Pseudogenization of the Umami Taste Receptor Gene Tas1r1 in the Giant Panda Coincided with its Dietary Switch to Bamboo (original) (raw)
Abstract
Although it belongs to the order Carnivora, the giant panda is a vegetarian with 99% of its diet being bamboo. The draft genome sequence of the giant panda shows that its umami taste receptor gene Tas1r1 is a pseudogene, prompting the proposal that the loss of the umami perception explains why the giant panda is herbivorous. To test this hypothesis, we sequenced all six exons of Tas1r1 in another individual of the giant panda and five other carnivores. We found that the open reading frame (ORF) of Tas1r1 is intact in all these carnivores except the giant panda. The rate ratio (ω) of nonsynonymous to synonymous substitutions in Tas1r1 is significantly higher for the giant panda lineage than for other carnivore lineages. Based on the ω change and the observed number of ORF-disrupting substitutions, we estimated that the functional constraint on the giant panda Tas1r1 was relaxed ∼4.2 Ma, with its 95% confidence interval between 1.3 and 10 Ma. Our estimate matches the approximate date of the giant panda's dietary switch inferred from fossil records. It is probable that the giant panda's decreased reliance on meat resulted in the dispensability of the umami taste, leading to Tas1r1 pseudogenization, which in turn reinforced its herbivorous life style because of the diminished attraction of returning to meat eating in the absence of Tas1r1. Nonetheless, additional factors are likely involved because herbivores such as cow and horse still retain an intact Tas1r1.
Keywords: giant panda, diet, pseudogenization, Tas1r1, umami taste
The giant panda Ailuropoda melanoleuca is an endangered species distributed in a few mountain ranges in central China. Its phylogenetic position was a long-standing puzzle; it had been assigned to Ursidae (bears), Procyonidae (raccoons), Ailuridae (red panda), and its own family Ailuropodidae based on different morphological characteristics (Bininda-Emonds 2004) before being conclusively classified into the bear family by overwhelming molecular evidence (O'Brien et al. 1985; Goldman et al. 1989; Yu et al. 2004; Arnason et al. 2007; Krause et al. 2008). The giant panda has been of interest to evolutionists because of several of its unusual traits such as the much reduced fecundity and the enlarged radial sesamoid that functions as a thumb. Most notably, in contrast to all other bears, the giant panda is herbivorous, with 99% of its diet being bamboo; the remaining 1% includes honey, eggs, fish, yams, shrub leaves, oranges, and bananas when available (Schaller et al. 1985; Schaller et al. 1989). Multiple lines of evidence support that the giant panda descended from a carnivorous ancestor. For example, the distribution, structure, and morphology of the lingual papillae in the giant panda are more similar to those of carnivores than herbivores (Pastor et al. 2008). The giant panda also has powerful jaws and teeth that are capable of tearing meat (Bininda-Emonds 2004), a carnivore-like digestive system (Li et al. 2010), and all the genetic components of a digestive system that carnivores possess (Bininda-Emonds 2004; Li et al. 2010). The fossil record provides key information about the timing of the giant panda's dietary switch. Dental remains of ancient pandas recovered from Yunnan, south China, exhibit a crushing dentition for bamboo eating, indicating that the giant panda already started eating bamboo at least 7 Ma (Jin et al. 2007). The premolar and molar teeth from a recently unearthed 2.0–2.4 MY old skull show a great resemblance to the living giant panda, suggesting that the giant panda had completed its dietary transition by that time (Jin et al. 2007). However, the molecular basis and consequences of the giant panda's dietary switch have been elusive.
Recently, it was discovered from the draft genome sequence of the giant panda that its Tas1r1 gene is inactivated due to two frame-shifting mutations in exon 3 and exon 6, respectively (Li et al. 2010). Tas1r1 belongs to the vertebrate Tas1r family that consists of three members in most species (Shi and Zhang 2006): Tas1r1 and Tas1r3 form a heterodimer that functions as the G-protein coupled receptor mediating the umami taste, the taste of glutamic acid and other amino acids, whereas Tas1r2 and Tas1r3 form a heterodimeric sweet receptor (Nelson et al. 2001; Nelson et al. 2002; Zhao et al. 2003). Because amino acids are much rarer in bamboo than in animal tissues (Kozukue et al. 1983), it was suggested that the pseudogenization of Tas1r1 explains why the giant panda does not eat meat (Li et al. 2010). Because Tas1r1 is intact in dog, Li et al. (2010) inferred that the pseudogenization of the giant panda Tas1r1 occurred after the giant panda diverged from the dog lineage ∼61 Ma. The lack of a precise date of the pseuodgenization event hinders a critical evaluation of the relationship between the Tas1r1 pseudogenization and the giant panda's dietary switch. By examining additional carnivores, we here show that the Tas1r1 pseudogenization not only is specific to the giant panda but also occurred approximately when the giant panda changed its diet.
We first sequenced all six coding exons of Tas1r1, totaling ∼2.5 kb, from a giant panda individual not used in the genome sequencing. We confirmed the previously reported 2-bp insertion in exon 3 and the 4-bp deletion in exon 6; these indels create multiple premature stop codons such that the resultant Tas1r1 lacks any of its seven transmembrane domains and is nonfunctional. The observation of these indels in the homozygous state in two unrelated giant pandas suggests that the null allele is likely fixed in the giant panda. In addition, we identified a 6-bp deletion in a region of exon 6 that is not covered in the draft genome sequence. The two alleles of Tas1r1 from the giant panda we sequenced differ at one nonsynonymous site (supplementary fig. S1, Supplementary Material online). Our two allelic Tas1r1 coding sequences differ from the genome sequence at 9 (five synonymous + four nonsynonymous) and 10 (five synonymous + five nonsynonymous) sites, respectively (supplementary fig. S1, Supplementary Material online). These differences may be due to sequencing errors in the genome sequence, although the possibility of polymorphism cannot be excluded.
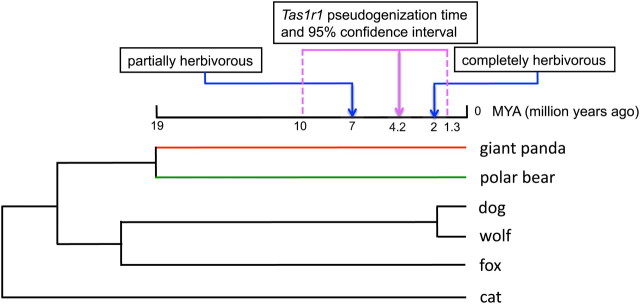
We also sequenced all coding exons of Tas1r1 from the polar bear Ursus maritimus, dog Canis familiaris, wolf Chrysocyon brachyurus, fox Vulpes lagopus, and cat Felis catus (fig. 1). In each of these five carnivores, we found Tas1r1 to be intact. Because the giant panda is the sister species to all other bears (O'Brien et al. 1985; Goldman et al. 1989; Yu et al. 2004; Arnason et al. 2007; Krause et al. 2008), our results indicate that the pseudogenization of Tas1r1 postdated the separation of the giant panda from all other bears (fig. 1), which has been dated to ∼19 Ma based on mitochondrial genome sequences, multiple nuclear genes, and multiple fossil calibrations (Krause et al. 2008; Eizirik et al. 2010).
FIG. 1.
A species tree of the carnivores studied in this work. Branch lengths are not drawn to scale except for the giant panda branch. Dates indicated by blue arrows are inferred from fossil records, whereas dates indicated with the purple arrow and dash lines are inferred from the molecular genetic evidence of this study.
To analyze the selective constraint on Tas1r1, we estimated the rate ratio (ω) of nonsynonymous to synonymous substitutions using PAML (Yang 2007). We aligned the Tas1r1 sequences from the six carnivores and removed the codons where indels are found. We then compared a series of evolutionary models in the likelihood framework, using the species tree shown in figure 1. We found that the average ω across the tree is 0.162, which is significantly lower than 1 (P < 10−50; Table 1), indicative of a generally strong purifying selection on Tas1r1 in carnivore evolution. Consistent with the fact that Tas1r1 became pseudogenized in the giant panda branch, a two-ratio model that allows a difference in ω between the giant panda branch (ω = 0.392) and all other branches (ω = 0.150) fits the data significantly better than a one-ratio model that assumes a single ω for all branches (P = 0.03; table 1). Interestingly, a more general model that allows the variation of ω across all branches does not fit significantly better than the above two-ratio model (P = 0.37; table 1), suggesting that ω does not vary significantly among the nonpanda branches. We further found that the ω for the giant panda branch in the two-ratio model is significantly lower than 1 (P = 0.05; table 1), suggesting that the relaxation of the functional constraint on Tas1r1 did not happen immediately after the giant panda diverged from other bears but rather more recently.
Table 1.
Likelihood Ratio Tests of Various Models on the Selective Pressures on Tas1r1.
| Models | ω | ln La | npb | Models Compared | 2Δ(ln L)c | _P_-Values |
|---|---|---|---|---|---|---|
| A. All branches have the same ω | 0.162 | −4753.63 | 11 | |||
| B. All branches have the same ω = 1 | 1 | −4880.22 | 10 | B vs. A | 253.17 | 5.3 × 10−57 |
| C. The panda branch has _ω_2, other branches have _ω_1 | _ω_1 = 0.150, _ω_2 = 0.392 | −4751.37 | 12 | A vs. C | 4.52 | 0.03 |
| D. The panda branch has _ω_2 = 1, other branches have _ω_1 | _ω_1 = 0.148, _ω_2 = 1 | −4753.24 | 11 | D vs. C | 3.74 | 0.05 |
| E. Each branch has one ω | Variable ω by branch | −4747.59 | 19 | C vs. E | 7.57 | 0.37 |
To estimate when the functional constraint on the giant panda Tas1r1 became relaxed, we assume a simple scenario in which a normal level of constraint as observed in other carnivores was suddenly and completely relaxed t Ma. Using the information of the rate of nucleotide substitution, especially the ω increase in the giant panda, we estimated a posterior probability distribution of t (fig. 2_A_; see Materials and Methods). Using the information that exactly two open reading frame (ORF)-disrupting substitutions are observed in the giant panda Tas1r1, we estimated another posterior probability distribution of t (fig. 2_B_; see Materials and Methods). Because these two distributions were estimated using independent information, we combined them to obtain the final posterior probability distribution of t (fig. 2_C_; see Materials and Methods). In this final distribution, t has a mode of 4.15, a median of 4.25, a mean of 4.59, and a 95% confidence interval of (1.25, 9.95) Ma. If the relaxation of the functional constraint was gradual rather than abrupt as assumed here, the starting date of the gradual relaxation is likely earlier than the above estimate.
FIG. 2.
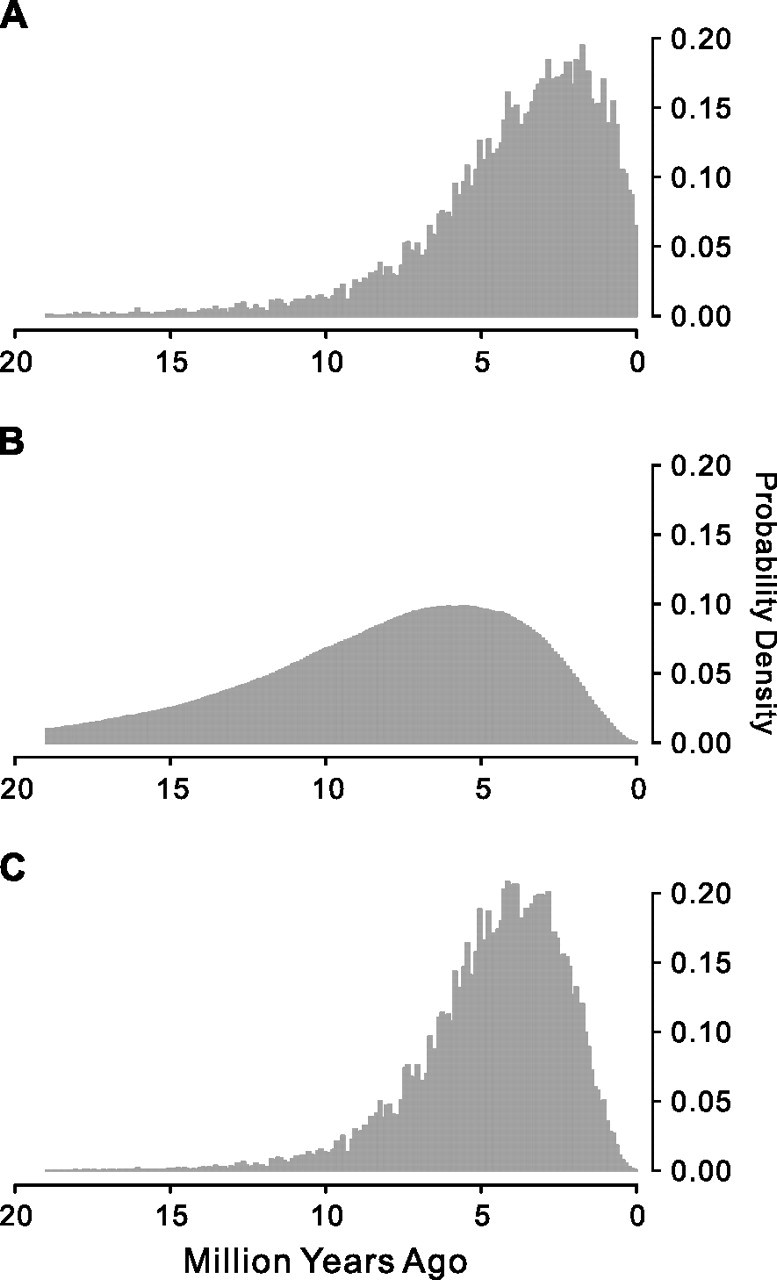
Posterior probability distributions of the time at which the functional constraint on the giant panda Tas1r1 was completely relaxed, based on the observations from (A) the rate of nucleotide substitutions, (B) the number of ORF-disrupting substitutions, and (C) both.
Because the fossil data suggested that the giant panda had started eating bamboo by 7 Ma and had completed this dietary change by 2 Ma, the functional relaxation of Tas1r1 began approximately when the dietary switch of the giant panda occurred (fig. 1). This concurrence provides additional support to the hypothesis that the loss of Tas1r1 is related to the giant panda's herbivorous feeding behavior (Li et al. 2010). Furthermore, Tas1r1 is functional in all other mammals examined (Shi and Zhang 2006), suggesting that its pseudogenization in the giant panda must be due to a relatively recent change unique to the giant panda; the dietary switch to bamboo appears to be the only plausible reason. Nonetheless, our molecular dating has a relatively wide 95% confidence interval, making it difficult to discern whether the functional relaxation of Tas1r1 started prior to, during, or after the giant panda's dietary switch. It is possible that ancient giant pandas started to change their diet to bamboo due to meat scarcity. Their less reliance on meat may have rendered the umami taste less important, leading to the pseudogenization of Tas1r1. The gene loss may have in turn reduced the attraction of returning to meat eating because of the lack of the umami perception. An alternative scenario is that the giant panda lost Tas1r1 prior to its change of diet, due to genetic drift in a small population. This scenario appears much less likely because, although the giant panda is endangered today, it may not have had a particularly small population in its evolutionary history. In fact, the nucleotide diversity of the giant panda is about twice that of humans and there is no apparent upsurge in the rate of gene loss in the giant panda compared that in other mammals (Li et al. 2010). It is worth noting that zebrafish Tas1r2/3 heterodimer detects amino acids rather than sugars in a heterologous expression system (Oike et al. 2007). If a similar situation happens to the giant panda, the pseudogenization of the giant panda Tas1r1 might be unrelated to the dietary switch. However, this scenario is exceedingly improbable, given the clear functional distinction between Tas1r1 and Tas1r2 in mammals (Chandrashekar et al. 2006).
The pseudogenization of the giant panda Tas1r1 prompted us to examine it in the draft genome sequences of two other herbivorous mammals: the cow Bos taurus and horse Equus caballus. Interestingly, Tas1r1 is intact in both species. Although ω is somewhat high in the horse (0.375), it appears normal in the cow (0.206) compared with other mammals. Thus, even though the loss of Tas1r1 in the giant panda may be related to its dietary switch to bamboo, additional factors must be involved. This observation echoes previous observations of the lack of a consistent correlation between the evolutionary patterns of sensory genes and sensory ecologies across lineages (Li et al. 2005; Young et al. 2010; Zhao et al. 2010), suggesting that our current understanding of the physiological roles of various sensory systems is still limited.
Materials and Methods
We used the complete coding sequence of the dog Tas1r1 (GenBank accession no. XM_546753) as the query to BLAT search the dog genome assembly (7.6× coverage) available at the University of California–San Cruz (http://genome.ucsc.edu/); the exon–intron junctions were determined by comparing the genomic sequence with the cDNA sequence using SPIDEY (http://www.ncbi.nlm.nih.gov). Using the dog Tas1r1 sequence as the query, we BlastN-searched the giant panda genome assembly (http://panda.genomics.org.cn). Based on the alignment of the dog and giant panda sequences, we designed 10 pairs of primers to amplify and sequence all 6 exons of Tas1r1 from the giant panda, polar bear, dog, wolf, fox, and cat. Polymerase chain reactions (PCRs) were performed with Go-Taq DNA polymerase (Promega) following the manufacturer's instructions. The PCR products were purified and sequenced by the Sanger method in both directions. The GenBank accession numbers of the newly determined sequences are HM468447–HM468452.
We used the DNA sequences thus obtained in the following analysis. When allelic variations exist in an individual that we sequenced (giant panda, dog, and cat), we used the allelic sequence that is more similar to the available genome sequence. We used two methods to estimate the time when the functional constraint on Tas1r1 was relaxed. In the first method, we used the information of nucleotide substitution rates. On the basis of our observations (see main text), we assume that the nonsynonymous/synonymous substitution rate ratio prior to the functional relaxation was constant among the carnivores examined but became 1 at t Ma in the giant panda. Let the total number of nonsynonymous substitutions per site for the giant panda branch (red branch in fig. 1) be _d_R. Then
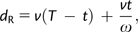 |
(1) |
|---|
where T = 19 MY is the time since the divergence between the giant panda and the polar bear, ω is the nonsynonymous/synonymous substitution rate ratio prior to the functional relaxation, and v is the nonsynonymous substitution rate per nonsynonymous site per MY before the functional relaxation. Equation 1 can be rewritten as
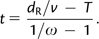 |
(2) |
|---|
We can estimate ω for the black and green branches in figure 1 using PAML and estimate v by _d_BG/t′, where _d_BG is the total number of nonsynonymous substitutions per site for black and green branches of the tree and t′ is the total evolutionary time that these branches represent. Thus,
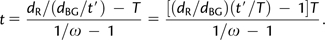 |
(3) |
|---|
We can estimate t′/T by the total number of synonymous and nonsynonymous substitutions on the black and green branches (_D_BG) divided by that on the green branch (_D_G). In other words, we estimated t by
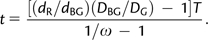 |
(4) |
|---|
By bootstrapping the codons of Tas1r1 104 times, we estimated the frequency distribution of t. We considered a t value to be valid when it is between 0 and 19 and subsequently derived a posterior distribution of t, or _P_1(t|Data 1) (fig. 2_A_), where Data 1 refers to the point substitutions in Tas1r1 used here.
In the second method, we estimated t by considering the fact that exactly two ORF-disrupting substitutions are found in the giant panda Tas1r1. Modifying the program PSEUDOGENE (Zhang and Webb 2003), we estimated the waiting times for two (_t_2) and three (_t_3) ORF-disrupting substitutions in Tas1r1, respectively. The actual time since the functional relaxation should be between _t_2 and _t_3. The PSEUDOGENE program requires the input of point and indel mutation rates. The genomic analysis showed that the point mutation rate in the giant panda is 1.3 × 10−9 per site per year (Li et al. 2010). Because this point mutation rate is almost identical to that in Old World primates (Yi et al. 2002), it is likely that the indel mutation rates of the two groups of organisms are also similar. We thus used the panda point mutation rate and the Old World monkey indel mutation rate (1.0 × 10−10 per site per year) (Podlaha and Zhang 2003) in the analysis. Using the polar bear Tas1r1 sequence as the input sequence, we simulated the pseudogenization process and acquired 104 pairs of _t_2 and _t_3. We thus obtained another posterior probability distribution for the starting time of the functional relaxation or _P_2(t|Data 2) (fig. 2_B_), where Data 2 refers to the observation of two ORF-disrupting substitutions. Because Data 1 and Data 2 are independent from each other, it can be shown that P(t|Data 1 and 2) = _P_1(t|Data 1) × _P_2(t|Data 1)/P(t), where P(t) is the prior distribution of t and is assumed to be uniform within (0, 19). We thus calculated _P_1(t|Data 1) × P_2(t|Data 2) and then rescaled the distribution such that the total probability over (0, 19) MY is 1 (fig. 2_C).
Supplementary Material
Supplementary fig. S1 is available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Supplementary Data
Acknowledgments
We thank Meg Bakewell, Ying Li, Wenfeng Qian, and three anonymous reviewers for valuable comments. This work was supported by a research grant from the U.S. National Institutes of Health to J.Z. J.R.Y. was supported in part by the 985 Project Fund from Sun Yat-sen University and X.L. was supported in part by a fellowship from the Chinese government.
References
- Arnason U, Gullberg A, Janke A, Kullberg M. Mitogenomic analyses of caniform relationships. Mol Phylogenet Evol. 2007;45:863–874. doi: 10.1016/j.ympev.2007.06.019. [DOI] [PubMed] [Google Scholar]
- Bininda-Emonds OR. Phylogenetic position of the giant panda. In: Lindburg D, Baragona K, editors. Giant panda: biology and conservation. Berkeley (CA): University of California Press; 2004. [Google Scholar]
- Chandrashekar J, Hoon MA, Ryba NJ, Zuker CS. The receptors and cells for mammalian taste. Nature. 2006;444:288–294. doi: 10.1038/nature05401. [DOI] [PubMed] [Google Scholar]
- Eizirik E, Murphy WJ, Koepfli KP, Johnson WE, Dragoo JW, Wayne RK, O'Brien SJ. Pattern and timing of diversification of the mammalian order Carnivora inferred from multiple nuclear gene sequences. Mol Phylogenet Evol. 2010;56:49–63. doi: 10.1016/j.ympev.2010.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman DP, Giri R, J O'Brien S. Molecular genetic-distance estimates among the Ursidae as indicated by one- and two-dimensional protein electrophoresis. Evolution. 1989;43:282–295. doi: 10.1111/j.1558-5646.1989.tb04228.x. [DOI] [PubMed] [Google Scholar]
- Jin C, Ciochon RL, Dong W, Hunt RM, Jr, Liu J, Jaeger M, Zhu Q. The first skull of the earliest giant panda. Proc Natl Acad Sci U S A. 2007;104:10932–10937. doi: 10.1073/pnas.0704198104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozukue E, Kozukue N, Kurosaki T. Organic acid, sugar and amino acid composition of bamboo shoots. J Food Sci. 1983;48:935–938. [Google Scholar]
- Krause J, Unger T, Nocon A, et al. (18 co-authors) Mitochondrial genomes reveal an explosive radiation of extinct and extant bears near the Miocene-Pliocene boundary. BMC Evol Biol. 2008;8:220. doi: 10.1186/1471-2148-8-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Fan W, Tian G, et al. (123 co-authors) The sequence and de novo assembly of the giant panda genome. Nature. 2010;463:311–317. doi: 10.1038/nature08696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Li W, Wang H, et al. (11 co-authors) Pseudogenization of a sweet-receptor gene accounts for cats' indifference toward sugar. PLoS Genet. 2005;1:27–35. doi: 10.1371/journal.pgen.0010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson G, Chandrashekar J, Hoon MA, Feng L, Zhao G, Ryba NJ, Zuker CS. An amino-acid taste receptor. Nature. 2002;416:199–202. doi: 10.1038/nature726. [DOI] [PubMed] [Google Scholar]
- Nelson G, Hoon MA, Chandrashekar J, Zhang Y, Ryba NJ, Zuker CS. Mammalian sweet taste receptors. Cell. 2001;106:381–390. doi: 10.1016/s0092-8674(01)00451-2. [DOI] [PubMed] [Google Scholar]
- O'Brien SJ, Nash WG, Wildt DE, Bush ME, Benveniste RE. A molecular solution to the riddle of the giant panda's phylogeny. Nature. 1985;317:140–144. doi: 10.1038/317140a0. [DOI] [PubMed] [Google Scholar]
- Oike H, Nagai T, Furuyama A, Okada S, Aihara Y, Ishimaru Y, Marui T, Matsumoto I, Misaka T, Abe K. Characterization of ligands for fish taste receptors. J Neurosci. 2007;27:5584–5592. doi: 10.1523/JNEUROSCI.0651-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor JF, Barbosa M, De Paz FJ. Morphological study of the lingual papillae of the giant panda (Ailuropoda melanoleuca) by scanning electron microscopy. J Anat. 2008;212:99–105. doi: 10.1111/j.1469-7580.2008.00850.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podlaha O, Zhang J. Positive selection on protein-length in the evolution of a primate sperm ion channel. Proc Natl Acad Sci U S A. 2003;100:12241–12246. doi: 10.1073/pnas.2033555100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller GB, Deng Q, Johnson KG, Wang X, Shen L, Hu J. The feeding ecology of giant pandas and asiatic black bears in the Tangjiahe reserve, China. In: Gittleman JL, editor. Carnivore behavior, ecology and evolution. Ithaca: Cornell University Press; 1989. pp. 212–241. [Google Scholar]
- Schaller GB, Hu J, Pan W, Zhu J. The giant pandas of Wolong. Chicago: University of Chicago Press; 1985. [Google Scholar]
- Shi P, Zhang J. Contrasting modes of evolution between vertebrate sweet/umami receptor genes and bitter receptor genes. Mol Biol Evol. 2006;23:292–300. doi: 10.1093/molbev/msj028. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yi S, Ellsworth DL, Li WH. Slow molecular clocks in Old World monkeys, apes, and humans. Mol Biol Evol. 2002;19:2191–2198. doi: 10.1093/oxfordjournals.molbev.a004043. [DOI] [PubMed] [Google Scholar]
- Young JM, Massa HF, Hsu L, Trask BJ. Extreme variability among mammalian V1R gene families. Genome Res. 2010;20:10–18. doi: 10.1101/gr.098913.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Li QW, Ryder OA, Zhang YP. Phylogeny of the bears (Ursidae) based on nuclear and mitochondrial genes. Mol Phylogenet Evol. 2004;32:480–494. doi: 10.1016/j.ympev.2004.02.015. [DOI] [PubMed] [Google Scholar]
- Zhang J, Webb DM. Evolutionary deterioration of the vomeronasal pheromone transduction pathway in catarrhine primates. Proc Natl Acad Sci U S A. 2003;100:8337–8341. doi: 10.1073/pnas.1331721100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao GQ, Zhang Y, Hoon MA, Chandrashekar J, Erlenbach I, Ryba NJ, Zuker CS. The receptors for mammalian sweet and umami taste. Cell. 2003;115:255–266. doi: 10.1016/s0092-8674(03)00844-4. [DOI] [PubMed] [Google Scholar]
- Zhao H, Zhou Y, Pinto CM, Charles-Dominique P, Galindo-González J, Zhang S, Zhang J. Evolution of the sweet taste receptor gene Tas1r2 in bats. Mol Biol Evol. 2010 doi: 10.1093/molbev/msq152. forthcoming. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data