Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells (original) (raw)
. Author manuscript; available in PMC: 2011 Aug 2.
Published in final edited form as: Nat Biotechnol. 2010 Jul 19;28(8):848–855. doi: 10.1038/nbt.1667
Abstract
Induced pluripotent stem cells (iPSCs) have been derived from various somatic cell populations through ectopic expression of defined factors. It remains unclear whether iPSCs generated from different cell types are molecularly and functionally similar. Here we show that iPSCs obtained from mouse fibroblasts, hematopoietic and myogenic cells exhibit distinct transcriptional and epigenetic patterns. Moreover, we demonstrate that cellular origin influences the in vitro differentiation potentials of iPSCs into embryoid bodies and different hematopoietic cell types. Notably, continuous passaging of iPSCs largely attenuates these differences. Our results suggest that early-passage iPSCs retain a transient epigenetic memory of their somatic cells of origin, which manifests as differential gene expression and altered differentiation capacity. These observations may influence ongoing attempts to use iPSCs for disease modeling and could also be exploited in potential therapeutic applications to enhance differentiation into desired cell lineages.
IPSCs are usually obtained from fibroblasts after infection with viral constructs expressing the four transcription factors Oct4, Sox2, Klf4 and c-Myc1–10. In addition, other cell types, including blood2,4,11, stomach and liver cells1, keratinocytes12,13, melanocytes14, pancreatic β cells7 and neural progenitors3,15–17 have been reprogrammed into iPSCs. Although these iPSC lines have been shown to express pluripotency genes and support the differentiation into cell types of all three germ layers, recent studies detected substantial molecular and functional differences among iPSCs derived from distinctive cell types. For example, iPSCs produced from various fibroblasts, stomach and liver cells showed different propensities to form tumors in mice, although the underlying molecular mechanisms remain elusive18. Another study identified persistent donor cell–specific gene expression patterns in human iPSCs produced from different cell types, suggesting an influence of the somatic cell of origin on the molecular properties of resultant iPSCs19. Whether cellular origin also affected the functional properties of iPSCs remained unexplored in that report. Of note, the findings of some of these studies may be confounded by the presence of different viral insertions in individual iPSC lines and by the fact that the analyzed iPSC lines were of different genetic background, which can affect both gene expression patterns20 and the functionality9,21 of cells. Indeed, we have recently shown that many mouse iPSC lines derived from different somatic cell types show aberrant silencing of a surprisingly small set of transcripts compared with embryonic stem cells (ESCs)22. However, our study did not investigate whether additional cell-of-origin–specific differences may exist in iPSC lines derived from different cell types.
Patient-specific iPSCs are a valuable tool for the study of disease and possibly for the development of therapies20,23–26. Thus, resolving the question of whether iPSCs produced from different cell types are molecularly and functionally equivalent is crucial for using these cells to model disease, which entails detecting subtle differences in the differentiation potential of patient-derived iPSCs24,27. Furthermore, the identification of somatic cells that influence the differentiation capacities of resultant iPSCs into desired cell lineages could be useful in a therapeutic setting.
To assess whether iPSCs derived from different somatic cell types are distinguishable, we compared here the transcriptional and epigenetic patterns, as well as the in vitro differentiation potentials, of iPSCs produced from four genetically identical adult mouse cell types that differed only in the lineage from which they were derived.
RESULTS
Genetically matched iPSCs derived from different cell types
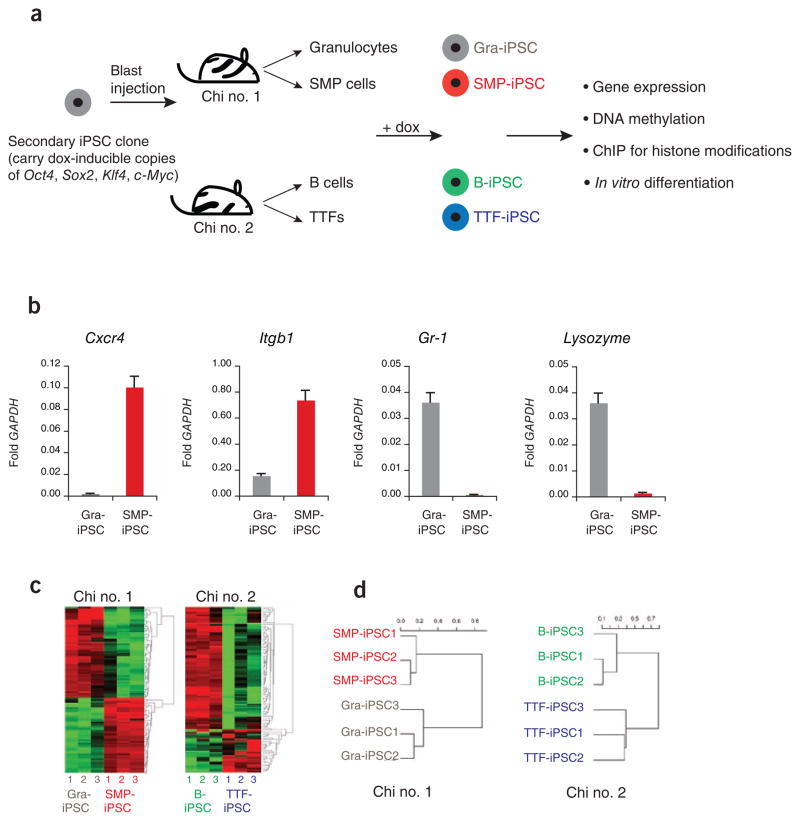
Because the genetic background of ESCs can influence their transcriptional and functional behaviors, we used a previously described ‘secondary system’ to generate genetically identical iPSCs2,28 (Fig. 1a). Briefly, iPSCs were generated from somatic cells using doxycycline-inducible lentiviruses expressing Oct4, Sox2, Klf4 and c-Myc29, and then injected into blastocysts to produce isogenic chimeric mice. Thus, isolation of different cell types from these chimeras and their subsequent exposure to doxycycline gave rise to iPSCs with the same genetic makeup. In this study, we focused on iPSCs derived from tail tip–derived fibroblasts (TTFs), splenic B cells (B), bone marrow–derived granulocytes and skeletal muscle precursors (SMPs)30, which were continuously cultured for 2–3 weeks (passage 4 to 6) after picking. The pluripotency of some of these cell lines has been previously documented2, or was analyzed in this study (Supplementary Table 1 and Supplementary Fig. 1). All cell lines grew at similar rates and independently of viral transgene expression (Supplementary Fig. 2) and upregulated the endogenous pluripotency genes Nanog, Sox2 and Oct4, indicating successful molecular reprogramming (Supplementary Table 1). Moreover, all lines gave rise to differentiated teratomas, and all tested lines supported the development of chimeric animals upon blastocyst injection, demonstrating their pluripotency (Supplementary Table 1). We therefore concluded that the cell lines analyzed here qualify as bona fide iPSC lines.
Figure 1.
iPSCs derived from different cell types are transcriptionally distinguishable. (a) Flow chart explaining the derivation and analysis of genetically matched iPSCs from different cell types. Secondary iPSCs were first injected into blastocysts to generate chimeric mice, from which the indicated somatic cell types were isolated. Exposure of these cells to doxycycline (dox) then gave rise to iPSCs. ChIP, chromatin immunoprecipitation. (b) Quantification of the expression levels of Cxcr4, Itgb1, Gr-1 and Lysozyme by quantitative PCR in SMP-iPSCs, in red, and Gra-iPSCs, in gray. The values were normalized to GAPDH expression; the error bars depict the s.e.m. (n = 3). (c) Heat map showing top 104 probes with highest variance in their expression levels. Left panel, SMP-iPSCs and Gra-iPSCs derived from chimera no. 1. Right panel, TTF-iPSCs and B-iPSCs derived from chimera no. 2. (d) Hierarchical, unsupervised clustering of iPSC expression profiles using the correlation distance and the Ward method. SMP-iPSCs and Gra-iPSCs were derived from chimera no. 1 (left panel), TTF-iPSCs and B-iPSCs originate from chimera no. 2 (right panel). Chi no. 1, chimera no. 1; chi no. 2, chimera no. 2.
iPSCs produced from different cell types are transcriptionally distinguishable
We first evaluated whether iPSCs derived from defined somatic cell types retain gene expression patterns indicative of their cells of origin. Specifically, we assessed the expression of cell lineage–specific candidate genes in iPSCs derived from granulocytes (Gra-iPSCs) and SMPs (SMP-iPSCs). As expected, the SMP markers Cxcr4 and Integrin B1 and the granulocyte markers Lysozyme (also known as Lyz1 and Lyz2) and Gr-1 (also known as Ly6g) were expressed at considerably higher levels in the somatic cells of origin than in resultant iPSCs (Supplementary Fig. 3). Moreover, SMP-iPSCs expressed substantially higher levels of Cxcr4 and Itgb1 than did Gra-iPSCs (Fig. 1b), and Gra-iPSCs showed higher expression levels of Lysozyme and Gr-1 compared with SMP-iPSCs (Fig. 1b). Together, these data suggest that iPSCs retain a transcriptional memory of their somatic cell of origin.
To test this notion globally, we compared the transcriptional profiles of iPSC lines originating from SMPs (n = 3) with those derived from granulocytes (n = 3), as well as expression profiles of iPSC lines originating from B cells (n = 3) with those produced from TTFs (n = 3). Note that iPSCs were compared with each other only if they originated from the same chimeric mouse (SMP-iPSCs versus Gra-iPSCs and B-iPSCs versus TTF-iPSCs) (Fig. 1a) to eliminate potential variability between different experiments and individual animals. All iPSC lines analyzed were between passage (p) 4 and 6. There were 1,388 genes differentially expressed (twofold, corrected P = 0.05) between SMP-iPSCs and Gra-iPSCs, and 1,090 genes between B-iPSCs and TTF-iPSCs (Supplementary Table 2). An analysis of the 100 genes with the greatest range of expression levels across all samples indicated that iPSCs with the same cell of origin clustered together (Fig. 1c). Consistent with this observation, unsupervised hierarchical clustering (Fig. 1d) as well as principal component analysis (Supplementary Fig. 4) of all genes placed SMP-iPSCs and Gra-iPSCs, as well as B-iPSCs and TTF-iPSCs, into different groups according to their cells of origin. Notably, Gene Ontology (GO) analysis of the 100 genes with the greatest range of expression between SMP-iPSCs and Gra-iPSCs indicated an enrichment for genes belonging to the categories ‘myofibril’ (7.6-fold enrichment), ‘contractile fiber’ (7.3-fold enrichment) and ‘muscle development’ (5.9-fold enrichment) as well as ‘B-cell activation’ (6.8-fold enrichment) and ‘leukocyte activation’ (3.7-fold enrichment) (when compared with the expected background). Together, these results show that genetically identical iPSCs obtained from four different somatic cell types are distinguishable from each other using genome-wide transcriptional analyses, further supporting the notion that the donor cell type influences the overall gene expression pattern of resultant iPSCs.
To determine the effect on gene expression patterns of deriving iPSCs from different animals in independent experiments, we compared the expression profiles of Gra-iPSCs derived from chimera no. 1 (n = 3) with Gra-iPSCs from chimera no. 2 (n = 3) as well as with SMP-iPSCs from chimera no. 1 and TTF-iPSCs from chimera no. 2 (Fig. 1a). Hierarchical clustering separated Gra-iPSCs according to their origin from different animals, suggesting a significant contribution of this experimental variable to gene expression patterns (Supplementary Fig. 5). However, when the expression data from TTF-iPSCs and SMP-iPSCs were included in the analysis, we found that differences due to cell of origin were stronger than those arising from variations in experimental conditions or animals. These data reinforce the observation that iPSCs derived from different somatic cell types are transcriptionally distinguishable, even when they originate from different animals.
To exclude the possibility that the observed gene expression differences were due to the specific secondary system used, we derived iPSCs from SMPs, granulocytes, B cells and peritoneal fibroblasts from reprogrammable mice31, which carry dox-inducible copies of all four reprogramming factors in a defined genomic locus. All iPSC lines grew independently of dox and gave rise to differentiated teratomas (Supplementary Fig. 6a). Analysis of gene expression profiles of these lines at p4 showed clustering according to their cells of origin, with the exception of peritoneal fibroblast–derived iPSCs, which may be a consequence of the heterogeneity of the starting population. Collectively, these results corroborate the notion that iPSCs generated from different cell types exhibit distinct transcriptional patterns (Supplementary Fig. 6b).
iPSCs derived from different cell types exhibit distinguishable epigenetic patterns
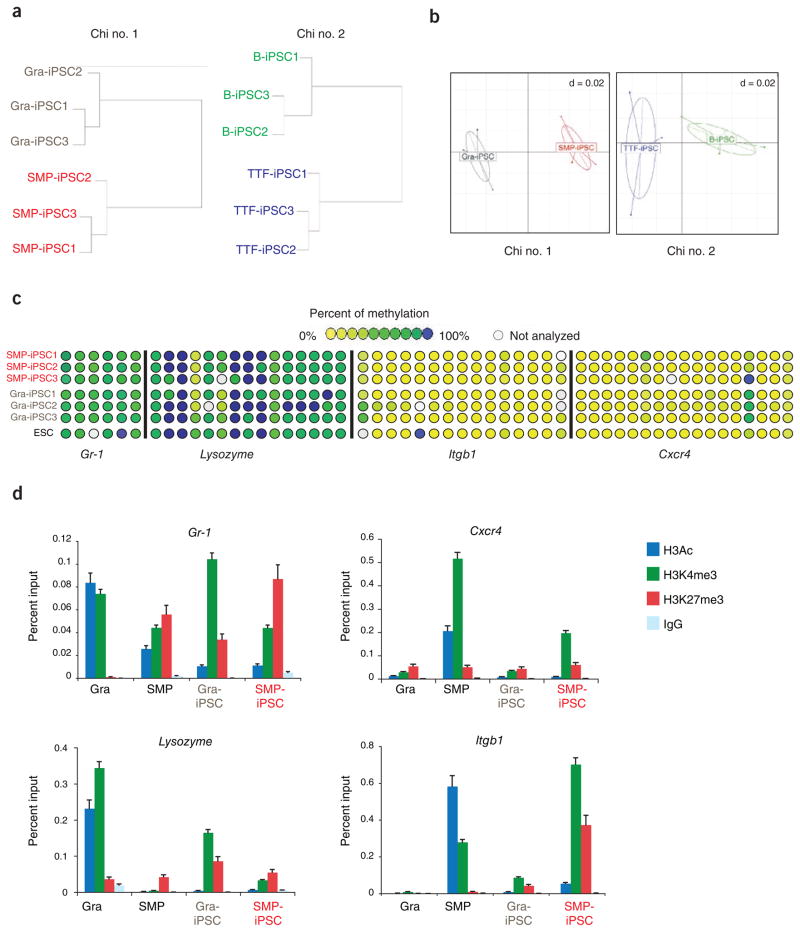
We next asked whether the differential gene expression patterns we observed correlated with differences in epigenetic marks. To this end, we performed a genome-wide, restriction enzyme–based methylation analysis of promoters termed ‘HpaII tiny fragment enrichment by ligation-mediated PCR’ (HELP) on the same cell lines we used for expression analysis. Unsupervised hierarchical clustering showed that Gra-iPSCs and SMP-iPSCs, as well as B-iPSCs and TTF-iPSCs, which clustered separately in the transcriptional assays, were also distinguishable based on their methylation patterns (Fig. 2a). Correspondence analysis of the same samples corroborated this finding (Fig. 2b), indicating that the donor cell type affects not only the overall transcriptional pattern but also the promoter methylation pattern of resultant iPSCs.
Figure 2.
iPSCs derived from different cell types exhibit distinguishable epigenetic signatures. (a) Hierarchical unsupervised clustering analysis of HELP genome-wide methylation data from indicated iPSC lines. (b) Correspondence analysis of SMP-iPSCs and Gra-iPSCs (left panel) from chimera no. 1, TTF-iPSCs and B-iPSCs (right panel) from chimera no. 2. (c) Graphic representation of DNA methylation quantification of specific CpGs (circles) in the promoter regions of the indicated candidate genes using EpiTYPER DNA methylation analyses. Yellow indicates 0% methylation and blue 100% methylation. (d) Chromatin immunoprecipitation (ChIP) for H3 pan-acetylated (H3Ac, in blue), H3K4 trimethylated (H3K4me3, in green), H3K27 trimethylated (H3K27me3, in red) and isotype control (IgG, in light blue) of granulocytes (Gra), SMPs, Gra-iPSCs and SMP-iPSCs. Chi no. 1, chimera no. 1; chi no. 2, chimera no. 2. The error bars depict the s.e.m. (n = 3).
Despite the separation of Gra-iPSCs from SMP-iPSCs and of TTF-iPSCs from B-iPSCs (Fig. 2a, b) by hierarchical clustering, we detected few loci that were differentially methylated with statistical significance using supervised analysis (69 genes between Gra-iPSCs and SMP-iPSCs and 0 genes between B-iPSCs and TTF-iPSCs; Supplementary Table 3). To complement these results, we interrogated the DNA methylation status at the promoter regions of the previously analyzed markers Cxcr4, Itgb1, Lysozyme and Gr-1 (Fig. 1b) using EpiTYPER DNA methylation analysis, which quantifies gene-specific CpG methylation. We failed to detect differences in the methylation levels of these candidate genes between SMP-iPSCs and Gra-iPSCs (Fig. 2c), further indicating that methylation differences are more subtle than the observed gene expression differences and raising the possibility that other chromatin marks may be responsible for the observed expression differences.
Indeed, we observed high levels of the activating marks H3Ac and H3K4me3 and low levels of the repressive marks H3K27me3 at the promoters of Cxcr4 and Itgb1 in SMPs and at the promoters of Lysozyme and Gr-1 in granulocytes, respectively, consistent with their abundant expression in these cell types (Fig. 2d). Notably, SMP-iPSCs, which showed higher expression levels of Cxcr4 and Itgb1 than did Gra-iPSCs (Fig. 1b), were enriched for H3K4me3 compared with Gra-iPSCs at these two genes. A similar pattern was observed for the granulocyte-specific genes in Gra-iPSCs compared with SMP-iPSCs, with Gr-1 and Lysozyme being elevated for H3K4me3 (Fig. 2d). These data show that the observed expression differences among iPSCs derived from different cell types may be predominantly the consequence of differences in histone marks, further suggesting that iPSCs retain an epigenetic memory of their cells of origin.
iPSCs derived from different cell types have distinctive in vitro differentiation potentials
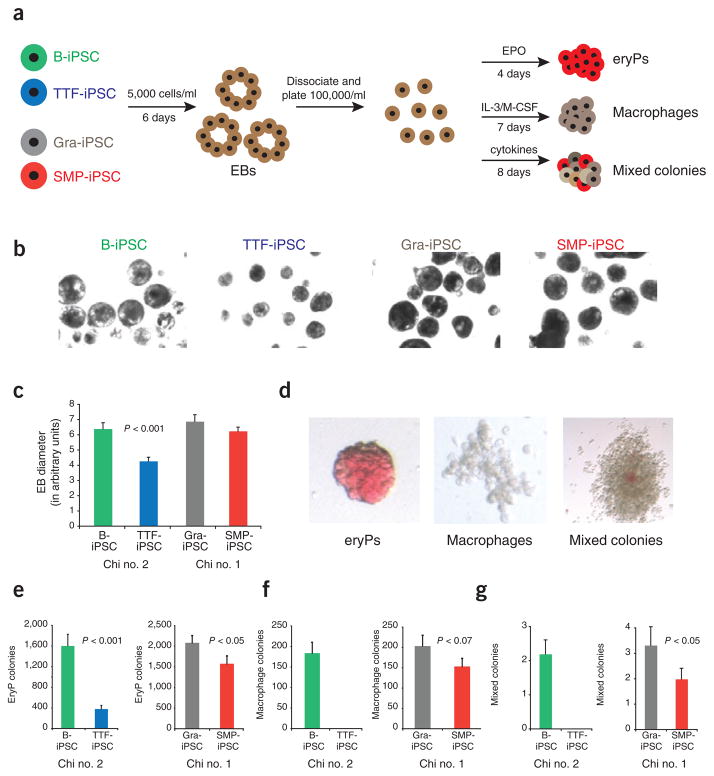
Because the gene expression differences we observed among different iPSC lines affected genes known to be involved in the lineage-specific differentiation and function of the somatic cell types from which they were derived, we reasoned that these differences might affect their capacity to differentiate into defined cell lineages. Thus, we evaluated the autonomous differentiation potential of the four types of iPSC lines by assessing their abilities to produce embryoid bodies, erythrocyte progenitors, macrophages and mixed hematopoietic colonies using established semiquantitative differentiation protocols (Fig. 3a). Most notably, TTF-iPSCs produced significantly smaller and fewer embryoid bodies compared with all the other iPSC lines (P < 0.001; Fig. 3b, c). Moreover, the embryoid bodies derived from TTF-iPSC generated relatively few erythrocyte, macrophage and mixed colony progenitors compared with B-iPSCs derived from the same animal despite equal numbers of input cells, indicating striking differences in the differentiation potentials of these iPSCs (Fig. 3d–g). In contrast, SMP-iPSCs and Gra-iPSCs showed equivalent abilities to produce embryoid bodies (Fig. 3d–g). However, Gra-iPSCs gave rise to erythrocyte, macrophages and mixed colonies at higher efficiencies than SMP-iPSCs, suggesting a pattern of differentiation that reflects their cells of origin. Together, these data show that the cell type of origin may bias the differentiation potential of resultant iPSC lines.
Figure 3.
iPSCs derived from different cell types have distinctive in vitro differentiation potentials. (a) experimental outline. iPSCs were first differentiated into embryoid bodies. At day 6, embryoid bodies were dissociated and plated in conditions to favor differentiation into erythrocyte progenitors (eryP) and macrophage and mixed hematopoietic colonies. (b) Phase contrast images showing embryoid bodies derived from B-iPSCs, TTF-iPSCs, Gra-iPSCs and SMP-iPSCs at same magnification. (c) Quantification of embryoid body sizes derived from B-iPSCs, TTF-iPSCs, Gra-iPSCs and SMP-iPSCs; the diameter of the embryoid bodies was measured using arbitrary units (AU). The error bars depict the s.e.m. (n = 30) (d) Representative images of erythrocyte progenitors (eryPs), macrophage colonies and mixed hematopoietic colonies. (e–g) Quantification of in vitro differentiation potentials of the different iPSCs into EryPs (e), macrophage colonies (f) and mixed hematopoietic colonies (g). Chi no. 1, chimera no. 1; chi no. 2, chimera no. 2. The error bars depict the s.e.m. (n = 12).
Continuous passaging of iPSCs abrogates transcriptional, epigenetic and functional differences
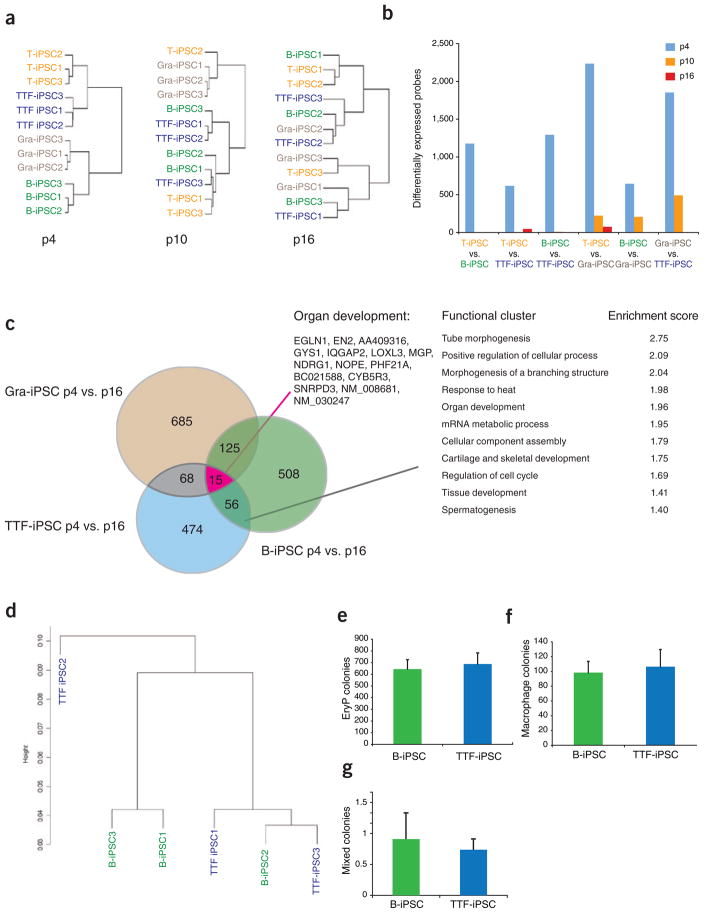
Previously published data suggest that early-passage, human iPSCs derived from fibroblasts are transcriptionally distinct from late-passage iPSCs32. However, that study did not examine the effect of passaging on the iPSC functionality. We therefore wondered whether continuous passaging of the various iPSC lines would eliminate the observed differences in gene expression and differentiation potential. For this analysis, we added to the B-iPSC/TTF-iPSC group, studied before (Figs. 1 and 2a, b), a new set of T cell– and granulocyte-derived iPSCs, which were all derived from chimera no. 2. These 12 iPSC lines were subjected to several additional rounds of passaging under identical culture conditions, and RNA was harvested at p10 and p16 for expression profiling. Whereas unsupervised hierarchical clustering of these cell lines at early passage (p4) clearly separated each of the different iPSC lines according to their cells of origin (Fig. 4a, left panel), unsupervised clustering of these lines at p10 showed that B-iPSCs, TTF-iPSCs and T-iPSCs were indistinguishable from each other, whereas the Gra-iPSCs still clustered together (Fig. 4a, middle panel). Further passaging of these cells until p16 entirely eliminated these differences (Fig. 4a, right panel). Together, these data indicate that continuous cell division resolves transcriptional differences among iPSC lines. Consistent with this observation, the total number of differentially expressed genes between various pairs of iPSC lines derived from different cellular origins was reduced from ~500–2,000 in early-passage cultures to only ~50 or even 0 in late-passage cultures, further demonstrating that after extensive in vitro propagation, these iPSC lines have become very similar to each other (Fig. 4b).
Figure 4.
Continuous passaging of iPSCs abrogates transcriptional, epigenetic and functional differences. (a) Hierarchical unsupervised clustering of expression profiles from B-iPSCs, T-iPSCs, TTF-iPSCs and Gra-iPSCs from chimera no. 2. Left panel shows clustering analysis of all iPSC samples at passage p4, the middle panel at p10 and the right panel at p16. (b) Number of differentially expressed probes between pairs of iPSC samples used in a; iPSCs at p4 are shown in blue bars, iPSCs at p10 are shown in orange bars and iPSCs at p16 are shown in red bars. The number of differently expressed probes between iPSCs was calculated using a pairwise analysis (twofold), with _t_-test P = 0.05, with Bejamini and Hochberg correction (n = 3). (c) Venn diagram and GO analysis showing overlap of genes that change from p4 to p16 in Gra-IPSCs, TTF-iPSCs and B-iPSCs. Red line marks functional GO cluster of genes shared between all three iPSC groups. Black line marks functional GO cluster of genes shared by at least two of the iPSC groups. Functional ontology cluster analysis was performed using the DAVIS algorithm. (d) Hierarchical unsupervised clustering using HELP genome-wide methylation profiles of B-iPSCs and TTF-iPSCs at p16. (e–g) Quantification of in vitro differentiation potentials of B-iPSCs and TTF-iPSCs at p16 into EryPs (e), macrophage colonies (f) and mixed hematopoietic colonies (g). The error bars depict the s.e.m. (n = 9).
Analysis of the genes whose expression changed between p4 and p16 in Gra-iPSCs, B-iPSCs and TTF-iPSCs showed 25% overlap with at least one of the other two groups of iPSC lines, suggesting that iPSCs undergo some common changes during passaging, irrespective of their cell of origin (Fig. 4c). GO analysis of these changes indicated a strong enrichment for developmental regulators. Moreover, the only GO cluster common to all three groups was ‘organ development’, indicating that the passaging of iPSCs results in a change of differentiation-associated gene expression patterns (Fig. 4c). The expression levels of the pluripotency genes Sox2 and Oct4, which are high already at early passage (Supplementary Table 1), increased even further during the passaging process, supporting the notion that the pluripotency network becomes increasingly solidified during culture (Supplementary Fig. 7), consistent with a previous report showing gradual upregulation of pluripotency-associated genes upon passaging of human iPSC lines32.
To evaluate whether the passaging of iPSCs attenuates the observed epigenetic differences, we performed HELP analysis on B-iPSCs and TTF-iPSCs at late passage. In contrast to early-passage iPSCs, the late-passage iPSCs could not be separated by hierarchical unsupervised clustering analysis based on their cells of origin (Fig. 4d). Accordingly, the methylation levels of histones at candidate genes in Gra-iPSCs and SMP-iPSCs became indistinguishable (Supplementary Fig. 8). Notably, several of the analyzed loci showed an enrichment for both H3K4me3 and H3K27me3, indicative of bivalent domains that are characteristic of pluripotent stem cells33. Thus, continuous passaging leads to an equilibration of the epigenetic differences detected in early-passage iPSCs.
Two possible mechanisms could account for the observed loss of epigenetic and transcriptional memory with increased passage number: (i) passive replication-dependent loss of somatic marks in the majority of iPSCs and (ii) selection of rare, preexisting, fully reprogrammed cells over time. Because the selection model predicts that such rare clones would have a growth or survival advantage, we would expect to see impaired growth rates of bulk iPSC cultures at early passage compared with late passage, which we did not observe (Supplementary Fig. 9a). We also did not detect significant differences when the growth rates of single-cell clones established from early and late passage iPSC lines were examined using a colorimetric assay (XTT assay) that detects metabolic activity (Supplementary Fig. 10) or by measuring the increase in cell numbers on three consecutive days (Supplementary Figs. 11 and 12). Similarly, an analysis of the colony formation efficiency of single cell-sorted iPSC from early- and late-passage cultures did not yield detectable differences (Supplementary Fig. 9b). Collectively, these data argue against the presence of rare subclones that become selected over time and are consistent with the notion that all iPSC lines gradually resolve transcriptional and epigenetic differences with increased passaging. However, our results do not exclude a combined model involving passive resolution of epigenetic marks as well as selection of multiple clones.
Finally, we asked whether the similar transcriptional and epigenetic patterns of late-passage iPSCs derived from distinct cells of origin would translate into an equalization of their differentiation potentials. We first performed an embryoid-body formation assay at different passages for TTF-iPSCs and B-iPSCs, which showed a strong difference at early passage. TTF-iPSCs gave rise to similarly-sized embryoid bodies as B-iPSCs around p10–p12 (Supplementary Fig. 13a, b) and were indistinguishable at p16 (Supplementary Fig. 13c, d). Moreover, embryoid bodies derived from TTF-iPSCs and B-iPSCs at p16 differentiated into similar numbers of erythrocyte (Fig. 4e), macrophage (Fig. 4f) and mixed-colony progenitors (Fig. 4g), thus proving that extensive cellular passaging eliminates differences in the differentiation potentials of these iPSCs.
DISCUSSION
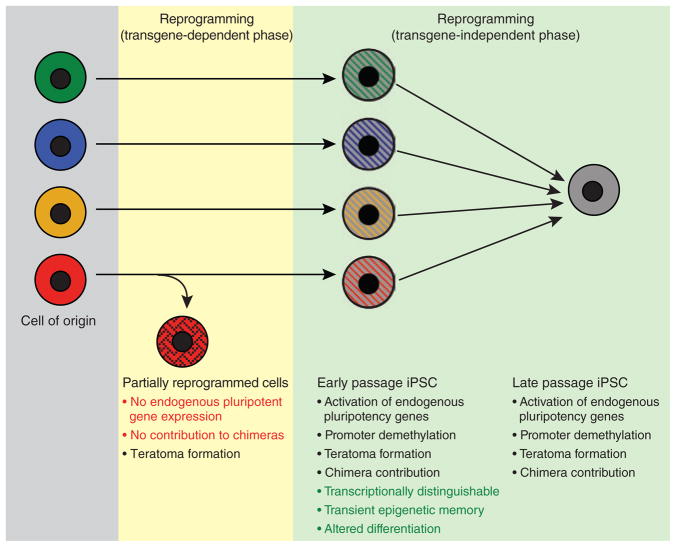
Our study shows that genetically matched iPSCs retain a transient transcriptional and epigenetic memory of their cell of origin at early passage, which can substantially affect their potential to differentiate into embryoid bodies and different hematopoietic cell types (Fig. 5). These molecular and functional differences are lost upon continuous passaging, however, indicating that complete reprogramming is a gradual process that continues beyond the acquisition of a bona fide iPSC state as measured by the activation of endogenous pluripotency genes, viral transgene–independent growth and the ability to differentiate into cell types of all three germ layers. Notably, the previously seen silencing of the Dlk1-Dio3 locus in many iPSC lines22 is not affected by the passaging of cells (data not shown). Of note, the early-passage iPSCs described here are different from “partially reprogrammed iPSCs”34,35, which depend on the continuous expression of viral transgenes and do not activate and demethylate pluripotency genes or contribute to the formation of viable chimeras (Fig. 5).
Figure 5.
Model summarizing the presented data. iPSCs derived from different somatic cell types retain a transient epigenetic and transcriptional memory of their cell type of origin at early passage, despite acquiring pluripotent gene expression, transgene-independent growth and the ability to contribute to tissues in chimeras. Continuous passaging resolves these differences, giving rise to iPSCs that are molecularly and functionally indistinguishable. Note the difference between early passage iPSCs and partially reprogrammed cells, which require continuous viral transgene expression and fail to activate endogenous pluripotency genes or support the development of viable mice.
The mechanism by which passaging eliminates the molecular and functional differences between iPSCs of different origins remains to be determined. Three key observations argue against the possibility of selective expansion of a rare subset of completely reprogrammed iPSCs: (i) both early- and late-passage iPSCs had similar proliferation rates; (ii) there was little variability in the growth rate of single-cell iPSC clones from early- and late-passage lines; and (iii) the number of passages required to resolve cell-of-origin differences was dependent upon the starting cell type. These observations suggest that the consolidation of the pluripotent transcriptional network upon passaging is a slow process, potentially facilitated by a positive feedback mechanism that gradually resolves the residual cell-of-origin–specific epigenetic marks and transcriptional patterns. In accordance with this idea is the finding that telomeres become gradually elongated with increased passage number of iPSCs36. Our results are also consistent with the previous observation that cloned embryos often retain donor cell–specific transcriptional patterns and do not efficiently activate embryonic genes over many cell divisions37–40, suggesting possible similarities in the mechanisms of reprogramming by nuclear transfer and induced pluripotency.
Because of the lack of ESC lines genetically matched to the secondary iPSC lines used here, we did not include ESC lines in our comparative analysis. Nevertheless, the present results may help to explain some of the previously reported differences between ESCs and iPSCs41,42. Some of these studies compared late-passage ESC lines with iPSC lines of undefined, but presumably earlier, passage that may not yet have reached an ESC-equivalent ground state. It should be informative to revisit these studies with genetically matched, trans-gene-free late-passage iPSCs to determine whether this abrogates such gene expression and differentiation differences.
The observed tendency of early-passage iPSC lines to differentiate preferentially into the cell lineage of origin could potentially be exploited in clinical settings to produce certain somatic cell types that have been difficult to obtain from ESCs thus far. However, these data also serve as a cautionary note for ongoing attempts to recapitulate disease phenotypes in vitro using patient-specific, early-passage iPSC lines, as the epigenetic, transcriptional and functional ‘immaturity’ of these cells might confound the data obtained from them. Further elucidation of the molecular indicators of fully reprogrammed iPSCs should help in the establishment of standardized iPSC lines that can be compared with confidence in basic biological and drug discovery studies.
METHODS
Methods and any associated references are available in the online version of the paper at http://www.nature.com/naturebiotechnology/.
Accession code. GEO: GSE22043, GSE22827, GSE22908.
Supplementary Material
Supp Data
Supp Data2
Supp Data3
Supp Data4
Supp Data5
Acknowledgments
We thank N. Maherali and R. Walsh for helpful suggestions and critical reading of the manuscript, B. Wittner for statistical advice, J. LaVecchio, G. Buruzula, K. Folz-Donahue and L. Prickett for expert cell sorting and K. Coser for technical assistance. J.M.P. was supported by an MGH ECOR fellowship, E.A. by a Jane Coffin Childs fellowship, M.S. by a Schering fellowship and K.Y.T. by the Agency of Science, Technology and Research Singapore. Support to A.M. was from the Lymphoma Society, SCOR no. 7132-08; to T.E. from National Institutes of Health (NIH) grant HL056182 and NYSTEM; to A.J.W. in part from the Burroughs Wellcome Fund, Harvard Stem Cell Institute, Peabody Foundation, and NIH 1 DP2 OD004345-01, and the Joslin Diabetes Center DERC (P30DK036836); to K.H. from Howard Hughes Medical Institute, the NIH Director’s Innovator Award and the Harvard Stem Cell Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Note: Supplementary information is available on the Nature Biotechnology website.
AUTHOR CONTRIBUTIONS
J.M.P. and K.H. conceived the study, interpreted results and wrote the manuscript; J.M.P. performed most of the experiments with help from W.K.; S.L. and T.E. performed and interpreted in vitro differentiation assays; M.E.F and A.M. performed and analyzed HELP methylation experiments; K.Y.T. and A.J.W. isolated SMPs and derived most SMP-iPSCs; T.S. and S.N. performed expression arrays; and S.E., E.A. and M.S. provided essential study material. All authors gave critical input to the manuscript draft.
References
- 1.Aoi T, et al. Generation of pluripotent stem cells from adult mouse liver and stomach cells. Science. 2008;321:699–702. doi: 10.1126/science.1154884. [DOI] [PubMed] [Google Scholar]
- 2.Eminli S, et al. Differentiation stage determines potential of hematopoietic cells for reprogramming into induced pluripotent stem cells. Nat Genet. 2009;41:968–976. doi: 10.1038/ng.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eminli S, Utikal J, Arnold K, Jaenisch R, Hochedlinger K. Reprogramming of neural progenitor cells into induced pluripotent stem cells in the absence of exogenous Sox2 expression. Stem Cells. 2008;26:2467–2474. doi: 10.1634/stemcells.2008-0317. [DOI] [PubMed] [Google Scholar]
- 4.Hanna J, et al. Direct reprogramming of terminally differentiated mature B lymphocytes to pluripotency. Cell. 2008;133:250–264. doi: 10.1016/j.cell.2008.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lowry WE, et al. Generation of human induced pluripotent stem cells from dermal fibroblasts. Proc Natl Acad Sci USA. 2008;105:2883–2888. doi: 10.1073/pnas.0711983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park IH, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 7.Stadtfeld M, Brennand K, Hochedlinger K. Reprogramming of pancreatic beta cells into induced pluripotent stem cells. Curr Biol. 2008;18:890–894. doi: 10.1016/j.cub.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 9.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 10.Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 11.Loh YH, et al. Generation of induced pluripotent stem cells from human blood. Blood. 2009;113:5476–5479. doi: 10.1182/blood-2009-02-204800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aasen T, et al. Efficient and rapid generation of induced pluripotent stem cells from human keratinocytes. Nat Biotechnol. 2008;26:1276–1284. doi: 10.1038/nbt.1503. [DOI] [PubMed] [Google Scholar]
- 13.Maherali N, et al. A high-efficiency system for the generation and study of human induced pluripotent stem cells. Cell Stem Cell. 2008;3:340–345. doi: 10.1016/j.stem.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Utikal J, Maherali N, Kulalert W, Hochedlinger K. Sox2 is dispensable for the reprogramming of melanocytes and melanoma cells into induced pluripotent stem cells. J Cell Sci. 2009;122:3502–3510. doi: 10.1242/jcs.054783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim JB, et al. Pluripotent stem cells induced from adult neural stem cells by reprogramming with two factors. Nature. 2008;454:646–650. doi: 10.1038/nature07061. [DOI] [PubMed] [Google Scholar]
- 16.Shi Y, et al. A combined chemical and genetic approach for the generation of induced pluripotent stem cells. Cell Stem Cell. 2008;2:525–528. doi: 10.1016/j.stem.2008.05.011. [DOI] [PubMed] [Google Scholar]
- 17.Silva J, et al. Promotion of reprogramming to ground state pluripotency by signal inhibition. PLoS Biol. 2008;6:e253. doi: 10.1371/journal.pbio.0060253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miura K, et al. Variation in the safety of induced pluripotent stem cell lines. Nat Biotechnol. 2009;27:743–745. doi: 10.1038/nbt.1554. [DOI] [PubMed] [Google Scholar]
- 19.Ghosh Z, et al. Persistent donor cell gene expression among human induced pluripotent stem cells contributes to differences with human embryonic stem cells. PLoS One. 2010;5:e8975. doi: 10.1371/journal.pone.0008975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soldner F, et al. Parkinson’s disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell. 2009;136:964–977. doi: 10.1016/j.cell.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 22.Stadtfeld M, et al. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature. 2010;465:175–181. doi: 10.1038/nature09017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dimos JT, et al. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science. 2008;321:1218–1221. doi: 10.1126/science.1158799. [DOI] [PubMed] [Google Scholar]
- 24.Ebert AD, et al. Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature. 2009;457:277–280. doi: 10.1038/nature07677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park IH, et al. Disease-specific induced pluripotent stem cells. Cell. 2008;134:877–886. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saha K, Jaenisch R. Technical challenges in using human induced pluripotent stem cells to model disease. Cell Stem Cell. 2009;5:584–595. doi: 10.1016/j.stem.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee G, et al. Modelling pathogenesis and treatment of familial dysautonomia using patient-specific iPSCs. Nature. 2009;461:402–406. doi: 10.1038/nature08320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wernig M, et al. A drug-inducible transgenic system for direct reprogramming of multiple somatic cell types. Nat Biotechnol. 2008;26:916–924. doi: 10.1038/nbt1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stadtfeld M, Maherali N, Breault DT, Hochedlinger K. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell. 2008;2:230–240. doi: 10.1016/j.stem.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cerletti M, et al. Highly efficient, functional engraftment of skeletal muscle stem cells in dystrophic muscles. Cell. 2008;134:37–47. doi: 10.1016/j.cell.2008.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stadtfeld M, Maherali N, Borkent M, Hochedlinger K. A reprogrammable mouse strain from gene-targeted embryonic stem cells. Nat Methods. 2010;7:53–55. doi: 10.1038/nmeth.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chin MH, et al. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell. 2009;5:111–123. doi: 10.1016/j.stem.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 34.Mikkelsen TS, et al. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sridharan R, et al. Role of the murine reprogramming factors in the induction of pluripotency. Cell. 2009;136:364–377. doi: 10.1016/j.cell.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marion RM, et al. Telomeres acquire embryonic stem cell characteristics in induced pluripotent stem cells. Cell Stem Cell. 2009;4:141–154. doi: 10.1016/j.stem.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 37.Boiani M, Eckardt S, Scholer HR, Mclaughlin KJ. Oct4 distribution and level in mouse clones: consequences for pluripotency. Genes Dev. 2002;16:1209–1219. doi: 10.1101/gad.966002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bortvin A, et al. Incomplete reactivation of Oct4-related genes in mouse embryos cloned from somatic nuclei. Development. 2003;130:1673–1680. doi: 10.1242/dev.00366. [DOI] [PubMed] [Google Scholar]
- 39.Ng RK, Gurdon JB. Epigenetic memory of active gene transcription is inherited through somatic cell nuclear transfer. Proc Natl Acad Sci USA. 2005;102:1957–1962. doi: 10.1073/pnas.0409813102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ng RK, Gurdon JB. Epigenetic memory of an active gene state depends on histone H3.3 incorporation into chromatin in the absence of transcription. Nat Cell Biol. 2008;10:102–109. doi: 10.1038/ncb1674. [DOI] [PubMed] [Google Scholar]
- 41.Feng Q, et al. Hemangioblastic derivatives from human induced pluripotent stem cells exhibit limited expansion and early senescence. Stem Cells. 2010;28:704–712. doi: 10.1002/stem.321. [DOI] [PubMed] [Google Scholar]
- 42.Hu BY, et al. Neural differentiation of human induced pluripotent stem cells follows developmental principles but with variable potency. Proc Natl Acad Sci USA. 2010;107:4335–4340. doi: 10.1073/pnas.0910012107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim K, et al. Epigenetic memory in induced pluripotent stem cells. Nature. 2010 July 19; doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supp Data
Supp Data2
Supp Data3
Supp Data4
Supp Data5