Genomic Study of Replication Initiation in Human Chromosomes Reveals the Influence of Transcription Regulation and Chromatin Structure on Origin Selection (original) (raw)
DNA replication in metazoans initiates from multiple chromosomal loci called origins. This study identifies 150 new origins of replication that were confirmed by two methods of nascent strand purification. We discern the role of transcription initiation and regulation, as well as chromatin signatures in determining origin selection in human genome.
Abstract
DNA replication in metazoans initiates from multiple chromosomal loci called origins. Currently, there are two methods to purify origin-centered nascent strands: lambda exonuclease digestion and anti-bromodeoxyuridine immunoprecipitation. Because both methods have unique strengths and limitations, we purified nascent strands by both methods, hybridized them independently to tiling arrays (1% genome) and compared the data to have an accurate view of genome-wide origin distribution. By this criterion, we identified 150 new origins that were reproducible across the methods. Examination of a subset of these origins by chromatin immunoprecipitation against origin recognition complex (ORC) subunits 2 and 3 showed 93% of initiation peaks to localize at/within 1 kb of ORC binding sites. Correlation of origins with functional elements of the genome revealed origin activity to be significantly enriched around transcription start sites (TSSs). Consistent with proximity to TSSs, we found a third of initiation events to occur at or near the RNA polymerase II binding sites. Interestingly, ∼50% of the early origin activity was localized within 5 kb of transcription regulatory factor binding region clusters. The chromatin signatures around the origins were enriched in H3K4-(di- and tri)-methylation and H3 acetylation modifications on histones. Affinity of origins for open chromatin was also reiterated by their proximity to DNAse I-hypersensitive sites. Replication initiation peaks were AT rich, and >50% of the origins mapped to evolutionarily conserved regions of the genome. In summary, these findings indicate that replication initiation is influenced by transcription initiation and regulation as well as chromatin structure.
INTRODUCTION
DNA replication is a highly orchestrated process that precisely duplicates the genome once every cell cycle and initiates from sites in the genome called origins of replication. A catalog of well-validated origins of replication in human chromosomes is absolutely essential to understand how the chromosomes are replicated in the normal S phase, how abnormalities in replication such as rereplication or delays in fork migration affect chromosomal stability, and how intra-S phase checkpoints induced by radiation and cancer chemotherapy impacts on chromosomal replication and fragility.
In simple eukaryotes such as Saccharomyces cerevisiae, origins of replication are located at multiple sites along each chromosome (Huberman and Riggs, 1968; Newlon et al., 1974) and are used reproducibly and relatively efficiently in successive cell cycles (Raghuraman et al., 2001; Bell and Dutta, 2002). These origins interact with specific initiator proteins such as the origin recognition complex (ORC) proteins (Bell and Stillman, 1992; Bell and Dutta, 2002). The majority of S. cerevisiae origins are small (∼150 base pairs), and most of them are characterized by an 11- to 17-base pair consensus element named autonomously replicating sequence.
In the fission yeast, Schizosaccharomyces pombe, origins are much larger and consist of multiple elements that each contribute partially to origin activity (Clyne and Kelly, 1995; Dubey et al., 1996; Chuang and Kelly, 1999). These elements are characterized by asymmetric AT stretches and are bound by ORC proteins (Clyne and Kelly, 1995; Dubey et al., 1996; Segurado et al., 2003), but a specific ORC binding sequence similar to the autonomously replicating sequence consensus sequence element in S. cerevisiae is not apparent.
The situation is even less defined for origins in metazoan chromosomes, which are estimated to be spaced ∼100 kb apart on average (Huberman and Riggs, 1968). To date, only 20 origins have been well characterized by multiple methods in different metazoans (12 in humans; Aladjem et al., 2006). Of these, only the lamin-B2 origin seems to correspond to a fixed initiation site (Abdurashidova et al., 1998). In contrast, the human b-globin, c-myc, and rDNA origins (Little et al., 1993; Waltz et al., 1996; Kamath and Leffak, 2001), as well as the Chinese hamster dihydrofolate reductase (DHFR) and rhodopsin (Dijkwel et al., 2000, 2002) origins, contain multiple inefficient initiation sites in zones that are ∼1 to >50 kb. There has not been a clear demonstration of an essential _cis_-acting genetic element (replicator) that requires ORC (initiator protein) binding for functional origin activity for mammalian origins. It is also not clear how many origins are reproducibly used during multiple cell cycles.
During the past 30 y, the most popular method to map origins of replication on mammalian chromosomes has been the polymerase chain reaction (PCR)-based quantification of origin-centered nascent strands (NS) peaks (Dijkwel et al., 1991; DePamphilis, 1993; Giacca et al., 1997). There are currently two different methods for purifying nascent strands. The first method is to pulse label nascent DNA with bromodeoxyuridine (BrdU), size select DNA strands of 0.5–2.5 kb on sucrose gradient, and purify BrdU-labeled nascent strands away from any unlabeled broken genomic DNA by immunoprecipitation with an anti-BrdU antibody. The second method takes advantage of the fact that nascent strands are resistant to digestion by lambda exonuclease (LExo) due to their 5′ RNA primers. Although both methods are acknowledged to have their own limitations, a head-to-head comparison of the two methods has not been published.
In the present study, we purified origin-centered nascent strands by both methods, each in duplicate and hybridized each preparation to high-resolution genome-tiling arrays covering 1% of the human genome, the subject of the National Institutes of Health ENCODE project. The studied area contains 0.5- to 2-Mb regions from 21 chromosomes and is sufficiently large and diverse to be a reliable sample of the whole genome (ENCODE Consortium, 2004). By comparing the two approaches, we identified 150 origins that initiate replication reproducibly across the methods and from well demarcated sites in multiple cells in the population. We also find that 93% of these origins associate with ORC subunits, Orc2 and Orc3. Interestingly, replication initiates close to transcription start sites and is also enriched around RNA polymerase (pol) II binding sites. We also found a link between origin activity and transcriptional regulation as these origins are preferentially seeded in the segments of the genome involved in recruiting transcription factors. Finally, the nucleosomes around the origins are rich in activating histone marks, suggesting a requirement for open chromatin to facilitate replication initiation. This careful survey of origins by two different methods of nascent strand purification filters out the efficient origins and characterizes their molecular determinants accurately.
MATERIALS AND METHODS
NS-BrdU Immunoprecipitation (BrIP) Nascent Strand Preparation
HeLa cells (108) were labeled with 100 μM BrdU for 30 min, and then genomic DNA was extracted. The BrdU-labeled DNA was separated from parental strands by boiling for 3 min and then size fractionated on a 5–20% sucrose gradient prepared in TNE (10 mM Tris, pH 8.0, 1 mM EDTA, and 0.3 M NaCl) for 20 h at 26,000 rpm in an SW55 rotor. BrdU-labeled nascent strands (0.5–2.5 kb) were then precipitated with a monoclonal anti-BrdU antibody (catalog no. 555627; BD Biosciences Pharmingen, San Diego, CA). The DNA recovered by immunoprecipitation was amplified in the linear range (14 cycles) by using a WGA2 kit from Sigma-Aldrich (St. Louis, MO) and was purified with a PCR cleanup kit (QIAGEN, Valencia, CA). Ten micrograms of amplified DNA was labeled and hybridized to the arrays as described previously (Karnani et al., 2007). A control sample representing total genomic DNA was similarly labeled and hybridized.
NS-LExo Nascent Strand Preparation
Genomic DNA was extracted from 108 HeLa cells in an RNase-free condition. Nascent DNA was released by boiling for 3 min, chilled on ice, and then loaded onto a neutral 5–20% sucrose gradient prepared in TNE. Gradients were centrifuged for 20 h at 26,000 rpm in an SW55 rotor at 4°C. Fractions corresponding to 0.5–2.5 kb were pooled, dialyzed against TE (10 mM Tris, pH 8.0 and 1 mM EDTA, pH 8.0), and precipitated with sodium acetate and ethanol. The DNA mixture was boiled for 3 min, chilled on ice, and phosphorylated with T4 polynucleotide kinase (New England Biolabs, Ipswich, MA). The reaction was stopped by the addition of 0.5% SDS/0.1 M EDTA. Proteinase K was added to 0.25 μg/ml, and the reaction was incubated at 50°C for 30 min. The reaction was diluted with 150 μl of TE, extracted once with phenol-chloroform, and sodium acetate and ethanol precipitated. The pellet was resuspended in 20 μl of sterile water. This DNA was digested overnight at 37°C in nuclease buffer with 2 μl of lambda exonuclease (10 U/μl; Epicenter Technologies, Madison, WI) as describe previously (Bielinsky and Gerbi, 1998). As a positive control for the completeness of the lambda exonuclease digestion, 100-base pair double-stranded, phosphorylated DNA (Supplemental Table 1) was spiked into nascent strands prep (Supplemental Figure 1D). Negative controls composed of either unphosphorylated 100-base pair double-stranded DNA or single-stranded RNA-DNA hybrid (first 17-nt RNA at the 5′ end and 83-nt DNA at the 3′ end) were independently spiked into nascent strands (Supplemental Figure 1, D and E). To test for RNase contamination in any of the enzymes and buffers, yeast tRNA was incubated independently with different enzyme/buffer for 12 h and analyzed on 2% agarose gel (Supplemental Figure 1A). The DNA recovered by lambda exonuclease digestion of nascent strands was amplified and hybridized to arrays as mentioned under NS-BrdU Immunoprecipitation (BrIP) Nascent Strand Preparation.
Microarray Hybridization
Nascent strands and genomic DNA were hybridized to ENCODE01-Forward (P/N 900543; Affymetrix, Santa Clara, CA) tiling arrays as described previously (Karnani et al., 2007). These arrays contain nonrepetitive, 25-mer oligonucleotide probe pairs (Perfect Match and Mis-Match control) spaced at an average distance of 22 base pairs from the central nucleotide. Each microarray was scanned and analyzed for signal intensities by GeneChIP Scanner 3000 and GeneChIP operating software software (Affymetrix). The primary data in the form of .cel files can be accessed at http://genome.bioch.virginia.edu/encode/origins/. Hybridization data were analyzed by Model-based analysis tool (MAT) for tiling arrays (Johnson et al., 2006) and genomic positions with a statistically significant enrichment (p ≤ 10−3, within a 1-kb window) of nascent strand signal over genomic control were flagged as nascent strand peaks. All the processed data have been generated using hg17 build (May 2004) of the Human genome assembly and can also be accessed at http://genome.bioch.virginia.edu/encode/origins/. The data will be made freely available through the ENCODE website at the time of publication of this article.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation (ChIP) assay was performed as per the protocol described previously (Trinklein et al., 2004), with a variation in the sonication step. Samples were sonicated (10 cycles of 15-s pulse at 50% amplitude and 45 s of cooling on ice) by using a microtip (3.2 mm; Branson Ultrasonics, Danbury, CT) and Model 500 Sonic Dismembrator (Thermo Fisher Scientific, Waltham, MA). The antibodies used for ChIP recognize Orc2 (BWH48) and Orc3 (BWH84) and have been validated extensively by us (Dhar et al., 2001). To determine the ChIP signal, 10 μl of ChIP DNA was amplified in a linear range (14 cycles) by using the WGA2 kit (Sigma-Aldrich) and cleaned by the PCR cleanup kit (QIAGEN). Two microliters of this purified DNA was used as template for semiquantitative PCR. The PCR reaction was set up in 20-μl reaction volume and subjected to 35 amplification cycles using LA Taq enzyme (Takara Bio USA, Madison, WI). Amplified fragments were analyzed on ethidium bromide stained 2% agarose gel. As a negative control, ChIP DNA from a rabbit immunoglobulin G (IgG) sample was amplified in a similar way. Ten percent input was used as template in the input lanes. The details on primers used for Orc-ChIP assay are provided in Supplemental Table 4.
Calculation of Interorigin Distances
For a given set of origins, the origins were segregated into the 44 different Encode regions. For each ENCODE region with n origins, interorigin distances were calculated for n − 1 distances (Table 1). The interorigin distance calculation had to ignore lengths of the ENCODE regions that were outside the outermost origins in a given segment. Thus 43 and 23% of base pairs within region interrogated were not covered by the interorigin intervals in NS-LExo and NS-BrIP, respectively.
Table 1.
Summary of replication origins identified by different origin mapping methods in HeLa cells
| Originmappingmethod | No. oforigins | Interorigindistance (bp) | Length oforigins (bp) | ||
|---|---|---|---|---|---|
| Mean | Median | Mean | Median | ||
| NS-LExo | 320 | 58,442 | 28,060 | 1,374 | 1,275 |
| NS-BrIP | 815 | 27,595 | 16,163 | 1,654 | 1,555 |
Calculation of AT Content
To treat all origins homogenously for AT content analysis, the peak position identified by MAT algorithm was selected to represent the location of the origin site and extended 100 bp on each side. The AT content was then calculated as the sum of As and Ts divided by the sum of As, Ts, Cs, and Gs and expressed as a percentage for each site.
A random model was generated for the NS-BrIP and NS-LExo data sets by the following method. The origin sites were randomly placed within the ENCODE regions such that no two origin sites actually overlap or lie within a specified distance of one another. This distance was approximately similar to the minimum interorigin distance observed in the original data set. Next, the AT content analysis described above was performed for this random origin set. This randomization was iterated 1000 times to produce a random null distribution. Then, the observed AT content for the given origin set was compared with the mean and SD of the random distribution, and a p value was determined that conveys the chance of observing this value by chance given the null background.
Comparative Analysis with Genome Features
Data sets for genomic features such as transcription start sites (TSSs), RNA polymerase II binding sites (RNA pol II), DNase I HS (DHS), CpG islands, regulatory factor binding region (RFBR), histone modification marks, and replication timing were downloaded from the UCSC genome browser (the ENCODE consortium, http://genome.ucsc.edu/ENCODE/). For comparative analysis, replication origins were treated as the source data set and the genomic feature to which it was compared was referred as the target data set. End points were determined for each origin in the source set and these were compared with the target set to find how many origins intersected/lay within a specified distance of the nearby target sites.
A random model was generated for each source/target comparison by the following method. The minimum distance between two origins within the source data set was determined. Then, the source sites were randomly placed within the ENCODE regions universe such that no two source sites lay within a distance of one another less than this minimum distance. The source/target analysis described above was then performed for this randomized source set against the fixed target set. This randomization was iterated 9999 times, and each iteration was checked to see how many source sites hit target sites. The p value reports the number of random iterations that achieved a higher number of hits than the actual source set. For example a p value <0.0005 indicates that there is a 5 in 10,000 observed occurrence of recovering a hit rate higher than the actual hit rate of the given source set by random chance.
Comparative Analysis with Conserved Elements (CEs)
The CEs correspond to three conservation algorithms (phastCons, binCons, and GERP) and three sequence alignment methods (TBA, MLAGAN, and MAVID) applied to the ENCODE region sequences of 28 vertebrate species (Margulies et al., 2007). For comparison with origins, we choose the moderate stringency data set of CE that were derived from bases shown to be constrained by at least two of the three conservation algorithms on at least two of the three alignments. For comparison with the conserved elements of genome, nascent strands were checked for any base pairs overlap with the conserved elements. The random model for this comparison was generated as mentioned for the source/target comparison described above.
RESULTS
Mapping Replication Origins Using Nascent Strands Purified by NS-LExo and NS-BrIP Methods
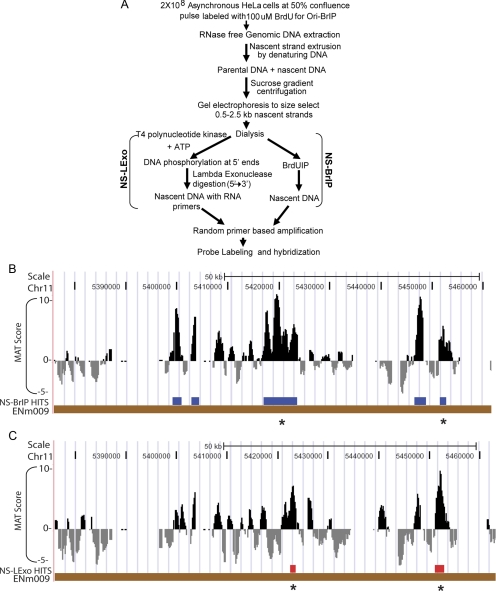
The two methods of purifying nascent strands are described in Figure 1A. In NS-BrIP, nascent strands are purified from contaminating genomic DNA by size selection (0.5–2.5 kb) of denatured DNA followed by BrdU immunoprecipitation (Pelizon et al., 1996). However, this method has the limitation of not completely removing contaminating nicked BrdU-labeled DNA. The NS-LExo method enriches for nascent strands because they are resistant to digestion by lambda exonuclease due to their 5′ RNA primers (Bielinsky and Gerbi, 1998). However, variable efficiency of the lambda exonuclease enzyme or contamination of RNAses in the buffers and enzymes used in this method introduce noise in the nascent strand preparations. Because each method has its own strengths and limitations, we isolated origin-centered nascent strands from asynchronous cells by both methods independently to get the most accurate view of the distribution and reproducibility of origins on a genome-wide scale. As a control for the completeness of the lambda exonuclease digestion, phosphorylated double-stranded 100-base pair DNA was spiked into nascent strands preparation before adding lambda exonuclease (Supplemental Table 1 and Supplemental Figure 1D). Complete digestion of this spiked DNA confirmed the lambda exonuclease activity in the reaction. Unphosphorylated 100-base pair double-stranded DNA or 100-base pair single-stranded RNA-DNA hybrid spiked controls were not digested by the exonuclease in parallel reactions (Supplemental Table 1 and Supplemental Figure 1, D and E). In addition, all buffers and enzymes were checked for any RNAase contamination by incubation with tRNA (Supplemental Figure 1A).
Figure 1.
(A) Schematic of NS-BrIP and NS-LExo methods used to purify nascent strands and map replication origins. (B and C) NS peaks from array hybridizations. UCSC browser image showing MAT score for enrichment of nascent strand signal relative to control, and nascent strand hit positions identified for NS-BrIP and NS-Lexo on chromosome 11 (ENCODE region ENm009). The nascent strand hits had signal intensity that was enriched above the background at p ≤ 10−3. Nascent strand hits that overlapped across the NS-Lexo and NS-BrIP methods are indicated by asterisk.
Enrichment of known origins b-globin and c-myc in nascent strands purified by both NS-BrIP and NS-LExo methods provided a check for the quality of the nascent strand preparations (Supplemental Figure 1, F and G). Two biological replicates of nascent strands prepared by a given method and total genomic DNA controls were hybridized independently to high-density tiling arrays (25-mer oligonucleotide probes with an average spacing of 22 base pairs) representing the nonrepetitive sequence of the 30-Mb ENCODE region (ENCODE Consortium, 2004; Karnani et al., 2007). Hybridization data were analyzed by using the Model-based Analysis of Tiling (MAT) arrays tool (Johnson et al., 2006) and genomic positions with a statistically significant enrichment (p ≤ 10−3) of nascent strand signal over genomic control (1-kb window) were flagged as origins (Figure 1, B and C). Hybridization with the HeLa NS-BrIP and NS-LExo identified 815 and 320 nascent strand peaks, respectively, with median lengths of 1275 and 1555 base pairs (Table 1).
The median interorigin distances calculated for NS-BrIP and NS-LExo were <30 kb (NS-BrIP, 16.1 kb and NS-LExo, 28.1 kb; Table 1). The shorter interorigin distance for NS-BrIP was due to the higher number of nascent stand peaks detected by this method. We also found interorigin distance to be >100 kb for 5–17% of NS-BrIP and NS-LExo peaks, respectively (Supplemental Figure 2, A and B).
Validation of Nascent Strand Peaks Identified by Microarray Hybridization
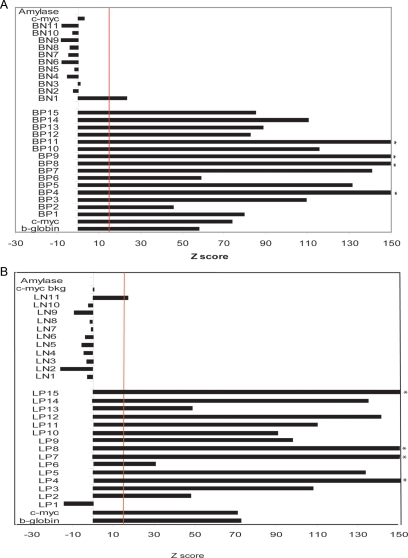
To assess the sensitivity and specificity of the array hybridizations, NS-BrIP and NS-LExo sites were validated by quantitative PCR (qPCR) by using two independent biological replicates of nascent strand preparations for each method (see primer details in Supplemental Tables 2 and 3). Each nascent strand preparation was checked for quality by performing qPCR for the b-globin and c-myc origins as well as their background control regions, amylase and c-myc background, respectively. Average Z scores of biological replicates for the known origins and their respective control background regions were calculated and the threshold for calling a site positive was set at ≥15 standard deviations higher than the background control regions (in this case amylase; Figure 2, A and B). Of the 15 microarray-positive calls tested (randomly selected from different chromosomes), the true positives for NS-BrIP and NS-LExo were 15 and 14, respectively. For the 11 randomly selected regions that did not show enrichment of nascent strand peaks by microarray analysis, the true negatives were 10 for NS-BrIP and 11 for NS-LExo (Figure 2, A and B). Based on these validation numbers the specificity and sensitivity for NS-BrIP method are 100 and 94%, whereas that for NS-LExo are 92 and 100%.
Figure 2.
Validation by qPCR confirms microarray data. Positive and negative sites detected by hybridization of nascent strands to microarrays were randomly selected from different chromosomal fragments and tested for enrichment (nascent strands over genomic) by qPCR by using two biological replicates of nascent strand preparations. qPCR for each biological replicate was performed in triplicate. Average Z scores of biological replicates were plotted for all the tested chromosomal regions as well as the known origins (b-globin and c-myc origins) and their respective background control regions (amylase and cymc background). The threshold for calling a microarray site positive was set at ≥15 standard deviations higher (vertical red line) than the background control regions (in this case amylase). Validation for NS-BrIP microarray data (primers to test array positives BP1 … BP15 and array negatives BN1 … BN11) (A) and validation of NS-LExo data set (primers to test array positives LP1 … LP15 and array negatives N1 LN11) (B). Asterisk, >150 Z score.
We also used a range of primers along a genomic region to check whether the signals from qPCR formed a peak at a position similar to that identified by the microarray results. Two primer pairs spanning the microarray peak and additional sets of primers (at 1-kb interval) covering 5 kb on either side of the peak were designed. As expected both NS-BrIP and NS-LExo methods gave specific peaks in the tested region and this peak corresponded with the peak position identified through microarray analysis (Supplemental Data 1, H–J).
Correlation with Genomic Features Indicates a Link between Replication Initiation and Transcription Initiation and Regulation
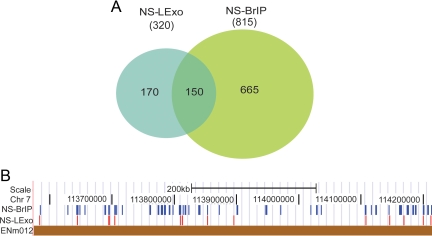
Because both the methods of nascent strand purification have their strengths and weaknesses we intersected the hybridization data from the two methods and identified 150 nascent strand peaks that reproducibly intersected at or within 2.5 kb (upper limit of nascent strand purification through sucrose gradient) across the methods (Figure 3, A and B). Note that in Figure 1, B and C, there is an example of a region from chromosome 11 where there were three peaks in the NS-BrIP panel that could score positive by NS-LExo method if we lowered the threshold for calling a positive site. We tested whether better concordance can be obtained between the two methods by relaxing the stringency for NS-LExo to p ≤ 0.01 and comparing it with BrIP peaks identified at p ≤ 0.001. Even though we gained on two of the three peaks in the chromosome 11 region shown in Figure 1C, the overall NS-LExo concordance with NS-BrIP dropped by 11% and the false discovery rate doubled, suggesting inclusion of more false positives. Hence, we proceeded with p ≤ 0.001 cut-off for both the methods. We named the intersection data set of the two methods as origins (ORIs) and explored their characteristics by comparing them with other functional features of the genome.
Figure 3.
Concordance of origins across the methods. (A) Venn diagram showing the overlap between NS-LExo and NS-BrIP methods. The number of fragments found in each group is shown between brackets and the number of overlapping segments is given in the intersected regions. (B) Browser image showing a comparison of the origins identified by NS-BrIP and NS-LExo methods within a 400-kb region of chromosome 7 (ENCODE region ENm012).
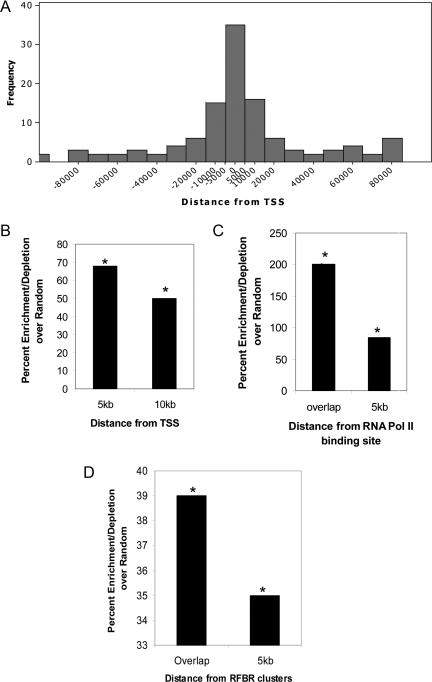
Studies in the past have proposed a connection between transcriptional machinery and replication initiation. The nonspecific initiation observed in Xenopus egg extracts can be localized by the assembly of a transcription domain (Hyrien et al., 1995; Danis et al., 2004). In addition transcription from the DHFR promoter in Chinese hamster ovary cells acts to regulate and define the boundaries of initiation zones (Saha et al., 2004). Finally, Drosophila ORC binding sites have been found to associate with RNA pol II binding sites (MacAlpine et al., 2004). These observations led us to check for any possible link between ORI activity and transcription initiation. Interestingly, these initiation peaks were enriched (68%) in the genomic segments 5 kb upstream or downstream of the TSS, and this correlation was highly significant compared with random (p < 0.0001; Figure 4, A and B). A similar distribution was also noted when the analysis was confined to active TSSs in HeLa cells (data not shown). Consistent with the proximity of ORIs to transcription start sites, these peaks were significantly enriched at or near (≤5 kb) RNA polymerase II binding sites (Figure 4C). The exact concordance was up to 31%, but the enrichments relative to a random model were large (75–175%) and significant (p < 0.0015).
Figure 4.
Correlation of ORI with transcription initiation and transcription factors. (A) Frequency-histograms showing distribution of replication initiation sites with respect to distance from TSS. (B) Enrichment relative to random model of the localization of ORI with respect to TSS at 5 and 10 kb. (C) Origin activity at or within 5 kb of RNA pol II binding sites. (D) Origin activity relative to RFBR clusters. Data in B–D are expressed as percentage of enrichment (or depletion) in experimental data minus the percentage of enrichment (or depletion) in the random model. Asterisk indicates statistically significant enrichment/depletion relative to the random model (p range < 0.01–0.0001).
Transcription factors recognize and bind to specific DNA sequences and hence play a major role in regulating transcription. In addition to transcriptional regulation, these factors directly or indirectly recruit histone-modifying enzymes and chromatin remodeling factors to alter the chromatin structure and hence influence multiple cellular processes including DNA replication. The β-globin locus control region (LCR), which is located >20 kb away from the replication origins and yet regulates its activity, is a classic example of such a link between transcriptional regulation and DNA replication (Forrester et al., 1990; Aladjem et al., 1995). Some of the metazoan origins that have been highly characterized have been shown to be located near a variety of transcription factor binding sites. The human c-Myc origin binds E2F proteins, Drosophila angiotensin-converting enzyme binds c-Myb homologues and Rb, and the LMNB2 origin associates with USF and SP1 (Biamonti et al., 1992; Dimitrova et al., 1996; Bosco et al., 2001; Maser et al., 2001; Beall et al., 2002). Recently, under ENCODE consortium 689 high-density transcription factor binding clusters were identified in 1% genome. These clusters were named RFBRs and were generated after pooling the ChIP-chip data for 29 different transcription factors. The distribution of RFBRs is nonrandom (ENCODE Project Consortium et al., 2007) and correlates with the positions of TSSs. We examined the positions of replication origins in the genome with respect to these clusters and found 20% of the ORIs (p < 0.04; Figure 4D) and ∼50% of early firing ORIs (p < 0.005, using early replicating genomic regions for generating random model) to initiate within 5 kb of such clusters. On comparison with genes transcribed in exponentially growing HeLa cells, we found >60% of RFBR associated ORIs to be located near actively transcribing genes (data not shown). These results indicate that these RFBRs might play a dual role of regulating transcription as well as replication initiation along the human genome.
Chromatin Signature Influences Replication Initiation
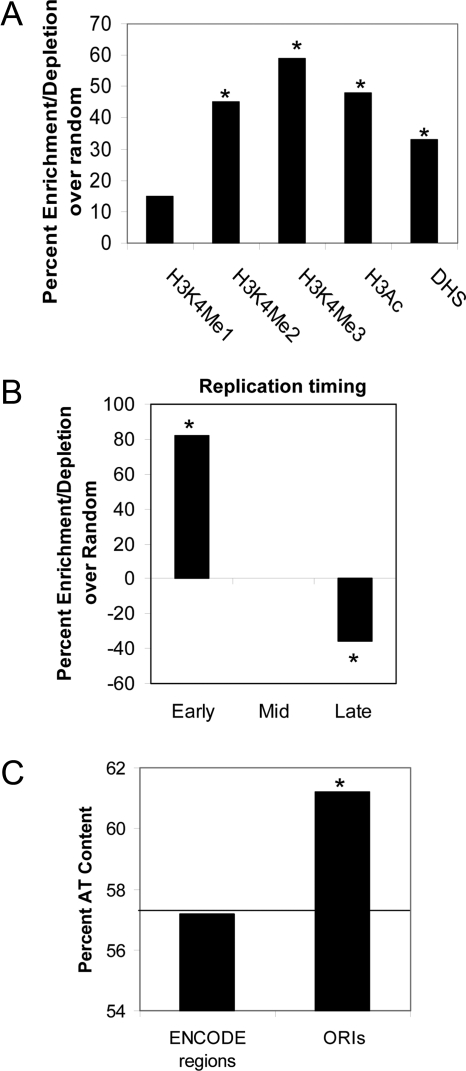
Due to the lack of consensus sequence, one of the biggest challenges in the replication field has been to determine how ORC binds to specific regions in the metazoan genome. One reasoning could be the involvement of chromatin structure. The chemical modifications on histones in the nucleosomes surrounding these origins would favor open chromatin and facilitate recruitment of ORC complex. To test this hypothesis, we analyzed the histone marks around the ORIs. The distribution of several of these modifications in HeLa have been mapped by the ENCODE consortium, allowing us to do this analysis (ENCODE Project Consortium et al., 2007; Karnani et al., 2007). As shown in Figure 5A, the origins were enriched (p < 0.0001) in three different active histone marks (H3K4 methylation and H3Ac). The preference for proximity to these modifications was higher for H3K4Me3, H3K4Me2, and H3Ac compared with H3k4Me1. This observation mirrors the chromatin signature requirement for transcription initiation and transcriptional regulation as both TSS and DNAse I hypersensitivity (DHS) are also known to tightly associate with H3K4Me3, H4K4Me2, and H3Ac modifications but have weak correlations with H3K4Me1 (ENCODE Project Consortium et al., 2007). Because all these functional elements are interconnected, we anticipated ORI activity to also associate with DHS sites. In support of this, we found ∼40% ORI to lie within 5 kb of DHS sites (p < 0.005; Figure 5A).
Figure 5.
(A) Distribution of histone marks and DHS sites around ORIs. (B) Histograms showing whether origins were selectively enriched or depleted in chromosomal segments known to replicate in different parts of S phase: early, mid, late as defined previously (Karnani et al., 2007). (C) Correlation between replication origins and AT content. The horizontal line indicates the percent AT content of the ENCODE region (1% human genome). Data in A and B are expressed as percentage of enrichment (or depletion) in the experimental data minus that in the random model. Asterisk indicated significant enrichment/depletion relative to random model (p < 0.01–0.0001).
Late Firing Origins Are Less Efficient
Replication initiates from a number of potential sites on a chromosomal locus, with the most efficient initiation events being triggered from the same origin across a large cell population. Efficient origins are therefore expected to be reproducibly identified using multiple nascent strand purification methods. ORIs represent this pool of origins as they were identified by their reproducibility/proximity across the two nascent strand purification methods. We compared the ORIs with our previously published replication timing data for the ENCODE regions to see whether efficient replication initiation occurs during any particular part of S phase. We had divided the synchronously replicating areas of the ENCODE regions into thirds, as replicating early, mid-, and late in S phase (Karnani et al., 2007). The temporal segregation of ORIs identified 49% of the origins to be located in the early replicating chromosomal segments (p < 0.0001 and 84% enrichment over random; Figure 5B). The mid- and late-replicating regions of the genome contained 30 and 21% of the ORIs, respectively, but the mid-S phase ORIs failed to show any enrichment relative to random expectation, whereas the late ORIs were disenriched (mid-ORIs: p < 0.5 and enrichment = 0% late ORIs: p < 0.004 and depletion = 36%, Figure 5B). This is similar to the observations from a genome-wide analysis of fission yeast origin efficiencies (Heichinger et al., 2006), although a contradicting study has also been published (Eshaghi et al., 2007).
Replication Initiation Peaks Are AT Rich and Show Evolutionary Conservation
Until now a metazoan DNA replication origin consensus sequence has not been identified, but most of the studied origins contain AT-rich regions (Aladjem et al., 2006). We checked whether the origins had any preference for AT abundance. To treat the origins homogenously, peak positions of all the origins were extended 100 bp on either side. The mean AT content of the origin sequences was 61% compared with 57% for all sequences in the ENCODE region (p < 10−6; Figure 5C). This AT enrichment does not dictate origin efficiency as this increase in AT content was also seen in NS-LExo– and NS-BrIP–specific nascent strand peaks (discussed in the following sections).
To determine whether sites of replication initiation were under any evolutionary selection, we performed an intersection between the origins and the CEs identified under ENCODE by using genomes of 28 vertebrate species (Margulies et al., 2007), (details under Materials and Methods). More than 50% of origins overlapped with CEs, and this intersection was significant compared with random (p < 0.001).
Orc2 and Orc3 Localize to ORI Sites
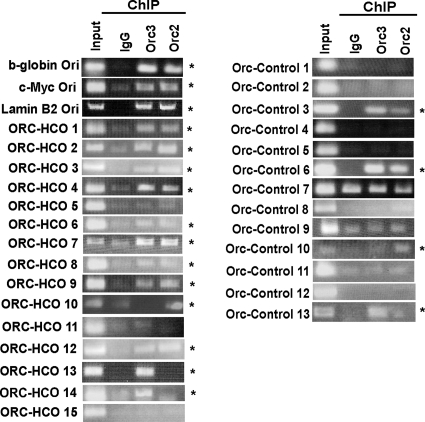
To validate the NS peaks by another origin-mapping method, we performed chromatin immunoprecipitation assay by using antibodies against ORC subunits. Orc2 and Orc3 are the core subunits of ORC complex and are known to associate with each other (Dhar et al., 2001; Vashee et al., 2003). ChIPs with antibodies against Orc2 or Orc3 were tested for enrichment of the site of replication initiation for 15 ORIs and for 13 nascent strand free sites. As positive controls for the assay, ORC binding was tested and found to be positive for the b-globin and c-myc origins.
Twelve of the 15 (80%) of the ORIs peaks had an ORC binding site (Figure 6). Of the three sites that were negative for ORC binding, an additional two sites had an ORC binding site within 1 kb of the nascent strand peak (Supplemental Figure 3). Thus, 93% of the tested ORIs identified in this study have ORC binding sites at or within 1 kb of the nascent strand peak. In contrast no Orc binding was observed for 70% of the nascent strand negative regions (Figure 6). The 30% of sites that did not have NS peaks but still bound ORC, suggesting that there are more ORC binding sites compared with initiation sites on a chromosome.
Figure 6.
Validation of ORIs by ORC ChIP. (A) Fifteen ORIs were tested for ORC binding by performing ChIP assays with Orc2 and Orc3 antibodies. Rabbit IgG was used as a measure of nonspecific chromatin precipitation. b-globin, c-myc, and lamin B2 origins were used as positive controls. Origins that were positive for one or both ORC subunits are indicated with an asterisk. (B) Thirteen sites that were not called as ORIs by microarray data were used as negative controls. The ethidium bromide gel documentation images were stretched or reduced in width to make all panels the same size.
Comparison of Two Methods of Nascent Strand Isolation Identifies Unique Properties of Each Method
As mentioned above, the two methods of nascent strand purification identified independent sets of nascent strand peaks on hybridization to the arrays. Forty-seven percent of NS-LExo sites either overlapped or were within 2.5 kb of NS-BrIP sites. We used these 150 overlapping nascent strand peaks/ORIs as the efficient origin pool of the total peaks identified by the two methods. However, there were still subsets of peaks that were specific to each method (Figure 3A). We classified these as NS-LExo–specific and NS-BrIP–specific sites.
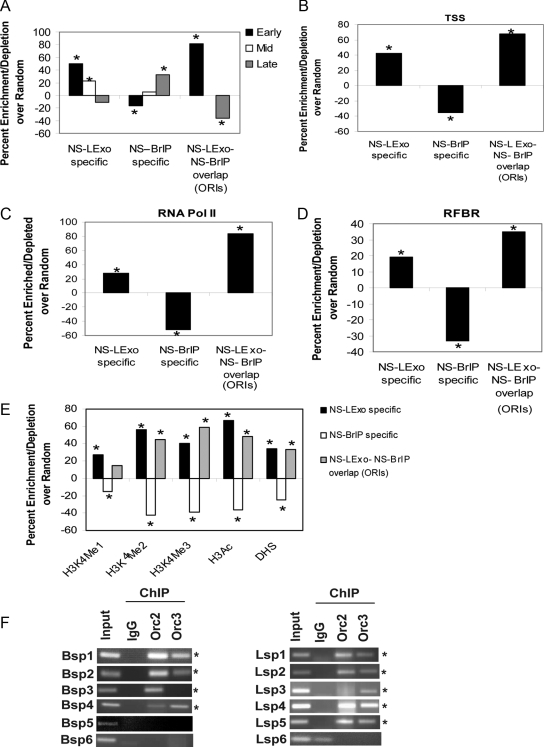
A very interesting feature appeared upon comparison of the NS-LExo– and BrIP–specific sites. The NS-LExo–specific peaks were very similar to ORIs as they primarily represented initiation events occurring in early S phase (Figure 7A) and preferred to localize in open parts of the genome that were close to transcription start sites, RNA polymerase II binding sites, RFBRs, activating histone marks, and DNAse I hypersensitivity sites (Figure 7, B–E). The enrichment of the experimental set in each of these correlations was statistically significant relative to the random, although the magnitude of the enrichment was lower compared with ORIs.
Figure 7.
Comparison of genomic features specific to two methods of nascent stand purification. (A) Segregation of NS-method specific peaks and ORIs into different temporal classes of S phase (early, mid, and late). (B) Localization of NS-method–specific peaks and ORIs with respect to TSS at 5 and 10 kb. (C) NS-method peaks and ORIs at or within 5 kb of RNA pol II binding sites. (D) RFBR clusters. (E) Histone marks and DHS sites. Data in A–E are expressed as percent enrichment (or depletion) minus that in the random model. Asterisk indicated significant enrichment/depletion relative to random model (p range < 0.01–0.0001). (F) Validation of NS-BrIP– and NS-LExo–specific peaks by ORC ChIP. Six nascent strand peaks each from NS-BrIP– (Bsp1–6) and NS-LExo–specific (Lsp1–6) categories were tested for ORC binding by performing ChIP assays with Orc2 and Orc3 antibodies. Rabbit IgG was used as a negative control. Nascent strand peaks that were positive for one or both ORC subunits are indicated with an asterisk. The ethidium bromide gel documentation images were stretched or reduced in width to make all panels the same size.
In contrast, NS-BrIP–specific sites initiated replication in late S phase and were significantly depleted in sites of transcription initiation and open chromatin structure (Figure 7, B–E). The reason for this unique property of NS-BrIP–specific sites is discussed in the following section.
Interestingly, both NS-LExo– and NS-BrIP–specific peaks were significantly AT rich just like ORIs (NS-LExo specific, 60%, p <0.0001; NS-BrIP specific, 63%, p <1015; ENCODE regions, 57%) and also had significant affinity for evolutionary conserved segments of the genome (NS-LExo specific, 59%, p < 0.0001; and NS-BrIP specific, 47%; p < 0.004).
Because the two classes of method-specific sites showed such different properties, we asked whether these were bona fide origins by exploring ORC occupancy on these sites. Six primer pairs were designed for six each of the NS-BrIP and NS-Lexo sites. These primers amplify ∼200-bp fragments spanning the nascent strand peak positions. As is evident from the Figure 7F, 67% of the tested BrIP-specific and 83% of the LExo-specific sites had ORC binding on the chromatin. Because nascent strands were in the size range of 0.5–2.5 kb, it is quite possible that ORC binding may have been missed at some of the tested sites. This can be checked in future by doing more elaborate experiments such as a primer walk around the nascent strand peaks or ChIP on chip assays.
DISCUSSION
This study is the first direct comparison of two methods of nascent strand purification for origin identification. As expected both the methods show overlapping and method-specific sites with nascent strand peaks. NS-LExo–specific peak show similar features as the overlapping data set (ORIs), but both these sets are depleted in late replicating origins. Conversely, NS-BrIP method captures both the early and late initiation events in the genome but the late firing nascent strand peaks dominate the NS-BrIP population. Both the method-specific peaks show ORC occupancy at 67–83% of nascent strand peak positions, so we believe that the majority of these sites are bona fide origins and yet are picked up by one method preferentially over the other. One explanation for the higher number of NS-BrIP sites is that the late firing origins are highly inefficient and BrdU-IP is a more sensitive assay to purify low abundance nascent strand peaks. This would suggest that an analysis that combines the two methods of nascent strand purification is useful in distinguishing efficient, reproducibly used replication origins of the human genome. By using this criterion, we have identified 150 new origins of replication in the ENCODE area and examined their molecular determinants on a genome-wide scale.
Our study suggests that early replication is a strong determinant of origin activity and efficiency. The preference of replication initiation for open chromatin and proximity to TSS and RFBR (dense transcription factor binding regions of genome) suggest that pre-replicative complexes bound at or near these functional elements of genome take advantage of the nucleosome-free local environment to initiate replication. This observation is in conformity with the findings from ORC binding studies in Drosophila and recent genome-wide nascent strand hybridization studies done in mouse embryonic stem cells (MacAlpine et al., 2004; Sequeira-Mendes et al., 2009).
Although replication initiation in metazoans is quite inefficient, there is an ongoing debate as to whether the DNA sequence is critical for origin selection. Sequence analysis of the DHFR-ori β, HBB, and laminB2 origins have revealed the existence of AT-rich stretches and asymmetric purine:pyrimidine tracks (AG) (Aladjem et al., 2006). Consistent with this, we find ORI peaks to be enriched in AT content even though most of these ORIs are located in GC-rich early replicating regions. Thus, an AT-rich local sequence may facilitate origin opening or helicase loading.
Recently, Lucas et al. (2007) investigated replication origins in the human lymphoblastoid cell line 11365. They hybridized nascent strands purified by BrIP method to the microarrays and identified 28 new origins that were reproducible in two biological replicates of BrIP preparation. The b-globin locus was the only genomic region that was covered by their and our studies. In the b-globin locus they identified two new origins (Chr11: 5209792-5211028 and Chr11: 5217893-5223314; Supplemental Figures S6, A and B). Both of these origins were identified by our NS-BrIP method in HeLa cells and were part of the same initiation zone (Supplemental Figure 4, A and B). Consistent with the idea that NS-BrIP alone is sensitive enough to identify low abundance, late-replicating origins, NS-LExo failed to detect a nascent strand peak in b-globin origin, known to fire late in S phase in HeLa cells.
Another study published by Cadoret et al. (2008) used the NS-LExo method in HeLa S3 cells (suspension cells that differ from HeLa adherent cells used here) and mapped NS peaks within ENCODE regions. We compared our data with their findings and found that <14% of our NS-LExo peaks were within 2.5 kb of nascent strand peaks in their findings (data not shown). To further investigate this low concordance between the two studies, we performed qPCR by using the primers Cadoret et al. (2008) used to validate their microarray results. Cadoret et al. (2008) plotted the qPCR enrichment of NS peaks (nascent strands/genomic ratio) relative to the c-myc origin. NS peaks that showed any qPCR signal enrichment relative to the c-myc background signal were considered to be validated origins of replication. Our qPCR analysis showed that only 2/15 (13.3%) of their origins were enriched in our NS-LExo nascent strand preparations even after using their threshold for a positive call (Supplemental Figure 5). Major causes of the low concordance rate between the two studies could be biological or technical. Of the biological differences, the most important is the difference in cell lines used: HeLa adherent by us versus HeLa S3 by them. The technical differences include the following. 1) Cadoret et al. (2008) used linear amplification of the nascent strand preparation using RNA polymerase, whereas we used 14 cycles of PCR amplification as done by most of the groups under the ENCODE consortium. 2) Because of the lower yield of nascent strands, a low cut-off had to be used by Cadoret et al. (2008) to validate the origins of replication by qPCR, i.e., any enrichment ≥ c-myc background sample, increasing the possibility of a higher false-positive rate. In our study, we find >90% of origins have nascent strand enrichment ≥15 standard deviations above the background site used by Cadoret et al. (2008) as the cut-off. 3) Differences stemming from the array platforms used (Affymetrix vs. Agilent Technologies, Santa Clara, CA). Clearly, the differences have to be explored further, but in our study we used two independent methods of nascent strand preparation (LExo and BrIP), each as two independent biological replicates for a total of four biological replicates, selected a more stringent criterion for making a positive call in qPCR, and validated a subset of our ORIs by ORC ChIP assays.
In summary, we have generated a catalogue of origins that contains both efficient and inefficient origins of the genome. A high level of inefficiency in initiation events dictates that origins have to be determined experimentally and so are very different from genes that happen to be fixed entities of the genome. This inventory of origins has been created for 1% of the genome in HeLa cells, but by applying this approach to the rest of the genome, to other cell lines, and to cells undergoing differentiation, we expect to delineate housekeeping origins from cell type-specific origins. Finally, we can now investigate how replication stress affects origin firing and which origins can act as hot spots for rereplication and be involved in events that lead to genomic instability.
Supplementary Material
[Supplemental Materials]
ACKNOWLEDGMENTS
We thank all the members of the Dutta laboratory for helpful discussions. This work was supported by National Institutes of Health grants HG-003157 and CA-60499 (to A. D.).
Footnotes
REFERENCES
- Abdurashidova G., Riva S., Biamonti G., Giacca M., Falaschi A. Cell cycle modulation of protein-DNA interactions at a human replication origin. EMBO J. 1998;17:2961–2969. doi: 10.1093/emboj/17.10.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aladjem M. I., Falaschi A., Kowalski D. Eukaryotic DNA replication origins. In: DePamphilis M.L., editor. DNA Replication Hum. Dis. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2006. pp. 31–61. [Google Scholar]
- Aladjem M. I., Groudine M., Brody L. L., Dieken E. S., Fournier R. E., Wahl G. M., Epner E. M. Participation of the human beta-globin locus control region in initiation of DNA replication. Science. 1995;270:815–819. doi: 10.1126/science.270.5237.815. [DOI] [PubMed] [Google Scholar]
- Beall E. L., Manak J. R., Zhou S., Bell M., Lipsick J. S., Botchan M. R. Role for a Drosophila Myb-containing protein complex in site-specific DNA replication. Nature. 2002;420:833–837. doi: 10.1038/nature01228. [DOI] [PubMed] [Google Scholar]
- Bell S. P., Dutta A. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- Bell S. P., Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- Biamonti G., Perini G., Weighardt F., Riva S., Giacca M., Norio P., Zentilin L., Diviacco S., Dimitrova D., Falaschi A. A human DNA replication origin: localization and transcriptional characterization. Chromosoma. 1992;102:S24–S31. doi: 10.1007/BF02451782. [DOI] [PubMed] [Google Scholar]
- Bielinsky A. K., Gerbi S. A. Discrete start sites for DNA synthesis in the yeast ARS1 origin. Science. 1998;279:95–98. doi: 10.1126/science.279.5347.95. [DOI] [PubMed] [Google Scholar]
- Bosco G., Du W., Orr-Weaver T. L. DNA replication control through interaction of E2F-RB and the origin recognition complex. Nat. Cell Biol. 2001;3:289–295. doi: 10.1038/35060086. [DOI] [PubMed] [Google Scholar]
- Cadoret J. C., Meisch F., Hassan-Zadeh V., Luyten I., Guillet C., Duret L., Quesneville H., Prioleau M. N. Genome-wide studies highlight indirect links between human replication origins and gene regulation. Proc. Natl. Acad. Sci. USA. 2008;105:15837–15842. doi: 10.1073/pnas.0805208105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang R. Y., Kelly T. J. The fission yeast homologue of Orc4p binds to replication origin DNA via multiple AT-hooks. Proc. Natl. Acad. Sci. USA. 1999;96:2656–2661. doi: 10.1073/pnas.96.6.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clyne R. K., Kelly T. J. Genetic analysis of an ARS element from the fission yeast Schizosaccharomyces pombe. EMBO J. 1995;14:6348–6357. doi: 10.1002/j.1460-2075.1995.tb00326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danis E., Brodolin K., Menut S., Maiorano D., Girard-Reydet C., Mechali M. Specification of a DNA replication origin by a transcription complex. Nat. Cell Biol. 2004;6:721–730. doi: 10.1038/ncb1149. [DOI] [PubMed] [Google Scholar]
- DePamphilis M. L. Origins of DNA replication in metazoan chromosomes. J. Biol. Chem. 1993;268:1–4. [PubMed] [Google Scholar]
- Dhar S. K., Delmolino L., Dutta A. Architecture of the human origin recognition complex. J. Biol. Chem. 2001;276:29067–29071. doi: 10.1074/jbc.M103078200. [DOI] [PubMed] [Google Scholar]
- Dijkwel P. A., Mesner L. D., Levenson V. V., d'Anna J., Hamlin J. L. Dispersive initiation of replication in the Chinese hamster rhodopsin locus. Exp. Cell Res. 2000;256:150–157. doi: 10.1006/excr.2000.4809. [DOI] [PubMed] [Google Scholar]
- Dijkwel P. A., Vaughn J. P., Hamlin J. L. Mapping of replication initiation sites in mammalian genomes by two-dimensional gel analysis: stabilization and enrichment of replication intermediates by isolation on the nuclear matrix. Mol. Cell. Biol. 1991;11:3850–3859. doi: 10.1128/mcb.11.8.3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkwel P. A., Wang S., Hamlin J. L. Initiation sites are distributed at frequent intervals in the Chinese hamster dihydrofolate reductase origin of replication but are used with very different efficiencies. Mol. Cell. Biol. 2002;22:3053–3065. doi: 10.1128/MCB.22.9.3053-3065.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova D. S., Giacca M., Demarchi F., Biamonti G., Riva S., Falaschi A. In vivo protein-DNA interactions at human DNA replication origin. Proc. Natl. Acad. Sci. USA. 1996;93:1498–1503. doi: 10.1073/pnas.93.4.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey D. D., Kim S. M., Todorov I. T., Huberman J. A. Large, complex modular structure of a fission yeast DNA replication origin. Curr. Biol. 1996;6:467–473. doi: 10.1016/s0960-9822(02)00514-6. [DOI] [PubMed] [Google Scholar]
- ENCODE Consortium. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- ENCODE Project Consortium et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshaghi M., Karuturi R. K., Li J., Chu Z., Liu E. T., Liu J. Global profiling of DNA replication timing and efficiency reveals that efficient replication/firing occurs late during S-phase in S. pombe. PLoS ONE. 2007;2:e722. doi: 10.1371/journal.pone.0000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrester W. C., Epner E., Driscoll M. C., Enver T., Brice M., Papayannopoulou T., Groudine M. A deletion of the human beta-globin locus activation region causes a major alteration in chromatin structure and replication across the entire beta-globin locus. Genes Dev. 1990;4:1637–1649. doi: 10.1101/gad.4.10.1637. [DOI] [PubMed] [Google Scholar]
- Giacca M., Pelizon C., Falaschi A. Mapping replication origins by quantifying relative abundance of nascent DNA strands using competitive polymerase chain reaction. Methods. 1997;13:301–312. doi: 10.1006/meth.1997.0529. [DOI] [PubMed] [Google Scholar]
- Heichinger C., Penkett C. J., Bahler J., Nurse P. Genome-wide characterization of fission yeast DNA replication origins. EMBO J. 2006;25:5171–5179. doi: 10.1038/sj.emboj.7601390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huberman J. A., Riggs A. D. On the mechanism of DNA replication in mammalian chromosomes. J. Mol. Biol. 1968;32:327–341. doi: 10.1016/0022-2836(68)90013-2. [DOI] [PubMed] [Google Scholar]
- Hyrien O., Maric C., Mechali M. Transition in specification of embryonic metazoan DNA replication origins. Science. 1995;270:994–997. doi: 10.1126/science.270.5238.994. [DOI] [PubMed] [Google Scholar]
- Johnson W. E., Li W., Meyer C. A., Gottardo R., Carroll J. S., Brown M., Liu X. S. Model-based analysis of tiling-arrays for ChIP-chip. Proc. Natl. Acad. Sci. USA. 2006;103:12457–12462. doi: 10.1073/pnas.0601180103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath S., Leffak M. Multiple sites of replication initiation in the human beta-globin gene locus. Nucleic Acids Res. 2001;29:809–817. doi: 10.1093/nar/29.3.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnani N., Taylor C. M., Malhotra A., Dutta A. Pan-S replication patterns and chromosomal domains defined by genome tiling arrays of ENCODE genomic areas. Genome Res. 2007;17:865–876. doi: 10.1101/gr.5427007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little R. D., Platt T. H., Schildkraut C. L. Initiation and termination of DNA replication in human rRNA genes. Mol. Cell. Biol. 1993;13:6600–6613. doi: 10.1128/mcb.13.10.6600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas I., Palakodeti A., Jiang Y., Young D. J., Jiang N., Fernald A. A., Le Beau M. M. High-throughput mapping of origins of replication in human cells. EMBO Rep. 2007;8:770–777. doi: 10.1038/sj.embor.7401026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlpine D. M., Rodriguez H. K., Bell S. P. Coordination of replication and transcription along a Drosophila chromosome. Genes Dev. 2004;18:3094–3105. doi: 10.1101/gad.1246404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies E. H., Cooper G. M., Asimenos Analyses of deep mammalian sequence alignments and constraint predictions for 1% of the human genome. Genome Res. 2007;17:760–774. doi: 10.1101/gr.6034307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maser R. S., Mirzoeva O. K., Wells J., Olivares H., Williams B. R., Zinkel R. A., Farnham P. J., Petrini J. H. Mre11 complex and DNA replication: linkage to E2F and sites of DNA synthesis. Mol. Cell. Biol. 2001;21:6006–6016. doi: 10.1128/MCB.21.17.6006-6016.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newlon C. S., Petes T. D., Hereford L. M., Fangman W. L. Replication of yeast chromosomal DNA. Nature. 1974;247:32–35. doi: 10.1038/247032a0. [DOI] [PubMed] [Google Scholar]
- Pelizon C., Diviacco S., Falaschi A., Giacca M. High-resolution mapping of the origin of DNA replication in the hamster dihydrofolate reductase gene domain by competitive PCR. Mol. Cell. Biol. 1996;16:5358–5364. doi: 10.1128/mcb.16.10.5358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghuraman M. K., Winzeler E. A., Collingwood D., Hunt S., Wodicka L., Conway A., Lockhart D. J., Davis R. W., Brewer B. J., Fangman W. L. Replication dynamics of the yeast genome. Science. 2001;294:115–121. doi: 10.1126/science.294.5540.115. [DOI] [PubMed] [Google Scholar]
- Saha S., Shan Y., Mesner L. D., Hamlin J. L. The promoter of the Chinese hamster ovary dihydrofolate reductase gene regulates the activity of the local origin and helps define its boundaries. Genes Dev. 2004;18:397–410. doi: 10.1101/gad.1171404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segurado M., de Luis A., Antequera F. Genome-wide distribution of DNA replication origins at A+T-rich islands in Schizosaccharomyces pombe. EMBO Rep. 2003;4:1048–1053. doi: 10.1038/sj.embor.7400008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sequeira-Mendes J., Diaz-Uriarte R., Apedaile A., Huntley D., Brockdorff N., Gomez M. Transcription initiation activity sets replication origin efficiency in mammalian cells. PLoS Genet. 2009;5:e1000446. doi: 10.1371/journal.pgen.1000446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinklein N. D., Chen W. C., Kingston R. E., Myers R. M. Transcriptional regulation and binding of heat shock factor 1 and heat shock factor 2 to 32 human heat shock genes during thermal stress and differentiation. Cell Stress Chaperones. 2004;9:21–28. doi: 10.1379/481.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashee S., Cvetic C., Lu W., Simancek P., Kelly T. J., Walter J. C. Sequence-independent DNA binding and replication initiation by the human origin recognition complex. Genes Dev. 2003;17:1894–1908. doi: 10.1101/gad.1084203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waltz S. E., Trivedi A. A., Leffak M. DNA replication initiates non-randomly at multiple sites near the c-myc gene in HeLa cells. Nucleic Acids Res. 1996;24:1887–1894. doi: 10.1093/nar/24.10.1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental Materials]