Regulation of photoreceptor gene expression by the retinal homeobox (Rx) gene product (original) (raw)
. Author manuscript; available in PMC: 2011 Mar 15.
Abstract
The retinal homeobox (Rx) gene product is essential for eye development. However little is known about its molecular function. It has been demonstrated that Rx binds to photoreceptor conserved element (PCE-1), a highly conserved element found in the promoter region of photoreceptor-specific genes such as rhodopsin and red cone opsin. We verify that Rx is co-expressed with rhodopsin and red cone opsin in maturing photoreceptors and demonstrate that Rx binds to the rhodopsin and red cone opsin promoters in vivo. We also find that Rx can cooperate with the Xenopus analogs of Crx and Nrl, otx5b and XLMaf (respectively), to activate a Xenopus opsin promoter-dependent reporter. Finally, we demonstrate that reduction of Rx expression in tadpoles results in decreases in expression of several PCE-1 containing photoreceptor genes, abnormal photoreceptor morphology, and impaired vision. Our data suggests that Rx, in combination with other transcription factors, is necessary for normal photoreceptor gene expression, maintenance, and function. This establishes a direct role for Rx in regulation of genes expressed in a differentiated cell type.
Keywords: retinal homeobox, Rx, photoreceptors, rhodopsin, red cone opsin, retinal development, shRNA, Argonaute 2
INTRODUCTION
The retinal homeobox (Rx) gene is essential for vertebrate eye development [reviewed in (Bailey et al., 2004)]. It is expressed in the prospective eye field at neural plate stages, prior to the appearance of morphological evidence of eye development (Casarosa et al., 1997; Chen and Cepko, 2002; Deschet et al., 1999; Furukawa et al., 1997a; Mathers et al., 1997; Ohuchi et al., 1999). Loss of Rx function results in severe reduction, or complete lack, of eye development in a variety of vertebrate species (Andreazzoli et al., 1999; Chen and Cepko, 2002; Chuang and Raymond, 2001; Loosli et al., 2003; Loosli et al., 2001; Mathers et al., 1997; Voronina et al., 2004) while over-expression of Rx results in development of extra retinal tissue (Andreazzoli et al., 1999; Chuang and Raymond, 2001; Mathers et al., 1997). Xenopus neural retinal cells forced to express Rx can develop into any retinal cell type, suggesting that Rx functions to maintain cells in a pluripotent state without biasing cell fate (Casarosa et al., 2003). In mouse, retinal cells expressing Rx tend to develop into Muller glial cells, a cell type that is thought to be capable of de-differentiating to provide a source of progenitors in the mature retina (Furukawa et al., 2000). Taken together, these results establish the importance of Rx in retinal development and suggest that Rx may be involved in regulating specification and proliferation of retinal progenitor cells. However, Rx is also expressed in photoreceptors in the mature retina (Perron et al., 1998). In zebrafish, knockdown of Rx1 and Rx2 during retinal maturation results in attenuation in photoreceptor development (Nelson et al., 2009). The molecular details of Rx function in these differentiated cells has not been determined.
The Rx gene product has been shown to function as a weak transcriptional activator. First, a constitutive repressor form of Rx (a fusion with the engrailed repression domain) functions as an antimorph, suggesting that Rx normally functions as a transcriptional activator (Andreazzoli et al., 1999). Second, Rx has been shown to bind a sequence element known as Photoreceptor Conserved Element 1 (PCE-1; also known as Ret1) (Kimura et al., 2000), a conserved sequence element found in promoters of many genes expressed in photoreceptors, including rhodopsin (rho), red cone opsin (RCO), rod arrestin, and interphotoreceptor retinoid-binding protein (IRBP) genes (Batni et al., 1996; Boatright et al., 1997; Kikuchi et al., 1993; Ma et al., 2001; Mani et al., 2001; Moritz et al., 2002). The rho promoter is highly conserved among vertebrates and has been analyzed in detail (Ma et al., 2001). Rx weakly activates synthetic gene reporter constructs containing multiple copies of PCE-1 (Chen and Cepko, 2002; Kimura et al., 2000). In addition to the PCE-1 site, rho promoters contain three additional conserved cis-acting elements, the BAT (Ret2), NRE (Ret3), and Ret4 sites. The BAT and Ret4 sites contain consensus core homeobox protein binding motifs. The BAT site primarily binds members of the orthodenticle (otx) family of transcription factors (Kimura et al., 2000), such as otx2 and cone-rod homeobox (crx) or its analog otx5b (Whitaker and Knox, 2004). Rx can also bind the BAT site, although it has greater affinity for the PCE-1 site (Kimura et al., 2000; Pan et al., 2006; Wang et al., 2004). NRE is the binding site for the transcription factor neural retina leucine zipper (Nrl) or its Xenopus analog XLmaf (Ishibashi and Yasuda, 2001). It has been well established that crx and Nrl synergize to activate rho promoters (Chen et al., 1997; Mitton et al., 2000), as do the Xenopus analogs otx5b and XLmaf (Whitaker and Knox, 2004). Additionally, transcription factors of the zinc finger (Sp4, KLF15) and nuclear hormone receptor (Nr1d1, Nr2e3) families are also involved in regulating rho promoters (Cheng et al., 2004; Lerner et al., 2005; Otteson et al., 2005; Otteson et al., 2004). The function of Rx has not been characterized in the context of an intact PCE-1-containing promoter such as the rho promoter.
Here we verify that Rx is expressed in rhodopsin-positive photoreceptors of the maturing retina. We report that Xenopus laevis Rx can specifically bind PCE-1-containing promoters in vivo, including rho and RCO. Further, we show that Rx can activate the Xenopus opsin promoter (XOP) in functional cooperation with XLmaf and otx5b. Finally, we demonstrate that expression of several photoreceptor-specific genes and photoreceptor development and function are dependent on Rx expression.
MATERIALS AND METHODS
Embryos
Embryos were produced by in vitro fertilization (Sive et al., 2000). Transgenic embryos were generated by intracytosolic sperm injection (ICSI) (Sparrow et al., 2000). For experiments involving injection of RNA into transgenic embryos, eggs were injected with sperm nuclei and transgene (ICSI). Dividing embryos were subsequently injected with RNA into one blastomere at 4-cell stage. Embryos were then screened for transgenesis markers and RNA lineage tracers at appropriate stages.
Plasmids
We have described the preparation of XOP-Luc and pCS2/RxL previously (Pan et al., 2006). To prepare MT-Rx, the Rx coding region (starting with the second codon) was amplified from pSP64T/Rx1A (Mathers et al., 1997) and subcloned into compatible sites in pCS2 + MT (Turner and Weintraub, 1994). DsRedExpress RNA was prepared from pCS2/HA-dsRedExpress (Seufert et al., 2005). The PCE-1 site was mutated in XOP-GFP (Mani et al., 2001) using the QuickChange XL Site Directed Mutagenesis Kit (Stratagene) and the primer 5’-CGTTTAAGGGAAGCCAGCTGACACTTTGCAATTTTAGCTTGG-3’ and its reverse complement, based on a previously described mutagenesis of the PCE-1 site (Kimura et al., 2000). XOP-dsRed was prepared by liberation of XOP from XOP-GFP (digested using BglII and BamHI) and insertion into pL-dsRed (digested using BamHI). pL-dsRed was prepared by liberation of the dsRed coding region and SV40 poly(A) signal from pCS2/HA-dsRedExpress and insertion into pLITMUS29 (New England Biolabs) (both digested using HinDIII and NsiI).
The Rx shRNA was designed using the RNAi OligoRetriever program with the Shagging-pSHAG1 BseRI-BamHI new design method (URL: http://katahdin.cshl.org:9331/RNAi/html/rnai.html) (Paddison and Hannon, 2002). The coding sequences for Rx1A or Rx2A (Mathers et al., 1997) were individually entered in the program to design an shRNA oligonucleotide that was identical for both Rx1A and Rx2A. The shRNA template was generated by PCR using the full-length oligonucleotide sequence 5’-GATCAAGCTTCTATACTGTGCAGCCTTGATGGGTTACCTTTGGAGGTAACCCATCAA GGCTGTACAGTATAGTTTTTTGGATCCAAGCTTGATC-3’. The shRNA second strand was generated and the double-stranded oligonucleotide was amplified for using Pfx (Invitrogen) and the primers shown in Table 1. The 94bp PCR product was TA cloned in pCR2.1 (Invitrogen) and sequenced to confirm that point mutations were not introduced during PCR. The double-stranded oligonucleotide was then subcloned into a modified pRNAT vector (Genscript) containing the X. laevis U6 promoter (Li and Rohrer, 2006) using HinDIII sites included in the oligonucleotide sequence (underlined). Orientation was determined using a BamHI site adjacent to one of the HinDIII sites (bold). As a control, an shRNA template encoding a reversed target sequence was similarly obtained and cloned. shRNA transgenes were prepared by restriction endonuclease digestion using BglII, PstI and SalI to release a 3 kb fragment.
TABLE 1.
PRIMERS USED FOR QUANTITATIVE PCR
| GENE | FORWARD PRIMER | REVERSE PRIMER |
|---|---|---|
| Rx1A | GCACAAGTTGGGCATAGTAGGG | GTTTTTGGGAAAGCTGGGC |
| Rx2A | CCGTGAGCCACATTCAAATCTA | AATACCAATCACAGCGCCCCT |
| L8 | AAGCTTCGGGCTATCGACTTT | ACGGCCTGGATCATGGATAA |
| Rhodopsin | TCCTGATCTGTTGGGTGCC | TGAAGACTGGGCCAAAGTCG |
| RCO | TTTCCGCAAAGCAAAATGG | GTAGTTACGCCCCTGGTGATG |
| silver | CAATAAATGAGCTGCCCGG | AATAACCACAACCTCCTCGGC |
| XOP | TCAGACCTGGATTTCTTCCTGT | ATTGCAAAGTGTTAATTGGCTTC |
| RCO pro | GGGTGGACGTTAAAACTCATATGC | GCTCAGCTGGATTAGCCTAGACAC |
| Rho exon5 | CTGCTTGATCACCACCCTGTG | TCTGTCTTGGAGGTGGCTGC |
| RCO UTR | GTAGTTACGCCCCTGGTGATG | TTTCCGCAAAGCAAAATGG |
| Rx shRNA | GATCAAGCTTCTATACTGTGCAGCCTTGATG | GATCAAGCTTGGATCCAAAAAACTATACTGTAC |
| IRBP | TCCTCGCCTGGTTGTCTCCTAT | AGCTGTTCCGGTGTTAGCTCCA |
| arrestin | TTCCCTGATTTCTTGCCATGC | TTCAAAATCCACACCACAGGC |
The X. laevis ortholog of the silver gene was amplified from X. laevis tadpole head cDNA by degenerate RT-PCR (accession number FJ643547).
The mRx rescue construct was prepared in several steps. First, the mRx coding region (Mathers et al., 1997) was amplified by PCR using appropriate primters and cloned into pCS2 (EcoRI and XhoI, contained in primers). Second, a 3 kb region of DNA adjacent to the Rx coding region was amplified from X. tropicalis genomic DNA (corresponding to scaffold_228:571,277–574,283 of Xenopus tropicalis genome August, 2005, assembly) using specific primers and cloned into pBlueScript II KS (BamHI and EcoRV, contained in primers) to make pBS/tRx3000. UCE2 (scaffold_228:579,112–579,379), was similarly amplified, digested (BamHI and BglII) and cloned into pBS/tRx3000 (BamHI) to make pBS/UCE2+tRx3000. Third, the dsRed expression cassette, including the CMV promoter, was excised from pCS2+HA/dsRedExpress in 2 fragments (a SalI and KpnI fragment and a KpnI fragment) and cloned into pBS/UCE2+tRx3000 (digested with SalI and KpnI) in two steps (first the SalI + KpnI fragment and then the KpnI fragment) to make UCE2+tRx3000 + dsRed. Finally, the mRx coding region and SV40 poly(A) signal were excised from pCS2/mRx (HinDIII and NotI and blunt-ended using the Klenow fragment of DNA polymerase) and introduced into UCE2+tRx3000 + dsRed (EcoRV) to make the mRx rescue transgene plasmid. The mRx rescue transgene was prepared by digestion of the plasmid with NotI.
A full-length Argonaute2 cDNA clone was identified as encoding Xl eIF2C (NCBI UniGene Xl.43287) in Sport6 was obtained from Open Biosystems (catalogue number MXL1736-9506800; accession number BC077863). The coding region was amplified by PCR using the following primers XL-eIF2C-F: 5’- GAATTCCATGTACTCCGGGGCC-3’; XL-eIF2C-R: 5’-GAATTCAAGTCACTGGATAAACAGCGG-3’ to introduce EcoRI sites (underlined) and subcloned into the TOPO 2.1 vector (Invitrogen) by TOPO TA cloning. Ago2 was then subcloned into the EcoRI site of pCS2 + MT. Transformants were checked for insertion and orientation by colony PCR and confirmed by sequencing. Synthetic mRNA was prepared by linearization with NotI and in vitro transcription using the mMessage mMachine SP6 in vitro transcription kit (Ambion).
Histology, in situ hybridization and immunohistochemistry
Antisense riboprobes were prepared by in vitro transcription as described previously for Rx, rhodopsin, and red cone opsin (Liu et al., 2001; Mathers et al., 1997; Mathers and Jamrich, 2000; Pan et al., 2006). In situ hybridization using whole or sectioned embryos was performed as described previously (Sive et al., 2000; Viczian et al., 2003). Staining of paraffin sections with antibodies was performed as previously described (El-Hodiri et al., 1997). Primary antibodies were used at the following dilutions: mouse anti-rhodopsin (RetP1, Biomeda) 1:50; mouse anti-islet1, 1:50 [39.4D5, Developmental Studies Hybridoma Bank (DSHB), University of Iowa]. Biotinylated-peanut agglutinin (Vector) was used at 5 µg/ml.
Chromatin immunoprecipitation
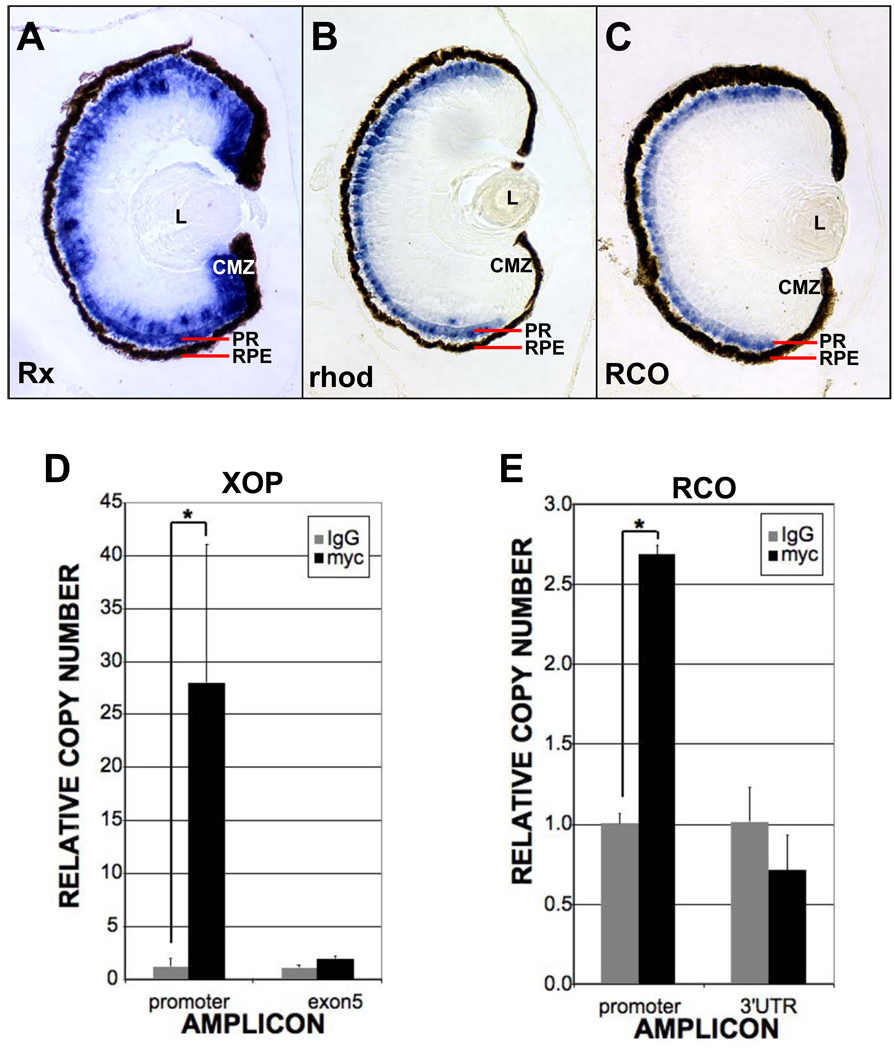
Chromatin immunoprecipitation (ChIP) was performed using a combination of protocols from the Farnham (URL: http://www.genomecenter.ucdavis.edu/farnham/protocols/tissues.html) (Wells et al., 2002) and Young labs (URL: http://jura.wi.mit.edu/young_public/hESregulation/ChIP.html) (Boyer et al., 2005). Briefly, 4-celled embryos were cultured from in vitro fertilized eggs (Sive et al., 2000) and injected with 10 pg myc-tagged Rx (MT-Rx) or 1 pg myc tag (MT) RNA into both prospective dorsal blastomeres. Abnormally developing embryos were discarded. Embryos were cultured to st 41 (Nieuwkoop and Faber, 1994) and anesthetized by immersion in MS222. Embryos were then collected and washed in Embryo Wash Buffer [0.03% (v/v) Triton X-100, 100 mM NaCl, 1 X Roche Mini-Complete Protease Inhibitors] and fixed for 15 minutes in Crosslinking Buffer [50 mM HEPES (pH 8), 1 mM EDTA, 0.5 mM EGTA, 100 mM NaCl, 1.8% (w/v) formaldehyde] at room temperature. Crosslinking was halted by addition of glycine to 0.125 M and incubation for 5 minutes at room temperature. Embryos were then washed in phosphate-buffered saline containing 10 mM EDTA and 0.1% (v/v) Tween-20 (PBSE-Tw). Embryo heads were isolated and homogenized in 2 ml Cell Lysis Buffer. Nuclei were pelleted by centrifugation. Pellets were resuspended in 2 ml Nuclei Lysis Buffer, transferred to 15 ml conical tubes fitted with ultrasound reflecting bars, and sonicated in a water bath sonicator (Diagenode Bioruptor) for 12 minutes (30 seconds on/30 seconds off) at “High” power setting. Lysates were incubated overnight at 4° C with rabbit IgG (Upstate) or polyclonal antibodies raised to the myc epitope tag (Sigma) prebound to Protein G magnetic beads (Dynal), prepared as described (Boyer et al., 2005). DNA was recovered following reversal of crosslinking, purified and used as template for real-time PCR. Real-time PCR was performed in a 25-µL amplification mixture containing 1 µL of ChIP product, 12.5 µL of 2x PCR master mix (SYBR Green; Applied Biosystems, Foster City, CA), and 100 nM forward and reverse primers, respectively (Table 1). The PCR conditions included a polymerase activation step at 95°C for 10 minutes followed by 50 cycles of 95°C for 15 seconds and 60°C for 60 seconds and run on a sequence detector (Model 7500; Applied Biosystems). The CT for XOP or RCO promoter was normalized by the CT of the rabbit IgG control. The statistical significance of relative differences in expression levels was determined by Student’s group t-test.
Quantitative RT-PCR
Total RNA was extracted from the isolated heads of individual st 41 tadpoles using Trizol according to manufacturer’s protocol and treated with DNase I to eliminate genomic DNA contamination. cDNA was prepared from RNA template by reverse transcription (RT) using qScript cDNA SuperMix (Quanta BioSciences) in a 20 µl reaction, according to manufacturer’s directions. Quantitative (real-time) PCR was performed in a 25 µL amplification mixture containing 1 µL of cDNA product, 12.5 µL of 2 X SYBR Green PCR master mix (Applied Biosystems), and 100 nM forward and reverse primers specific to each gene and PCR conditions as described for ChIP, above. A housekeeping gene, L8, was used as an internal control. The statistical significance of relative differences in expression levels was determined by Student’s group t-test.
Luciferase assays
Embryos were injected at 2-cell stage with 25 pg XOP-Luc DNA (Pan et al., 2006), pRLtkLuc (encoding Renilla luciferase as an internal control) and RNAs encoding effectors as indicated in the figure legends. Embryo lysates were prepared at st 11 and assayed for luciferase activity using the Stop n Glo Dual Luciferase Assay Kit (Promega) as described previously (Kelly et al., 2006; Pan et al., 2006). Firefly luciferase activity values were normalized against Renilla luciferase activity values and presented as fold activation compared to the baseline activity of the reporter (without co-injected effectors).
Transgenesis
Transgenes were digested with appropriate restriction endonucleases and purified from an agarose gel (GeneClean Kit, Bio101), unless indicated otherwise. Transgene concentration was determined by direct comparison of purified transgene to a DNA mass ladder (Invitrogen) resolved by agarose gel electrophoresis. Transgenic Xenopus laevis embryos were prepared by intracytosolic sperm injection (ICSI) as described previously (Sparrow et al., 2000) using digitonin-treated sperm nuclei (Huang et al., 1999). Embryos transgenic for the pRNAT plasmid (shRNA transgenics) were identified by observation of blue fluorescence (from the cGFP expression cassette). Embryos transgenic for the mRx rescue plasmid were identified by observation of red fluorescence (from the dsRed Express expression cassette). Fluorescence in live embryos was observed using a Leica MZFLIII fluorescent microscope with a GFP2 filter. Digital images were captured using a SPOT RT SE 7.2 Color Mosaic digital camera (Diagnostic Instruments).
Quantitation of fluorescence
Fluorescence in photomicrographs was quantified essentially as described previously (Ghai et al., 2009). Tadpoles were generated cotransgenic for XOP-dsRed containing a wild-type or mutated PCE-1 site and Rx or control shRNA. As discussed above, the shRNA transgene also contains a cGFP expression cassette. Tadpoles were cultured to st 41, fixed, and cryopreserved. Sections were cut at 10 µM thickness and mounted using Vectashield Hard Set with DAPI (Vector Labs). All photomicrographs were captured by a SPOT RT digital camera connected to a Nikon Eclipse E800 microscope using identical exposure times (10 sec). Fluorescence was quantified using IMAGEPRO 6.2 software (Media Cybernetics, Bethesda, MD, USA). For each image, the whole retina area was selected. Green or red cells or cell clusters were picked automatically by program by adjusting intensity value for each color, threshold (0 = black, 255 = saturated green or red). Threshold was set to cover all the labeled cells in the retina. The average pixel intensity was calculated for all pixels within threshold regions. These calculations were determined for each retinal region sampled from two to three different embryos for each experiment conditions. Red fluorescence values (XOP-dsRed readout) were normalized to green fluorescent values (as a measure of the shRNA transgene). Values are presented as average normalized fluorescence values ± standard deviation. Statistical analysis was performed using Student’s group t-test.
Tadpole visual function assay
Tadpoles were tested for visual system function essentially as described previously (Moriya et al., 1996). Tadpoles were raised to st 45 or 50 and kept on a white background for at least 3 days. Tadpoles were then placed into a 4” X 6” X 3” clear colorless plastic container containing approximately 500 ml 0.1 X MMR. This container was placed inside a slightly larger container, half covered with black paper and half covered with white paper. Tadpoles were introduced singly into the container. Their position (white half or black half) was recorded after two minutes. This test was repeated 5 – 10 times on each of two consecutive days.
RESULTS
It has been reported that Rx binds the highly conserved photoreceptor conserved element (PCE-1) found in the regulatory regions of many genes expressed in photoreceptors (Boatright et al., 1997; Kikuchi et al., 1993; Ma et al., 2001; Mani et al., 2001). Previous reports have focused on the striking expression of Rx in the anterior neural plate and during early phases of eye specification and retinal development, prior to differentiation of photoreceptors. To determine if PCE-1 site-containing promoters may be targets of Rx, we first asked if Rx expression overlaps the expression of photoreceptor-specific genes, such as rhodopsin (rho) and red cone opsin (RCO). There are two Rx genes in X. laevis, Rx1A and Rx2A (Mathers et al., 1997) with apparently identical expression patterns (unpublished data). Rx1A has been reported to be expressed weakly in the photoreceptor layer of the maturing Xenopus retina (Perron et al., 1998). Additionally, a X. laevis Rx1A promoter-GFP transgene is expressed in both rods and cones and zebrafish Rx1 is expressed in rods and cones (Chuang et al., 1999; Nelson et al., 2008; Zhang et al., 2003). We verified that Rx1A is expressed in the mature Xenopus retina (st 41), primarily in the marginal zone and photoreceptor layer (Fig 1A). Importantly, genes regulated by PCE-1-dependent promoters are also expressed in the photoreceptor layer, such as rho and RCO (Fig 1B, C). These results confirm the expression of Rx1A in photoreceptor cells where PCE-1-dependent genes are expressed. Thus, genes regulated by promoters containing PCE-1 sites are biologically relevant potential Rx targets.
Figure 1.
Rhodopsin and red cone opsin (RCO) are Rx targets. A – C. Rx is co-expressed with rhodopsin and RCO in photoreceptors. In situ hybridization on sections from paraffin-embedded st 41 tadpoles using probes for Rx (A), rhodopsin (B) or RCO (C). D, E. Chromatin immunoprecipitation (ChIP) results indicating that myc-tagged Rx (MT-Rx) can bind to the rhodopsin (D) and RCO (E) promoters in vivo. Results are presented as the CT of each sample normalized to the CT of a “no antibody” control. * - p < 0.003 (XOP), p < 0.002 (RCO).
The Xenopus opsin promoter (XOP) regulates transcription of the rho gene in rod photoreceptors and contains a highly conserved PCE-1 element (Batni et al., 1996; Mani et al., 2001). Since Rx1A is expressed in photoreceptors and can bind the PCE-1 site, we selected XOP as a putative Rx1A target. We asked if Rx1A can bind to XOP in vivo by chromatin immunoprecipitation (ChIP). We have not found an antibody that reliably immunoprecipitates Xenopus Rx (YP, SN, LEK, and HME, unpublished). Therefore, we expressed a myc-tagged Rx1A (MT-Rx) in Xenopus embryos and performed ChIP using an antibody raised against the myc tag. As a control, we expressed myc-tag (MT) alone. XOP chromatin was not precipitated from embryos expressing MT only (Fig 1D). We found that the anti-myc antibody specifically precipitated chromatin containing XOP when MT-Rx was expressed (Fig 1D). A rho coding exon was not efficiently precipitated from embryos expressing MT-Rx (Fig 1D). Additionally, a non-PCE-1 containing promoter, the Rx2A promoter, was not precipitated by MT-Rx (not shown). Taken together, these results demonstrate that Rx can specifically bind the rho promoter in vivo. Further, the antibody could precipitate chromatin that included another PCE-1-containing gene regulatory region, the promoter of the red cone opsin (RCO) gene (Fig 1E) (Moritz et al., 2002). These ChIP results demonstrate that Rx1A can bind to PCE-1 containing promoters in vivo. Specifically, these results demonstrate that Rx can bind regulatory regions of photoreceptor-specific genes in the differentiating retina and retina-derived cells in vivo.
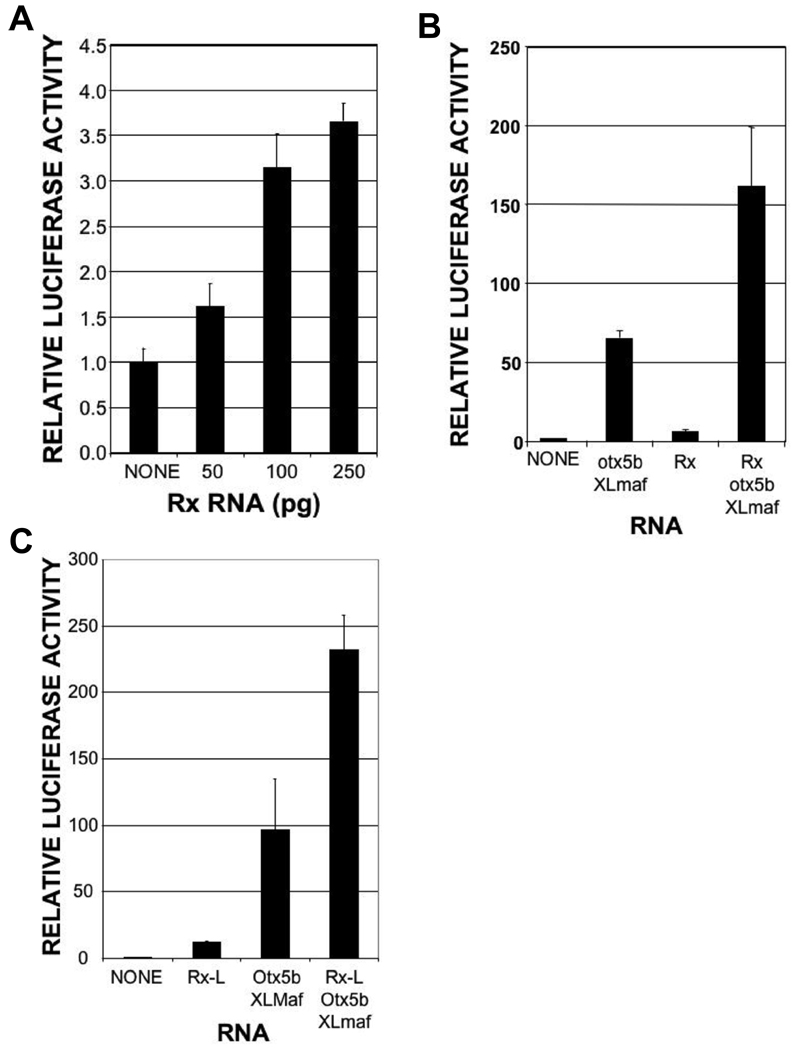
To investigate the function of Rx1A in regulating transcription, we tested its ability to activate a luciferase reporter gene construct containing XOP. We found that Rx1A could indeed activate this reporter in a dose-dependent manner, albeit weakly (Fig 2A). Rho promoters are activated, in part, by synergistic interactions between the crx and Nrl transcription factors (Chen et al., 1997; Mitton et al., 2000; Whitaker and Knox, 2004). Binding sites for these proteins (BAT and NRE, respectively) are well conserved in rho promoters (Chen et al., 1997; Ma et al., 2001; Mani et al., 2001; Mears et al., 2001), including XOP. XOP can be activated by mammalian crx and Nrl and by the analogous Xenopus proteins, otx5b and XLmaf (Ishibashi and Yasuda, 2001), also known as XLNrl. To investigate the possibility that Rx can cooperatively activate XOP, we tested the activity of Rx1A on XOP in the presence of otx5b and XLmaf. We found that otx5b and XLmaf synergistically activated XOP in our experimental system and that Rx1A cooperated with these factors to further activate XOP (Fig 2B).
Figure 2.
Rx and RxL can cooperate with other factors to activate the Xenopus opsin promoter (XOP). Luciferase assay performed using lysates from embryos co-injected with XOP-Luc reporter plasmid and Rx, Rx-L, otx5b, and/or XLmaf RNAs as shown. A. Rx activates XOP-Luc in a dose-dependent manner. B. Rx cooperates with otx5b and XLmaf to activate the XOP-Luc reporter. C. Rx-L cooperates with otx5b and XLmaf to activate the XOP-Luc reporter.
We have previously characterized Rx-like gene product, Rx-L, that contains a homeodomain that is nearly identical to the Rx homeodomain but does not contain an octapeptide motif (OP) (Pan et al., 2006). It has been demonstrated that the chicken and human Rx-like gene products, cRaxL and QRX, bind the PCE-1 site and are stronger transcriptional activators than Rx gene products, presumably because they lack an OP (Chen and Cepko, 2002; Wang et al., 2004). It has also been shown that QRX can cooperate with CRX and NRL to activate a rhodopsin promoter-based reporter gene (Wang et al., 2004). To determine whether Rx-L functioned similarly, we tested its ability to activate the XOP-Luc reporter in our experimental system. We found that the combination of Rx-L, otx5b, and XLmaf activated the XOP-Luc reporter to a greater degree than either Rx-L alone or the combination of otx5b and XLmaf (Fig 2C), suggesting that Rx-L can also functionally cooperate with otx5b and XLmaf to activate XOP.
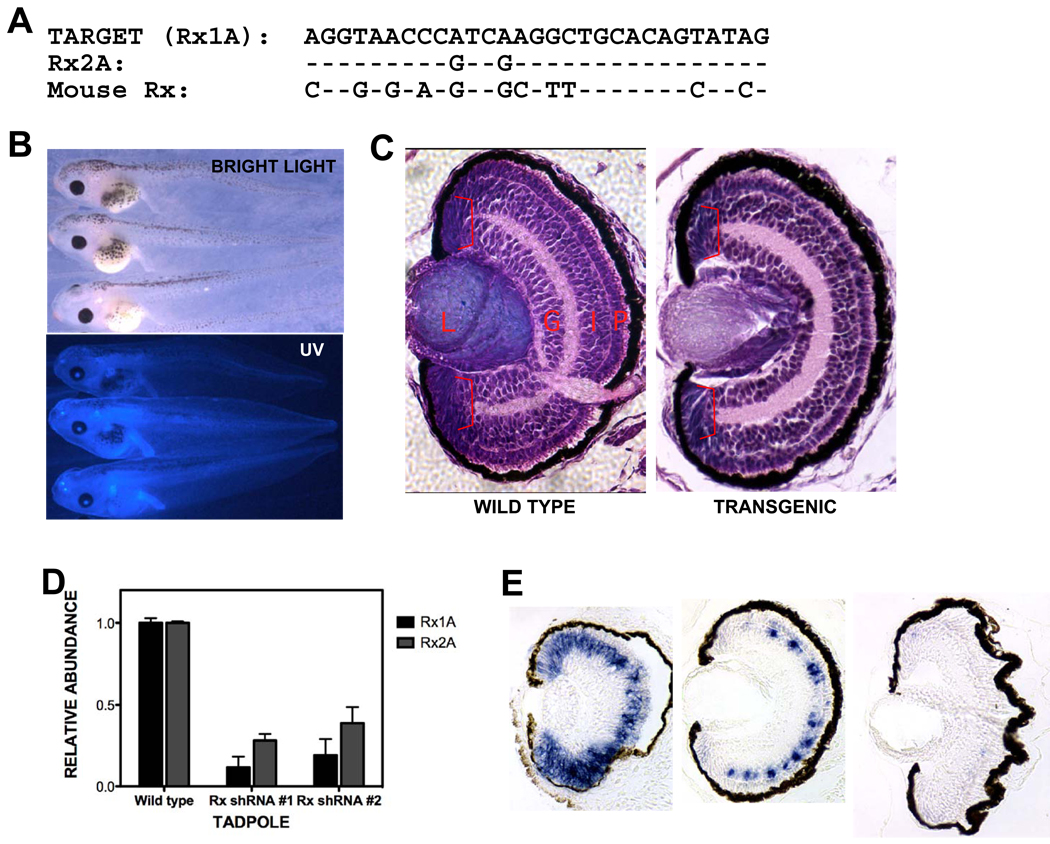
We next investigated the involvement of Rx gene products in regulating rho and RCO gene expression in vivo. We used a shRNA-based approach to knock down Rx expression during embryonic development. Oligonucleotides encoding a Rx-specific shRNA were produced and introduced into a modified version of the pRNAT plasmid. This allowed us to prepare transgenic embryos ubiquitously expressing the Rx shRNA under the control of the Xenopus tropicalis U6 promoter. The Rx shRNA matched 29 nucleotides of the Rx1A transcript and 26 of 29 nucleotides of the Rx2A transcript (Fig 3A). Transgenic embryos were identified by blue fluorescence generated by a cGFP expression cassette that was included in the shRNA transgene (Fig 3B). We were surprised to observe no apparent external or histological phenotype in eye development of embryos transgenic for the Rx shRNA construct at st 41 (Fig 3B,C). However, we found that embryos transgenic for the Rx shRNA construct exhibited reduced expression of both Rx1A and Rx2A at st 41 (Fig 3D). Both Rx1A and Rx2A were knocked down approximately 60 – 90%. Additionally, we found that Rx1A expression in the retina is essentially normal at st 38 but disappears from the CMZ and photoreceptor layer by st 41 (Fig 3E). These results demonstrate that Rx shRNA transgenic tadpoles can serve as a model in which Rx expression is reduced in the maturing retina.
Figure 3.
Generation of Rx knockdown embryos. A. A portion of X. laevis Rx1A was selected as a target for development of an shRNA. Alignment of the shRNA target sequence with corresponding regions of X. laevis Rx2A and mouse Rx. B. Tadpoles transgenic for the Rx shRNA plasmid appeared normal. Bright light or UV-light views of transgenic tadpoles at st 41. C. Retinas from Rx shRNA transgenic tadpoles appeared histologically normal. Hematoxylin and eosin stained sections of paraffin-embedded wild type (left panel) or Rx shRNA transgenic tadpole. D. Rx shRNA transgenic tadpoles have reduced levels of both Rx1A and Rx2A as determined by quantitative RT-PCR (qRT-PCR) performed using total RNA purified from isolated tadpole heads (st 41). E. Rx1A is expression is reduced by in situ hybridization performed using 8 µM sections of paraffin-embedded tadpoles at st 38, 41, and 45 (from left to right). The reduction in Rx expression appears to increase as development progresses.
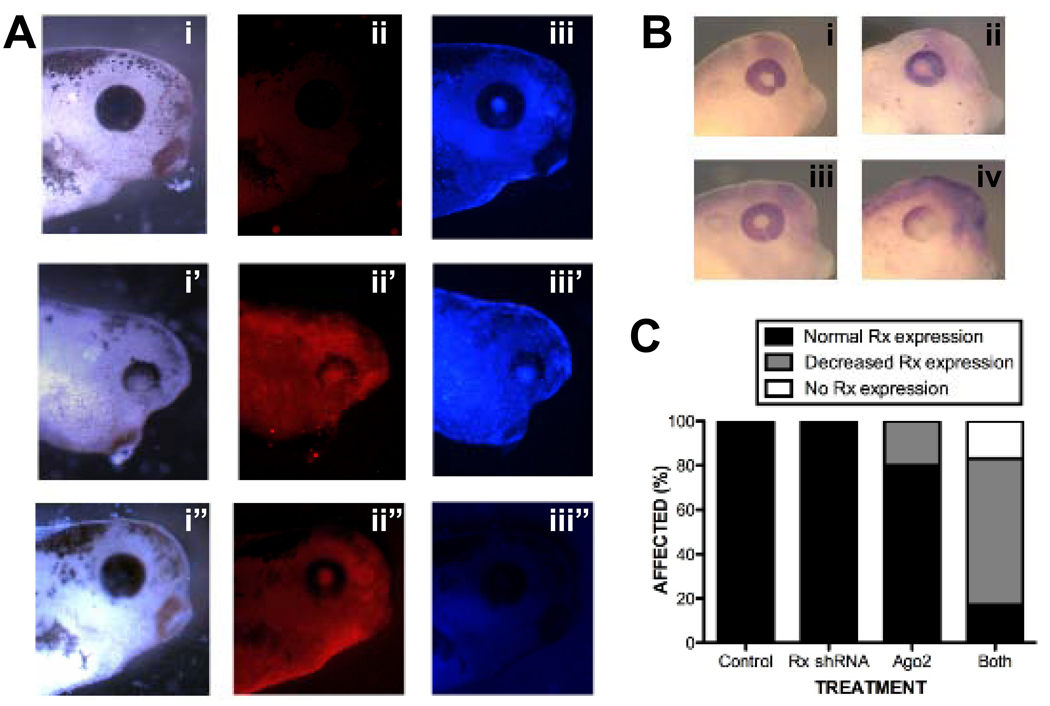
As mentioned above, it was curious that shRNA-mediated Rx knockdown resulted in a late phenotype. We observed that expression of the cGFP was not readily detectable before st 28 and intensified as the tadpoles developed (not shown). These results suggest that the transgene products may accumulate with time, resulting in effective knockdown of Rx1A and Rx2A at tadpole stages, but not before. A second possibility is that the Rx shRNA was generated during early development but was not functional until later. One possibility is that the biomolecules involved in either shRNA processing or function might be in relatively limited supply. The processing and function of shRNAs is mediated by Argonaute (Ago) gene products [reviewed in (Faehnle and Joshua-Tor, 2007; Hock and Meister, 2008; Hutvagner and Simard, 2008)]. Ago proteins bind shRNAs and mediate shRNA-target interactions, including target RNA degradation. It is possible that expression of the Rx shRNA does not result in a discernable eye phenotype because the shRNA overwhelms the system. It has been demonstrated that co-expression of Ago2 enhances the effects of shRNAs in tissue culture (Diederichs et al., 2008) and in Xenopus tadpoles (Chen, 2009). We found that overexpression of X. laevis Ago2 in Rx shRNA transgenic tadpoles results in defects in eye development, including anophthalmia and microophthalmia (Fig 4A). Eye development defects were not observed in non-transgenic control tadpoles overexpressing Ago2. Further, overexpression of Ago2 resulted in decreased Rx expression in tailbud stage Rx shRNA transgenic embryos (Fig 4B,C). We previously did not observe reduction of Rx expression until st 41 (Fig 3E). These results demonstrate that the effects of the Rx shRNA can be induced earlier in development by co-expression of the Ago2 gene product.
Figure 4.
Exogenous Argonaute2 (Ago2) exacerbates the effects of shRNA-mediated Rx knockdown. A. Exogenous Ago2 exacerbates the Rx shRNA knockdown phenotype. Embryos were generated by intra-cytosolic sperm injection (ICSI) with Rx shRNA transgene and injected with RNA encoding X. laevis Ago2 RNA. Embryos were co-injected with dsRed Express RNA as a lineage tracer. Embryos were photographed under white light (i, ii, iii), red fluorescence to visualize dsRed lineage tracer (i’, ii’, iii’), or blue fluorescence to visualize Rx shRNA trasngene (i”, ii”, iii”). Embryos receiving only the Rx shRNA (panels i – iii) or Ago2 RNA (panels i” – iii”) have apparently normal eyes while embryos transgenic for the Rx shRNA and receiving Ago2 RNA in the developing eye exhibited abnormally developed eyes (panels i’ – iii’). B. Exogenous Ago2 exacerbates the effects of Rx shRNA on Rx expression. Wholemount in situ hybridization using a Rx antisense riboprobe and embryos receiving either the Rx shRNA transgene (ii), Ago2 RNA (iii), or both (iv). Control embryos (i) received neither. These results are presented in graph form in C.
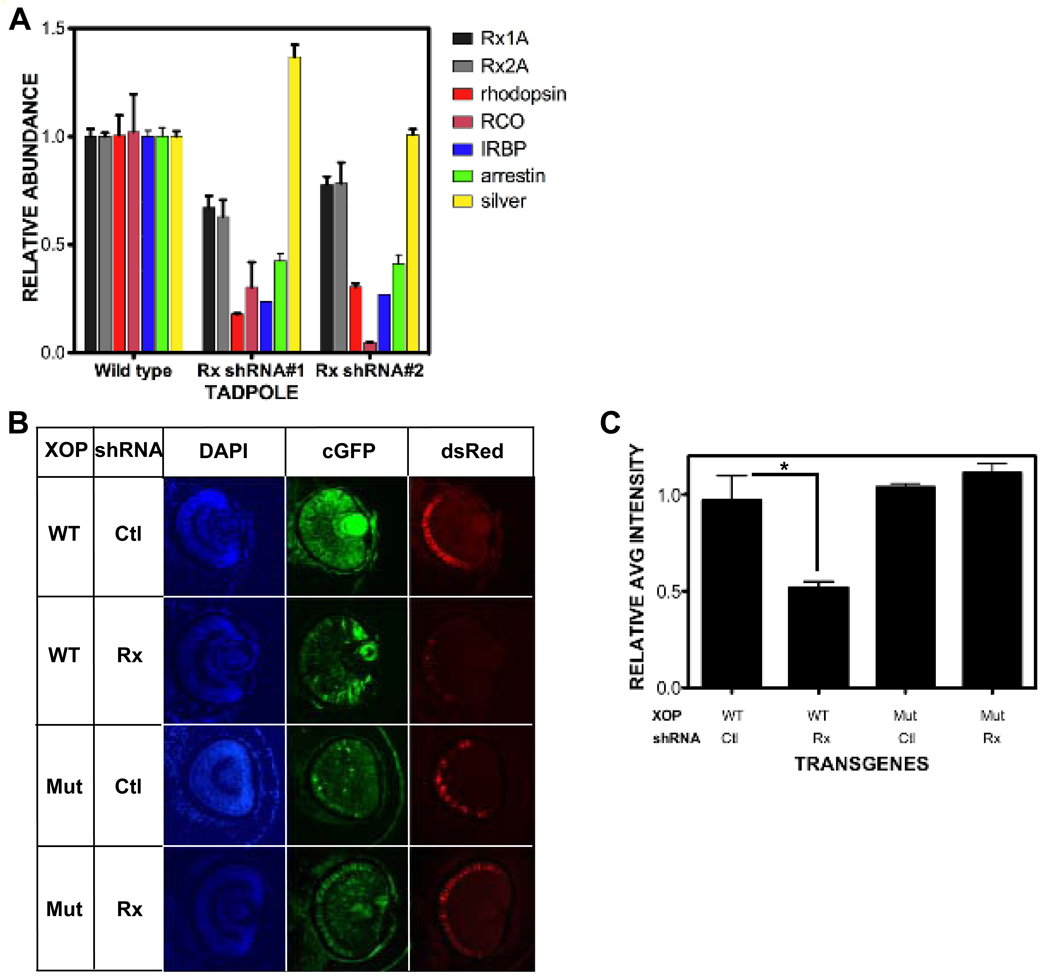
We next investigated the effect of decreasing Rx mRNA levels on putative target gene expression. Rx expression was knocked down late in development using the Rx shRNA trasngene alone. We found that rho expression was reduced by approximately 50% in Rx shRNA embryos (Fig 5A). Additionally, we found that RCO expression was also reduced by approximately 50% in Rx shRNA transgenic tadpoles. IRBP and arrestin are additional examples of genes expressed in photoreceptors under the control of transcriptional regulatory regions that contain PCE-1 sites. We found that IRBP and arrestin expression levels are reduced by approximately 75% and 60%, respectively (Fig 5A). As a control, we analyzed the expression of silver, a gene expressed specifically in the RPE and not the neural retina (Fig S1). Silver expression was not reduced in Rx shRNA embryos (Fig 5A). Therefore knockdown of Rx expression results in specific reduction of expression of photoreceptor genes with regulatory regions that contain PCE-1 elements.
Figure 5.
Photoreceptor gene expression is reduced by Rx knockdown. A. Expression of the photoreceptor genes rhodopsin, red cone opsin, IRBP, and arrestin are reduced in Rx shRNA transgenic tadpoles by qRT-PCR, as are Rx1A and Rx2A. Expression of silver is not reduced in Rx shRNA transgenic embryos by qRT-PCR. B. Rx knockdown results in PCE-1 site-dependent reduction in rhodopsin promoter activity. Co-transgenic tadpoles were generated using a Rx or control shRNA transgene and XOP-dsRed containing a wild type or mutated PCE-1 site, as indicated on the left side of the figure. Left column: retinal cell nuclei visualized using DAPI. Middle column: shRNA transgene visualized by the coral GFP (cGFP) expression cassette. Right column: XOP-dsRed trasngene expression. C. Quantification of results shown in B as described in Methods. Rx shRNA results in reduction of XOP-dsRed as compared to control (reversed sequence) shRNA. Introduction of the Rx shRNA transgene does not result in decreased XOP-dsRed expression level when the PCE-1 site is mutated. * - p < 0.025. Abbreviations: WT – wild type; Ctl – control.
The results presented above demonstrate that expression of several genes with PCE-1-containing transcriptional regulatory regions is dependent on Rx expression. Among these was the rhodopsin gene, which contains a well-characterized conserved PCE-1 site in its promoter. We have demonstrated that the Xenopus rhodopsin promoter (XOP) can be activated by Rx and can physically interact with Rx in vivo (Fig 1D, Fig 2A,B). We next asked if the PCE-1 site was necessary for regulation of XOP on Rx expression. We prepared embryos co-transgenic for control or Rx shRNA and a XOP-dsRed reporter and determined the level of dsRed reporter gene expression relative to transgene abundance. Consistent with the results presented above, we found that expression of the dsRed reporter was reduced by approximately 50% in embryos co-transgenic for Rx shRNA as compared to control shRNA (Fig 5B,C). This indicated that the XOP-dsRed transgene was dependent on Rx for full activity. We then repeated the experiment using XOP-dsRed containing a mutated PCE-1 site. We found that expression of the mutated reporter was similar in embryos co-transgenic for either control or Rx shRNA (Fig 5B,C). These results demonstrate that the Rx is necessary for normal levels of rhodopsin promoter activity and that the PCE-1 site is necessary for this effect. Further, these results indicate that the effects of Rx knockdown on photoreceptor gene expression are dependent on the PCE-1 site.
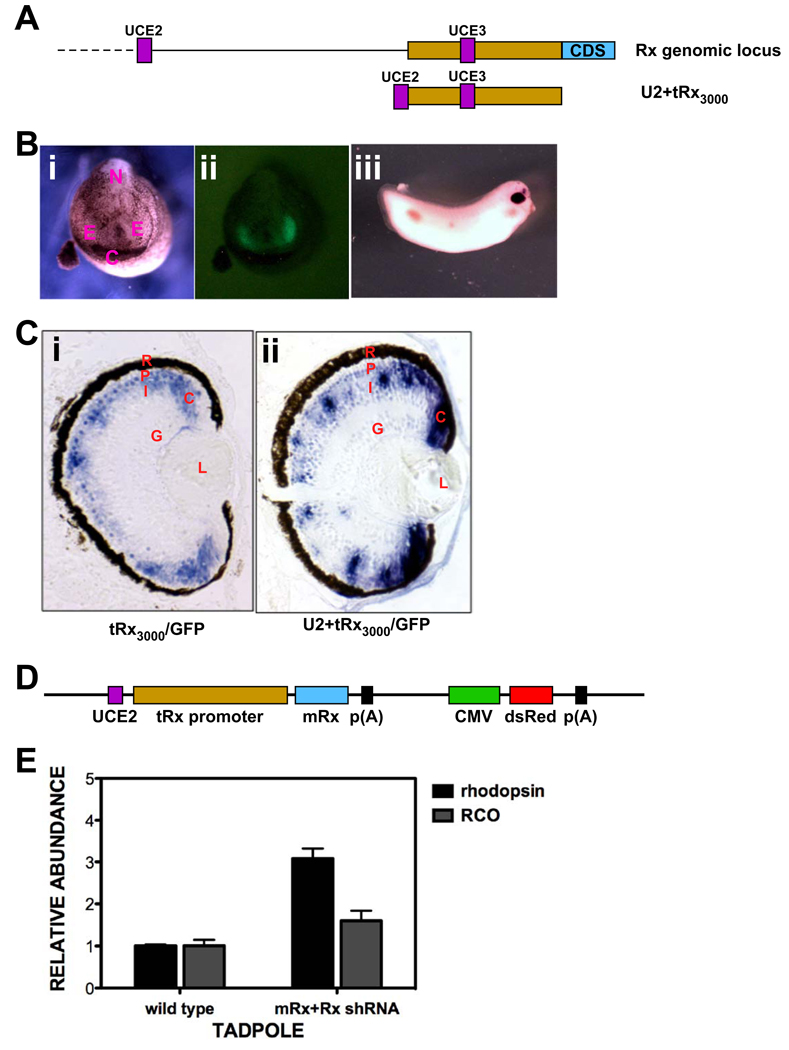
To further investigate the specificity of shRNA-mediated Rx knockdown, we undertook a rescue experiment where exogenous Rx was co-expressed with the Rx shRNA. We chose to express mouse Rx (mRx) since it contains 12 mismatches in the 32-nucleotide shRNA target region (Fig 3A), allowing it to escape knockdown by the Rx shRNA. We used X. tropicalis Rx regulatory regions to drive specific expression of mRx. We combined 3 kb of genomic DNA flanking the 5’-end of the X. tropicalis Rx coding region (tRx3000) with an additional genomic ultra-conserved element (UCE2) (Fig 6A). We found that tRx3000 alone was capable of driving expression of a GFP transgene in a similar pattern to endogenous Rx, including the anterior neural plate and developing eyes at tailbud stages and the photoreceptor layer, INL, and most of the CMZ of the maturing tadpole retina (Fig 6B, C). However, expression was absent from the distal tip of the CMZ, where Rx is expressed in retinal stem cells (compare Fig 6Ci and Fig 1A). We used comparative genomics to identify conserved genomic elements that may function regulating gene expression. The tRx3000 promoter contains three UCEs, previously reported as conserved noncoding sequences (Danno et al., 2008). One of these is located approximately 6 kb upstream of the UCE contained in tRx3000 (UCE3, Fig 6A). We amplified it from genomic DNA and named it UCE2. Addition of UCE2 to tRx3000 (U2+tRx3000) resulted in additional transgene expression in the distal tip of the CMZ (Fig 6Cii), similar to the expression of the endogenous Rx gene. Further characterization of the X. tropicalis Rx promoter and UCEs will be described elsewhere.
Figure 6.
Knockdown of photopigment gene expression can be rescued by expression of mouse Rx. A. Top: Schematic diagram of the X. tropicalis Rx genomic locus. Shown are: two of the three ultra-conserved elements (UCEs) (UCE2 and UCE3, purple), 3 kb promoter (yellow), first coding exon (blue). Bottom: Schematic diagram of Rx regulatory region construct UCE2+tRx3000, containing the 3 kb Rx promoter and UCE2. B. Expression of tRx3000/GFP transgene in tailbud embryos. i: white light image of st 20 neural tube stage embryo; ii: fluorescent image of the same embryo shown in (i); iii: wholemount in situ hybridization of tailbud stage (st 28) tRx3000/GFP transgenic embryo using an antisense riboprobe to GFP. C. In situ hybridization of sections of paraffin-embedded st 41 tadpoles using an antisense riboprobe to GFP. i: The tRx3000/GFP transgene is expressed in the photoreceptor layer, in the INL, and the CMZ. It is not expressed in the distal portion of the CMZ where retinal stem cells are found. ii: Addition of UCE2 to the tRx3000/GFP transgene drives expression throughout the CMZ. D. Schematic diagram of rescue construct. The construct includes the mRx coding region driven by UCE2+tRx3000 and a dsRed expression cassette driven by the CMV promoter for selection of transgenic embryos. E. Expression of photopigment genes rhodopsin and red cone opsin is not reduced in embryos transgenic for both Rx shRNA and the mRx by qRT-PCR.
We next prepared transgenic embryos using both the Rx shRNA and U2+tRx3000-mRx transgenes (Fig 6D). These embryos did not exhibit reduced levels of rho or RCO gene expression (Fig 6E). These results demonstrate that the effects of Rx knockdown by shRNA can be restored by expression of mRx and suggest that the effects of Rx shRNA expression are specifically a result of reduction or loss of Rx expression.
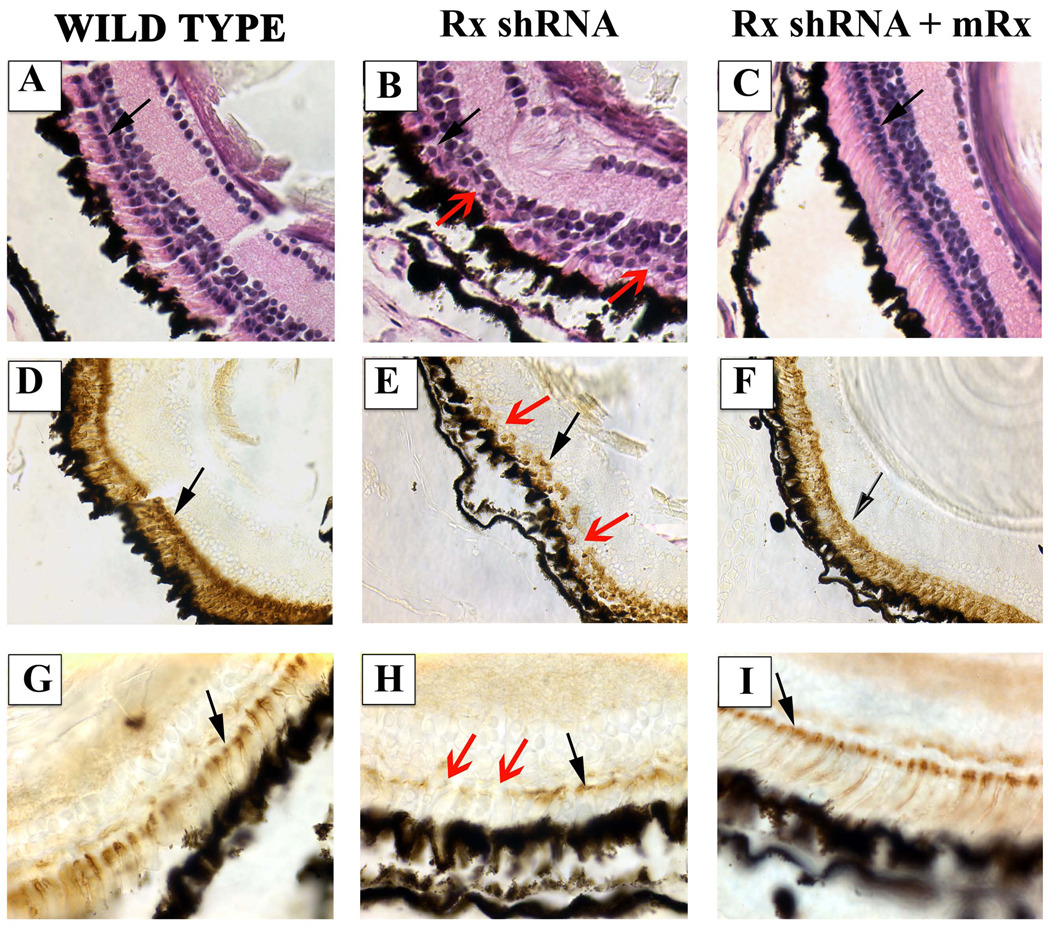
Rod development, morphology, and function are dependent on normal expression, subcellular localization, and function of rhodopsin and other gene products necessary for photoreceptor function (Wilson and Wensel, 2003). We next investigated the downstream effects of Rx knockdown on photoreceptor histology and function. To test photoreceptor function, we employed a vision-dependent behavioral assay that depends on the finding that tadpoles conditioned to a light colored background prefer a light substratum to a dark one (Moriya et al., 1996). We found that 5 tadpoles transgenic for the Rx shRNA exhibited normal visual function in this assay at st 45 (Table 2). However, a few days later (st 50), 3 of 5 of these tadpoles exhibited no preference for substratum color (Table 2), indicating that vision was impaired in these shRNA transgenic tadpoles. This result suggests that visual function developed essentially normal and then deteriorated later. Further, embryos transgenic for both the Rx shRNA and mRx chose the white substratum, indicating that visual function was “rescued”. We found that the vision-impaired tadpoles had abnormal photoreceptors. Photoreceptors exhibited abnormal organization and morphology in these retinas (Fig 7B). The outer segments of these photorecptotrs were extremely short and no longer arranged in a parallel manner (compare Fig 7B to 7A). Additionally, the outer nuclear layer was discontinuous (Fig 7B, E, H - red arrows). Rods appeared normal in retinas of Rx shRNA tadpoles that performed normally in the visual function assay (not shown). We also observed that RPE appeared wavy in affected embryos, perhaps indicating that the neural retina was reduced relative to the RPE. Further, rhodopsin expression was reduced in these tadpoles (Fig 7E). We also observed abnormalities in cone development, as visualized by peanut agglutinin (PNA) staining (Fig 7H). PNA staining was decreased and discontinuous in Rx shRNA transgenics relative to nontransgenic controls. Both rods and cones appeared normal in Rx shRNA mRx co-transgenics (Fig 7A, D, G). Visual function, photoreceptor histology, and rod and cone markers all appeared normal in tadpoles transgenic for both Rx shRNA and U2+tRx3000-mRx (Table 2 and Fig 7C, F, I). Interestingly, some tadpoles receiving the Rx shRNA and mRx transgenes had normal cones but elongated rod photoreceptors (Fig 7C), suggesting that overexpression of mRx can result in hyperdevelopment of photoreceptors. Taken together, these results demonstrate that the development of normal photoreceptor structure and function require normal Rx expression.
TABLE 2.
VISUAL FUNCTION TEST
| TADPOLE | ST 45 | ST 50+ |
|---|---|---|
| Wild type | 100% (10/10) | 100% (10/10) |
| Wild type | 100% (10/10) | 100% (10/10) |
| Wild type | 100% (10/10) | 100% (10/10) |
| Rx shRNA 1 | 100% (10/10) | 80% (8/10) |
| Rx shRNA 2 | 100% (10/10) | 40% (4/10) |
| Rx shRNA 3 | 100% (10/10) | 100% (10/10) |
| Rx shRNA 4 | 100% (10/10) | 60% (12/20) |
| Rx shRNA 5 | 100% (10/10) | 60% (12/20) |
| Rx shRNA + rescue 1 | N.D. | 90% (18/20) |
| Rx shRNA + rescue 2 | N.D. | 90% (17/19) |
| Rx shRNA + rescue 3 | N.D. | 95% (18/19) |
Figure 7.
Specific degeneration of photoreceptors in visually impaired Rx shRNA tadpoles. A – C. Hematoxylin and eosin staining of sections prepared from paraffin-embedded st 50 tadpoles. Black arrows indicate nuclei in the outer nuclear layer (ONL). Red arrow indicates a gap in the ONL. D – F. Immunohistochemical staining of sections from paraffin-embedded tadpoles using an antibody raised against rhodopsin (RetP1). Black arrows indicate RetP1-positive cells in the photoreceptor layer. Red arrow indicates a gap in RetP1 staining in the photoreceptor layer. G – I. Staining of sections prepared from paraffin-embedded tadpoles using peanut agglutinin (PNA). Black arrows indicate PNA-positive cells in the photoreceptor layer. Red arrow indicates a gap in PNA staining in the photoreceptor layer. Wild type (A, D, G), Rx shRNA transgenic (B, E, H), or Rx shRNA + mRx rescue construct cotransgenic (C, F, I) tadpoles were raised to st 50 and tested for visual function (Table 1). Rx shRNA transgenic tadpoles with impaired visual function exhibited abnormal photoreceptor histology, including missing nuclei (red arrow) compared to a nontransgenic control (A, B). Photoreceptor histology was normal in a tadpole transgenic for both Rx shRNA and mRx (C). Rod and cone photoreceptors of Rx shRNA tadpoles exhibited reduced staining with rhodopsin and PNA (red arrows in E and H) compared to nontransgenic controls (D, E, G, H). Rhodopsin and PNA staining appeared normal in tadpoles transgenic for both the Rx shRNA and mRx (F, I).
DISCUSSION
In this paper we presented data demonstrating that Rx gene products play a role in regulating PCE-1-containing promoters. It has been demonstrated that zebrafish Rx gene products are necessary for normal photoreceptor development and expression of normal levels of markers of differentiated photoreceptors (Wilson and Wensel, 2003). We have provided evidence that Rx may directly regulate expression of PCE-1-dependent genes.
It is not entirely surprising that Rx may play a role in promoting transcription of photoreceptor genes. We and others have previously characterized the activity of Rx-like gene products (Ohuchi et al., 1999; Pan et al., 2006; Wang et al., 2004; Wu et al., 2009). These gene products are expressed during photoreceptor differentiation and function as stronger transcriptional activators than Rx. However, mice do not have a Rx-like gene (Wang et al., 2004). This suggests that Rx may play the role of Rx-like gene products in mice in regulating promoters containing PCE-1 sites. If this is the case, then Rx may also play a role in regulating those target genes in species that express a Rx-like gene product. However, it is not clear how Rx function is modified by the co-expression of a Rx-like gene product in photoreceptors.
It has been suggested that Rx may bind to homeobox response elements in the rhodopsin promoter in addition to the PCE-1 site (Wang et al., 2004). It has been demonstrated that Rx can bind the BAT site with reduced affinity (Kimura et al., 2000). We have demonstrated that Rx can bind both the BAT and Ret4 sites, albeit with lower affinity than the PCE-1 site (Pan et al., 2006). In this work, we demonstrated that knockdown of Rx expression results in reduced expression of rhodopsin and a rhodopsin promoter reporter (XOP-dsRed). However, Rx knockdown did not affect expression of the reporter when the PCE-1 site was mutated. This result indicates that Rx does not regulate transcription appreciably through the BAT or Ret4 sites in vivo. Therefore Rx regulates rhodopsin promoter activity primarily through the PCE-1 site.
Expression of mRx in Rx knockdown rescued retinal development, maintenance, and function. Interestingly, these embryos exhibited normal-appearing cones but elongated rods. This suggests that mRx and endogenous Rx1A/Rx2A may not be functionally equivalent. As discussed above, mRx must functionally replace Rx-like transcriptional activators that are absent from the mouse genome. Therefore, it is possible that this dual functionality results in hyperdeveloped rod photoreceptors. Alternatively, there may be other functional differences between mRx and frog Rx orthologs aside from strength of transactivation. Mammalian and Xenopus Rx proteins are highly conserved in the major domains shared among aristaless-related paired type homeobox proteins (octapeptide, homeodomain, OAR domain), but also contain regions of relative divergence. It is possible that these less well-conserved domains contribute to putative functional differences between mouse and frog Rx.
Our results suggest that Rx may have a role in promoting photoreceptor differentiation in the maturing retina in cooperation with crx/otx5b and Nrl/XLmaf. Additional factors are involved in rho promoter regulation in vivo. For example, mammalian rho promoters are also regulated by the zinc finger proteins Sp4 (Lerner et al., 2005) and KLF15 (Otteson et al., 2004) and the nuclear hormone receptors Nr2e3 and Nr1d1 (Cheng et al., 2004). It will be interesting to investigate whether Rx can also cooperate with these additional factors or how Rx interaction with otx5b/Crx and XLmaf/Nrl might be affected by their inclusion.
Rx is already well known for its essential role during retinal specification very early in eye development. Thus the intriguing question arises: does Rx function differ during fundamentally different phases of retinal specification and maturation? It is likely that Rx interacts with different transcription factors when regulating target genes early vs late in retinal development, especially since transcription factors associated with photoreceptor differentiation, such as Crx and Nrl, are not expressed during early phases of mammalian retinal development (Chen et al., 1997; Furukawa et al., 1997b; Swaroop et al., 1992). However, the definitive answer to this question awaits fine characterization of bona fide Rx target promoters that regulate transcription prior to the onset of expression of photoreceptor-specific genes such as opsins.
It has been suggested that the transcription factor that binds the PCE-1 site must function as a transcriptional repressor since deletion of the site results in an increase in opsin promoter activity (Mani et al., 2001). This is in apparent contradiction to the function of Rx as a transcriptional activator, discussed above. It has been proposed that Rx may repress opsin promoter activity in vivo since it is primarily expressed early in development, before the onset of opsin gene expression. However, Rx expression has been reported in the maturing Xenopus retina (Perron et al., 1998 and this work) and in mature photoreceptors in the zebrafish retina (Chuang et al., 1999). Additionally, Rx-like gene products are expressed in developing photoreceptors. QRX and Erx have been identified as additional gene products that bind to PCE-1 sites in cDNA library screens (Martinez and Barnstable, 1998; Wang et al., 2004). QRX and other Rx-like gene products, chicken cRaxL and Xenopus Rx-L, share a conserved homeodomain and OAR domain with Rx but differ elsewhere (Chen and Cepko, 2002; Ohuchi et al., 1999; Pan et al., 2006; Wang et al., 2004). Little is known about Erx aside from its partial sequence and expression in photoreceptors. Recently, it has been demonstrated that another retinal paired-type homeobox protein, Chx10/Vsx2, also binds the PCE-1 site (Dorval et al., 2006). Thus, it seems that the results from previous XOP PCE-1 mutagenesis experiments might represent the regulation of reporters by multiple PCE-1-binding proteins (Rx, QRX/RxL, Erx, Vsx). While Rx and Rx-like gene products are expressed in photoreceptors, Chx10/Vsx2 is expressed in a subset of bipolar cells. This is consistent with Rx and Rx-like gene products playing a role in enhancing photoreceptor-specific gene expression. Additionally, Chx10/Vsx2 and other PCE-1 binding gene products may lend specificity to PCE-1-dependent gene expression by repressing photoreceptor-specific gene expression outside of the photoreceptor layer.
Knockdown of Rx expression resulted in a decrease in photoreceptor gene expression and led to a reduction in vision and abnormal photoreceptor arrangement and morphology. These deficits were observed later in tadpole development (st 50+ as opposed to st 45), reminiscent of retinal degeneration. Several retinal degeneration diseases are caused by mutations in rhodopsin that result in its mislocalization and eventual interference in normal photoreceptor function but not a reduction in rhodopsin expression levels (Wilson and Wensel, 2003). Additionally, mutations in IRBP or arrestin genes are found in patients with retinitis pigmentosa, a pregressive retinal dystrophy (den Hollander et al., 2009; Nakazawa et al., 1998; Valverde et al., 1998). However, in the case of our Rx shRNA tadpoles, expression levels of many photoreceptor-specific genes (potentially every gene with a PCE-1-dependent promoter/enhancer) are being reduced simultaneously. The effects of reduction or loss of expression of every PCE-1-dependent gene product is unknown. However, since many, if not most, photoreceptor-specific genes contain conserved PCE-1 sites within regulatory regions, it is easy to imagine that knockdown of these gene products would be catastrophic to the development, maintenance, function, and integrity of photoreceptor cells.
Knockdown of Rx expression using translation-inhibitory antisense morpholino oligonucleotides results in anophthalmia (Pan and El-Hodiri, unpublished and (Bailey et al., 2004)), similar to the phenotype of Rx knock-out mice and Rx mutant fish (Loosli et al., 2003; Loosli et al., 2001; Mathers et al., 1997). We expect the Rx shRNA transcript to be present from mid-blastula stages but did not observe significant reduction in Rx1A transcript levels by Rx shRNA until retinal maturation stages (after st 38). This explains how the retina is able to form essentially normally in these embryos. It is possible that the Rx shRNA is produced but does not accumulate to sufficiently high concentrations to affect Rx expression until relatively late stages allowing early eye development to proceed normally. Alternatively, it is possible that shRNA is produced but is not functional until later stages. It has been reported that knockdown of dicer expression results perturbs retinal development beginning at approximately st 30 (Decembrini et al., 2008). The lack of functional Rx shRNA would allow early eye development to escape the effects of knockdown and only later development would be affected. Indeed, we demonstrated that Rx shRNA embryos exhibit eye defects earlier in development when exogenous Argonaute 2 (Ago2) is also expressed. This suggests that RxshRNA is produced early but is not functional until later stages without additional Argonaute expression. As discussed by Chen and coworkers (Chen, 2009), manipulation of spatial and temporal expression of shRNA and Ago2 allows specific spatial and temporal control of gene knockdown. shRNA and Ago2-mediated knockdown can be regulated by the use of tissue- or developmental stage-specific promoters or inducible forms of these reagents.
Supplementary Material
01
Figure S1 - The silver gene is specifically expressed in the developing eye at tailbud stages. A. Wholemount in situ hybridization of late tailbud stage embryo (st 34) using silver antisense riboprobe. Silver is expressed in the eye. B. The embryos shown in (A) was embedded in paraffin and sectioned. The in situ hybridization signal is localized to the outer layer of the eye, the retinal pigmented epithelium (RPE). C. In situ hybridization performed using sections of a paraffin-embedded st 41 tadpole. Expression of the silver gene (blue color) is restricted to the RPE (brown color). L – lens.
ACKNOWLEDGEMENTS
This work was supported by NIH/NEI grant EY015480 to HME. The authors are grateful to Anne Griep and Kate Hyde for providing hands-on instruction in chromatin immunoprecipitation, Mike Zuber for advice regarding the visual function assay, and Melissa Scott and Andy Fischer for help with quantification of fluorescence. We also thank Rob Orford for sharing the Xenopus ChIP protocol before publication. We thank Carla Green, Barry Knox, Orson Moritz, and Barb Roehrer for providing plasmids and Orson Moritz and Deborah Parris for helpful suggestions regarding shRNA design and function. Finally, we thank Dawn Chandler, Andy Fischer, Brenda Lilly, and Holly Moose for critical reading of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Andreazzoli M, Gestri G, Angeloni D, Menna E, Barsacchi G. Role of Xrx1 in Xenopus eye and anterior brain development. Development. 1999;126:2451–2460. doi: 10.1242/dev.126.11.2451. [DOI] [PubMed] [Google Scholar]
- Bailey TJ, El-Hodiri H, Zhang L, Shah R, Mathers PH, Jamrich M. Regulation of vertebrate eye development by Rx genes. Int J Dev Biol. 2004;48:761–770. doi: 10.1387/ijdb.041878tb. [DOI] [PubMed] [Google Scholar]
- Batni S, Scalzetti L, Moody SA, Knox BE. Characterization of the Xenopus rhodopsin gene. J Biol Chem. 1996;271:3179–3186. doi: 10.1074/jbc.271.6.3179. [DOI] [PubMed] [Google Scholar]
- Boatright JH, Borst DE, Peoples JW, Bruno J, Edwards CL, Si JS, Nickerson JM. A major cis activator of the IRBP gene contains CRX-binding and Ret-1/PCE-I elements. Mol Vis. 1997;3:15. [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, Gifford DK, Melton DA, Jaenisch R, Young RA. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casarosa S, Amato MA, Andreazzoli M, Gestri G, Barsacchi G, Cremisi F. Xrx1 controls proliferation and multipotency of retinal progenitors. Mol Cell Neurosci. 2003;22:25–36. doi: 10.1016/s1044-7431(02)00025-8. [DOI] [PubMed] [Google Scholar]
- Casarosa S, Andreazzoli M, Simeone A, Barsacchi G. Xrx1, a novel Xenopus homeobox gene expressed during eye and pineal gland development. Mech Dev. 1997;61:187–198. doi: 10.1016/s0925-4773(96)00640-5. [DOI] [PubMed] [Google Scholar]
- Chen C-M, Chiu S-L, Shen W, Cline HT. Co-expression of Argonaute2 enhances short hairpin RNA-induced RNA interference in Xenopus CNS neurons in vivo. Frontiers in Neuromethods. 2009;1:1–13. doi: 10.3389/neuro.17.001.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CM, Cepko CL. The chicken RaxL gene plays a role in the initiation of photoreceptor differentiation. Development. 2002;129:5363–5375. doi: 10.1242/dev.00114. [DOI] [PubMed] [Google Scholar]
- Chen S, Wang QL, Nie Z, Sun H, Lennon G, Copeland NG, Gilbert DJ, Jenkins NA, Zack DJ. Crx, a novel Otx-like paired-homeodomain protein, binds to and transactivates photoreceptor cell-specific genes. Neuron. 1997;19:1017–1030. doi: 10.1016/s0896-6273(00)80394-3. [DOI] [PubMed] [Google Scholar]
- Cheng H, Khanna H, Oh EC, Hicks D, Mitton KP, Swaroop A. Photoreceptor-specific nuclear receptor NR2E3 functions as a transcriptional activator in rod photoreceptors. Hum Mol Genet. 2004;13:1563–1575. doi: 10.1093/hmg/ddh173. [DOI] [PubMed] [Google Scholar]
- Chuang JC, Mathers PH, Raymond PA. Expression of three Rx homeobox genes in embryonic and adult zebrafish. Mech Dev. 1999;84:195–198. doi: 10.1016/s0925-4773(99)00077-5. [DOI] [PubMed] [Google Scholar]
- Chuang JC, Raymond PA. Zebrafish genes rx1 and rx2 help define the region of forebrain that gives rise to retina. Dev Biol. 2001;231:13–30. doi: 10.1006/dbio.2000.0125. [DOI] [PubMed] [Google Scholar]
- Danno H, Michiue T, Hitachi K, Yukita A, Ishiura S, Asashima M. Molecular links among the causative genes for ocular malformation: Otx2 and Sox2 coregulate Rax expression. Proc Natl Acad Sci U S A. 2008;105:5408–5413. doi: 10.1073/pnas.0710954105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decembrini S, Andreazzoli M, Barsacchi G, Cremisi F. Dicer inactivation causes heterochronic retinogenesis in Xenopus laevis. Int J Dev Biol. 2008;52:1099–1103. doi: 10.1387/ijdb.082646sd. [DOI] [PubMed] [Google Scholar]
- den Hollander AI, McGee TL, Ziviello C, Banfi S, Dryja TP, Gonzalez-Fernandez F, Ghosh D, Berson EL. A homozygous missense mutation in the IRBP gene (RBP3) associated with autosomal recessive retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2009;50:1864–1872. doi: 10.1167/iovs.08-2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschet K, Bourrat F, Ristoratore F, Chourrout D, Joly JS. Expression of the medaka (Oryzias latipes) Ol-Rx3 paired-like gene in two diencephalic derivatives, the eye and the hypothalamus. Mech Dev. 1999;83:179–182. doi: 10.1016/s0925-4773(99)00037-4. [DOI] [PubMed] [Google Scholar]
- Diederichs S, Jung S, Rothenberg SM, Smolen GA, Mlody BG, Haber DA. Coexpression of Argonaute-2 enhances RNA interference toward perfect match binding sites. Proc Natl Acad Sci U S A. 2008;105:9284–9289. doi: 10.1073/pnas.0800803105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorval KM, Bobechko BP, Fujieda H, Chen S, Zack DJ, Bremner R. CHX10 targets a subset of photoreceptor genes. J Biol Chem. 2006;281:744–751. doi: 10.1074/jbc.M509470200. [DOI] [PubMed] [Google Scholar]
- El-Hodiri HM, Shou W, Etkin LD. xnf7 functions in dorsal-ventral patterning of the Xenopus embryo. Dev Biol. 1997;190:1–17. doi: 10.1006/dbio.1997.8692. [DOI] [PubMed] [Google Scholar]
- Faehnle CR, Joshua-Tor L. Argonautes confront new small RNAs. Curr Opin Chem Biol. 2007;11:569–577. doi: 10.1016/j.cbpa.2007.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa T, Kozak CA, Cepko CL. rax, a novel paired-type homeobox gene, shows expression in the anterior neural fold and developing retina. Proc Natl Acad Sci U S A. 1997a;94:3088–3093. doi: 10.1073/pnas.94.7.3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa T, Morrow EM, Cepko CL. Crx, a novel otx-like homeobox gene, shows photoreceptor-specific expression and regulates photoreceptor differentiation. Cell. 1997b;91:531–541. doi: 10.1016/s0092-8674(00)80439-0. [DOI] [PubMed] [Google Scholar]
- Furukawa T, Mukherjee S, Bao ZZ, Morrow EM, Cepko CL. rax, Hes1, and notch1 promote the formation of Muller glia by postnatal retinal progenitor cells. Neuron. 2000;26:383–394. doi: 10.1016/s0896-6273(00)81171-x. [DOI] [PubMed] [Google Scholar]
- Ghai K, Zelinka C, Fischer AJ. Serotonin released from amacrine neurons is scavenged and degraded in bipolar neurons in the retina. J Neurochem. 2009;111:1–14. doi: 10.1111/j.1471-4159.2009.06270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hock J, Meister G. The Argonaute protein family. Genome Biol. 2008;9:210. doi: 10.1186/gb-2008-9-2-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Marsh-Armstrong N, Brown DD. Metamorphosis is inhibited in transgenic Xenopus laevis tadpoles that overexpress type III deiodinase. Proc Natl Acad Sci U S A. 1999;96:962–967. doi: 10.1073/pnas.96.3.962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- Ishibashi S, Yasuda K. Distinct roles of maf genes during Xenopus lens development. Mech Dev. 2001;101:155–166. doi: 10.1016/s0925-4773(00)00585-2. [DOI] [PubMed] [Google Scholar]
- Kelly LE, Carrel TL, Herman GE, El-Hodiri HM. Pbx1 and Meis1 regulate activity of the Xenopus laevis Zic3 promoter through a highly conserved region. Biochem Biophys Res Commun. 2006;344:1031–1037. doi: 10.1016/j.bbrc.2006.03.235. [DOI] [PubMed] [Google Scholar]
- Kikuchi T, Raju K, Breitman ML, Shinohara T. The proximal promoter of the mouse arrestin gene directs gene expression in photoreceptor cells and contains an evolutionarily conserved retinal factor-binding site. Mol Cell Biol. 1993;13:4400–4408. doi: 10.1128/mcb.13.7.4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura A, Singh D, Wawrousek EF, Kikuchi M, Nakamura M, Shinohara T. Both PCE-1/RX and OTX/CRX interactions are necessary for photoreceptor-specific gene expression. J Biol Chem. 2000;275:1152–1160. doi: 10.1074/jbc.275.2.1152. [DOI] [PubMed] [Google Scholar]
- Lerner LE, Peng GH, Gribanova YE, Chen S, Farber DB. Sp4 is expressed in retinal neurons, activates transcription of photoreceptor-specific genes, and synergizes with Crx. J Biol Chem. 2005;280:20642–20650. doi: 10.1074/jbc.M500957200. [DOI] [PubMed] [Google Scholar]
- Li M, Rohrer B. Gene silencing in Xenopus laevis by DNA vector-based RNA interference and transgenesis. Cell Res. 2006;16:99–105. doi: 10.1038/sj.cr.7310013. [DOI] [PubMed] [Google Scholar]
- Liu Y, Lupo G, Marchitiello A, Gestri G, He RQ, Banfi S, Barsacchi G. Expression of the Xvax2 gene demarcates presumptive ventral telencephalon and specific visual structures in Xenopus laevis. Mech Dev. 2001;100:115–118. doi: 10.1016/s0925-4773(00)00505-0. [DOI] [PubMed] [Google Scholar]
- Loosli F, Staub W, Finger-Baier KC, Ober EA, Verkade H, Wittbrodt J, Baier H. Loss of eyes in zebrafish caused by mutation of chokh/rx3. EMBO Rep. 2003;4:894–899. doi: 10.1038/sj.embor.embor919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loosli F, Winkler S, Burgtorf C, Wurmbach E, Ansorge W, Henrich T, Grabher C, Arendt D, Carl M, Krone A, Grzebisz E, Wittbrodt J. Medaka eyeless is the key factor linking retinal determination and eye growth. Development. 2001;128:4035–4044. doi: 10.1242/dev.128.20.4035. [DOI] [PubMed] [Google Scholar]
- Ma GC, Wang TM, Su CY, Wang YL, Chen S, Tsai HJ. Retina-specific cis-elements and binding nuclear proteins of carp rhodopsin gene. FEBS Lett. 2001;508:265–271. doi: 10.1016/s0014-5793(01)03058-7. [DOI] [PubMed] [Google Scholar]
- Mani SS, Batni S, Whitaker L, Chen S, Engbretson G, Knox BE. Xenopus rhodopsin promoter. Identification of immediate upstream sequences necessary for high level, rod-specific transcription. J Biol Chem. 2001;276:36557–36565. doi: 10.1074/jbc.M101685200. [DOI] [PubMed] [Google Scholar]
- Martinez JA, Barnstable CJ. Erx, a novel retina-specific homeodomain transcription factor, can interact with Ret 1/PCEI sites. Biochem Biophys Res Commun. 1998;250:175–180. doi: 10.1006/bbrc.1998.9261. [DOI] [PubMed] [Google Scholar]
- Mathers PH, Grinberg A, Mahon KA, Jamrich M. The Rx homeobox gene is essential for vertebrate eye development. Nature. 1997;387:603–607. doi: 10.1038/42475. [DOI] [PubMed] [Google Scholar]
- Mathers PH, Jamrich M. Regulation of eye formation by the Rx and pax6 homeobox genes. Cell Mol Life Sci. 2000;57:186–194. doi: 10.1007/PL00000683. [DOI] [PubMed] [Google Scholar]
- Mears AJ, Kondo M, Swain PK, Takada Y, Bush RA, Saunders TL, Sieving PA, Swaroop A. Nrl is required for rod photoreceptor development. Nat Genet. 2001;29:447–452. doi: 10.1038/ng774. [DOI] [PubMed] [Google Scholar]
- Mitton KP, Swain PK, Chen S, Xu S, Zack DJ, Swaroop A. The leucine zipper of NRL interacts with the CRX homeodomain. A possible mechanism of transcriptional synergy in rhodopsin regulation. J Biol Chem. 2000;275:29794–29799. doi: 10.1074/jbc.M003658200. [DOI] [PubMed] [Google Scholar]
- Moritz OL, Peck A, Tam BM. Xenopus laevis red cone opsin and Prph2 promoters allow transgene expression in amphibian cones, or both rods and cones. Gene. 2002;298:173–182. doi: 10.1016/s0378-1119(02)00923-x. [DOI] [PubMed] [Google Scholar]
- Moriya T, Kito K, Miyashita Y, Asami K. Preference for background color of the Xenopus laevis tadpole. J Exp Zool. 1996;276:335–344. doi: 10.1002/(SICI)1097-010X(19961201)276:5<335::AID-JEZ4>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Nakazawa M, Wada Y, Tamai M. Arrestin gene mutations in autosomal recessive retinitis pigmentosa. Arch Ophthalmol. 1998;116:498–501. doi: 10.1001/archopht.116.4.498. [DOI] [PubMed] [Google Scholar]
- Nelson SM, Frey RA, Wardwell SL, Stenkamp DL. The developmental sequence of gene expression within the rod photoreceptor lineage in embryonic zebrafish. Dev Dyn. 2008;237:2903–2917. doi: 10.1002/dvdy.21721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson SM, Park L, Stenkamp DL. Retinal homeobox 1 is required for retinal neurogenesis and photoreceptor differentiation in embryonic zebrafish. Dev Biol. 2009;328:24–39. doi: 10.1016/j.ydbio.2008.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. Garland Publishing, Inc. New York: 1994. Normal Table of Xenopus laevis (Daudin) [Google Scholar]
- Ohuchi H, Tomonari S, Itoh H, Mikawa T, Noji S. Identification of chick rax/rx genes with overlapping patterns of expression during early eye and brain development. Mech Dev. 1999;85:193–195. doi: 10.1016/s0925-4773(99)00094-5. [DOI] [PubMed] [Google Scholar]
- Otteson DC, Lai H, Liu Y, Zack DJ. Zinc-finger domains of the transcriptional repressor KLF15 bind multiple sites in rhodopsin and IRBP promoters including the CRS-1 and G-rich repressor elements. BMC Mol Biol. 2005;6:15. doi: 10.1186/1471-2199-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otteson DC, Liu Y, Lai H, Wang C, Gray S, Jain MK, Zack DJ. Kruppel-like factor 15, a zinc-finger transcriptional regulator, represses the rhodopsin and interphotoreceptor retinoid-binding protein promoters. Invest Ophthalmol Vis Sci. 2004;45:2522–2530. doi: 10.1167/iovs.04-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddison PJ, Hannon GJ. RNA interference: the new somatic cell genetics? Cancer Cell. 2002;2:17–23. doi: 10.1016/s1535-6108(02)00092-2. [DOI] [PubMed] [Google Scholar]
- Pan Y, Nekkalapudi S, Kelly LE, El-Hodiri HM. The Rx-like homeobox gene (Rx-L) is necessary for normal photoreceptor development. Invest Ophthalmol Vis Sci. 2006;47:4245–4253. doi: 10.1167/iovs.06-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perron M, Kanekar S, Vetter ML, Harris WA. The genetic sequence of retinal development in the ciliary margin of the Xenopus eye. Dev Biol. 1998;199:185–200. doi: 10.1006/dbio.1998.8939. [DOI] [PubMed] [Google Scholar]
- Seufert DW, Prescott NL, El-Hodiri HM. Xenopus aristaless-related homeobox (xARX) gene product functions as both a transcriptional activator and repressor in forebrain development. Dev Dyn. 2005;232:313–324. doi: 10.1002/dvdy.20234. [DOI] [PubMed] [Google Scholar]
- Sive HL, Grainger RM, Harland RM. Early Development of Xenopus laevis: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2000. [Google Scholar]
- Sparrow DB, Latinkic B, Mohun TJ. A simplified method of generating transgenic Xenopus. Nucleic Acids Res. 2000;28:E12. doi: 10.1093/nar/28.4.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaroop A, Xu JZ, Pawar H, Jackson A, Skolnick C, Agarwal N. A conserved retina-specific gene encodes a basic motif/leucine zipper domain. Proc Natl Acad Sci U S A. 1992;89:266–270. doi: 10.1073/pnas.89.1.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner DL, Weintraub H. Expression of achaete-scute homolog 3 in Xenopus embryos converts ectodermal cells to a neural fate. Genes Dev. 1994;8:1434–1447. doi: 10.1101/gad.8.12.1434. [DOI] [PubMed] [Google Scholar]
- Valverde D, Vazquez-Gundin F, del Rio E, Calaf M, Fernandez JL, Baiget M. Analysis of the IRBP gene as a cause of RP in 45 ARRP Spanish families. Autosomal recessive retinitis pigmentosa. Interstitial retinol binding protein. Spanish Multicentric and Multidisciplinary Group for Research into Retinitis Pigmentosa. Ophthalmic Genet. 1998;19:197–202. doi: 10.1076/opge.19.4.197.2312. [DOI] [PubMed] [Google Scholar]
- Viczian AS, Vignali R, Zuber ME, Barsacchi G, Harris WA. XOtx5b and XOtx2 regulate photoreceptor and bipolar fates in the Xenopus retina. Development. 2003;130:1281–1294. doi: 10.1242/dev.00343. [DOI] [PubMed] [Google Scholar]
- Voronina VA, Kozhemyakina EA, O'Kernick CM, Kahn ND, Wenger SL, Linberg JV, Schneider AS, Mathers PH. Mutations in the human RAX homeobox gene in a patient with anophthalmia and sclerocornea. Hum Mol Genet. 2004;13:315–322. doi: 10.1093/hmg/ddh025. [DOI] [PubMed] [Google Scholar]
- Wang QL, Chen S, Esumi N, Swain PK, Haines HS, Peng G, Melia BM, McIntosh I, Heckenlively JR, Jacobson SG, Stone EM, Swaroop A, Zack DJ. QRX, a novel homeobox gene, modulates photoreceptor gene expression. Hum Mol Genet. 2004;13:1025–1040. doi: 10.1093/hmg/ddh117. [DOI] [PubMed] [Google Scholar]
- Wells J, Graveel CR, Bartley SM, Madore SJ, Farnham PJ. The identification of E2F1-specific target genes. Proc Natl Acad Sci U S A. 2002;99:3890–3895. doi: 10.1073/pnas.062047499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitaker SL, Knox BE. Conserved transcriptional activators of the Xenopus rhodopsin gene. J Biol Chem. 2004;279:49010–49018. doi: 10.1074/jbc.M406080200. [DOI] [PubMed] [Google Scholar]
- Wilson JH, Wensel TG. The nature of dominant mutations of rhodopsin and implications for gene therapy. Mol Neurobiol. 2003;28:149–158. doi: 10.1385/MN:28:2:149. [DOI] [PubMed] [Google Scholar]
- Wu HY, Perron M, Hollemann T. The role of Xenopus Rx-L in photoreceptor cell determination. Dev Biol. 2009;327:352–365. doi: 10.1016/j.ydbio.2008.12.017. [DOI] [PubMed] [Google Scholar]
- Zhang L, El-Hodiri HM, Ma HF, Zhang X, Servetnick M, Wensel TG, Jamrich M. Targeted expression of the dominant-negative FGFR4a in the eye using Xrx1A regulatory sequences interferes with normal retinal development. Development. 2003;130:4177–4186. doi: 10.1242/dev.00626. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01
Figure S1 - The silver gene is specifically expressed in the developing eye at tailbud stages. A. Wholemount in situ hybridization of late tailbud stage embryo (st 34) using silver antisense riboprobe. Silver is expressed in the eye. B. The embryos shown in (A) was embedded in paraffin and sectioned. The in situ hybridization signal is localized to the outer layer of the eye, the retinal pigmented epithelium (RPE). C. In situ hybridization performed using sections of a paraffin-embedded st 41 tadpole. Expression of the silver gene (blue color) is restricted to the RPE (brown color). L – lens.