Non-Mendelian determinant [ISP+] in yeast is a nuclear-residing prion form of the global transcriptional regulator Sfp1 (original) (raw)
Abstract
Four protein-based genetic determinants or prions—[SWI+], [_MCA_], [OCT+], and [_MOT3+_]—are recent additions to the list of well-known Saccharomyces cerevisiae prions, [PSI+], [_URE3_], and [PIN+]. A rapid expansion of this list may indicate that many yeast proteins can convert into heritable prion forms and underscores a problem of prion input into cellular physiology. Here, we prove that the global transcriptional regulator Sfp1 can become a prion corresponding to the prion-like determinant [ISP+] described earlier. We show that SFP1 deletion causes an irreversible [ISP+] loss, whereas increased SFP1 expression induces [ISP+] appearance. Cells that display the [ISP+] phenotype contain the aggregated form of Sfp1. Indeed, these aggregates demonstrate a nuclear location. We also show that the phenotypic manifestation of Sfp1 prionization differs from the manifestation of SFP1 deletion. These properties and others distinguish [ISP+] from yeast prions described to date.
Keywords: protein inheritance, nonsense suppression
The list of protein-based genetic determinants, or prions, in yeast is continuously expanding, especially during the last 2 y. In addition to the classic yeast prions [PSI+] and [_URE3_] that have been studied since the 1960s and the subsequently discovered [PIN+] (1), several new prions have been identified since 2008. These are [SWI +] (2), [_MCA_] (3), [OCT +] (4), and [_MOT3+_] (5). They correspond to amyloid forms of Swi1, Mca1, Cyc8, and Mot3 proteins, respectively. Besides these amyloid-based prions, two self-perpetuating determinants of nonamyloid nature, [β] and [GAR+], were recently described (6, 7). Thus, it is evident that protein inheritance is a widespread phenomenon, at least in lower eukaryotes.
The discovery of prions in yeast occurred in different ways. Some (i.e., [PSI+] and [_URE_3]) were long known as genetic determinants of mysterious nature until their prion nature was proposed (8). The others were revealed by purposeful screening of potentially prionogenic proteins and corresponding determinants. The prion-like determinant [ISP+], described in our earlier work (9), belongs to the first group, because it was detected as a nonchromosomal antisuppressor in strains containing specific sup35 nonsense suppressor mutations and the nonsense mutations his7-1 (UAA) and lys2-87 (UGA). Consequently, strains that contain [ISP+] express the nonsuppressor (Sup- or His-Lys-) phenotype, whereas their [_isp_-] derivatives display the suppressor (Sup+ or His+Lys+) phenotype. These strains also contain sup45 cryptic mutations necessary for phenotypic manifestation of antisuppression (10). The Sup- phenotype that is determined by [ISP+] is dominant. The [ISP+] can be eliminated in the presence of guanidine chloride (GuHCl), and it reappears at high frequency after elimination. However, GuHCl-mediated elimination of [ISP+] is less efficient than for known yeast prions and is caused not only by [ISP+] curing but also by the selection of [_isp_-] cells more resistant to GuHCl than are [ISP+] cells (9). Contrary to known yeast prions, [ISP+] maintenance does not depend on the Hsp104p chaperone protein, and efficiency of [ISP+] cytoduction is much lower than for known yeast prions. Thus, the [ISP+] combines properties of typical yeast prions and its own specific properties.
To establish the prion nature of [ISP+], it is required to identify its host gene and characterize prion-related features of the corresponding protein. Recently, we recognized the SFP1 gene as a candidate gene encoding [ISP+] (11). Here, we corroborate this finding and demonstrate some properties of the prion form of Sfp1.
Results
SFP1 Deletion Causes Irreversible [ISP+] Loss.
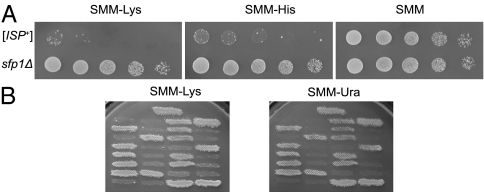
In a large-scale screen of the insertion gene library, we have shown that insertion of a minitransposon into the SFP1 gene altered the phenotype of the [ISP+] strain from Sup- to Sup+ (11). The same effect was observed in the _sfp1_Δ derivative of [ISP+] strain 25–25-2V-P3982 obtained in this work by URA3 replacement of SFP1 (Fig. 1_A_). The Sup+ phenotype cosegregated with Ura+ in tetrads of the diploid that was obtained by crossing the _sfp1_Δ and [ISP+] strains (Fig. 1_B_). These findings indicate either that [ISP+] is a prion form of Sfp1 or that the change in phenotype was caused by an independent manifestation of the _SFP1_-null allele.
Fig. 1.
Deletion of the SFP1 changes phenotype of [ISP+] strains from Sup− to Sup+. (A) Growth of [ISP+] strain 25–25-2V-P3982 and _sfp1_Δ derivative of this strain on supplemented minimal medium (SMM)-Lys and SMM-His media allowed monitoring of lys2-87 and his7-1 nonsense suppression. SMM medium was used as a growth control. (B) Cosegregation of Ura+ and Sup+ phenotypes on tetrads of the diploid obtained by crossing the _sfp1_Δ derivative of 25–25-2V-P3982 and [ISP+] strain 5B-P4513. This diploid is heterozygous for SFP1 deletion and homozygous for lys2-87, and it contains the nonchromosomal determinant [ISP+].
To distinguish between these two possibilities, the _sfp1_Δ strain was transformed with the centromeric vector pRS315-SFP1. Introduction of the wild-type SFP1 allele did not change the phenotype of the _sfp1_Δ strain [i.e., the absolute majority (556 of 559) of transformants has retained the Sup+ phenotype]. This fact suggests that the change of phenotype in the _sfp1_Δ strain was caused by [ISP+] loss rather than phenotypic effects of the SFP1 deletion; otherwise, restoration of the Sup− phenotype would be observed. Notably, this loss was irreversible, because we have not observed a single example of Sup- clones appearing in the mitotic progeny of _sfp1_Δ strains in contrast to [_isp_-] strains obtained by GuHCl treatment, which produced Sup- clones at a high frequency (9). These results confirmed that SFP1 could be considered as a likely host gene for [ISP+].
Increased Expression of SFP1 Induces [ISP+] Appearance.
Increased production of prionogenic proteins induces the de novo appearance of corresponding prions (12). To determine effects of SFP1 overexpression, the [_isp−_] variant of 25–25-2V-P3982, obtained by GuHCl treatment, was transformed with the high copy-number plasmid pRS426-SFP1. Contrary to the recipient Sup+ strain, nearly 100% of transformants showed the Sup− phenotype. In the empty vector-control transformation, Sup− transformants appeared at a frequency that was typical of spontaneous [ISP+] appearance (Table 1).
Table 1.
Expression of SFP1 from the high copy-number plasmid changes phenotype of [_isp_−] strain 25–25-2V-P3982 from Sup+ to Sup-
| No. from total | ||||
|---|---|---|---|---|
| Plasmid | No. of transformants | Sup+ | Sup− | % of Sup− transformants* |
| pRS426-SFP1 | 22,997 | 97 | 22,900 | 99.6 ± 0.04 |
| pRS426 | 2,530 | 2,441 | 89 | 3.5 ± 0.37 |
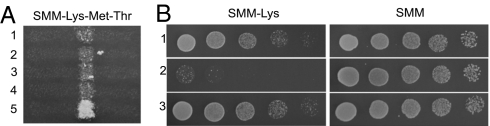
Next, effects of plasmid elimination in 100 independent Sup- transformants obtained by SFP1 overexpression were examined. Of 1,946 clones that were isolated in mitotic progeny of these transformants grown on YPD, 1,389 clones (∼71%) retained the Sup− phenotype, despite the fact that they lost the plasmid marker URA3. Retention of the Sup− phenotype after elimination of the _SFP1_-overexpressing plasmid by the majority of clones indicates that the Sup- phenotype of these clones was caused not by phenotypic manifestation of Sfp1 overproduction but by induction of an [ISP+]-like determinant. This determinant is dominant, because all diploids obtained by crossing the [_isp_−] strain to 120 randomly selected clones, which retain the Sup- phenotype after plasmid loss, displayed the Sup- phenotype (examples of these crosses are presented in Fig. 2_A_). Notably, GuHCl treatment converts these Sup− clones to Sup+ (Fig. 2_B_).
Fig. 2.
Expression of the SFP1 from a high-copy plasmid induces the dominant, GuHCl-curable nonsuppressor phenotype. (A) The Sup- phenotype induced by SFP1 overexpression is dominant. Crosses of the [_isp_−] strain 5B-P4513 to three strains retaining the Sup- phenotype after loss of _SFP1_-expressing plasmid are shown in lines 1–3. Control crosses of 5B-P4513[_isp_−] to [ISP+] and [_isp_−] variants of 25-25-2V-P3982 are in line 4 and line 5, correspondingly. The SMM-Lys medium does not also contain methionine and threonine, because met13-A1 and thr4-B15 were used as selective markers in this cross. (B) The Sup- phenotype induced by transient SFP1 overexpression changes for Sup+ after GuHCl treatment. The original [_isp_−] strain is shown in line 1, one of the strains retaining Sup- phenotype after plasmid loss is shown in line 2, and subsequent 5 mM GuHCl treatment is shown in line 3.
Earlier, we showed that neither HSP104 overexpression nor deletion caused [ISP+] loss (9), whereas propagation of yeast prions usually depends on the level of Hsp104 production (13). We examined the consequences of HSP104 expression from the high copy-number plasmid pLH105 in strains containing the determinant induced by SFP1 overexpression. Among 1,687 transformants studied, 1,680 did not change their Sup− phenotype despite Hsp104 overproduction, similar to the control [ISP+]-containing strain. The appearance of exceptional Sup+ clones among transformants may be explained by spontaneous [ISP+] loss. Concurrently, the construct efficiently eliminated another yeast prion, [PSI+].
Then, effects of SFP1 overexpression in the _hsp104_Δ derivative of the [_isp_−] strain 25–25-2V-P3982 were studied. All 120 selected transformants displayed the Sup− phenotype in contrast to the recipient Sup+ strain. After plasmid loss on YPD medium, the majority of clones (154 of 160 examined) retained the Sup− phenotype. Thus, Hsp104 absence does not prevent effects of transient SFP1 overexpression. Collectively, these data indicate that the determinant induced by transient Sfp1 overproduction is identical to [ISP+].
Nuclei of [ISP+] Cells Contain the Aggregated Form of Sfp1-GFP.
Prion conversion of a protein leads to formation of amyloid-like aggregates (14). To monitor the presence of Sfp1 aggregates in [ISP+] cells, the Sfp1-GFP fusion protein was used. First, we have demonstrated that the fused protein allows [ISP+] to appear. To this end, the _sfp1_Δ-25–25-2V-P3982 strain, which had a stable Sup+ phenotype, was transformed with the centromeric vector pMT3453, expressing the SFP1-GFP from the native SFP1 promoter. Notably, the Sup− clones appeared in the mitotic progeny of transformants at a frequency similar to that of spontaneous [ISP+] appearance (i.e., ∼1.0 × 10−4/cell/generation) (9).
We also examined the [ISP+]-inducing effect of the SFP1-GFP fusion. The [_isp_−] variant of 25–25-2V-P3982 was transformed with pSFP1-GFP, which expressed SFP1-GFP from the GAL1/10 promoter. Of the 674 clones that were isolated from the mitotic progeny of 10 transformants, 577 clones (∼85.6%) switched phenotypes from Sup+ to Sup− on galactose-containing medium and retained this phenotype after transfer to glucose-containing medium. The Sup− phenotype that was induced by SFP1-GFP expression was dominant, which was shown in the cross to the [_isp_−] strain, and GuHCl treatment restored the suppression. Thus, overproduction of Sfp1-GFP induces the appearance of [ISP+].
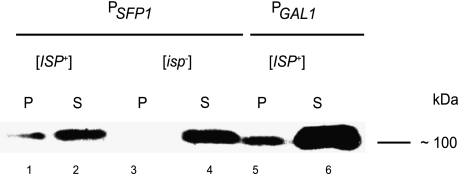
The aggregation of Sfp1-GFP was analyzed by centrifugation of cell extracts obtained from [ISP+] and [_isp_−] cells and subsequent Western-blotting with anti-GFP antibody. [_isp_−] cells contained the Sfp1-GFP only in supernatant, whereas [ISP+] cells expressing _SFP1_-GFP both from the galactose-induced promoter and the native SFP1 promoter contained the protein not only in the supernatant but also in the pellet (Fig. 3).
Fig. 3.
Sfp1-GFP hybrid protein forms aggregates in [_ISP+_] cells but not in [_isp_−] cells. Detection of Sfp1-GFP by Western blot with anti-GFP antibody in the pellet and supernatant fractions of cell lysates of [ISP+] cells expressing _SFP1_-GFP from the native SFP1 promoter (lines 1 and 2), Gal1/10 promoter (lines 5 and 6), and [_isp_−] cells (lines 3 and 4).
The intracellular distribution of Sfp1-GFP in [ISP+] and [_isp_−] cells was studied by fluorescence microscopy. In six independent isolates of the [_isp_−] strain, we observed a weak signal that was distributed between the cytoplasm and nucleus, although the signal was occasionally stronger in the nucleus (Fig. 4_A_). A similar pattern of Sfp1-GFP fluorescence has been observed by other authors (15, 16).
Fig. 4.
Fluorescent assay of Sfp1-GFP's ability to aggregate. [_isp_−] cells (A) and [_ISP+_] cells (B) producing the Sfp1-GFP from the native SFP1 promoter are shown. [_ISP+_] cells obtained by Sfp1-GFP overproduction in [_isp_−] strain are shown in C; [_ISP+_] cells obtained by Sfp1-GFP overproduction in _sfp1_Δ strain are shown in D.
In [ISP+] strains, ∼5–7% of cells contained brightly fluorescing foci. Notably, these foci resided both in cells that expressed the Sfp1-GFP from the native SFP1 promoter (Fig. 4_B_) and in cells overproducing Sfp1p-GFP (Fig. 4 C and D). Additionally, in Sfp1-GFP–overexpressing cells, the foci assumed a granular structure (Fig. 4 C and D), similar to that formed by yeast prions. Distinct granules were not visible, however, in cells that expressed Sfp1-GFP from the SFP1 promoter (Fig. 4_B_). Remarkably, analysis of foci location revealed that they were located in the nuclei of [ISP+] cells (Fig. 5).
Fig. 5.
Nuclear location of Sfp1-GFP aggregates in [_ISP+_] cells. [_ISP+_] cells overproducing the Sfp1-GFP in _sfp1_Δ strain are shown in A, and [_ISP+_] cells producing the Sfp1-GFP from the native SFP1 promoter are shown in B.
Prionization Does Not Cause a Loss of Sfp1 Functions.
Yeast prions usually represent an inactive form of the corresponding protein; therefore, prionization causes the same phenotype as a whole or partial inactivation of a gene encoding this protein (12). Comparison of phenotypes of [ISP+] and _sfp1_Δ strains, however, shows that, in the case of [ISP+], we certainly have a distinct situation. The Sfp1 prionization causes antisuppression, whereas the absence of Sfp1 does not (strains containing the SFP1 deletion display the Sup+ phenotype) (Fig. 1 A and B).
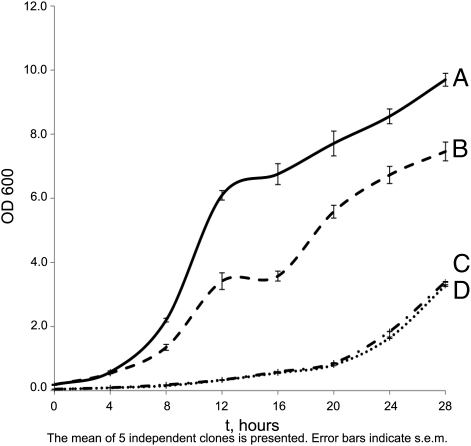
In an earlier study, we showed that [ISP+] strains grew faster than isogenic [_isp_−] strains (9). In this study, we expounded on this comparison with two variants of _sfp1_Δ strain. One was obtained in an [ISP+] background, and the other was obtained in an [_isp_−] background. We showed that both variants of _sfp1_Δ strain demonstrateded the slowest growth (Fig. 6). Thus, the influence of Sfp1 prionization on growth rate opposes the effect of SFP1 deletion.
Fig. 6.
The influences of Sfp1 prionization and Sfp1 absence on the strain growth are opposite. Growth of [ISP+] strain 25–25-2V-P3982 is shown in A. The [_isp_−] derivative of this strain obtained by GuHCl treatment is shown in B; the _sfp1_Δ derivative of [ISP+] is in C, and _sfp1_Δ derivative of [_isp_−] strain is in D. YPD medium was used.
It is known that SFP1 is one of the key genes controlling cell size in yeast (17–19); therefore, cells that lack Sfp1 show a reduced size. At the same time, comparison of cell area (Materials and Methods) showed that [ISP+] cells are significantly larger than _sfp1_Δ cells (Table 2).
Table 2.
Comparison of cell size in [ISP+], [_isp_−], and _sfp1_Δ strains
| Strain | Size of cell area (μm2) |
|---|---|
| [ISP+] | 32.3 ± 0.49 |
| [_isp_-] | 31.8 ± 0.57 |
| _sfp1_Δ* | 14.5 ± 0.37 |
| _sfp1_Δ† | 14.4 ± 0.21 |
Also, taking into account that sfp1 mutations manifest an increased sensitivity to drugs that target translation, such as cycloheximide and paromomycin (20), we compared the [ISP+], [_isp_−], and _sfp1_Δ strains with regard to their sensitivity to these drugs. It was shown that the [ISP+] strain was more resistant than the [_isp_−] strain, whereas both variants of _sfp1_Δ strain were more sensitive (Table 3).
Table 3.
Different resistance to translational drugs shown by [ISP+], [_isp_−], and _sfp1_Δ strains
| Zone of inhibition (mm) | ||||
|---|---|---|---|---|
| Drug | [ISP+] | [_isp_−] | _sfp1_Δ* | _sfp1_Δ† |
| Cycloheximide | 23.2 ± 0.28 | 26.3 ± 0.40 | 31.7 ± 0.13 | 31.6 ± 0.11 |
| Paromomycin | 11.4 ± 0.23 | 14.2 ± 0.20 | 23.3 ± 0.07 | 23.4 ± 0.01 |
Taken together, our findings suggest that consequences of Sfp1 prion conversion are not equivalent to the loss of Sfp1 function. It should also be noted that the similarity of properties displayed by _sfp1_Δ derivatives of [ISP+] and [_isp_−] strains shows once more that the distinction of [ISP+] and [_isp_−] strains is determined by the status of Sfp1 protein.
Discussion
The data obtained in this work indicate that the non-Mendelian determinant [ISP+] represents a prion form of the Sfp1 protein. We have shown that deletion of SFP1 in an [ISP+] strain caused the irreversible loss of the determinant, whereas increased SFP1 expression caused the appearance of an antisuppressor determinant, which is similar in properties to [ISP+]. Furthermore, cells of the [ISP+] strain transformed with _SFP1_-_GFP_–bearing plasmid contain the aggregated form of the Sfp1-GFP hybrid protein, although [_isp_−] cells do not.
Sfp1 is a transcription factor that contains three Cys2His2 zinc-finger domains (17); at least two of them, located in the C terminus, are functional (19). It is the global regulator of transcription that positively controls the expression of ∼10% of all yeast genes, including genes that encode ribosomal proteins and other components of the translational machinery as well as genes that control ribosome biogenesis (15, 16, 19, 20). It is a component of the target of rapamycin (TOR) signaling pathway, and phosphorylation of Sfp1 by TORC1 kinase regulates its function, particularly nuclear targeting (21).
Importantly, Sfp1 belongs to a group of asparagine-enriched proteins and was revealed as a potential prion by several computational surveys (5, 22, 23). We suggest that the prion domain is located in the central region of the protein restricted roughly by positions 230 and 430. This Asn-rich region does not contain functional domains, such as zinc fingers or phosphorylation sites (21). Location of the prion domain in this region was also predicted by the method developed by Alberti et al. (5). An exact identification of the prion domain is certainly a separate task for the future.
As already mentioned, some features distinguish [ISP+] from known yeast prions. For instance, the frequency of spontaneous [ISP+] appearance is 1.0 × 10−4/cell/generation (9), whereas switching rates of other prions can be as low as 10−6 to 10−7 /cell/generation (24–26). The efficiency of [ISP+] induction by Sfp1 overexpression approximates 70%, whereas it is much lower for other prions. This difference possibly reflects a higher ability of Sfp1 to nucleation and polymerization than other known prion proteins. At the same time, the amount of aggregated protein in pellets of [ISP+] cells is less than in supernatants, even when _SFP1_-GFP is expressed from galactose-inducible promoter (Fig. 3). It is evident that further study is necessary to resolve this contradiction.
Specific properties of Sfp1 aggregates may underlie the independence of [ISP+] propagation on Hsp104p. At the same time, we register the curing effect of GuHCl to [ISP+], although it was weaker than for other yeast prions. These data contradict a well-established mechanism of prion curing by GuHCl through inhibition of Hsp104p ATPase activity (27) and support the idea that an Hsp104-independent pathway of prion shearing may exist (28). Whether [ISP+] requires other chaperones for its propagation should be a subject of future studies.
The additional distinction of [ISP+] from yeast prions characterized at present is its nuclear location. The nuclear location of [ISP+] was suggested earlier in our work (9) to explain a low efficiency of [ISP+] transfer by cytoduction compared with other yeast prions. We have also proposed that a protein corresponding to [ISP+] is a shuttle protein, and its conversion into prion form should hamper its export from the nucleus. These suggestions are now confirmed, at least partially, because Sfp1 is a shuttle protein that operates in the nucleus under normal growth conditions and exits the cytoplasm under stress (15, 19, 21). Indeed, a nuclear location of its prion form was shown by fluorescence microscopy.
Another specific property of [ISP+]-bearing strains is the distinction of their phenotype from the phenotype of the _sfp1_Δ strain. The SFP1 deletion does not cause antisuppression, whereas [ISP+] does, the _sfp1_Δ strain consists of cells that are significantly smaller than cells of the [ISP+] strain, and it grows much more slowly and is more sensitive to antibiotics than the [ISP+] strain. Although it is believed that prion switching causes a phenotype similar to genetic inactivation of the corresponding gene (1), exclusions from this rule are documented. In the case of the [Het-s] prion of Podospora anserina, the protein acquires a property that triggers vegetative incompatibility by interaction with the product of another allele of the same gene in heterokaryotic mycelia (29). However, the exact explanation of this phenomenon is complicated by gaps in our knowledge of the molecular mechanisms of incompatibility in fungi. Formally, the [PIN+] prion in yeast may also be attributed as one of such exclusions, because prion switching transforms the corresponding protein, Rnq1, into a template for the polymerization of some other prion proteins (30, 31). It is unclear, however, how Rnq1 prionization influences its own functioning, because Rnq1 functions, besides its influence on induction of other prions, are not known.
In the case of [ISP+], we observe a more obvious situation, because Sfp1 functions are generally recognized and [ISP+] has a clear phenotypic manifestation in the system used. A possibility of retention of protein functions after prion switching exists, as was shown with the [_URE3_] model (32). Also, diverse consequences of Sfp1 prionization and SFP1 deletion may be explained by the influence of the prion form of Sfp1 on the status of some other proteins. This influence may include either induction of their prionization or stimulation of nonprion polymerization or sequestration because of inclusion into prion aggregates (33, 34). Thus, the nonsuppressor phenotype of [ISP+] strains may be determined by numerous changes in proteome. This possibility remains, despite the fact that the _sfp1_Δ derivatives of the [ISP+] and [_isp_−] strains display similar properties (Fig. 6 and Table 3).
Interestingly, from seven yeast prions described to date, four proteins—Ure2, Swi1, Cyc8, and Mot3—participate in regulation of gene expression. Sfp1 should be added to this list. Whether the ability of these proteins to switch into prion form is related to their functions remains to be established.
Materials and Methods
Yeast Strains.
We used [ISP+] and [_isp_−] variants of the strain 25–25-2V-P3982, which is the MATa derivative of 25–2V-P3982, MATα ade1-14 his7-1 lys2-87 ura3 Δ thr4-B15 leu2-1sup35-25 sup45-400 (9, 10). [ISP+] and [_isp_−] variants of 5B-P4513, _MAT_α ade1-14 his7-1 lys2-87 ura3 Δ met13-A1 leu2-1 sup35-25 sup45-400 (10) were used as tester strains. hsp104Δ-25–2V-P3982 contains deletion of HSP104 (9). sfp1Δ-25–25-2V-P3982 was obtained in this study and contained URA3 replacement of SFP1 in 25–25-2V-P3982.
Plasmids.
pRS425-SFP1 and pRS426-SFP1, the derivatives of pRS425 and pRS426 containing SFP1, were generated by cloning SFP1 by PCR of the chromosomal SFP1 copy from 2V-P3982. pMT3193 contained SFP1 under the control of its own promoter, and pMT3453 expressed GFP-SFP1 under the control of the SFP1 promoter (16). Both pMT3193 and pMT3453 were provided by M. Tyers (Toronto, ON, Canada). The ORF plus 799 bp of upstream and 300 bp of downstream sequence of SFP1 from pMT3193 was cloned into pRS315 to generate pRS315-SFP1. pSFP1-GFP, obtained from A. Vershon's laboratory (Piscataway, NJ), contained the entire SFP1 gene, fused in frame to the 5′ end of the GFP gene, under control of the GAL1/GAL10 promoter (19). A 2-μm plasmid, pLH105, containing HSP104 under control of the constitutively activated GPD promoter, was obtained from the Y. Chernoff laboratory (Atlanta, GA) (35). The pCORE plasmid (36) was used to replace SFP1 with URA3.
Yeast Culture and Media.
Yeast cultures were grown at 26 °C in rich medium YPD, supplemented minimal medium (SMM), or SMM lacking one or more supplements (e.g., SMM-lysine). All media were prepared as described (37). Five millimolar GuHCl (Sigma) was added where indicated.
Standard yeast genetics methods were used (37). To monitor Sup- and Sup+ phenotypes corresponding to [ISP+] and [_isp_−] status, strains were replica plated on SMM-Lys and SMM-His media. Sometimes, manifestation of [ISP+]-based antisuppression was clearer for lys2-87 than for his7-1; in these cases, results that were obtained using SMM-Lys are presented.
GuHCl treatment and subsequent examination of the clones were performed as described (38). The drug-sensitivity test was carried out as described (19) using paromomycin and cycloheximide (Sigma).
Fluorescence Microscopy.
A fluorescence assay of GFP fusion proteins was performed using a Leica DM6000B Leica Microsystems GmBH with Leica QWin Standard software (version 3.2.0). DAPI (Invitrogen) staining was performed according to the manufacturer's protocol.
Preparation, Fractionation, and Analysis of Yeast-Cell Lysates.
Protein isolation and differential centrifugation were performed accordingly to protocol described in ref. 39. Fractionation of cell lysates was performed at 13,000 × g for 20 min. After SDS/PAGE in 12% gel, proteins were transferred onto a PVDF membrane. Western blotting was performed with primary monoclonal antibody 3A9 against GFP obtained from the Institute of Bioorganic Chemistry in Moscow, Russia. Reactions with the secondary anti-mouse antibodies as well as chemiluminescent detection were performed with the use of ECL detection kit (General Electric). Staining of gel with Coomassie Blue was used for normalization of the total protein amount.
Cell-Size Evaluation.
Strains were grown in YPD up to the early stationary phase. Then, cells were placed in a counting chamber (hemocytometer) and examined in transmitted light under a Leica DM LS microscope equipped with FLUOTAR objective 20×/0.40 photoadapter Leica DFC 320 camera. Images were captured with Leica DFC Twain software and processed with Adobe Photoshop CS2. The photographed cell area was estimated using the ImageJ 1.34s program (http://rsb.info.nih.gov/ij/). Student t test was used for statistical evaluation of the data.
Acknowledgments
We thank M. Tyers (University of Toronto), A. Vershon (Rutgers University), and Y. Chernoff (Georgia Institute of Technology) for plasmids. We are grateful to S. G. Inge-Vechtomov for helpful discussion of our data and M. D. Ter Avanesyan for critical review of the manuscript and important suggestions. We thank A. P. Galkin and A. F. Saifitdinova for help with fluorescence microscopy and A. V. Svitin for help with manuscript preparation. This work was supported by Russian Foundation of Basic Research Grant 08-04-00353 and Federal Agency for Education (Russia) Grant P799.
Footnotes
The authors declare no conflict of interest.
References
- 1.Wickner RB, Edskes HK, Shewmaker F, Nakayashiki T. Prions of fungi: Inherited structures and biological roles. Nat Rev Microbiol. 2007;5:611–618. doi: 10.1038/nrmicro1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Du Z, Park KW, Yu H, Fan Q, Li L. Newly identified prion linked to the chromatin-remodeling factor Swi1 in Saccharomyces cerevisiae. Nat Genet. 2008;40:460–465. doi: 10.1038/ng.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nemecek J, Nakayashiki T, Wickner RB. A prion of yeast metacaspase homolog (Mca1p) detected by a genetic screen. Proc Natl Acad Sci USA. 2009;106:1892–1896. doi: 10.1073/pnas.0812470106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 4.Patel BK, Gavin-Smyth J, Liebman SW. The yeast global transcriptional co-repressor protein Cyc8 can propagate as a prion. Nat Cell Biol. 2009;11:344–349. doi: 10.1038/ncb1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alberti S, Halfmann R, King O, Kapila A, Lindquist S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 2009;137:146–158. doi: 10.1016/j.cell.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roberts BT, Wickner RB. Heritable activity: A prion that propagates by covalent autoactivation. Genes Dev. 2003;17:2083–2087. doi: 10.1101/gad.1115803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown JC, Lindquist S. A heritable switch in carbon source utilization driven by an unusual yeast prion. Genes Dev. 2009;23:2320–2332. doi: 10.1101/gad.1839109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wickner RB. [URE3] as an altered URE2 protein: Evidence for a prion analog in Saccharomyces cerevisiae. Science. 1994;264:566–569. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- 9.Volkov KV, et al. Novel non-Mendelian determinant involved in the control of translation accuracy in Saccharomyces cerevisiae. Genetics. 2002;160:25–36. doi: 10.1093/genetics/160.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aksenova AIu, Volkov KV, Rovinsky NS, Svitin AV, Mironova LN. Phenotypic expression of epigenetic determinant [ISP+] in Saccharomyces cerevisiae depends on the combination of sup35 and sup45 mutations. Mol Biol (Mosk) 2006;40:844–849. [PubMed] [Google Scholar]
- 11.Rogoza TM, et al. Search for the genes critical for propagation of the prion-like antisuppressor determinant [ISP+] in yeast using insertion library. Mol Biol (Mosk) 2009;43:392–399. [PubMed] [Google Scholar]
- 12.Wickner RB, et al. Prion genetics: New rules for a new kind of gene. Annu Rev Genet. 2004;38:681–707. doi: 10.1146/annurev.genet.38.072902.092200. [DOI] [PubMed] [Google Scholar]
- 13.Romanova NV, Chernoff YO. Hsp104 and prion propagation. Protein Pept Lett. 2009;16:598–605. doi: 10.2174/092986609788490078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wickner RB, et al. Yeast prions act as genes composed of self-propagating protein amyloids. Adv Protein Chem. 2001;57:313–334. doi: 10.1016/s0065-3233(01)57026-6. [DOI] [PubMed] [Google Scholar]
- 15.Marion RM, et al. Sfp1 is a stress- and nutrient-sensitive regulator of ribosomal protein gene expression. Proc Natl Acad Sci USA. 2004;101:14315–14322. doi: 10.1073/pnas.0405353101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jorgensen P, et al. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 2004;18:2491–2505. doi: 10.1101/gad.1228804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blumberg H, Silver P. A split zinc-finger protein is required for normal yeast growth. Gene. 1991;107:101–110. doi: 10.1016/0378-1119(91)90302-r. [DOI] [PubMed] [Google Scholar]
- 18.Sudbery P. Cell biology. When wee meets whi. Science. 2002;297:351–352. doi: 10.1126/science.1073042. [DOI] [PubMed] [Google Scholar]
- 19.Fingerman I, Nagaraj V, Norris D, Vershon AK. Sfp1 plays a key role in yeast ribosome biogenesis. Eukaryot Cell. 2003;2:1061–1068. doi: 10.1128/EC.2.5.1061-1068.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cipollina C, et al. Saccharomyces cerevisiae SFP1: At the crossroads of central metabolism and ribosome biogenesis. Microbiology. 2008;154:1686–1699. doi: 10.1099/mic.0.2008/017392-0. [DOI] [PubMed] [Google Scholar]
- 21.Lempiäinen H, et al. Sfp1 interaction with TORC1 and Mrs6 reveals feedback regulation on TOR signaling. Mol Cell. 2009;33:704–716. doi: 10.1016/j.molcel.2009.01.034. [DOI] [PubMed] [Google Scholar]
- 22.Michelitsch MD, Weissman JS. A census of glutamine/asparagine-rich regions: Implications for their conserved function and the prediction of novel prions. Proc Natl Acad Sci USA. 2000;97:11910–11915. doi: 10.1073/pnas.97.22.11910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harrison PM, Gerstein M. A method to assess compositional bias in biological sequences and its application to prion-like glutamine/asparagine-rich domains in eukaryotic proteomes. Genome Biol. 2003;4:R40. doi: 10.1186/gb-2003-4-6-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lund PM, Cox BS. Reversion analysis of [psi-] mutations in Saccharomyces cerevisiae. Genet Res. 1981;37:173–182. doi: 10.1017/s0016672300020140. [DOI] [PubMed] [Google Scholar]
- 25.Liu JJ, Lindquist S. Oligopeptide-repeat expansions modulate ‘protein-only’ inheritance in yeast. Nature. 1999;400:573–576. doi: 10.1038/23048. [DOI] [PubMed] [Google Scholar]
- 26.Tuite MF, Cox BS. Propagation of yeast prions. Nat Rev Mol Cell Biol. 2003;4:878–890. doi: 10.1038/nrm1247. [DOI] [PubMed] [Google Scholar]
- 27.Grimminger V, Richter K, Imhof A, Buchner J, Walter S. The prion curing agent guanidinium chloride specifically inhibits ATP hydrolysis by Hsp104. J Biol Chem. 2004;279:7378–7383. doi: 10.1074/jbc.M312403200. [DOI] [PubMed] [Google Scholar]
- 28.Crist CG, Nakayashiki T, Kurahashi H, Nakamura Y. [PHI+], a novel Sup35-prion variant propagated with non-Gln/Asn oligopeptide repeats in the absence of the chaperone protein Hsp104. Genes Cells. 2003;8:603–618. doi: 10.1046/j.1365-2443.2003.00661.x. [DOI] [PubMed] [Google Scholar]
- 29.Saupe SJ. A short history of small s: A prion of the fungus Podospora anserina. Prion. 2007;1:110–115. doi: 10.4161/pri.1.2.4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Derkatch IL, Bradley ME, Hong JY, Liebman SW. Prions affect the appearance of other prions: The story of [PIN+] Cell. 2001;106:171–182. doi: 10.1016/s0092-8674(01)00427-5. [DOI] [PubMed] [Google Scholar]
- 31.Vitrenko YA, Gracheva EO, Richmond JE, Liebman SW. Visualization of aggregation of the Rnq1 prion domain and cross-seeding interactions with Sup35NM. J Biol Chem. 2007;282:1779–1787. doi: 10.1074/jbc.M609269200. [DOI] [PubMed] [Google Scholar]
- 32.Baxa U, Speransky V, Steven AC, Wickner RB. Mechanism of inactivation on prion conversion of the Saccharomyces cerevisiae Ure2 protein. Proc Natl Acad Sci USA. 2002;99:5253–5260. doi: 10.1073/pnas.082097899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Derkatch IL, Liebman SW. Prion-prion interactions. Prion. 2007;1:161–169. doi: 10.4161/pri.1.3.4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Urakov VN, et al. Interdependence of amyloid formation in yeast: Implications for polyglutamine disorders and biological functions. Prion. 2010;4:45–52. doi: 10.4161/pri.4.1.11074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chernoff YO, Newnam GP, Kumar J, Allen K, Zink AD. Evidence for a protein mutator in yeast: Role of the Hsp70-related chaperone ssb in formation, stability, and toxicity of the [PSI] prion. Mol Cell Biol. 1999;19:8103–8112. doi: 10.1128/mcb.19.12.8103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Storici F, Lewis LK, Resnick MA. In vivo site-directed mutagenesis using oligonucleotides. Nat Biotechnol. 2001;19:773–776. doi: 10.1038/90837. [DOI] [PubMed] [Google Scholar]
- 37.Kaiser C, Michaelis S, Mitchell A. Methods in Yeast Genetics. Plainview, NY: Cold Spring Harbor Laboratory Press; 1994. p. 234. [Google Scholar]
- 38.Derkatch IL, Bradley ME, Zhou P, Chernoff YO, Liebman SW. Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics. 1997;147:507–519. doi: 10.1093/genetics/147.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chernoff YO, Uptain SM, Lindquist SL. Analysis of prion factors in yeast. Methods Enzymol. 2002;351:499–538. doi: 10.1016/s0076-6879(02)51867-x. [DOI] [PubMed] [Google Scholar]