncoding Xist RNA along the future inactive X chromosome (Xi), triggering long-range transcriptional silencing and the deposition of repressive chromatin marks such as histone H3 lysine 27 trimethylation (H3K27me3) in cis (27, 28). Although decoration of the Xi with Xist is not significantly affected upon telomere shortening (Fig. S2 A and B), we observed a significant decrease of Xi-associated H3K27me3 in skin sections of K5TRF2/_G2 Terc_−/− E12 embryos as well as in adult skin from _G3 Terc_−/− and K5TRF2/_G3 Terc_−/− mice, as detected by confocal immunofluorescence (Fig. 3 A–E). Exclusion of RNA polymerase II from the H3K27me3 mark at the Xi in K5TRF2/G2 _Terc_−/− embryos indicates that the establishment of _Xist_-mediated silencing is normal in the epidermis of E12 mice cells (Fig. 3 A and C). These findings suggest that increased DNA damage originated from short telomeres leads to a defective maintenance of X inactivation, marked by a reduced frequency of H3K27me3 at the Xi and the reexpression of the Xi-linked K5TRF2 transgene.
Fig. 3.
Critical telomere shortening results in impaired H3K27me3 of the Xi in vivo. (A) Combined immunostaining for H3K27me3 and RNA polymerase II in E12 embryo skin sections. (B) Reduced deposition of H3K27me3 on the Xi in K5TRF2/_G2 Terc_−/− skin sections. (C) Quantification of RNA polymerase II exclusion at the Xi (decorated with H3K27me3). (D) Immunostaining for H3K27me3 in adult tail-skin sections. (E) Reduced frequency of Xi-coupled H3K27me3 in adult skin sections of K5TRF2/_G2 Terc_−/− females. N, number of mice; n, nuclei analyzed. Error bars, standard error. An unpaired Student's t test was used to calculate statistical significance.
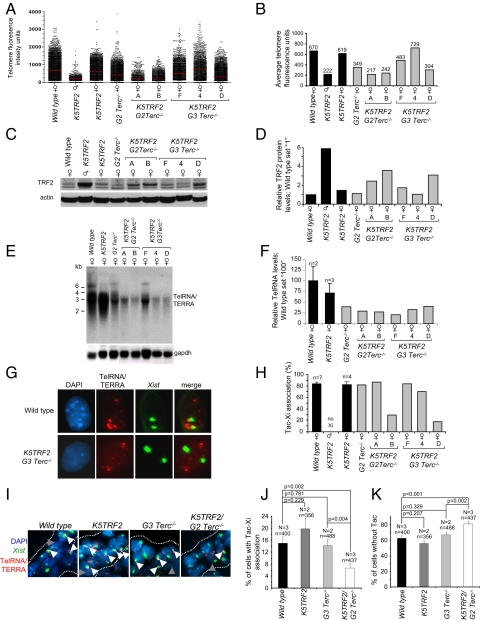
Vertebrate telomeres are transcribed by RNA polymerase II, giving rise to UUAGGG repeat-containing, noncoding RNAs (TelRNA/TERRA) that localize to mammalian telomeres but also form accumulations (Tacs, _T_elRNA ac_cumulation_s) in the vicinity of the inactive X chromosome (Xi) in female cells (29, 30). To test a possible link with XCI, we determined telomere length, TelRNA/TERRA expression, TRF2 protein levels, and Tac-Xi localization pattern in primary keratinocytes derived from female wild-type, K5TRF2, and K5TRF2/_G2–G3 Terc_−/− newborn mice. _G2 Terc_−/− cells displayed telomere shortening that was further augmented in K5TRF2/_G2 Terc_−/− keratinocytes (Fig. 4 A and B) (26). Selection for cells that rescue telomere length by recombination-based mechanisms explains the heterogeneity of telomere lengths in K5TRF2/_G3 Terc_−/− keratinocytes (see cultures F and 4 in Fig. 4 A and B) (26). Telomere attrition in K5TRF2/_G2 Terc_−/− (cultures A and B) and K5TRF2/_G3 Terc_−/− (culture D) cells is accompanied by loss of silencing of the X-linked K5TRF2 transgene, resulting in increased TRF2 protein levels (see cultures A, B, and D in Fig. 4 A–D), further confirming our in vivo data (Fig. 2A). Previous reports suggested a positive correlation between telomere length and TelRNA/TERRA levels (29, 30). In agreement with this, we observed reduced TelRNA/TERRA levels in keratinocyte cultures with short telomeres (Fig. 4 E and F). Furthermore, _Xist_–TelRNA/TERRA double-RNA FISH on female keratinocytes revealed an impaired association of Tacs with the inactive X in female K5TRF2/_G2 Terc_−/− (culture B) and PM K5TRF2/_G3 Terc_−/− (culture D) cells, which was confirmed in vivo in the epidermis of K5TRF2/_G2 Terc_−/− E12 embryos (Fig. 4 G–J). Tac-Xi association was normal in cells and embryos with sufficiently long telomeres (Fig. 4 G–J). Of notice, the frequency of Tacs was decreased in skin sections from E12 K5TRF2/_G2 Terc_−/− embryos (Fig. 4K), suggesting that TelRNA/TERRA expression levels influence Tac formation. To test whether a DNA damage signal elicited from dysfunctional telomeres could directly contribute to Tac-Xi dissociation, we exposed primary MEF (pMEF) to ionizing radiation. Thirty minutes after irradiation, cells exhibited an efficient DDR activation, as indicated by the appearance of abundant γH2AX foci, which decreased in the following hours postirradiation (Fig. S3_A_). Treatment with ionizing radiation caused only minimal alterations in TelRNA/TERRA expression levels and Tac frequency in the experimental time window used here, as determined both by Northern blotting and RNA FISH (Fig. S2 C–G). Importantly, we observed a severe reduction in Tac-Xi associations 3 h postirradiation, suggesting that these associations are disrupted as a consequence of DNA damage (Fig. S3_B_). These findings suggest that DNA damage signaling originating from dysfunctional telomeres alters the maintenance of epigenetic gene silencing on the Xi but also affects nuclear organization of chromatin territories, as exemplified by Tac-Xi dissociation in cells with critically short telomeres. These findings also raise the possibility that defective Xi maintenance may be part of the global chromatin defects triggered by telomere shortening.
Fig. 4.
Critical telomere shortening results in a loss of _Xist_-Tacs association. (A) Distributions of individual telomere lengths in metaphases from primary keratinocytes. a.f.u., arbitrary fluorescence units; red bars, medium telomere fluorescence. (B) Average telomere length per genotype. Average values are shown on top of the bars. (C) TRF2 protein expression. (D) Quantification of TRF2 levels normalized against β-actin. (E) TelRNA/TERRA levels as detected by Northern blotting. (F) Quantification of TelRNA/TERRA levels normalized against gapdh. n, cell lines analyzed; standard error is indicated. (G) Combined _Xist_-TelRNA/TERRA RNA FISH to detect _Xist_-Tac association. Cells shown are polyploid and present 2 Xist signals. DNA was stained with DAPI. (H) _Xist_-TelRNA association is lost in most cultures with short telomeres and high TRF2. n, cell lines analyzed; standard error is indicated. (I) _Xist_-TelRNA RNA FISH in E12 embryonic skin sections. Green, Xist RNA; red, TelRNA/TERRA; blue, DAPI. Gray arrowheads, telomere-associated TelRNA/TERRA; white arrowheads, TelRNA accumulations (Tacs) in the vicinity of the _Xist_-coated inactive X chromosome. (J) K5TRF2/_G2 Terc_−/− E13 embryos display reduced Xi-Tac association. (K) Quantification of Tac frequency in E12 embryos. N, number of mice; n, nuclei analyzed. Error bars, standard error. An unpaired Student's t test was used to calculate statistical significance.
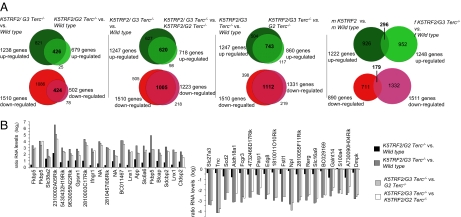
A recent report indicated that the accumulation of DNA damage during mammalian aging results in altered gene expression patterns due to the redistribution of the histone deacetylase SIRT on chromatin (22). To investigate whether progressive telomere shortening selectively affects transcriptional memory on the Xi or is associated with a global deregulation of gene expression, we performed comparative transcriptome analyses using primary skin keratinocytes from wild-type, K5TRF2, _G2 Terc_−/−, K5TRF2/_G2 Terc_−/−, and K5TRF2/_G3 Terc_−/− PM females. To control for sex-specific gene expression differences, we included wild-type and K5TRF2 PM males in the analysis (23). Differences in transcript levels of individual genes obtained in pairwise transcriptome comparisons were blotted along cytogenetic maps of all mouse chromosomes and revealed that progressive telomere shortening is associated with increasing transcriptome alterations, affecting all mouse chromosomes to a similar extent (Fig. S5 A–J and Table S1). Genome-wide alterations reached high statistical significance in female K5TRF2/_G3 Terc_−/− keratinocytes (P < 0.001) when compared with other genotypes with longer telomeres (Fig. S4_A_). We conclude that telomere attrition leads to a global impairment in the accomplishment of gene expression programs. Focusing our analyses on individual genes, we found 213 affected genes in female _G2 Terc_−/− keratinocytes [false detection rate (FDR) <0.05], and this number was further increased in K5TRF2/_Terc_−/− cells reaching a maximum of 2,757 gene expression changes in female K5TRF2/_G3 Terc_−/− keratinocytes (Fig. S4_B_). We note that highly significant gene expression changes (FDR <0.001) were detected only in K5TRF2/_G3 Terc_−/− keratinocytes (Fig. S4_B_). Progressive telomere shortening results in cumulative transcriptome changes affecting 15.1% of all genes (6.7% up-regulated; 8.4% down-regulated) in K5TRF2/_G3 Terc_−/− mice (Table S1). Importantly, transcriptome alterations were reproduced in male K5TRF2 keratinocytes, ruling out a sex-specific effect on transcriptome alterations associated with critically short telomeres (Fig. S4_B_, Fig. S5 A–J, and Table S1). We next addressed whether genes altered (FDR <0.05) in female K5TRF2/_G3 Terc_−/− keratinocytes were also affected in cells with less severe telomere shortening. Venn diagrams demonstrated a highly overlapping pattern of affected genes (FDR <0.05) between K5TRF2/_G3 Terc_−/− and K5TRF2/_G2 Terc_−/− keratinocytes when compared with wild-type or _G2 Terc_−/− transcriptomes (Fig. 5A). An independent set of gene expression-profiling analyses revealed that 24% of genes up-regulated and 20% of genes down-regulated in male K5TRF2 keratinocytes (FDR <0.05) showed the same regulation in female K5TRF2/_G3 Terc_−/− samples. This indicates that telomere attrition affects the expression of an overlapping set of genes in male and female cells (Fig. 5A). Focusing on individual probe sets displaying most robust alterations, we confirmed that robust gene expression changes in K5TRF2/_G3 Terc_−/− cells (FDR <1.00E-07) are gradually diminished in K5TRF2/_G2 Terc_−/− and G2 _Terc_−/− cells (Fig. 5B and Tables S2–S4).
Fig. 5.
Deregulation of the mouse transcriptome is linked to progressive telomere shortening. (A) Venn diagrams showing overlapping gene expression patterns between the indicated pairwise comparisons of transcriptomes. Telomere shortening affects the expression of a similar group of genes between different pairwise comparisons. Red circles, down-regulated genes; green circles, up-regulated genes. (B) Transcript levels ratios of 20 probe sets robustly up-regulated or down-regulated in K5TRF2/_G3 Terc_−/− versus wild-type comparisons. Probe sets are ranked according to the false detection rate (FDR <1.00E-07; also see Tables S2 and S3). Changes in transcript levels are tendentially reduced when the difference in telomere length between experimental samples is reduced. K5TRF2/_G2 Terc_−/− show only limited transcriptome changes when compared with wild-type keratinocytes.
Importantly, despite showing slightly elevated TRF2 expression, no significant alterations in gene expression were detected in female K5TRF2 keratinocytes (Fig. S4 and Table S1). In addition, high TRF2 levels in female K5TRF2/_G1–G2 Terc_−/− keratinocytes did not significantly alter the global transcriptome (Fig. S4 and Table S1). These findings indicate that short telomeres and not increased TRF2 expression are responsible for the transcriptome alterations described here. We conclude that progressive telomere shortening is linked with a continuous loss of stringency of global transcriptional control that affects the expression of a defined set of genes in a nonstochastic manner.
Next, we investigated whether transcriptome alterations were biased toward particular pathways, which could provide an explanation for the premature aging phenotypes in K5TRF2/_G2–G3 Terc_−/− females and K5TRF2 males. Pathways analysis revealed an overrepresentation (FDR <0.25) of genes involved in the mTOR, Akt, eIF4 and ErbB pathways in K5TRF2/_G3 Terc_−/− females and to a lesser extent in male K5TRF2 keratinocytes, presumably reflecting the activation of a cell survival program to escape cell cycle arrest/cellular senescence induced by critically short telomeres (Table S5 A–D). In this regard, we found an underrepresentation of genes involved in cell cycle regulation (FDR <0.25) in female K5TRF2/_G3 Terc_−/− and male K5TRF2 keratinocytes (Table S5 A–D). Interestingly, despite the presence of an increased DNA damage load, genes involved in DNA damage repair pathways, such as base excision, nucleotide excision, and mismatch repair, were underrepresented in female K5TRF2/_G3 Terc_−/− and male K5TRF2 keratinocytes (Table S5 A–D). Mutations in components of nucleotide excision repair are responsible for the development of premature aging syndromes such as xeroderma pigmentosa, Cockayne syndrome, or trichothiodystrophy (31). These findings suggest that the decreased ability of male K5TRF2 and female K5TRF2/_G2–G3 Terc_−/− keratinocytes to up-regulate DNA damage-response pathways in a context of severe telomere dysfunction may contribute to the known hypersensitivity to UV irradiation and premature appearance of skin-aging phenotypes in these mice (23).
Interestingly, we found that ≈10% of the gene expression changes detected in mouse models for aging, such as nucleotide excision repair-deficient mice (_Ercc1_−/− mice vs. wild-type mice) and very old mice (130 vs. 8 weeks old) (13), overlapped with alterations in K5TRF2/_G3 Terc_−/− females (Fig. S6 and Tables S6–S8). However, this overlap was less evident when comparing K5TRF2/_G3 Terc_−/− female keratinocytes with ES cells treated with oxidating agents (22) or with the aging mouse neocortex (22) (Fig. S6 and Tables S6–S8).
Discussion
Identifying sources of DNA damage associated with aging and analyzing their effect on cellular metabolism is essential to understanding the aging process. Accumulation of dysfunctional, critically short telomeres is associated with the development of aging pathologies (6–9). Telomere shortening is paralleled by activation of a DDR (4, 5). Recent studies show that oxidative stress as well as mutations in DNA repair pathways provoke epigenetic alterations and transcriptional changes paralleling those observed in aged animals (13, 22, 32). Here we show that accumulation of dysfunctional telomeres, associated with broad alterations of the mouse transcriptome and impaired maintenance of epigenetic silencing during X inactivation.
Gene expression changes accumulate with progressive telomere shortening and are detected along all chromosomes and are unbiased toward gene location, excluding a telomere position effect as a causative event for the observed transcriptome alterations (33, 34). Comparative transcriptome analyses further revealed that progressive telomere shortening gradually affects a defined set of genes driving the up-regulation of the mTOR, ErbB, and Akt survival pathways and down-regulation of cell cycle-promoting genes and DNA damage pathways. The observed induction of a cellular stress response promoting growth repression and cell survival is in line with previous findings in yeast (17–20). In particular, Sir relocation causes gene expression changes involving the up-regulation of survival genes and stress response factors and the down-regulation of ribosomal biogenesis, thus limiting proliferation and promoting cell survival (20, 21). Interestingly, yeast mTOR antagonizes Sir complex relocation upon the induction of cellular stress (20). Together with our current data, these findings support the view of mTOR as a major integrator of stress signals, including severe telomere dysfunction. Finally, the activation of survival mechanisms via the mTOR and Akt pathways is in line with previous findings from our group showing that mouse spermatocytes with critically short telomeres up-regulate the PI3-kinase survival pathway (35). Interestingly, we also detected an underrepresentation of genes involved in DNA damage repair pathways in cells with critically short telomeres. Conceivably, this could lead to an impaired response to DNA damage, which is in line the reported hypersensitivity of these mice to UV irradiation, and could contribute to genomic instability and premature tissue deterioration (23, 26).
Alterations in epigenetic regulators have been previously reported to cause gene expression patterns related to aging (14–16, 22). We show here that DNA damage originating from critically short telomeres causes the re-expression of an X-linked K5TRF2 transgene that is subjected to X chromosome inactivation in cells with sufficient telomere length. Importantly, loss of silencing is accompanied by a reduced deposition of H3K27me3 at the Xi and by a dissociation of TelRNA accumulations (Tacs) from the Xi. Interestingly, Tac-Xi dissociation is also induced by exogenously induced DNA damage. Together, this indicates an impact of telomere-originated DNA damage on the organization of chromatin territories, such as that of the Xi. The failure of transgene silencing in cells with short telomeres provides further evidence for the hypothesis of an age-related decrease in the stability of the X-inactivation mechanism (36, 37).
Genome deregulation caused by telomere attrition and telomere dysfunction is in line with a current model proposing that the accumulation of DNA damage causes the deregulation of epigenetic control and altered gene expression during organismal aging (13, 22). Considering that progressive telomere shortening is observed in elderly mice and humans (38, 39), our study indicates that DNA damage checkpoint activation by dysfunctional telomeres is sufficient to induce epigenetic alterations and gene expression changes that antagonize proliferation and promote the activation of survival pathways. These findings support the notion that dysfunctional telomeres represent a unique and cumulative source of DNA damage, which can promote organismal aging by impairing tissue regeneration.
Materials and Methods
Mice.
PM K5TRF2 and K5TRF2/_G1–G3 Terc_−/− mice were generated and maintained as previously described (23, 26).
Histopathology and Immunohistochemistry.
Immunohistochemistry was performed on deparaffinated adult skin sections or frozen OCT sections (E12) using a mouse monoclonal anti-phospho-histone H2AX antibody (1:500; Upstate Biotechnology), a polyclonal rabbit anti-H3K27me3 antibody (07–449; Upstate Biotechnology), and a monoclonal anti-RNA polymerase II antibody (ab5408; Abcam). Xi-linked H3K27me3 and exclusion of RNA polymerase II from the inactive X (marked by focal H3K27me3) was analyzed by visual appearance using confocal microscopy. Images were obtained using a confocal ultra-spectral microscope (TCS-SP2-A-OBS-UV; Leica) or captured at 100× magnification using a Leica CTR MIC microscope and a Cohu High-Performance CCD camera (Cohu, Inc.).
Telomere Length Analyses on Skin Sections and Skin Keratinocyte.
Q-FISH on keratinocytes and tissue sections was carried out as described (23). Nuclei or metaphases were captured at 100× magnification using a Leica CTR MIC microscope and a Cohu High-Performance CCD camera. Telomere fluorescence was determined as described (40).
TRF-Based Telomere Length Analysis of Adult Keratinocytes.
Adult epidermal keratinocytes were isolated as described (23) and included in agarose plugs following instructions provided by the manufacturer (Bio-Rad). TRF analysis was performed as described (3).
Isolation and Culture of Keratinocytes from Newborn Mouse Skin.
Newborn skin keratinocytes were isolated as described (23).
γ Irradiation of Primary Mouse Embryonic Fibroblast (pMEFs).
pMEFs were exposed to 3 Gy of γ irradiation, and 30 min and 3 h postirradiation induction of DNA damage was monitored using an anti-phospho-histone H2AX antibody (1:500; Upstate Biotechnology). In parallel, Xist and TelRNA/TERRA RNA FISH stainings were performed.
Western Blotting.
Western blots of primary keratinocytes were carried out as described (23). Twenty-five micrograms to 70 μg were used per condition. A polyclonal antibody to TRF2 (1:1,000; SF08, provided by E. Gilson, Lyon, France) and a monoclonal antibody to β-actin (1:10,000; Sigma) were used.
RNA Analysis.
Northern blot analysis for TelRNA/TERRA was performed as described (30).
RNA Fluorescence in Situ Hybridizations (RNA FISH).
RNA FISH analysis was carried out as previously described (30). RNA FISH analysis of embryos was performed on frozen OCT sections (5 μm) (30).
Microarray Analyses.
Total RNA was prepared from adult female tail-skin keratinocytes (23, 35) using the RNAeasy kit (Invitrogen): PM wild-type (n = 4), PM K5TRF2 (n = 4), _G2 Terc_−/− (n = 2), PM K5TRF2/_G2 Terc_−/− (n = 3), and PM K5TRF2/_G3 Terc_−/− (n = 3). RNA quality was tested using the Agilent 2100 Bioanalyzer (Agilent Technologies). RNA was labeled in a one-color format and hybridized to 44K Whole Mouse Genome Oligo microarrays (G4122F; Agilent Technologies). Details on gene expression profiling and analysis are in SI Text.
Supplementary Material
Supporting Information
Acknowledgments.
M.A.B.'s laboratory is funded by the Spanish Ministry of Science and Technology, the European Union, European Research Council Advanced Grants, the Spanish Association Against Cancer, and the Körber European Science Award 2008.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Chan SW, Blackburn EH. New ways not to make ends meet: Telomerase, DNA damage proteins and heterochromatin. Oncogene. 2002;21:553–563. doi: 10.1038/sj.onc.1205082. [DOI] [PubMed] [Google Scholar]
- 2.de Lange T. Protection of mammalian telomeres. Oncogene. 2002;21:532–540. doi: 10.1038/sj.onc.1205080. [DOI] [PubMed] [Google Scholar]
- 3.Blasco MA, et al. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997;91:25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- 4.d'Adda di Fagagna F, et al. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426:194–198. doi: 10.1038/nature02118. [DOI] [PubMed] [Google Scholar]
- 5.de Lange T. Shelterin: The protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 6.Goytisolo FA, Blasco MA. Many ways to telomere dysfunction: In vivo studies using mouse models. Oncogene. 2002;21:584–591. doi: 10.1038/sj.onc.1205085. [DOI] [PubMed] [Google Scholar]
- 7.Herrera E, et al. Disease states associated with telomerase deficiency appear earlier in mice with short telomeres. EMBO J. 1999;18:2950–2960. doi: 10.1093/emboj/18.11.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee HW, et al. Essential role of mouse telomerase in highly proliferative organs. Nature. 1998;392:569–574. doi: 10.1038/33345. [DOI] [PubMed] [Google Scholar]
- 9.Rudolph KL, et al. Longevity, stress response, and cancer in aging telomerase-deficient mice. Cell. 1999;96:701–712. doi: 10.1016/s0092-8674(00)80580-2. [DOI] [PubMed] [Google Scholar]
- 10.de Boer J, et al. Premature aging in mice deficient in DNA repair and transcription. Science. 2002;296:1276–1279. doi: 10.1126/science.1070174. [DOI] [PubMed] [Google Scholar]
- 11.Martin GM. Genetic modulation of senescent phenotypes in Homo sapiens. Cell. 2005;120:523–532. doi: 10.1016/j.cell.2005.01.031. [DOI] [PubMed] [Google Scholar]
- 12.Martin GM, Oshima J. Lessons from human progeroid syndromes. Nature. 2000;408:263–266. doi: 10.1038/35041705. [DOI] [PubMed] [Google Scholar]
- 13.Niedernhofer LJ, et al. A new progeroid syndrome reveals that genotoxic stress suppresses the somatotroph axis. Nature. 2006;444:1038–1043. doi: 10.1038/nature05456. [DOI] [PubMed] [Google Scholar]
- 14.Imai S, Kitano H. Heterochromatin islands and their dynamic reorganization: A hypothesis for three distinctive features of cellular aging. Exp Gerontol. 1998;33:555–570. doi: 10.1016/s0531-5565(98)00037-0. [DOI] [PubMed] [Google Scholar]
- 15.Vijg J. Impact of genome instability on transcription regulation of aging and senescence. Mech Ageing Dev. 2004;125:747–753. doi: 10.1016/j.mad.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 16.Villeponteau B. The heterochromatin loss model of aging. Exp Gerontol. 1997;32:383–394. doi: 10.1016/s0531-5565(96)00155-6. [DOI] [PubMed] [Google Scholar]
- 17.Martin SG, et al. Relocalization of telomeric Ku and SIR proteins in response to DNA strand breaks in yeast. Cell. 1999;97:621–633. doi: 10.1016/s0092-8674(00)80773-4. [DOI] [PubMed] [Google Scholar]
- 18.McAinsh AD, Scott-Drew S, Murray JA, Jackson SP. DNA damage triggers disruption of telomeric silencing and Mec1p-dependent relocation of Sir3p. Curr Biol. 1999;9:963–966. doi: 10.1016/s0960-9822(99)80424-2. [DOI] [PubMed] [Google Scholar]
- 19.Mills KD, Sinclair DA, Guarente L. MEC1-dependent redistribution of the Sir3 silencing protein from telomeres to DNA double-strand breaks. Cell. 1999;97:609–620. doi: 10.1016/s0092-8674(00)80772-2. [DOI] [PubMed] [Google Scholar]
- 20.Ai W, et al. Regulation of subtelomeric silencing during stress response. Mol Cell. 2002;10:1295–1305. doi: 10.1016/s1097-2765(02)00695-0. [DOI] [PubMed] [Google Scholar]
- 21.Taddei A, et al. The functional importance of telomere clustering: Global changes in gene expression result from SIR factor dispersion. Genome Res. 2009;19:611–625. doi: 10.1101/gr.083881.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oberdoerffer P, et al. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell. 2008;135:907–918. doi: 10.1016/j.cell.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Munoz P, Blanco R, Flores JM, Blasco MA. XPF nuclease-dependent telomere loss and increased DNA damage in mice overexpressing TRF2 result in premature aging and cancer. Nat Genet. 2005;37:1063–1071. doi: 10.1038/ng1633. [DOI] [PubMed] [Google Scholar]
- 24.Celli GB, de Lange T. DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat Cell Biol. 2005;7:712–718. doi: 10.1038/ncb1275. [DOI] [PubMed] [Google Scholar]
- 25.Smogorzewska A, et al. Control of human telomere length by TRF1 and TRF2. Mol Cell Biol. 2000;20:1659–1668. doi: 10.1128/mcb.20.5.1659-1668.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blanco R, et al. Telomerase abrogation dramatically accelerates TRF2-induced epithelial carcinogenesis. Genes Dev. 2007;21:206–220. doi: 10.1101/gad.406207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heard E. Delving into the diversity of facultative heterochromatin: The epigenetics of the inactive X chromosome. Curr Opin Genet Dev. 2005;15:482–489. doi: 10.1016/j.gde.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 28.Payer B, Lee JT. X chromosome dosage compensation: How mammals keep the balance. Annu Rev Genet. 2008;42:733–772. doi: 10.1146/annurev.genet.42.110807.091711. [DOI] [PubMed] [Google Scholar]
- 29.Azzalin CM, et al. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318:798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- 30.Schoeftner S, Blasco MA. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat Cell Biol. 2008;10:228–236. doi: 10.1038/ncb1685. [DOI] [PubMed] [Google Scholar]
- 31.Garinis GA, van der Horst GT, Vijg J, Hoeijmakers JH. DNA damage and ageing: New-age ideas for an age-old problem. Nat Cell Biol. 2008;10:1241–1247. doi: 10.1038/ncb1108-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bahar R, et al. Increased cell-to-cell variation in gene expression in ageing mouse heart. Nature. 2006;441:1011–1014. doi: 10.1038/nature04844. [DOI] [PubMed] [Google Scholar]
- 33.Baur JA, Zou Y, Shay JW, Wright WE. Telomere position effect in human cells. Science. 2001;292:2075–2077. doi: 10.1126/science.1062329. [DOI] [PubMed] [Google Scholar]
- 34.Koering CE, et al. Human telomeric position effect is determined by chromosomal context and telomeric chromatin integrity. EMBO Rep. 2002;3:1055–1061. doi: 10.1093/embo-reports/kvf215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Franco S, Canela A, Klatt P, Blasco MA. Effectors of mammalian telomere dysfunction: A comparative transcriptome analysis using mouse models. Carcinogenesis. 2005;26:1613–1626. doi: 10.1093/carcin/bgi107. [DOI] [PubMed] [Google Scholar]
- 36.Bennett-Baker PE, Wilkowski J, Burke DT. Age-associated activation of epigenetically repressed genes in the mouse. Genetics. 2003;165:2055–2062. doi: 10.1093/genetics/165.4.2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wareham KA, Lyon MF, Glenister PH, Williams ED. Age related reactivation of an X-linked gene. Nature. 1987;327:725–727. doi: 10.1038/327725a0. [DOI] [PubMed] [Google Scholar]
- 38.Flores I, et al. The longest telomeres: A general signature of adult stem cell compartments. Genes Dev. 2008;22:654–667. doi: 10.1101/gad.451008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harley CB, Futcher AB, Greider CW. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 40.Samper E, et al. Mammalian Ku86 protein prevents telomeric fusions independently of the length of TTAGGG repeats and the G-strand overhang. EMBO Rep. 2000;1:244–252. doi: 10.1093/embo-reports/kvd051. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information