Insights into ubiquitin transfer cascades from a structure of a UbcH5B~Ubiquitin-HECTNEDD4L complex (original) (raw)
. Author manuscript; available in PMC: 2010 Jun 25.
Summary
In E1-E2-E3 ubiquitin (Ub) conjugation cascades, the E2 first forms a transient E2~Ub covalent complex, and then interacts with an E3 for Ub transfer. For cascades involving E3s in the HECT class, Ub is transferred from an associated E2 to the acceptor Cys in the HECT domain C-lobe. To gain insights into this process, we determined the crystal structure of a complex between the HECT domain of NEDD4L and the E2 UbcH5B bearing a covalently-linked Ub at its active site (UbcH5B~Ub). Noncovalent interactions between UbcH5B and the HECT N-lobe and between Ub and the HECT domain C-lobe lead to an overall compact structure, with the Ub C-terminus sandwiched between UbcH5B and HECT domain active sites. The structure suggests a model for E2-to-HECT Ub transfer, in which interactions between a donor Ub and an acceptor domain constrain upstream and downstream enzymes for conjugation.
Keywords: Ubiquitin, HECT, E3, Ubiquitin ligase, UbcH5B, NEDD4L, NEDD4-2
Introduction
A key question in understanding ubiquitin (Ub) conjugation is how Ub is transferred between enzymes in E1-E2-E3 cascades. For E3s in the HECT (Homologous to E6AP C-Terminus) class, the ~40 kDa C-terminal HECT domain binds a reactive thioester-linked E2~Ub (here “~” refers to thioester or thioester-like covalent linkage). Then a transthiolation reaction ensues, whereby Ub is transferred from the E2 catalytic Cys to the HECT domain catalytic Cys (Huibregtse et al., 1995; Scheffner et al., 1993). Thus, the Ub C-terminus and the active sites of the E2 and HECT domain must all be juxtaposed for E2-to-E3 Ub transfer.
In humans, nearly 30 HECT E3s (Rotin and Kumar, 2009) become charged by selective interactions with distinct E2s, and subsequently catalyze target ubiquitination. For example, the HECT E3 NEDD4L has been shown to bind and receive Ub from a subset of E2s including UbcH5B and Ube2E3 (Debonneville and Staub, 2004; Fotia et al., 2006). A well-recognized downstream function of NEDD4L is regulation of blood pressure through ubiquitination of the Epithelial Sodium Channel (ENaC) (Kamynina et al., 2001; Rotin, 2008).
Despite important physiological roles of NEDD4L and other HECT E3s (Scheffner and Staub, 2007), their fundamental enzymatic mechanisms remain incompletely understood. A particularly vexing question is how a HECT domain and a specific Ub-loaded E2 interact to promote Ub transfer. Prior studies showed that HECT domains have two structural “lobes” tethered by a flexible linker (Huang et al., 1999; Verdecia et al., 2003). The N-terminal “N-lobe” binds part of an E2 distal from the E2 catalytic Cys. The C-terminal “C-lobe” contains the HECT catalytic Cys, which receives Ub from the E2 to form a thioester-linked E3~Ub complex. In the only crystal structure of an E2-HECT domain complex, containing UbcH7 and the HECT domain of E6AP (hereafter referred to as HECTE6AP), a 41 Å gap separates the E2 and E3 cysteines (Huang et al., 1999). Subsequent structures of isolated HECT domains revealed a range of relative orientations for E2-binding N-lobes and catalytic Cys containing C-lobes (Ogunjimi et al., 2005; Verdecia et al., 2003). However, detailed knowledge of how E2, Ub, and HECT domains may work together is also critical for understanding transfer cascades. To address this problem, we report here the crystal structure of a UbcH5B~Ub-HECTNEDD4L complex. The structure reveals combinatorial interactions between individual components of the E2~Ub intermediate and the HECT N- and C-lobes, respectively, important for E2-to-HECT Ub transfer.
Results and Discussion
An overall compact structure involving HECT N- and C-lobe interactions with E2 and Ub
Wild-type transthiolation complexes containing a thioester-linked E2~Ub and a HECT domain are unstable due to Ub transfer between E2 and E3 catalytic cysteines. Indeed, Ub is readily transferred from the Cys of UbcH5B to that of HECTNEDD4L (the isolated HECT domain from NEDD4L) (Fotia et al., 2006). This is also true for a L3S/T98K mutant UbcH5B with high affinity for some HECT domains (Eletr and Kuhlman, 2007). Ub linked via an oxy-ester bond to a Ser in place of the E2 catalytic Cys85 of the high-affinity UbcH5B (UbcH5B~Ub) is also susceptible to decomposition via a HECTNEDD4L catalytic Cys (Fig S1). Thus, we determined crystal structures of complexes between high-affinity UbcH5B~Ub and inactive versions of HECTNEDD4L (C922S and C922A, 3.1 and 3.3 Å resolution respectively) (Fig 1, S2, S3, Table S1), which superimpose with 0.25 Å root mean square deviation (rmsd). In both, the two UbcH5B~Ub-HECTNEDD4L complexes per asymmetric unit superimpose with ~1.0 Å rmsd. Minor differences between the two complexes result mainly from <4° rotations about hinges between the HECTNEDD4L N- and C-lobes, and between N-lobe subdomains. For simplicity, only one complex in the higher resolution structure containing the HECTNEDD4L C-to-S mutation is generally discussed below.
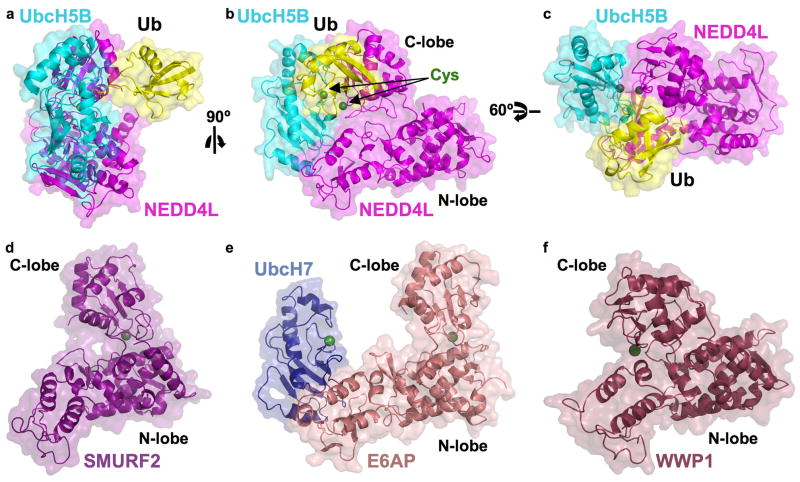
Fig 1. Structure of UbcH5B Ub-HECTNEDD4L.
a–c. 3 views, separated by 90° in y (b) and then 60° in x (c) of UbcH5B (cyan) Ub (yellow)-HECTNEDD4L (magenta). Catalytic residues (here serines, but normally cysteines as indicated) - green spheres.
d–e. Structures of HECTSMURF2 (violet, d), UbcH7-HECTE6AP (navy, pink, e), and HECTWWP1 (maroon), oriented as UbcH5B Ub-HECTNEDD4L in b. Catalytic cysteines - green spheres.
The individual structures of UbcH5B, Ub, and the HECTNEDD4L N- and C-lobes resemble prior E2, Ub, and HECT domain structures (Huang et al., 1999; Ogunjimi et al., 2005; Ozkan et al., 2005; Verdecia et al., 2003). Furthermore, the backbone of residues involved in contacts between UbcH5B with the HECTNEDD4L N-lobe superimpose on the corresponding portions of the earlier UbcH7-HECTE6AP structure (Huang et al., 1999) with 1.2 Å rmsd (Fig 2).
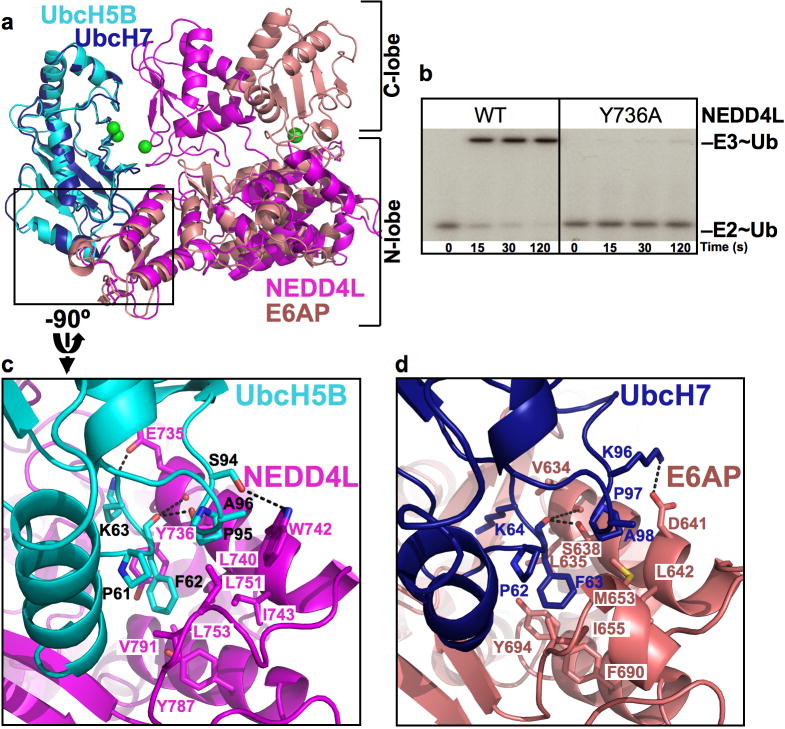
Fig 2. UbcH5B-HECTNEDD4L N-lobe interactions.
a. Structural superposition of the E2s in UbcH5B (cyan) Ub (not shown for clarity)-HECTNEDD4L (magenta) and UbcH7-HECTE6AP (navy, pink). Catalytic residues are green spheres.
b. Autoradiogram showing time course of pulse-chase transfer of 32P-Ub from UbcH5B (E2~Ub) to wild-type (WT) and the Y736A mutant HECTNEDD4L (E3~Ub).
c. Close-up view of interactions between UbcH5B (cyan, black labels) and HECTNEDD4L N-lobe (magenta), oriented by 90° rotation in y relative to a.
d. Close-up view of interactions between UbcH7 (navy) and HECTE6AP (salmon), in same orientation as UbcH5B-HECTNEDD4L in c. Oxygens are red, nitrogens blue, and salt-bridges/H-bonds dashes.
Ub is located in the center of the complex due to two major structural features that differ from those observed for UbcH7-HECTE6AP and other HECT domains (Fig 1, S4) (Huang et al., 1999; Ogunjimi et al., 2005; Verdecia et al., 2003). First, the HECT C-lobe displays a striking relative rotation in comparison to published HECT domain structures (Fig 1, S4). Second, the HECTNEDD4L C-lobe interacts noncovalently with the Ub globular domain (Fig 1, 3a). Thus, interactions between individual proteins in the UbcH5B~Ub complex and individual HECT domain N- and C-lobes, respectively, result in an overall compact structure for the UbcH5B~Ub-HECTNEDD4L complex. Interactions between the Ub-loaded UbcH5B and HECTNEDD4L bury ~4000 Å2.
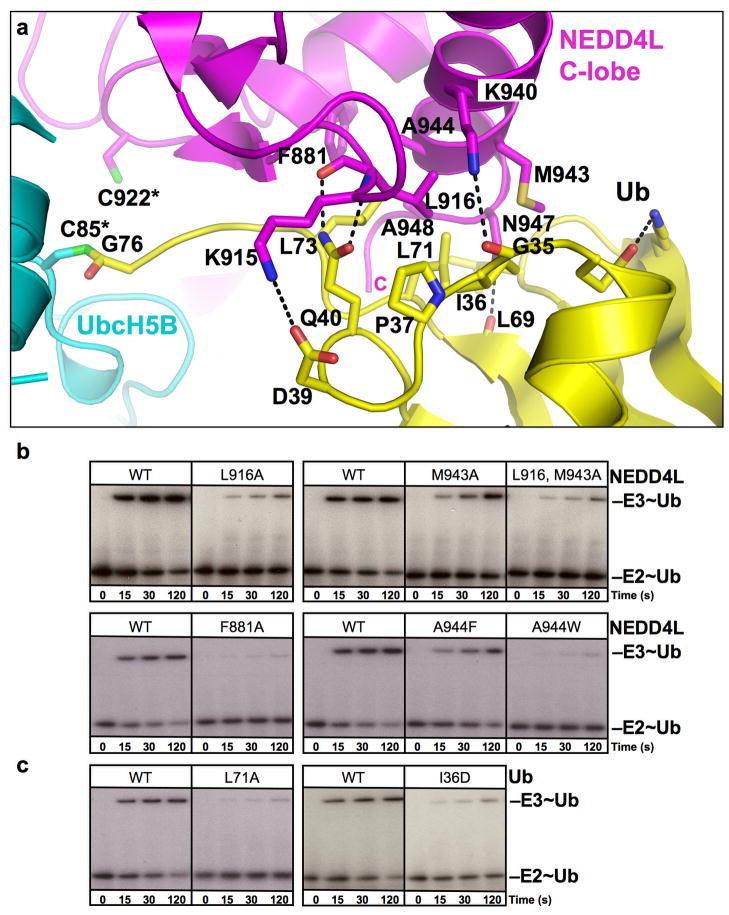
Fig 3. Ub-HECTNEDD4L C-lobe interface.
a. Close-up view of Ub (yellow)-HECTNEDD4L (magenta). In the structure, the active site cysteines are substituted with serines, but are labeled C85* and C922* from UbcH5B and NEDD4L, respectively, to indicate native sequences. Oxygens are red, nitrogens blue, and salt-bridges/H-bonds dashes.
b, c. Autoradiograms showing time course of pulse-chase transfer of 32P-Ub from UbcH5B (E2~Ub) to GST-tagged HECT domain of NEDD4L (E3~Ub), for WT and the indicated mutant of NEDD4L (b) or Ub (c).
In the UbcH5B~Ub-HECTNEDD4L structure, the E2 and HECT domain cysteines face each other, and are separated by <8 Å (Fig 1a–c). The HECT Cys-to-Ub C-terminus gap is ~8Å, which is relatively close but slightly longer than the distance required for Ub transfer to the HECT Cys. The Ub C-terminal tail is sandwiched between the catalytic centers of the E2 and HECT domain C-lobe (Fig 1c, 3a). Thus, the structure provides insights into how the HECT C-lobe, E2, and Ub may approach each other for Ub transfer.
E2-HECT specificity
Comparison of the UbcH5B-HECTNEDD4L interface with that observed in the prior structure of the UbcH7-HECTE6AP complex (Huang et al., 1999) reveals how distinct HECT domain sequences recognize the conserved E2 loop4 Phe (UbcH5B Phe62) absolutely required for binding to HECT E3s (Fig 2) (Nuber and Scheffner, 1999). Parallel surfaces from HECTNEDD4L and HECTE6AP interact with the loop4 Phe in UbcH5B and UbcH7, respectively. However, the identities of loop4 Phe-interacting hydrophobic side-chains differ between HECT domains. As a result, the relative roles of corresponding side-chains in E2-binding may be distributed divergently for the different HECT E3s. For example, HECTNEDD4L residue Tyr736 from Helix7 makes key contacts to the UbcH5B Phe62 (Fig 2). By contrast, in HECTE6AP, this role appears to be partially carried out by Tyr694, which occupies a similar side-chain position even though it comes from Helix8 (Huang et al., 1999). Hydrophobic interactions are generally malleable in nature (Lim and Sauer, 1989), so the wide range of hydrophobic loop4 Phe-binding surfaces displayed by HECT domain sequences may explain how some E2s can interact with numerous E3s, and vice-versa, with a range of affinities that can be regulated via additional protein-protein interactions (Ogunjimi et al., 2005).
The structure also visualizes determinants of E2-HECT selectivity. Known residues influencing specificity are UbcH5B Ser94 and HECTNEDD4L Trp742, which interact via an H-bond (Fig 2c). Trp742 is conserved among NEDD4-family E3s, which may explain their preference for UbcH4/5 and Ube2E E2s E2s that bear a Ser in a position corresponding to UbcH5B Ser94 (Figs S5, S6) (Debonneville and Staub, 2004; Fotia et al., 2006). The parallel interaction from UbcH7-HECTE6AP is a Lys96-Asp641 salt-bridge (Fig 2d). Like E6AP, some other HECT domains (e.g. HECT6, HECTD1, HECTD2, HERC3 and HERC6) have an acidic residue corresponding to HECTNEDD4L Trp742, and may preferentially interact with UbcH7, UbcH8, or Ube2T, potentially via their basic residue corresponding to UbcH5B Ser94. Mutational data support the notion that these polar interactions can influence E2-HECT specificity. A UbcH7 Lys96-to-acidic substitution decreased binding to HECTE6AP, and swapping charges between UbcH7 and HECTE6AP resulted in a high-affinity complex between UbcH7(K96E) and HECTE6AP(D641K) (Eletr and Kuhlman, 2007). In agreement with prior data that HECTNEDD4L is active with UbcH7 (Fotia et al., 2006), the UbcH5B S94K mutation has little effect on its own toward HECTNEDD4L (Fig S7). However, a HECTNEDD4L W742D mutant is inactivated toward UbcH5B, and this can be partially rescued by a UbcH5B S94K mutation (Fig S7).
Interactions between the HECTNEDD4L C-lobe and the UbcH5B-linked Ub
The UbcH5B~Ub-HECTNEDD4L structure provides insights into how Ub that is covalently linked to an E2 may be positioned for transfer to the HECT catalytic Cys. The HECT domain C-lobe makes extensive noncovalent contacts with the UbcH5B-linked Ub to bury ~1400 Å2 of surface area (Fig 3a). The interface is anchored by hydrophobic contacts from Ub residues Ile36, Leu71, and Leu73, and HECTNEDD4L residues Leu916 and Met943, and is further supported by Ub Pro37 and NEDD4L Phe881, Ala944, and Ala948. The interaction also involves a salt-bridge between Ub Asp39 and HECTNEDD4L Lys915, and by hydrogen bonds between the backbone oxygen of Ub Gly35 and HECTNEDD4L Lys940, Ub Gln40 and the backbone of HECTNEDD4L Leu916, and the backbone oxygen of Ub Leu69 and Asn947. We tested the importance of the Ub interactions with the C-lobe using a pulse-chase protocol to exclusively monitor effects of mutations on Ub transfer from UbcH5B to a GST-tagged version of the NEDD4L HECT domain. This assay excludes any mutagenic effects on the earlier step of forming the thioester intermediate between UbcH5B and Ub. Indeed, mutations in key Ub-HECTNEDD4L interface residues are deleterious (Fig 3b, c).
To gain insights into the roles of Ub hydrophobic patch interactions, we wished to perform kinetic analyses. The rapid velocity of UbcH5B-to-HECTNEDD4L transthiolation precluded kinetic analysis of wild-type proteins. Thus, we crippled the reaction with mutations at or near the active sites and distal from the interface with the Ub hydrophobic patch (UbcH5B N81A, or HECTNEDD4L T921A or C922S). We used multiple backgrounds since the crippling mutations themselves may alter catalytic mechanism in different ways (Fig S8). Although we were unable to saturate the rates for many mutants, we were able to obtain data for the single Ub I36D and L71A mutants. Whereas the I36D mutant, at the heart of the interface, had little effect on kcat, the Ub L71A mutant displayed either decreased or increased kcat values depending on the background, which might reflect its relative proximity to the active sites in the structure and differences in catalytic mechanisms among the crippled backgrounds. In all backgrounds, the Ub mutations increase _K_m (Fig S8), which is consistent with a role in binding the C-lobe during the transthiolation reaction.
Many Ub-interacting NEDD4L residues are conserved as hydrophobic among some HECT E3s (Fig S6), raising the possibility that other HECT domains could bind E2~Ub via the Ub hydrophobic patch. Consistent with this notion, Ub transfer is impaired by mutations in the corresponding hydrophobic patch in the yeast HECTRsp5 (Fig S9). Furthermore, the Ub I36D and L71A mutations essentially eliminate transfer from UbcH5B to HECT domains from Rsp5 and ITCH under our assay conditions (Fig S9). By contrast, minor effects of the Ub mutations on transfer to HECTE6AP raise the possibility that the structurally-observed interactions may occur for a subset of HECT E3s, such as the NEDD4 family. This difference might also reflect mechanistic distinctions for E6AP (Wang and Pickart, 2005).
The functional importance of the Ub surface that interacts with HECTNEDD4L is substantiated by the previous finding that individual Ala substitutions for Ile36, Leu71, or Leu73 are all lethal to yeast, and that mutation of Asp39 to Ala results in temperature-sensitivity (Sloper-Mould et al., 2001). We also find that mutations in Rsp5 corresponding to the Ub-hydrophobic binding surface fail to complement the temperature-sensitivity of rsp5-1 mutant cells (Fig S9).
Model of an E2-to-HECT transthiolation complex
Although the ~8Å gap between the HECTNEDD4L residue 922 (here a Ser or Ala, but normally the catalytic Cys) and the C-terminus of Ub linked to UbcH5B is still greater than the distance required for nucleophilic attack and Ub transfer, it is significantly closer than E2-to-Ub distances predicted from modeling an E2 onto structures of other HECT E3s. Only an additional relative rotation of ~4° about the flexible hinge linking the HECTNEDD4L lobes is required to generate a structural model of a transthiolation complex (Fig 4). This rotation is in-range with the differences observed between the two complexes in the asymmetric unit of the crystal (Fig S10).
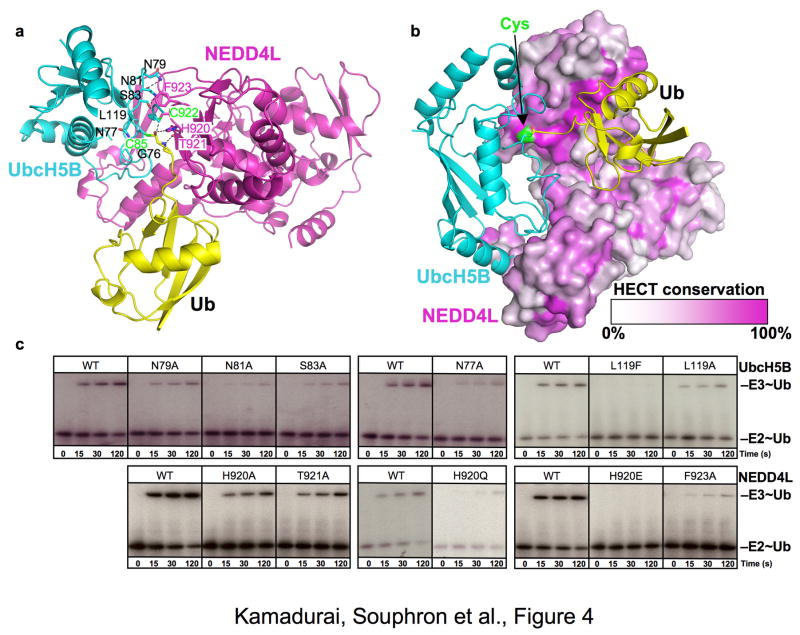
Fig 4. Model for E2-to-HECT E3 transthiolation.
a. Structural model of UbcH5B (cyan, catalytic Cys85 green) Ub (yellow)-NEDD4L HECT domain (magenta) in a putative conformation for the transthiolation reaction. ~4° rotation about the N- and C-lobes allows NEDD4L catalytic Cys922 (green) to approach the Ub C-terminus. Intermolecular contact residues poised to influence transthiolation are represented as sticks.
b. Conservation among HECTs displayed on HECTNEDD4L surface (white – no conservation, magenta – 100% identity except catalytic Cys that is green), in model of transthiolation complex with UbcH5B~Ub.
c. Autoradiograms showing time course of pulse-chase transfer of 32P-Ub from UbcH5B (E2~Ub) to GST-tagged HECT domain of NEDD4L (E3~Ub) for WT and indicated mutants in UbcH5B (top) or NEDD4L (bottom).
Our rotated model for E2-to-HECT transthiolation is supported by several features of the structure and by mutational analysis. First, a key feature of the model is that the HECT domain C-lobe maintains its interaction with the E2-linked Ub, which we find important for Ub transfer from UbcH5B to HECTNEDD4L (Fig 3, S11). Second, the portion of the C-lobe forming noncovalent interactions with Ub in the transthiolation model is conserved (Fig 4b, S6). Finally, our model predicts interactions between residues from UbcH5B and HECTNEDD4L during transthiolation, and mutational analyses agree with these predictions. The Ub C-terminal tail is flanked by HECTNEDD4L His920 and Thr921 and by UbcH5B Asn77 and Leu119. In turn, UbcH5B Leu119 makes van der Waals interactions with HECTNEDD4L His920 and Phe923 and also with the catalytic Cys922. Structural complementarity between UbcH5B L119 and HECTNEDD4L Phe923 is supported by mutational analysis, and conservation of these residues among many E2s and HECTs may reflect their importance (Fig S5, S6, S12). Furthermore, the HECTNEDD4L catalytic Cys922 is linked to an electrostatic network involving UbcH5B Asn79, Asn81, and Ser83. In support of the model, substitution of any of these residues with Ala impairs Ub transfer from UbcH5B to HECTNEDD4L (Fig 4c), although a definitive understanding of the role of each side chain in E2-to-HECT transthiolation will require additional higher resolution structures of catalytic complexes.
Implications for E1-E2-HECT cascades
The UbcH5B~Ub-HECTNEDD4L structure supports the notion that formation of the HECT~Ub thioester requires cycling of the E2 between E1 for its own loading with Ub, and then transferring Ub to the HECT Cys. Prior studies indicated that E1 and HECT E3 binding to an E2 is mutually exclusive (Eletr et al., 2005; Huang et al., 2005; Ozkan et al., 2005). Our data suggest that the extent of structural overlap is even greater than previously described, as the same catalytic Cys face of the E2 is occupied during receipt of a Ubl from E1 (Huang et al., 2007), and during transfer to HECTNEDD4L via anchoring from the covalently attached Ub (Fig 1). Based on the prior UbcH7-HECTE6AP structure (Huang et al., 1999), it seems likely that a Ub-loaded E2 would first engage a HECT domain N-lobe, and that conformational searching involving rotation between the HECT N- and C-lobes (Verdecia et al., 2003), and between the E2 and its covalently-bound Ub would allow E2-to-HECT Ub transfer.
The UbcH5B~Ub-HECTNEDD4L structure may also provide insights into downstream steps in E1-E2-HECT cascades. Following formation of a thioester-linked Ub~HECT intermediate, an isopeptide-linked polyUb chain is built on the target. A study of Rsp5 indicated that this occurs via a “sequential addition” mechanism, with Ubs transferred one-at-a-time from the HECT Cys first to a target acceptor residue, then to a specific Ub Lys of a growing polyUb chain (Kim and Huibregtse, 2009). It was proposed that HECT domain C-lobes bind noncovalently and position the thioester-bound Ub during these latter steps of transfer (Kim and Huibregtse, 2009). Although isolated Ub has not been found to bind to HECT domain C-lobes (French et al., 2009; Kim and Huibregtse, 2009)(also our unpublished data), roles for a C-lobe binding noncovalently to its thioester-bound Ub during ligation may be indicated by the findings that (1) swapping a HECT E3 C-lobe can swap specificity for a Ub acceptor Lys (Kim and Huibregtse, 2009), and (2) mutation of the conserved Phe near HECT domain C-termini (“-4F” – not visible in any HECT structure to date) almost entirely blocks Ub ligation from a HECT E3 to a target (Salvat et al., 2004). It is tempting to speculate that Rsp5 C-lobe-Ub interactions may resemble the UbcH5B~Ub-HECTNEDD4L structure, as polyUb chain specificity requires both the Rsp5 β-strand region and α-helix (Kim and Huibregtse, 2009), which corresponds to the location of the NEDD4L hydrophobic patch. Also, the -4F immediately follows the most C-terminally observed amino acid in UbcH5B~Ub-HECTNEDD4L. The NEDD4L C-lobe contacts observed in our structure would position Ub such that the -4F could access either the Ub C-terminal tail or globular domain to exert its catalytic function (Fig 3a). C-lobe-Ub interactions could also minimize potential clashing with other NEDD4L domains and/or the substrate during rotations en route to the site of Ub attachment during polyubiquitination. Future studies will be required to provide dynamic details of both upstream and downstream steps in HECT cascades.
Emerging roles for the “2nd hydrophobic patch” and other Ub surfaces
Despite its small size, Ub is recognized by a plethora of distinct partners that together utilize a remarkable fraction of the Ub surface. Early studies focused on interactions with the canonical “Leu8/Ile44/Val70 Ub hydrophobic patch” (Beal et al., 1998) that is recognized by the 26S Proteasome and Ub-binding domains such as UIM, UBA, NZF, MIU, and others (Harper and Schulman, 2006; Hicke et al., 2005; Hurley et al., 2006). However, a comprehensive analysis of the effects of surface Ala mutations on yeast growth revealed two other essential hydrophobic surfaces (Sloper-Mould et al., 2001), including the Ile36/Leu71/Leu73 patch we observe to bind the NEDD4L C-lobe. The third Ub surface essential for yeast viability is centered around Gln2/Phe4/Thr12 (Sloper-Mould et al., 2001). This latter patch was recently shown to bind the NEMO UBAN motif (Rahighi et al., 2009), and genetic data indicate other interactions (Sloper-Mould et al., 2001). Thus, future studies will undoubtedly reveal additional roles for Gln2/Phe4/Thr12, and most likely the Ile36/Leu71/Leu73 hydrophobic patch and other surfaces of Ub (Lee et al., 2006; Penengo et al., 2006). In support of this notion, the Ile36/Leu71/Leu73 hydrophobic patch is also responsible for high-affinity interactions with the Vps9 CUE domain (Prag et al., 2003), and with the ZnF-UBP domain as found in IsoT (Reyes-Turcu et al., 2006). Comparison to the UbcH5B~Ub-HECTNEDD4L structure reveals how three distinct folds make related complementary interactions with this common “2nd hydrophobic patch” on Ub (Fig S13).
Comparison to other acceptors of Ub/Ubls from E2
An important question in Ub/Ubl casades is how upstream enzymes and their thioester-bound Ub/Ubls come together with downstream enzymes or targets that accept Ub. The UbcH5B~Ub-HECTNEDD4L structure provides insights as to how the HECT C-lobe might be placed relative to E2~Ub for the transthiolation reaction. This is reminiscent of characteristics recently observed in other, distinct E2-target structures in Ub/Ubl pathways (Fig S14). For example, Mms2-Ubc13-mediated generation of Lys63-linked polyUb chains involves positioning of the acceptor Ub acceptor via noncovalent interactions with Mms2, which in turn interacts with and thus positions the E2 Ubc13 (Eddins et al., 2006). A distinct mechanism was observed for positioning of the SUMO acceptor Lys in RanGAP1, by direct noncovalent interactions with the SUMO E2 Ubc9 (Bernier-Villamor et al., 2002; Reverter and Lima, 2005; Yunus and Lima, 2006). Indeed, UbcH5B residues that influence Ub transfer to HECTNEDD4L partially correspond to those important for Ub transfer from Ubc13 and SUMO transfer from Ubc9. For example, UbcH5B Asn77 corresponds to the “catalytic Asn” in the conserved E2 “HPN motif” implicated in (1) stabilizing the structure of the E2 loop that contains UbcH5B Asp117 and Leu119 (Yunus and Lima, 2006), (2) suppressing the pKa of a target Lys (Yunus and Lima, 2006), and (3) stabilizing the oxyanion intermediate during RING E3-mediated Ub conjugation (Wu et al., 2003) (Fig S14). Although a conservative Asn-to-Gln substitution did not impair Ub transfer from an E2 to HECTE6AP (Wu et al., 2003), the kcat and Km defects of an Ala mutation for Ub transfer to variants of HECTNEDD4L may be most consistent with a structural role for Asn77 in promoting transfer to a HECT domain catalytic Cys (Fig S8). However, we did not observe any effect of an Ala substitution for Asp117 (not shown), which corresponds to a key catalytic residue in Ubc9 (Asp127) (Yunus and Lima, 2006). Therefore, it appears that a subset of common E2 features establish different catalytic functions associated with Ub/Ubl transfer from an E2 to a HECT Cys or a target or Ub acceptor Lys.
General themes in Ub interactions involved in transfer from E2s
An emerging theme from structural studies is that a Ub/Ubl thioester linked to an E2 is optimally oriented by noncovalent interactions that are important for transfer reactions. Although this has not been studied previously for either HECT or RING E3s, such an interaction was first revealed for the unique SUMO E3 RanBP2, which both binds Ubc9 and has a SUMO-interacting motif that also positions the Ubc9-linked SUMO for transfer to a target Lys (Reverter and Lima, 2005). Also, the A20_ZnF domain of Rabex-5 was shown to bind to a distinct Asp58 surface of Ub, presumably to position the E2-linked Ub for the ligation reaction (Lee et al., 2006; Penengo et al., 2006). Furthermore, E3-independent modification of Ub-binding proteins involve their binding noncovalently to the Ub that is thioester-linked to an E2 (Hoeller et al., 2007).
The UbcH5B~Ub-HECTNEDD4L structure reveals a variation on this theme for HECT E3s. By virtue of the HECT domain N-lobe recruiting UbcH5B, and interactions between the Ub (donor) and HECTNEDD4L C-lobe (acceptor), the relative locations of E2, its thioester-bound Ub, and the HECTNEDD4L Cys are all localized to promote conjugation. Downstream steps in target polyubiquitination involve conformational changes about the HECT domain N- and C-lobes (Verdecia et al., 2003). We speculate that access of the different HECT N- and C-lobe sites, whether for loading the HECT Cys, or unloading onto a target or Ub, helps establish coordination of the numerous reactions in the stepwise transfer mechanisms that have been proposed for some HECT transfer cascades (Kim and Huibregtse, 2009; Verdecia et al., 2003).
Experimental Procedures
Crystallography
UbcH5B~Ub was made by colysing cells expressing GST-TEV-GGS-Ub, untagged UbcH5B (L3S/T98K/C85S (Eletr and Kuhlman, 2007) and UBA1. Following glutathione affinity purification and overnight TEV digestion, UbcH5B~Ub was purified by cation exhange and sizing chromatography. GST-HECTNEDD4L was purified similarly, except anion rather than cation exchange was performed, and residual GST removed by affinity. UbcH5B~Ub:HECTNEDD4L were mixed 2:1-1.5, purified by sizing, flash-frozen and stored at −80 °C. Crystals were grown in hanging drops and structures determined as described in Supplemental Data.
Biochemical assays
[32P]-Ub was prepared as in (Eletr et al., 2005). For the pulse (WT and mutants), UbcH5B was [32P]-Ub-loaded and quenched on ice by 10-fold dilution with EDTA to 50 mM to block further E2-loading. [32P]-Ub was chased onto GST-HECT (WT and mutant) after dilution. Reactions were stopped with 2xSDS-buffer and products visualized from dried 4–12% NuPage Bis-Tris gels by autoradiography and/or Storm Phosphorimager. Details are in Supplemental Data.
Supplementary Material
01
Table 1.
Data collection and refinement statistics
| UbcH5B~Ub-NEDD4L Cys922Ser | UbcH5B~Ub-NEDD4L Cys922Ala | |
|---|---|---|
| Data Collection | ||
| Beamline | SERCAT (ID-22 APS) | NECAT (ID-24 APS) |
| Space group | C2221 | C2221 |
| Cell dimensions | ||
| a, b, c (Å) | 174.17, 200.57, 109.82 | 173.98, 199.89, 109.71 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 3.1 | 3.3 |
| _R_sym or _R_merge | 17.3 (38.2) | 12.5 (56.8) |
| I/σ_I_ | 23.4 (3.5) | 14.6 (2.4) |
| Completeness (%) | 98.6 (87.5) | 87.5 (89.4) |
| Refinement | ||
| Resolution (Å) | 50.0-3.1 | 50.0-3.3 |
| No. reflections for refinement | 33799 | 25449 |
| _R_work/_R_free | 25.6/29.0 | 21.7/26.7 |
| R.m.s deviations | ||
| Bond lengths (Å) | 0.009 | 0.009 |
| Bond angles (°) | 1.5 | 1.4 |
| Ramachandran plot statistics | ||
| Most favored regions | 83.8 | 81.7 |
| Additional allowed regions | 15.8 | 17.3 |
| Generously allowed regions | 0.4 | 1.0 |
| Disallowed regions | 0.0 | 0.0 |
Acknowledgments
We thank the Schulman lab for helpful discussions. This work was supported in part by ALSAC, NIH R01GM077053, and the Howard Hughes Medical Institute. BAS is an Investigator of the Howard Hughes Medical Institute. HBK and DS are supported by fellowships from the American Heart Association. We thank staff at SERCAT and NECAT. NECAT is supported by RR-15301 from the NIH NCRR. APS is supported by the US DOE under Contract No. W-31-109-Eng-38.
Footnotes
Accession codes
UbcH5B~Ub-HECTNEDD4L (C922S): 3JW0.pdb
UbcH5B~Ub-HECTNEDD4L (C922A): 3JVZ.pdb
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Beal RE, Toscano-Cantaffa D, Young P, Rechsteiner M, Pickart CM. The hydrophobic effect contributes to polyubiquitin chain recognition. Biochemistry. 1998;37:2925–2934. doi: 10.1021/bi972514p. [DOI] [PubMed] [Google Scholar]
- Bernier-Villamor V, Sampson DA, Matunis MJ, Lima CD. Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell. 2002;108:345–356. doi: 10.1016/s0092-8674(02)00630-x. [DOI] [PubMed] [Google Scholar]
- Debonneville C, Staub O. Participation of the ubiquitin-conjugating enzyme UBE2E3 in Nedd4-2-dependent regulation of the epithelial Na+ channel. Mol Cell Biol. 2004;24:2397–2409. doi: 10.1128/MCB.24.6.2397-2409.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddins MJ, Carlile CM, Gomez KM, Pickart CM, Wolberger C. Mms2-Ubc13 covalently bound to ubiquitin reveals the structural basis of linkage-specific polyubiquitin chain formation. Nat Struct Mol Biol. 2006;13:915–920. doi: 10.1038/nsmb1148. [DOI] [PubMed] [Google Scholar]
- Eletr ZM, Huang DT, Duda DM, Schulman BA, Kuhlman B. E2 conjugating enzymes must disengage from their E1 enzymes before E3-dependent ubiquitin and ubiquitin-like transfer. Nat Struct Mol Biol. 2005;12:933–934. doi: 10.1038/nsmb984. [DOI] [PubMed] [Google Scholar]
- Eletr ZM, Kuhlman B. Sequence determinants of E2-E6AP binding affinity and specificity. J Mol Biol. 2007;369:419–428. doi: 10.1016/j.jmb.2007.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fotia AB, Cook DI, Kumar S. The ubiquitin-protein ligases Nedd4 and Nedd4-2 show similar ubiquitin-conjugating enzyme specificities. Int J Biochem Cell Biol. 2006;38:472–479. doi: 10.1016/j.biocel.2005.11.006. [DOI] [PubMed] [Google Scholar]
- French ME, Kretzmann BR, Hicke L. Regulation of the RSP5 ubiquitin ligase by an intrinsic ubiquitin-binding site. J Biol Chem. 2009;284:12071–12079. doi: 10.1074/jbc.M901106200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JW, Schulman BA. Structural complexity in ubiquitin recognition. Cell. 2006;124:1133–1136. doi: 10.1016/j.cell.2006.03.009. [DOI] [PubMed] [Google Scholar]
- Hicke L, Schubert HL, Hill CP. Ubiquitin-binding domains. Nat Rev Mol Cell Biol. 2005;6:610–621. doi: 10.1038/nrm1701. [DOI] [PubMed] [Google Scholar]
- Hoeller D, Hecker CM, Wagner S, Rogov V, Dotsch V, Dikic I. E3-independent monoubiquitination of ubiquitin-binding proteins. Mol Cell. 2007;26:891–898. doi: 10.1016/j.molcel.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Huang DT, Hunt HW, Zhuang M, Ohi MD, Holton JM, Schulman BA. Basis for a ubiquitin-like protein thioester switch toggling E1–E2 affinity. Nature. 2007;445:394–398. doi: 10.1038/nature05490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DT, Paydar A, Zhuang M, Waddell MB, Holton JM, Schulman BA. Structural basis for recruitment of Ubc12 by an E2 binding domain in NEDD8’s E1. Mol Cell. 2005;17:341–350. doi: 10.1016/j.molcel.2004.12.020. [DOI] [PubMed] [Google Scholar]
- Huang L, Kinnucan E, Wang G, Beaudenon S, Howley PM, Huibregtse JM, Pavletich NP. Structure of an E6AP-UbcH7 complex: insights into ubiquitination by the E2–E3 enzyme cascade. Science. 1999;286:1321–1326. doi: 10.1126/science.286.5443.1321. [DOI] [PubMed] [Google Scholar]
- Huibregtse JM, Scheffner M, Beaudenon S, Howley PM. A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc Nat Acad Sci USA. 1995;92:2563–2567. doi: 10.1073/pnas.92.7.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JH, Lee S, Prag G. Ubiquitin-binding domains. Biochem J. 2006;399:361–372. doi: 10.1042/BJ20061138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamynina E, Debonneville C, Bens M, Vandewalle A, Staub O. A novel mouse Nedd4 protein suppresses the activity of the epithelial Na+ channel. Faseb J. 2001;15:204–214. doi: 10.1096/fj.00-0191com. [DOI] [PubMed] [Google Scholar]
- Kim HC, Huibregtse JM. Polyubiquitination by HECT E3s and the Determinants of Chain Type Specificity. Mol Cell Biol. 2009;29:3307–3318. doi: 10.1128/MCB.00240-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Tsai YC, Mattera R, Smith WJ, Kostelansky MS, Weissman AM, Bonifacino JS, Hurley JH. Structural basis for ubiquitin recognition and autoubiquitination by Rabex-5. Nat Struct Mol Biol. 2006;13:264–271. doi: 10.1038/nsmb1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim WA, Sauer RT. Alternative packing arrangements in the hydrophobic core of lambda repressor. Nature. 1989;339:31–36. doi: 10.1038/339031a0. [DOI] [PubMed] [Google Scholar]
- Nuber U, Scheffner M. Identification of determinants in E2 ubiquitin-conjugating enzymes required for hect E3 ubiquitin-protein ligase interaction. J Biol Chem. 1999;274:7576–7582. doi: 10.1074/jbc.274.11.7576. [DOI] [PubMed] [Google Scholar]
- Ogunjimi AA, Briant DJ, Pece-Barbara N, Le Roy C, Di Guglielmo GM, Kavsak P, Rasmussen RK, Seet BT, Sicheri F, Wrana JL. Regulation of Smurf2 ubiquitin ligase activity by anchoring the E2 to the HECT domain. Mol Cell. 2005;19:297–308. doi: 10.1016/j.molcel.2005.06.028. [DOI] [PubMed] [Google Scholar]
- Ozkan E, Yu H, Deisenhofer J. Mechanistic insight into the allosteric activation of a ubiquitin-conjugating enzyme by RING-type ubiquitin ligases. Proc Nat Acad Sci USA. 2005;102:18890–18895. doi: 10.1073/pnas.0509418102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penengo L, Mapelli M, Murachelli AG, Confalonieri S, Magri L, Musacchio A, Di Fiore PP, Polo S, Schneider TR. Crystal structure of the ubiquitin binding domains of rabex-5 reveals two modes of interaction with ubiquitin. Cell. 2006;124:1183–1195. doi: 10.1016/j.cell.2006.02.020. [DOI] [PubMed] [Google Scholar]
- Prag G, Misra S, Jones EA, Ghirlando R, Davies BA, Horazdovsky BF, Hurley JH. Mechanism of ubiquitin recognition by the CUE domain of Vps9p. Cell. 2003;113:609–620. doi: 10.1016/s0092-8674(03)00364-7. [DOI] [PubMed] [Google Scholar]
- Rahighi S, Ikeda F, Kawasaki M, Akutsu M, Suzuki N, Kato R, Kensche T, Uejima T, Bloor S, Komander D, et al. Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation. Cell. 2009;136:1098–1109. doi: 10.1016/j.cell.2009.03.007. [DOI] [PubMed] [Google Scholar]
- Reverter D, Lima CD. Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature. 2005;435:687–692. doi: 10.1038/nature03588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Turcu FE, Horton JR, Mullally JE, Heroux A, Cheng X, Wilkinson KD. The ubiquitin binding domain ZnF UBP recognizes the C-terminal diglycine motif of unanchored ubiquitin. Cell. 2006;124:1197–1208. doi: 10.1016/j.cell.2006.02.038. [DOI] [PubMed] [Google Scholar]
- Rotin D. Role of the UPS in Liddle syndrome. BMC biochemistry . 2008;9(Suppl 1):S5. doi: 10.1186/1471-2091-9-S1-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotin D, Kumar S. Physiological functions of the HECT family of ubiquitin ligases. Nat Rev Mol Cell Biol. 2009;10:398–409. doi: 10.1038/nrm2690. [DOI] [PubMed] [Google Scholar]
- Salvat C, Wang G, Dastur A, Lyon N, Huibregtse JM. The -4 phenylalanine is required for substrate ubiquitination catalyzed by HECT ubiquitin ligases. Journal of biological chemistry. 2004;279:18935–18943. doi: 10.1074/jbc.M312201200. [DOI] [PubMed] [Google Scholar]
- Scheffner M, Huibregtse JM, Vierstra RD, Howley PM. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- Scheffner M, Staub O. HECT E3s and human disease. BMC Biochemistry. 2007;8(Suppl 1):S6. doi: 10.1186/1471-2091-8-S1-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloper-Mould KE, Jemc JC, Pickart CM, Hicke L. Distinct functional surface regions on ubiquitin. J Biol Chem. 2001;276:30483–30489. doi: 10.1074/jbc.M103248200. [DOI] [PubMed] [Google Scholar]
- Verdecia MA, Joazeiro CA, Wells NJ, Ferrer JL, Bowman ME, Hunter T, Noel JP. Conformational flexibility underlies ubiquitin ligation mediated by the WWP1 HECT domain E3 ligase. Mol Cell. 2003;11:249–259. doi: 10.1016/s1097-2765(02)00774-8. [DOI] [PubMed] [Google Scholar]
- Wang M, Pickart CM. Different HECT domain ubiquitin ligases employ distinct mechanisms of polyubiquitin chain synthesis. EMBO J. 2005;24:4324–4333. doi: 10.1038/sj.emboj.7600895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu PY, Hanlon M, Eddins M, Tsui C, Rogers RS, Jensen JP, Matunis MJ, Weisman AM, Wolberger C, Pickart CM. A conserved catalytic residue in the ubiquitin-conjugating enzyme family. EMBO J. 2003;22:5241–5250. doi: 10.1093/emboj/cdg501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yunus AA, Lima CD. Lysine activation and functional analysis of E2-mediated conjugation in the SUMO pathway. Nat Struct Mol Biol. 2006;13:491–499. doi: 10.1038/nsmb1104. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01