Ancestral Roles of Small RNAs: An Ago-Centric Perspective (original) (raw)
Argonaute proteins lie at the heart of all RNAi-related effector complexes. Their PAZ domain recognizes small RNA targets; the PIWI domain is related to RNase H and responsible for target cleavage.
Abstract
RNAi has existed at least since the divergence of prokaryotes and eukaryotes. This collection of pathways responds to a diversity of “abberant” RNAs and generally silences or eliminates genes sharing sequence content with the silencing trigger. In the canonical pathway, double-stranded RNAs are processed into small RNAs, which guide effector complexes to their targets by complementary base pairing. Many alternative routes from silencing trigger to small RNA are continuously being uncovered. Though the triggers of the pathway and the mechanisms of small RNA production are many, all RNAi-related mechanisms share Argonaute proteins as the heart of their effector complexes. These can act as self-contained silencing machines, binding directly to small RNAs, carrying out homology-based target recognition, and in some cases cleaving targets using an endogenous nuclease domain. Here, we discuss the diversity of Argonaute proteins from a structural and functional perspective.
1. INTRODUCTION
The evolution of genomes capable of sustaining their own replication was almost certainly followed by the rapid emergence of parasites capable of exploiting this innovation. Hosts able to control or combat such parasites would have a strong selective advantage. Thus, the deeply rooted conflict between hosts and those seeking to exploit them has driven the development of a panoply of immune strategies that permit discrimination of self from nonself at the genomic level and provide strategies for selectively negating the propagation of pathogens at the expense of their host.
Small regulatory RNAs are pervasive throughout eukaryotes and components of small RNA pathways are also found in select archea and eubacteria (Hock and Meister 2008; Chen 2009; Karginov and Hannon 2010). In multiple kingdoms of life, small RNAs play key roles in responses to both exogenous nucleic acid pathogens, such as viruses, and to resident genomic parasites, namely transposons (Aravin et al. 2007). Though the precise architecture of these small RNA-based immune systems varies, their sharing of core components and of general regulatory strategies strongly suggests genome defense as an ancestral function of small RNAs. Numerous observations suggest later diversification of the biological roles of small RNAs, extending these to gene regulation at nearly every conceivable level and even to control of genomic architecture and content.
The diversity of small RNA families seems to be growing at an alarming rate, catalyzed in large part by the application of increasingly deep next-generation sequencing. At times, it seems as if we will find that nearly every position in every eukaryotic genome will be soon be reflected within at least one small RNA species. However, here we will confine our discussion to the classes of small RNAs that are united under the broad umbrella of RNA interference pathways. Though small RNAs may enter these pathways through a surprising number of different avenues (Kim et al. 2009), all pathways falling under the RNAi rubric share one common and immutable feature. They exert their functions through an effector complex, called RISC or the RNA-induced silencing complex, that has an Argonaute-family protein at its core.
Argonaute proteins were first linked to RNAi through genetic studies in Caenorhabditis elegans. A screen for RNAi-resistant mutants uncovered RDE-1 (RNAi-defective-1) as being essential for gene silencing in response to exogenously introduced double-stranded RNA (Tabara et al. 1999). This was a representative of a little-studied family of proteins, which had previously only been noted because of developmental phenotypes that arose upon their inactivation. In fact, the Argonaute designation came from the appearance of an Arabidopsis mutant whose morphology was reminiscent of a squid (Bohmert et al. 1998). However, even from the beginning, it was clear that Argonaute proteins were members of a highly conserved and quite large family of proteins, with multiple members in nearly every eukaryotic genome (Cerutti et al. 2000). Later studies placed Ago proteins within the emerging biochemical framework of RNA silencing mechanisms, showing that Argonaute was the component of RISC, which directly interacted with the small RNA species that contribute target specificity to the complex (Hammond et al. 2001).
2. ARGONAUTES IN THREE CLADES
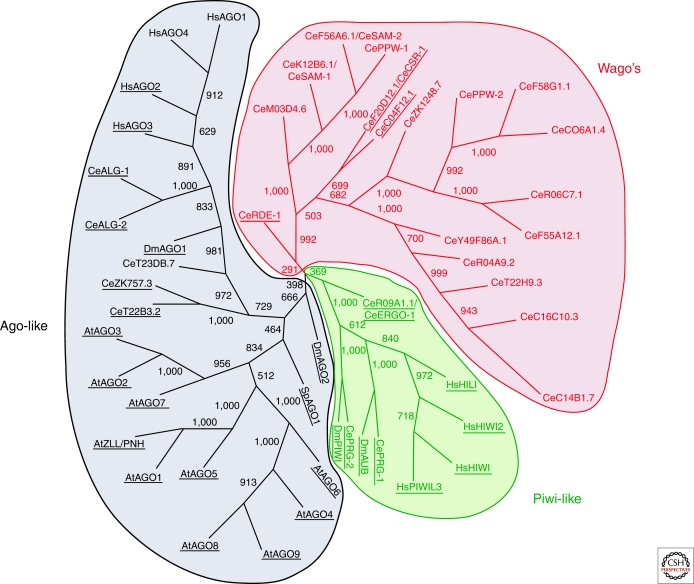
A phylogenic analysis of Argonautes from Homo sapiens (Hs), Arabidopsis thaliana (At), C. elegans (Ce), C. briggsae (Cb), Drosophila melanogaster (Dm), and Schizosaccharomyces pombe (Sp), representatives of the plant, animal, and fungal kingdoms, suggested that these proteins can be divided into three clades (Fig. 1) (Tolia and Joshua-Tor 2007). In many ways, this diversity mirrors the breadth of biological processes, which Argonaute proteins can impact and mechanisms through which they can act. The Ago-like subfamily proteins, defined based on their similarity to AtAgo1, are present in animals, plants, and fission yeast and are involved in transcriptional and posttranscriptional silencing and in genome organization (Joshua-Tor 2006). They show broad expression patterns being present in nearly every tissue and cell type examined in these representative species. They most often interact with what have become known as canonical small RNAs, which based upon their mechanisms of biogenesis average around 22 nucleotides in length and have 5′ phosphate and 3′ hydroxyl groups (Zamore et al. 2000; Bernstein et al. 2001). These include the well-known microRNAs and small interfering RNAs, both derived from double-stranded RNA precursors. The Piwi subfamily is an animal-specific clade that is expressed almost exclusively in gonadal tissues, often only in germline cells, where they form the core of animal transposon silencing pathways (Aravin et al. 2007). Piwi proteins interact with piRNAs (Piwi-interacting) RNAs, which are generated via a pathway distinct from canonical small RNAs. They have 5′ P and 3′ OH ends, suggesting nucleolytic processing from longer precursors, but are often larger than RNAs of the canonical classes (23–29 nt). The Wago clade is constituted entirely of worm Argonautes. It is unusually diverse, contributing 18 of the 27 family members in the C. elegans genome (Tolia and Joshua-Tor 2007). Many that have been characterized so far bind small RNAs generated by direct synthesis from RNA templates (Yigit et al. 2006). Thus, Wago proteins are generally occupied by “secondary siRNAs” which bear 5′ triphosphate and 3′ hydroxyl termini. The biological roles of Wago proteins are still emerging; however, it is clear that they are important for silencing responses to exogenously delivered dsRNAs, for small RNA-based regulation of endogenous genes, and chromosome structure and segregation (Tabara et al. 1999; Yigit et al. 2006; Claycomb et al. 2009; Conine et al. 2010; Gu et al. 2009).
Figure 1.
The three clades of the Argonautes. The Ago-like clade, shown in black, is based on similarity to AtAgo1; the Piwi-like clade, shown in green based on similarity to DmPiwi and the Wago's, which is a worm-specific clade is shown in red. Argonautes that contain a complete catalytic motif are underlined (Adapted from Tolia and Joshua-Tor 2007).
3. AGO STRUCTURE REVEALS FUNCTION
A continuous accumulation of genetic and biochemical evidence pointed to central roles for Ago proteins in virtually all RNAi-related processes. Characterization of the RISC, particularly from Drosophila and mammals demonstrated that it acted as a multiple turnover enzyme that could cleave RNA substrates in a manner determined by the sequence of its bound RNA, an activity termed “Slicer” (Martinez and Tuschl 2004; Schwarz et al. 2004). How Argonaute proteins contributed to this activity, however, remained a mystery.
Sequence conservation of Ago proteins pointed to the presence of two discrete domains, which were termed PAZ and PIWI (Cerutti et al. 2000). Unfortunately, sequence alone provided no insight into the functions of these regions nor could their roles be inferred by comparisons to other better-characterized proteins. A true understanding of their function and consequently the functions of the Ago proteins themselves came only after structures of PAZ domains and then full-length Argonautes emerged.
The first PAZ domain structures were accompanied by biochemical studies demonstrating that it formed an RNA binding module, which specifically interacts with the 3′ end of the small RNA that guides Ago target specificity (Lingel et al. 2003; Song et al. 2003; Yan et al. 2003). Most of the contacts to the RNA are made by highly conserved aromatic residues—two tyrosines and a histidine—that contact the phosphate that bridges the two terminal bases of the small RNA (Lingel et al. 2004; Ma et al. 2004).
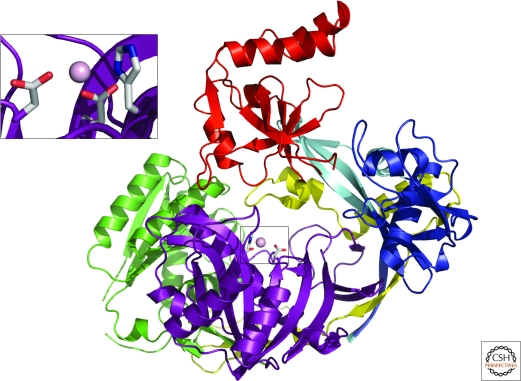
The determination of the first full-length Argonaute protein structure from P. furiosus (PfAgo) illustrated that Argonautes also contain three additional domains: the amino-terminal, middle (Mid), and PIWI domains (Fig. 2) (Song et al. 2004). The big surprise came upon examination of the PIWI domain located at the carboxyl terminus of the protein. The structure revealed that Argonautes clearly belong to the RNase H family of enzymes. Apart from having the RNase H fold at its core, it also has the two highly conserved aspartates that are invariably present on adjacent β-strands in this class of enzymes. Modeling of an siRNA guide strand and target mRNA further showed this to be consistent with what was known from biochemical experiments regarding the characteristics of the slicing activity of the RNA-Induced Silencing Complex, RISC (Song et al. 2004). These include the dependence on the presence of Mg++, the generation of products similar to those produced by RNase H family enzymes, and the position of the scissile phosphate, or the endonucleolytic cut, opposite the phosphate between the 10th and 11th nucleotide from the 5′ end of the small RNA guide. Thus the identity of the previously unknown Slicer activity was determined to reside in the Argonaute protein itself, defining this protein as a self-contained silencing machine (Liu et al. 2004; Song et al. 2004).
Figure 2.
The crystal structure of Argonaute from P. furiosus shown as a ribbon diagram. The amino-terminal domain is in blue, the PAZ domain is red, the middle domain is green, and the PIWI domain is in purple. The interdomain connector is shown in yellow. A close-up of the active site residues coordinating a Mn++ ion (pink ball) is shown in the inset.
The PAZ domain, which resembles an OB (Oligonucleotide Binding) fold domain with a slightly different topology (Lingel et al. 2003; Song et al. 2003; Yan et al. 2003), appears to be rather mobile as evident from the two structures of Aquifex aeolicus Argonaute (Yuan et al. 2005; Rashid et al. 2007). In one case the PAZ domain is in a similar orientation to that of the PAZ domain in the P. furiosus structure and in the other there is a shift of approximately 24°. Even though crystal contacts could play a role, this disparity does illustrate the conformational space that the PAZ domain may assume. The hypothesis of a flexible PAZ is underscored by molecular dynamics simulations (Rashid et al. 2007) and normal mode analyses of these structures (Ming et al. 2007) showing low frequency motions, especially implicating movement of the PAZ domain in opening the putative binding groove for RNA.
4. MULTIPLE MODES OF RNA BINDING
Within RISC, Ago proteins must exist in a large number of RNA bound configurations. Initially, the small RNA guide must be loaded to prime the complex. In the canonical pathway, it still remains unclear whether all guides are loaded initially as double-stranded RNAs. However, biochemical studies clearly demonstrate that following cleavage into discrete segments, double-stranded guides can enter RISC (Matranga et al. 2005). These can be converted by cleavage or by simple dissociation of the nonguide strand into an active complex, with a single-stranded guide ready to search for targets. In most instances, Ago clade proteins preferentially use a portion of the guide to identify targets. This region, known as the “seed” encompasses nucleotides two to eight of the small RNA (Bartel 2009). Although the remainder of the small RNA and its degree of pairing to its substrate can certainly contribute to both the efficiency of silencing and the mode of RISC action, structural studies have provided clues to the dominant role of the seed in target selection. They have also suggested that cycles of target cleavage and substrate release are accompanied not only by conformational changes in the Ago proteins themselves but also by changes in the way Ago engages its small RNA guide.
Much of this insight has been gained through a series of structures of the T.thermophilus Argonaute bound to various combinations of DNA guide strands and RNA and DNA target strands (Wang et al. 2008a; Wang et al. 2008b; Wang et al. 2009). The thermophilus protein actually prefers DNA as both target and guide; however the latter fact became apparent over time, causing initial studies to be done with DNA guide/RNA target combinations.
Bound to a 21-mer DNA guide in the absence of target, Argonaute anchors both ends of the small DNA (Wang et al. 2008b). The 3′ end resides within the PAZ domain as predicted based upon prior structural and biochemical studies of other family members (Lingel et al. 2003; Song et al. 2003; Ma et al. 2004). The 5′ end of the guide lies within a binding site in the mid domain that was identified previously through structures of a PIWI-domain protein with RNA, though the trajectory of the strand differs in the two complexes (Ma et al. 2005; Parker et al. 2005).
The existence of a 5′ end-binding pocket within the Ago protein itself is consistent with the many indications that different family members have different 5′ end preferences. In Arabidopsis, the 12 Ago proteins each show a different spectrum of preferences for the 5′ nucleotide of the bound small RNA (Mi et al. 2008; Montgomery et al. 2008). In fact, individual species can be redirected from one family member to another simply by changing the terminal base. In worms, different Ago proteins likely alter this pocket to engage small RNAs with mono versus triphosphate termini, an accommodation that is also likely made in certain fungal Argonautes. Some Piwi proteins bind almost exclusively to small RNAs beginning with U (Aravin et al. 2007), though it is presently unclear whether this preference is established during biogenesis or small RNA selection by members of this clade. Though the pyrococcus, thermophilus, and aquifex structures form a basis from which to build hypotheses regarding small RNA selection by Ago proteins, what is clearly required as a future step is to define binding pockets within family members whose natural small RNA preferences are clear.
A second correlate between structural, biochemical, and genetic studies extends to the way in which the seed is used for target selection. There appears to be no pressure on natural targets to pair with the first base of the small RNA (Lewis et al. 2003). This is in accord with its limited availability within the structure (Wang et al. 2008b). Moreover, the structure reveals that the bases of the first and second nucleotides are destacked and that the edges of the bases for nucleotides 2–6 are exposed for base pairing. The middle part of the guide (nucleotides 12–17) is disordered in the structure lacking substrate. However, one can see an arginine intercalating between nucleotides 10 and 11 resulting in a catalytically incompetent conformation.
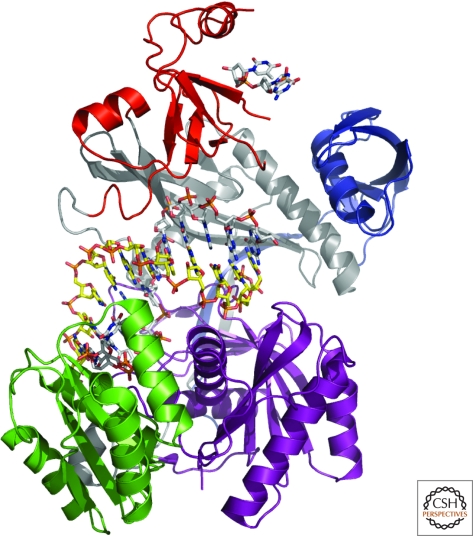
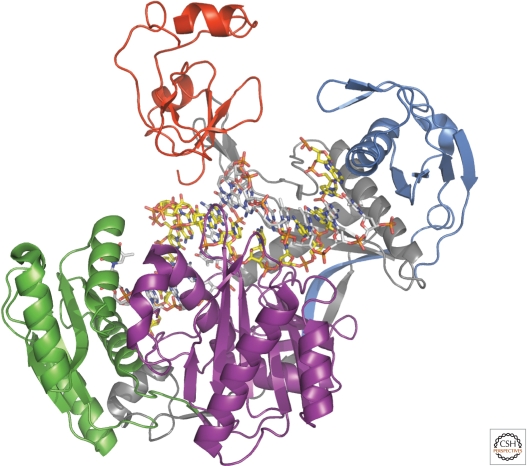
The exploitation of active site mutants of the thermophilus enzyme, substituting one of the active site aspartates with asparagine, enabled the examination of structures with target RNAs (Wang et al. 2009). In a complex containing a quite short, 12-mer target, the guide remained anchored at both ends as in the binary enzyme (Fig. 3). The 5′-phosphate was bound to several conserved residues and a Mg++ ion, underscoring the importance of the presence of this phosphate for selectivity in guide binding and for establishing fidelity in cleavage sites selection (Ma et al. 2005; Rivas et al. 2005). The first guide-strand base remained destacked from the next as was shown previously. The DNA-RNA duplex that follows at positions 2 through 12 are reported to resemble an A-form conformation and the scissile bond is placed at the active site of the enzyme in a helical and catalytically competent conformation in contrast to their orthogonal arrangement in the binary complex. The following nucleotides of the guide strand are presumably unstructured, because they are not observed. The final two nucleotides again appear anchored within the PAZ domain. The placement of the guide is a bit different in the ternary structure, but most notable is the substantial movement of the PAZ domain with respect to the binary complex. One must keep in mind that the PAZ domain is involved in significant crystal contacts that might affect these conformational changes, however, it does illustrate the range of possible motions it could sample. A different active site mutant with a longer 15-mer target RNA resulted in a smaller movement of the PAZ domain. However, in this case, the guide strand appears to have exited the PAZ domain to pair with the RNA and its 3′ end is now disordered. It is worth noting that guide-strand exiting the PAZ during the catalytic cycle had been previously predicted by careful kinetic analysis of Drosophila RISC (Schwarz et al. 2004). The 3′ end pairing between the passenger and guide is accompanied by a widening of the channel between the PIWI and N-domains. An even longer RNA target, 19-nucleotides in length (Fig. 4), helps localize the 16th nucleotide of the target RNA and in this case the N-domain appears to block simple propagation of the DNA-RNA duplex, though additional conformational changes in the protein might occur with an even longer target. Throughout the seed pairing, Argonaute contacts only the guide strand, though contacts to the target are observed around nucleotides 10–13.
Figure 3.
The crystal structure of Argonaute from Thermus thermophilus in complex with a 21-nucleotide DNA guide (in stick with gray carbons) and a 12-nucleotide RNA target (in stick with yellow carbons). The amino-terminal domain is in blue, the PAZ domain in red, the mid domain in green and the PIWI domain in purple. The two linkers are shown in gray. The first 12 nucleotides and the two 3′-nucleotides of the DNA guide are observed in the structure. The 3′ nucleotides are bound to the PAZ domain.
Figure 4.
The crystal structure of Argonaute from T. thermophilus in complex with a 21-nucleotide DNA guide (in stick with gray carbons) and a 19-nucleotide RNA target (in stick with yellow carbons). Domains are colored as in Figure 3. The first 16 nucleotides of the DNA guide and 15 nucleotides of the RNA target strand are observed in the structure. The 3′-end nucleotides of the guide are no longer bound to the PAZ domain in this structure.
The reloading of an RNA target into the complex during the catalytic cycle is a problem worth examining. In some of the aforementioned structures, the DNA guide appears to be blocking easy exit of the RNA. It appears to be nestled well between the guide and the protein. Would two turns of a duplex need to be unwound and rewound for each cycle to allow entrance of a new target RNA, or does the protein “help” distort the duplex so that there is no hindrance for exit and entrance of successive RNA target strands to be cleaved? This question is clearly relevant for achieving multiple catalytic cycles by each RISC but also likely for the conversion of RISC loaded with dsRNA guides to those complexes primed with ssRNA and ready for action.
5. SMALL RNA GUIDED CLEAVAGE
Though Argonaute proteins can impact their targets in many ways, one of the most critical and conserved is endonucleolytic cleavage (Song et al. 2004). The substrate specificity of Argonaute is determined by the sequence of the bound guide RNA, as the target is recognized via hybridization to the guide strand. As described earlier, Argonaute is an RNase H-like enzyme creating a 5′ product with a 3-OH and a 3′ product carrying a 5′ phosphate, with the DNA strand generally being replaced by a guide RNA. The RNAse H protein family consists of well-characterized enzymes such as retroviral integrases and transposases (Nowotny 2009). Escherichia coli RNase H1 catalyzes a single reaction resulting in substrate cleavage. Integrases and transposases catalyze two consecutive reactions, donor-end processing and nucleotidyl transfer, resulting in strand transfer. The nucleophile in these reactions is either a water molecule or a nucleotide 3′-OH. They involve a two-metal ion catalysis mechanism, with one metal activating the nucleophile and the second stabilizing the intermediate (Nowotny et al. 2005). Two metals have been observed in crystal structures of substrate-bound complexes of Tn5 transposase and both human and B. halodurans RNase H1 (Lovell et al. 2002; Nowotny et al. 2005; Nowotny et al. 2007). So far, the presence of at least one metal ion is clear in some of the Argonaute structures (Rivas et al. 2005; Yuan et al. 2005; Wang et al. 2008a; Wang et al. 2009). A second metal ion was reported in one of the ternary complexes, largely based on possible coordination with neighboring oxygen atoms (Wang et al. 2009).
A combination of structural and mutational analyses identified the two invariant aspartates characteristic of RNase H enzymes as well as a third residue that in most cases is a histidine rather than another carboxylate, critical for catalysis (Fig. 2) (Joshua-Tor 2006). The presence of all three residues is not sufficient to predict whether a particular Argonaute has catalytic activity. For example, the four human Argonaute clade proteins, HsAgo1, HsAgo2, HsAgo3, and HsAgo4 have been tested for activity, with HsAgo2 alone having slicer activity, even though HsAgo3 has an intact catalytic triad (Liu et al. 2004; Meister et al. 2004). The remaining Piwi clade proteins also conserve catalytic potential, and are presumed active based upon their conserved mechanism of transposon recognition and silencing (Faehnle and Joshua-Tor 2007).
The presence of the Argonaute protein and the guide RNA in a minimal recombinant RISC, is sufficient for target cleavage, but does not show ATP-stimulated product release and turns over rather slowly. However, other factors likely also contribute to the function of a complete RISC. In one report, a complex containing Argonaute, Dicer, and human immunodeficiency virus-1 trans-activating response (TAR) RNA-binding protein (TRBP) is capable of guide-strand loading and multiple rounds of target cleavage, stimulated by nucleotides (Gregory et al. 2005). Another report indicated that Dicer dissociates once Ago2 is loaded with the guide RNA (Maniataki and Mourelatos 2005). More recent studies support the dissociation of both Dicer and TRBP from Ago2 after loading, and these appear to not participate in target cleavage (MacRae et al. 2008).
Many questions still remain to be answered to fully understand target cleavage by the Argonautes. How is multiple turnover achieved? Does PAZ domain movement assist in substrate engagement and placement, and in product release? What is essential to constitute an active Slicer in addition to the active site requirements?
6. COMPARATIVE STUDIES OF ARGONAUTE BIOLOGY HINT AT ANCESTRAL FUNCTION
In essence, RISC is a programmable homology search engine, which can confer a variety of properties upon a target. Some of these depend upon the catalytic activity of Ago proteins, whereas others do not. This is reflected in a diversification of the Ago family into multiple clades and into groups of family members, which either retain or have lost catalytic potential. Given the staggering breadth of Ago functions, it seems difficult to infer the ancestral roles of the RNAi pathway. In both plants and animals, Argonautes are programmed by microRNAs to regulate gene expression, mainly at the posttranscriptional level (Bartel 2009). However, differences in microRNA biogenesis and effector mechanisms and the lack of any shared microRNAs between plant and animal kingdoms strongly suggests that gene regulatory functions of small RNAs may have evolved several times independently. Plants, animals, and fungi have a variety of small RNA-based pathways which impact chromatin structure and organization and which in some cases are essential for chromosome function or even control chromosome content (Mochizuki et al. 2002; Volpe et al. 2002; Mathieu and Bender 2004; Alexander et al. 2008). Although these pathways play different roles in different species, one shared feature is that all are driven by the recognition of repetitive sequences. In fact, recognition and control of repeated elements, transposons, and viruses—generally foreign nucleic acids—seems the one theme that unifies small RNA pathways wherever they are found.
The first hints of immune roles for RNAi came from plants, where canonical RNAi mechanisms—involving Dicer, siRNAs and Ago-clade proteins—proved a key mediator of antiviral defense (Hamilton and Baulcombe 1999; Mlotshwa et al. 2008). Here, simply the double-stranded nature of replication intermediates seemed sufficient to indicate that an RNA was “foreign” and to tag it for nucleolytic destruction. Similar observations in Drosophila and C. elegans extended the antiviral paradigm to animals (Lu et al. 2005; Wang et al. 2006). In mammals, where an adaptive immune system has evolved to fight viral infection, the roles of RNAi in antiviral immunity are less clear. However, mammals clearly use small RNA pathways to recognize and control endogenous nucleic acid parasites.
An animal-specific clade of Ago proteins, the Piwis, forms the basis of an elegant, RNA-based innate immune system that selectively silences transposons in the germ cells of animals (Aravin et al. 2007). This incorporates two key elements, a programmable evolutionary record of transposon exposure and control and an adaptive mechanism to optimize the response to ongoing transposon challenge.
Small RNAs that enter the piRNA pathway are produced by at least two distinct biogenesis mechanisms. So-called “primary piRNAs” are generated from specific loci within animal genomes, called piRNA clusters. These are often highly enriched for fragmented, diverged transposon remnants, and in some cases represent the most transposon-rich areas of the host genome. These loci appear to be transcribed as large single-stranded precursors, which are parsed into individual small RNAs to create the equivalent of an immune repertoire. In order to gain control over a new element, it must be incorporated into the piRNA pathway by integration into a piRNA cluster, and some clusters are known to act as hotspots for insertion of the elements that they control.
Once made, primary piRNA/Piwi protein complexes can recognize targets much as in the canonical RNAi pathway, catalyzing small RNA-dependent cleavage, in this case of transposon mRNAs. Here, the canonical and piRNA pathways diverge in the sense that target cleavage by Piwi can create the 5′ end of a new small RNA (Brennecke et al. 2007; Gunawardane et al. 2007). This target-derived RNA can catalyze the production of further small RNAs through the same mechanism. If target RNAs can recognize antisense transposon content synthesized as part of the precursors generated from piRNA clusters, then an amplification loop can be formed where interactions between the immune repertoire and their targets can increase populations of small RNAs to meet a particular transposon challenge. In this way, the adaptive amplification loop, called the ping-pong cycle, is analogous to the expansion of adaptive immune cells triggered by antigen-receptor interactions (Aravin et al. 2007).
piRNA pathways in animals clearly use post-transcriptional mechanisms as part of their regulatory arsenal. However, there is clear evidence that piRNA pathways direct DNA methylation of repeats in mammals (Carmell et al. 2007) and that posttranscriptional controls may also figure prominently in silencing of mobile elements in flies (Pal-Bhadra et al. 2004; Brower-Toland et al. 2007). In plants, a 24-nucleotide class of small RNAs directs the methylation of repeat elements via binding to an Ago-clade protein (Zilberman et al. 2003). These depend upon a Dicer for their production, implicating dsRNA precursors that are absent from the piRNA pathway. Unlike the piRNA pathway in animals where we are at least starting to gain glimpses into the mechanism of self-non-self recognition, there are no concrete hypotheses regarding how repeat elements are singled out for silencing in plants. Two distinct RNA polymerases, polIV and polV, appear essential for RNA-directed DNA methylation (Herr et al. 2005; Onodera et al. 2005) suggesting at least one construction for a reinforcing loop similar to the one in animals. If transcription by these polymerases triggers small RNA production, which in turn directs methylation, one could close a cycle if that same methylation also helped recruit the polymerase variants. The question, however, would be how the cycle would be initiated in the first place.
In ciliates, small RNAs are critical for repeat management through strategies that show some conceptual similarities to the piRNA pathway but also some remarkable differences (Mochizuki and Gorovsky 2004). Tetrahymena thermophila is unusual in many respects. Among these are that somatic and germ nuclei, the macronucleus and micronucleus respectively, are propagated within a single vegetatively growing cell. The nuclei differ not only in their transcriptional activity but also in their genomic content. During the sexual cycle when a new somatic nucleus is produced following fusion of the meiotic products of the micronucleus, a significant fraction of the genome is discarded through process that is guided by small RNAs. The small RNA population is adjusted based on the content of the old macronucleus through mechanisms that are not precisely clear. However the result is that through a process of genome comparison, small RNA populations targeting repeats are enriched, and these direct the physical elimination of sequences from the developing somatic genome. Small RNAs that direct elimination reside in a Piwi family protein but are Dicer-dependent (Malone et al. 2005; Mochizuki and Gorovsky 2005), a strange convergence of pathways that seem distinct in other species.
Examples of small RNA and repeat management can also be drawn from the fungal kingdom. In fission yeast, small RNAs are essential for the functional integrity of centromeres (Folco et al. 2008; White and Allshire 2008). Their effect in this case is on the organization of the centromeric heterochromatin domain, which is in turn essential for proper segregation. In S. pombe, a series of centromere-associated repeats becomes periodically transcribed during the S-phase of the cell cycle (Chen et al. 2008; Kloc et al. 2008). These are targeted by the RNAi machinery, in this case via a pathway that requires Dicer, an RdRP, and an Ago-clade protein, resulting in the deposition of histone methyl marks characteristic of heterochromatin (White and Allshire 2008). Here, the conserved theme of targeting repeats seems to have been co-opted to serve a positive role in the host cell. A second example can be seen in the phenomenon of unpaired silencing (MSUD, meiotic silencing of unpaired DNA) in Neurospora (Shiu et al. 2001). During MSUD, sequences that differ between two homologs are selectively and heritably silenced during the sexual reproductive cycle. Such a comparison would presumably pick up new transposon insertions in addition to other types of copy number variation between synapsed chromosomes. Though no small RNAs driving this process have yet been identified, it does require the same types of RNAi-related components required for the creation of centromeric heterochromatin in pombe (Alexander et al. 2008).
Though RNAi-related pathways are not the universal mechanism by which hosts battle genomic parasites, the conceptual framework that emerged particularly by studies of the piRNA pathway also underlies phage resistance in both archea and eubacteria. Here the CRISPR-CAS system uses small RNAs (crRNAs) to identify and cleave phage mRNAs or genomic DNAs (Jore et al. 2010; Karginov and Hannon 2010). There is also analogy in the mechanism by which invading elements are incorporated into a genetic memory of resistance. Fragments of invading phage become integrated into a locus (CRISPR), which serves as the source of phage-targeting crRNAs. Upon challenge with an unrecognized phage, the clusters can clearly evolve by the acquisition of new sequence information near the 5′ end of the CRISPR unit. Fragments can also be lost from this heritable resistance determinant in the absence of pressure to maintain them.
Thus, through all kingdoms of life, small RNAs underlie the ability of genomes to selectively propagate in the presence of endogenous and exogenous invaders that seek to hijack replicative potential. However, the flexible nature of small RNAs themselves and the protein complexes, which use these to guide target selection, has resulted in an explosion of regulatory function that now pervades almost every aspect of biology.
REFERENCES
- Alexander WG, Raju NB, Xiao H, Hammond TM, Perdue TD, Metzenberg RL, Pukkila PJ, Shiu PK 2008. DCL-1 colocalizes with other components of the MSUD machinery and is required for silencing. Fungal Genet Biol 45: 719–727 [DOI] [PubMed] [Google Scholar]
- Aravin AA, Hannon GJ, Brennecke J 2007. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science 318: 761–764 [DOI] [PubMed] [Google Scholar]
- Bartel DP 2009. MicroRNAs: Target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366 [DOI] [PubMed] [Google Scholar]
- Bohmert K, Camus I, Bellini C, Bouchez D, Caboche M, Benning C 1998. AGO1 defines a novel locus of Arabidopsis controlling leaf development. Embo J 17: 170–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089–1103 [DOI] [PubMed] [Google Scholar]
- Brower-Toland B, Findley SD, Jiang L, Liu L, Yin H, Dus M, Zhou P, Elgin SC, Lin H 2007. Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes Dev 21: 2300–2311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmell MA, Girard A, van de Kant HJ, Bourc'his D, Bestor TH, de Rooij DG, Hannon GJ 2007. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell 12: 503–514 [DOI] [PubMed] [Google Scholar]
- Cerutti L, Mian N, Bateman A 2000. Domains in gene silencing and cell differentiation proteins: The novel PAZ domain and redefinition of the Piwi domain. Trends Biochem Sci 25: 481–482 [DOI] [PubMed] [Google Scholar]
- Chen X 2009. Small RNAs and their roles in plant development. Annu Rev Cell Dev Biol 25: 21–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ES, Zhang K, Nicolas E, Cam HP, Zofall M, Grewal SI 2008. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature 451: 734–737 [DOI] [PubMed] [Google Scholar]
- Claycomb JM, Batista PJ, Pang KM, Gu W, Vasale JJ, van Wolfswinkel JC, Chaves DA, Shirayama M, Mitani S, Ketting RF, et al. 2009. The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell 139: 123–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conine CC, Batista PJ, Gu W, Claycomb JM, Chaves DA, Shirayama M, Mello CC 2010. Argonautes ALG-3 and ALG-4 are required for spermatogenesis-specific 26G-RNAs and thermotolerant sperm in Caenorhabditis elegans. Proc Natl Acad Sci 107: 3588–3593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faehnle CR, Joshua-Tor L 2007. Argonautes confront new small RNAs. Curr Opin Chem Biol 11: 569–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folco HD, Pidoux AL, Urano T, Allshire RC 2008. Heterochromatin and RNAi are required to establish CENP-A chromatin at centromeres. Science 319: 94–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R 2005. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell 123: 631–640 [DOI] [PubMed] [Google Scholar]
- Gu W, Shirayama M, Conte D Jr, Vasale J, Batista PJ, Claycomb JM, Moresco JJ, Youngman EM, Keys J, Stoltz MJ, et al. 2009. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol Cell 36: 231–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC 2007. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science 315: 1587–1590 [DOI] [PubMed] [Google Scholar]
- Hamilton AJ, Baulcombe DC 1999. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286: 950–952 [DOI] [PubMed] [Google Scholar]
- Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ 2001. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293: 1146–1150 [DOI] [PubMed] [Google Scholar]
- Herr AJ, Jensen MB, Dalmay T, Baulcombe DC 2005. RNA polymerase IV directs silencing of endogenous DNA. Science 308: 118–120 [DOI] [PubMed] [Google Scholar]
- Hock J, Meister G 2008. The Argonaute protein family. Genome Biol 9: 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jore MM, Brouns SJJ, van der Oost J 2010. RNA in defense: CRISPRs protect prokaryotes against alien nucleic acids. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a003772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshua-Tor L 2006. The Argonautes. Cold Spring Harb Symp Quant Biol 71: 67–72 [DOI] [PubMed] [Google Scholar]
- Karginov FV, Hannon GJ 2010. The CRISPR system: Small RNA-guided defense in bacteria and archaea. Mol Cell 37: 7–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC 2009. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 10: 126–139 [DOI] [PubMed] [Google Scholar]
- Kloc A, Zaratiegui M, Nora E, Martienssen R 2008. RNA interference guides histone modification during the S phase of chromosomal replication. Curr Biol 18: 490–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB 2003. Prediction of mammalian microRNA targets. Cell 115: 787–798 [DOI] [PubMed] [Google Scholar]
- Lingel A, Simon B, Izaurralde E, Sattler M 2003. Structure and nucleic-acid binding of the Drosophila Argonaute 2 PAZ domain. Nature 426: 465–469 [DOI] [PubMed] [Google Scholar]
- Lingel A, Simon B, Izaurralde E, Sattler M 2004. Nucleic acid 3′-end recognition by the Argonaute2 PAZ domain. Nat Struct Mol Biol 11: 576–577 [DOI] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ 2004. Argonaute2 is the catalytic engine of mammalian RNAi. Science 305: 1437–1441 [DOI] [PubMed] [Google Scholar]
- Lovell S, Goryshin IY, Reznikoff WR, Rayment I 2002. Two-metal active site binding of a Tn5 transposase synaptic complex. Nat Struct Biol 9: 278–281 [DOI] [PubMed] [Google Scholar]
- Lu R, Maduro M, Li F, Li HW, Broitman-Maduro G, Li WX, Ding SW 2005. Animal virus replication and RNAi-mediated antiviral silencing in Caenorhabditis elegans. Nature 436: 1040–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma JB, Ye K, Patel DJ 2004. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature 429: 318–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma JB, Yuan YR, Meister G, Pei Y, Tuschl T, Patel DJ 2005. Structural basis for 5′-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature 434: 666–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacRae IJ, Ma E, Zhou M, Robinson CV, Doudna JA 2008. In vitro reconstitution of the human RISC-loading complex. Proc Natl Acad Sci U S A 105: 512–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Anderson AM, Motl JA, Rexer CH, Chalker DL 2005. Germ line transcripts are processed by a Dicer-like protein that is essential for developmentally programmed genome rearrangements of Tetrahymena thermophila. Mol Cell Biol 25: 9151–9164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniataki E, Mourelatos Z 2005. A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Genes Dev 19: 2979–2990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J, Tuschl T 2004. RISC is a 5′ phosphomonoester-producing RNA endonuclease. Genes Dev 18: 975–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu O, Bender J 2004. RNA-directed DNA methylation. J Cell Sci 117: 4881–4888 [DOI] [PubMed] [Google Scholar]
- Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD 2005. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell 123: 607–620 [DOI] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T 2004. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell 15: 185–197 [DOI] [PubMed] [Google Scholar]
- Mi S, Cai T, Hu Y, Chen Y, Hodges E, Ni F, Wu L, Li S, Zhou H, Long C, et al. 2008. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133: 116–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming D, Wall ME, Sanbonmatsu KY 2007. Domain motions of Argonaute, the catalytic engine of RNA interference. BMC Bioinformatics 8: 470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlotshwa S, Pruss GJ, Vance V 2008. Small RNAs in viral infection and host defense. Trends Plant Sci 13: 375–382 [DOI] [PubMed] [Google Scholar]
- Mochizuki K, Gorovsky MA 2004. Small RNAs in genome rearrangement in Tetrahymena. Curr Opin Genet Dev 14: 181–187 [DOI] [PubMed] [Google Scholar]
- Mochizuki K, Gorovsky MA 2005. A Dicer-like protein in Tetrahymena has distinct functions in genome rearrangement, chromosome segregation, and meiotic prophase. Genes Dev 19: 77–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki K, Fine NA, Fujisawa T, Gorovsky MA 2002. Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in Tetrahymena. Cell 110: 689–699 [DOI] [PubMed] [Google Scholar]
- Montgomery TA, Howell MD, Cuperus JT, Li D, Hansen JE, Alexander AL, Chapman EJ, Fahlgren N, Allen E, Carrington JC 2008. Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell 133: 128–141 [DOI] [PubMed] [Google Scholar]
- Nowotny M 2009. Retroviral integrase superfamily: The structural perspective. EMBO Rep 10: 144–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowotny M, Gaidamakov SA, Crouch RJ, Yang W 2005. Crystal structures of RNase H bound to an RNA/DNA hybrid: Substrate specificity and metal-dependent catalysis. Cell 121: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Nowotny M, Gaidamakov SA, Ghirlando R, Cerritelli SM, Crouch RJ, Yang W 2007. Structure of human RNase H1 complexed with an RNA/DNA hybrid: Insight into HIV reverse transcription. Mol Cell 28: 264–276 [DOI] [PubMed] [Google Scholar]
- Onodera Y, Haag JR, Ream T, Nunes PC, Pontes O, Pikaard CS 2005. Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell 120: 613–622 [DOI] [PubMed] [Google Scholar]
- Pal-Bhadra M, Leibovitch BA, Gandhi SG, Rao M, Bhadra U, Birchler JA, Elgin SC 2004. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science 303: 669–672 [DOI] [PubMed] [Google Scholar]
- Parker JS, Roe SM, Barford D 2005. Structural insights into mRNA recognition from a PIWI domain-siRNA guide complex. Nature 434: 663–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid UJ, Paterok D, Koglin A, Gohlke H, Piehler J, Chen JC 2007. Structure of Aquifex aeolicus argonaute highlights conformational flexibility of the PAZ domain as a potential regulator of RNA-induced silencing complex function. J Biol Chem 282: 13824–13832 [DOI] [PubMed] [Google Scholar]
- Rivas FV, Tolia NH, Song JJ, Aragon JP, Liu J, Hannon GJ, Joshua-Tor L 2005. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat Struct Mol Biol 12: 340–349 [DOI] [PubMed] [Google Scholar]
- Schwarz DS, Tomari Y, Zamore PD 2004. The RNA-induced silencing complex is a Mg2+-dependent endonuclease. Curr Biol 14: 787–791 [DOI] [PubMed] [Google Scholar]
- Shiu PK, Raju NB, Zickler D, Metzenberg RL 2001. Meiotic silencing by unpaired DNA. Cell 107: 905–916 [DOI] [PubMed] [Google Scholar]
- Song JJ, Liu J, Tolia NH, Schneiderman J, Smith SK, Martienssen RA, Hannon GJ, Joshua-Tor L 2003. The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nat Struct Biol 10: 1026–1032 [DOI] [PubMed] [Google Scholar]
- Song JJ, Smith SK, Hannon GJ, Joshua-Tor L 2004. Crystal structure of Argonaute and its implications for RISC slicer activity. Science 305: 1434–1437 [DOI] [PubMed] [Google Scholar]
- Tabara H, Sarkissian M, Kelly WG, Fleenor J, Grishok A, Timmons L, Fire A, Mello CC 1999. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell 99: 123–132 [DOI] [PubMed] [Google Scholar]
- Tolia NH, Joshua-Tor L 2007. Slicer and the argonautes. Nat Chem Biol 3: 36–43 [DOI] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA 2002. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297: 1833–1837 [DOI] [PubMed] [Google Scholar]
- Wang XH, Aliyari R, Li WX, Li HW, Kim K, Carthew R, Atkinson P, Ding SW 2006. RNA interference directs innate immunity against viruses in adult Drosophila. Science 312: 452–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Juranek S, Li H, Sheng G, Tuschl T, Patel DJ 2008a. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature 456: 921–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Juranek S, Li H, Sheng G, Wardle GS, Tuschl T, Patel DJ 2009. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature 461: 754–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Sheng G, Juranek S, Tuschl T, Patel DJ 2008b. Structure of the guide-strand-containing argonaute silencing complex. Nature 456: 209–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White SA, Allshire RC 2008. RNAi-mediated chromatin silencing in fission yeast. Curr Top Microbiol Immunol 320: 157–183 [DOI] [PubMed] [Google Scholar]
- Yan KS, Yan S, Farooq A, Han A, Zeng L, Zhou MM 2003. Structure and conserved RNA binding of the PAZ domain. Nature 426: 468–474 [DOI] [PubMed] [Google Scholar]
- Yigit E, Batista PJ, Bei Y, Pang KM, Chen CC, Tolia NH, Joshua-Tor L, Mitani S, Simard MJ, Mello CC 2006. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell 127: 747–757 [DOI] [PubMed] [Google Scholar]
- Yuan YR, Pei Y, Ma JB, Kuryavyi V, Zhadina M, Meister G, Chen HY, Dauter Z, Tuschl T, Patel DJ 2005. Crystal structure of A. aeolicus argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol Cell 19: 405–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore PD, Tuschl T, Sharp PA, Bartel DP 2000. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101: 25–33 [DOI] [PubMed] [Google Scholar]
- Zilberman D, Cao X, Jacobsen SE 2003. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299: 716–719 [DOI] [PubMed] [Google Scholar]