T Cell Receptor (TCR)-induced Tyrosine Phosphorylation Dynamics Identifies THEMIS as a New TCR Signalosome Component (original) (raw)
Abstract
Stimulation of the T cell antigen receptor (TCR) induces formation of a phosphorylation-dependent signaling network via multiprotein complexes, whose compositions and dynamics are incompletely understood. Using stable isotope labeling by amino acids in cell culture (SILAC)-based quantitative proteomics, we investigated the kinetics of signal propagation after TCR-induced protein tyrosine phosphorylation. We confidently assigned 77 proteins (of 758 identified) as a direct or indirect consequence of tyrosine phosphorylation that proceeds in successive “signaling waves” revealing the temporal pace at which tyrosine kinases activate cellular functions. The first wave includes thymocyte-expressed molecule involved in selection (THEMIS), a protein recently implicated in thymocyte development but whose signaling role is unclear. We found that tyrosine phosphorylation of THEMIS depends on the presence of the scaffold proteins Linker for activation of T cells (LAT) and SH2 domain-containing lymphocyte protein of 76 kDa (SLP-76). THEMIS associates with LAT, presumably via the adapter growth factor receptor-bound protein 2 (Grb2) and with phospholipase Cγ1 (PLC-γ1). RNAi-mediated THEMIS knock-down inhibited TCR-induced IL-2 gene expression due to reduced ERK and nuclear factor of activated T cells (NFAT)/activator protein 1 (AP-1) signaling, whereas JNK, p38, or nuclear factor κB (NF-κB) activation were unaffected. Our study reveals the dynamics of TCR-dependent signaling networks and suggests a specific role for THEMIS in early TCR signalosome function.
Keywords: Immunology, Kinetics, Phosphotyrosine Signaling, Proteomics, Signal Transduction, ERK Activation, SILAC, TCR Signaling, THEMIS
Introduction
Signals generated by ligation of the T cell antigen receptor (TCR)6 to peptide-major histocompatibility complex (MHC) agonists induce dramatic changes in cell morphology and gene expression pattern and play a central role in initiating the adaptive immune response. TCR ligation triggers phosphorylation of tyrosine residues in the immunoreceptor tyrosine-based activation motifs of the CD3/ζ-chain complex by the Src-family protein-tyrosine kinase LCK followed by recruitment and activation of ζ-chain-associated protein kinase of 70 kDa (ZAP-70). ZAP-70 in turn activates a “canonical pathway” by phosphorylating the transmembrane scaffold linker for activation of T cells (LAT), which associates with the SLP-76/Grb2-related adapter protein (Gads) complex to form a platform for the assembly of crucial signaling proteins, including phospholipase Cγ1 (PLC-γ1), the adhesion and degranulation promoting adapter protein (ADAP)/Src kinase-associated phosphoprotein of 55 kDa (SKAP55) adapter module, the Tec-family kinase ITK, and the Rho-family guanine nucleotide exchange factor (GEF) Vav (1). Several signaling pathways are activated from and tuned by reactions taking place within this multiprotein complex and contribute to specify T cell fate during thymus development and exposure to antigen (2). TCR-induced gene expression is dependent on elevation of intracellular calcium and activation of small GTPases and PKCs and is essentially controlled by a number of key transcription factors, including nuclear factor κB (NF-κB), nuclear factor of activated T cells (NFAT), and activator protein 1 (AP-1) (1). A global view of temporal changes in TCR signaling components and complexes is crucial to improve our understanding of T cell physiology and pathophysiology. A powerful means to achieve these goals is to monitor large-scale protein phosphorylation dynamics using quantitative mass spectrometry (MS)-based proteomics (3). Recent technological improvements in MS instrumentation (4, 5), development of new methods for relative (6–9), and absolute quantitation (10, 11) and refined computing resources (7, 12, 13) have enabled the monitoring of phosphorylation changes of large numbers of signaling components in response to external stimuli with great accuracy (3, 6, 14). Previous MS-based investigations of TCR-proximal signaling (e.g. tyrosine phosphorylation) compared single activation time points and/or only two different activation conditions (15–17). However, the complexity and dynamics of signaling networks and their individual or integrated contributions to cellular functions can be more completely revealed by monitoring the kinetic unfolding of molecular activation events at higher temporal resolution. Recent investigations have used label-free or stable isotope labeling by amino acids in cell culture (SILAC) based approaches coupled to phosphopeptide enrichment techniques for assessing signaling events (18, 19). However, the limited number of phosphopeptides recovered for most proteins often lowers statistical confidence of protein identification and quantitation, a hurdle that can be circumvented by analyzing the peptide mixture before and after phosphopeptide enrichment (20) or increasing technical and biological replicas. These limitations can be overcome by quantitative enrichment for phosphorylated proteins rather than phosphopeptides and rigorous statistics provided by technical and biological replicas, thus, relying on a larger set of peptides for MS identification and quantitation.
Here we combined anti-phosphotyrosine immunoisolation with SILAC-based MS quantitation to examine the dynamics of TCR-induced activation events at a higher temporal resolution than previous studies. This analysis revealed not only the profiles of activation kinetics of individual proteins after TCR stimulation but also hinted at likely partnerships of phosphorylated proteins with functional complexes. We identified kinetic profiles as “waves” of activation controlling diverse cellular functions such as receptor-proximal signal transduction and propagation, cytoskeleton rearrangement, membrane trafficking, ubiquitination, and surprisingly, activation of nucleic acid-associated proteins. Importantly, we identified a novel signaling component, C6orf190, and characterized its signaling function. The murine ortholog of C6orf190, “thymocyte-expressed molecule involved in selection” (Themis), has recently been reported to play a crucial role in thymocyte development (21–25). However, its precise function remains unclear, and it is at present disputed whether Themis has a role in TCR signaling. With this study we aimed to elucidate the potential signaling function of human THEMIS. We provide strong support for the notion that THEMIS is a component of the early TCR signaling machinery by showing that its tyrosine phosphorylation depends on LCK, LAT, and SLP-76 and that it binds to LAT, Grb2, and PLC-γ1. Finally, we present evidence that THEMIS is required for effective ERK and NFAT/AP-1 activation but dispensable for c-Jun amino-terminal kinase (JNK), p38, and NF-κB signaling.
EXPERIMENTAL PROCEDURES
Mass spectrometry and SILAC-related experimental procedures can be found in the supplemental text.
Plasmids and Antibodies
Full-length cDNA encoding human THEMIS was obtained from Open Biosystems and used as the PCR template to generate THEMIS-OST, carrying a C-terminal One-STrEP-Tag (IBA BioTAGnology). THEMIS-OST was cloned into the lentiviral expression vector pHR-SIN-BX-IRES-Emerald (kindly provided by Dr. V. Cerundolo, WIMM, Oxford) and verified by bidirectional sequencing. Plasmids pGL4-NF-κB-luc (firefly luciferase) and RLuc (renilla luciferase) were kind gifts of Dr. Tomas Mustelin (Burnham Institute for Medical Research, La Jolla, CA). The lentiviral helper plasmids psPAX2 (Addgene 10703) and pMD2.G (Addgene 12259) were provided by Dr. Didier Trono (Ecole Polytechnique Fédérale de Lausanne, Lausanne, Switzerland) via Addgene. Lentiviral pLKO.1 plasmid targeting the LCK 3′-UTR (TRCN0000001598) was from Open Biosystems. Mouse monoclonal antibodies (Abs) used included: anti-phosphotyrosine (Tyr(P)) (4G10, Millipore, PY99, Santa Cruz Biotechnology, and PY20, BD Transduction Laboratories); anti-LCK (lymphocyte-specific kinase; 3A5, Santa Cruz); anti-ZAP-70 (2F3.2), anti-LAT (2E9), anti-phospho-LAT (Tyr226) (Millipore); anti-CD3ζ (Tyr142, BD Biosciences); anti-phospho-ERK1/2 (E10, Cell Signaling Technology); anti-GAPDH (6C5, Calbiochem); anti-One-STrEP-Tag mAb (StrepMAB Classic, IBA bioTAGnology); anti-human CD3ϵ (UCHT-1) and anti-SLP-76 (SLP-76/03, AbD Serotec); anti-human CD28 (CD28.2, BioLegend). Rabbit polyclonal Abs used were: anti-PLC-γ1 and anti-Grb2 (C-23, Santa Cruz); anti-actin (Sigma); anti-BRCC3 (BRCA1/BRCA2-containing complex subunit 36, ProSci Inc.); anti-ABRO1 (Abraxas brother protein 1, Bethyl Laboratories); anti-HGS (hepatocyte growth factor-regulated tyrosine kinase substrate), anti-GIT1 (Arf GTPase-activating protein), and anti-ELMO-1 (engulfment and cell motility 11) (Atlas Antibodies); anti-ARAP1 (Arf-GAP with Rho-GAP domain, ANK repeat, and pleckstrin homology domain-containing protein 1, a gift from Dr. P. Randazzo, NCI, NIH, Bethesda, MD); anti-Cool2/alphaPix, anti-ERK1/2 (137F5), anti-phospho-SAPK/JNK (Thr-183/Tyr-185), anti-phospho-p38 MAPK (3D7), anti-phospho-PLC-γ1 (Tyr-783), anti-phospho-ZAP-70 (Tyr-493), and anti-phospho-Src (Tyr-416, Cell Signaling Technology). Anti-THEMIS antiserum was obtained by immunizing rabbits with a peptide corresponding to the C-terminal 15 amino acids of the human C6orf190 sequence and affinity-purified using the same peptide.
Cells and Transfections
CD4+ Jurkat subclone 20, SLP-76-deficient Jurkat cells (J14), and Jurkat cells expressing luciferase reporter plasmids were maintained in RPMI 1640 (PAA Laboratories, Inc.) medium supplemented with 10% fetal calf serum (FCS, Perbio). Human embryonic kidney epithelial cells (HEK293) were maintained in DMEM, 10% FCS. HEK293 cells were transfected by standard calcium phosphate precipitation. Lentiviral particles were produced in HEK293 cells by co-transfection of the pHR-SIN-BX-IRES-Emerald vector with the packaging plasmids psPAX2 and pMD2.G. Forty-eight hours after transfection, viral supernatants were harvested, filtered, and used for the transduction of cells.
T Cells Isolation from Themis−/− and Control Mice and IL-2 ELISA
Conventional CD4+ T cells were purified from spleens of wild-type and Themis knock-out mice using a CD4 negative selection kit (Stemcell) supplemented with anti-CD25-biotin (7D4, eBioscience) for depletion of CD4+CD25+ Tregs and effector T cells. Cell purity was controlled with anti-CD25-PE (PC61, eBioscience) and was 90–95%. Purified cells were stimulated with plate-bound anti-CD3 Ab (BD BioCoatTM T cell activation plate, BD Biosciences) and soluble anti-CD28 antibody (1 μg/ml) for 48 h. Supernatants were collected, and IL-2 concentration was measured using ELISA (eBioscience, IL-2 ELISA kit, catalog #88-7024).
Silencing of Protein Expression
MISSION® shRNA lentiviral transduction particles specific for human C6orf190 and human LAT were purchased from Sigma. As a negative control MISSION®, non-target shRNA control transduction particles (SHC002V) were used, delivering a short hairpin that does not target any human or mouse protein. Stable knock-down cell lines were generated by lentiviral transduction of wild-type Jurkat cells or Jurkat cells stably expressing either a NFAT/AP-1-luciferase reporter construct (kindly provided by Prof. M. Tremblay, McGill University, Montreal, Canada) or a IL-2-luciferase reporter construct (a gift from Dr. H. Stockinger, Medical University of Vienna, Austria) and subsequent 2 μg/ml puromycin selection. Silencing efficiency was controlled by immunoblot. For stimulations we used Jurkat cells treated with non-targeting control shRNA (shControl), two shRNAs targeting THEMIS (shTHEMIS1 (TRCN0000130264) and shTHEMIS2 (TRCN0000128476), 60 and 90% knockdown efficiency, respectively), or shRNA targeting LAT (shLAT, TRCN0000029734, 60% knockdown efficiency).
Immunoprecipitations and Pulldown Assays
Resting or anti-CD3-activated wild-type Jurkat cells or Jurkat cells expressing THEMIS-OST were lysed with lysis buffer containing 0.5% dodecyl-β-d-maltoside (see above). THEMIS IPs were carried out on cleared lysates for 60–90 min at 4 °C with purified THEMIS antibody coupled to Sepharose protein G beads or Streptactin-Sepharose beads (IBA BioTAGnology, Göttingen, Germany), respectively. As controls, anti-THEMIS beads were incubated with a 1 mm solution of the peptide against which the antibody was raised, and Streptactin beads were incubated with 50 mm biotin before IP. After IP, beads were washed three times with lysis buffer and boiled in reducing SDS sample buffer.
ELISAs and Luciferase Assays
For IL-2 ELISAs, Jurkat cells transduced with the indicated shRNA constructs were either stimulated with plate-bound CD3 mAb UCHT-1 (concentration as indicated) and soluble CD28 mAb CD28.2 (1 μg/ml) in flat-bottom plates or with Raji B cells pre-pulsed with the indicated concentrations of staphylococcal enterotoxin E LPS-reduced, Toxin Technology) in round-bottom plates. All stimulations were done in triplicate and carried out in RPMI, 10% FCS at 37 °C in a 5% CO2 atmosphere for 24 h. Human IL-2 ELISA was performed from culture supernatants following the manufacturer's instructions (human IL-2 DuoSet, R&D Systems).
For IL-2 and NFAT/AP-1-luciferase assays, stable shRNA-transduced IL-2 and NFAT/AP-1-Jurkat luciferase cells were stimulated for 6 h as described above including an additional stimulation with 12 ng/ml phorbol 12-myristate 13-acetate and 500 ng/ml ionomycin. Luciferase activity was assayed according to the manufacturer's protocol (luciferase assay system E3040, Promega) on a Berthold Tristar LB941. Relative light units were normalized by the maximum phorbol 12-myristate 13-acetate/ionomycin stimulation for each cell line and for cell numbers in each sample by Bradford assay. For NF-κB-luciferase assays, shRNA-transduced Jurkat cells were transfected 24 h before the assay with NF-κB luciferase (firefly) and renilla luciferase plasmids by standard electroporation (GenePulser, Bio-Rad) and stimulated as described. Firefly and renilla luciferase activities were assayed according to the manufacturer's protocol (Dual-Luciferase Reporter Assay System, Promega). Relative light units of the firefly reporter were normalized for transfection efficiency by the renilla relative light units and by the maximum phorbol 12-myristate 13-acetate/ionomycin stimulation for each cell line.
RESULTS
Quantitative Kinetic Analysis of TCR-induced Tyrosine Phosphorylated Proteins
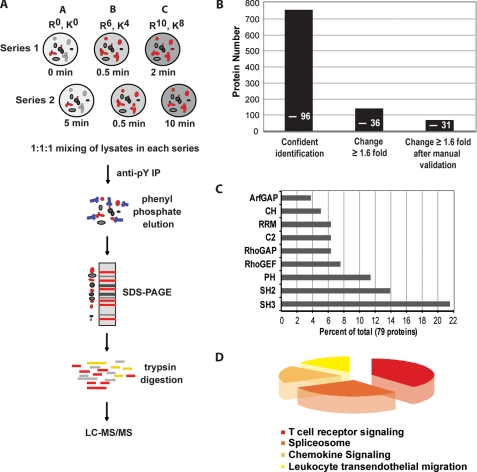
To obtain high resolution kinetic profiles of tyrosine phosphorylation after TCR triggering, we metabolically labeled three separate cultures of Jurkat cells with distinct isotopic forms of arginine and lysine (Fig. 1A). Cells were divided into two series and activated with anti-CD3 Ab for the indicated times, with one common time point to link and normalize the two stimulation series and obtain a continuous activation profile over five time points (Fig. 1A). Cell lysates of each series were mixed in 1:1:1 ratio and incubated with a mixture of anti-Tyr(P) Abs that routinely allowed >90% depletion of tyrosine-phosphorylated proteins (data not shown). The vast excess of anti-Tyr(P) Abs used likely out-competed phosphotyrosine-based interactions (e.g. Src homology 2 and phosphotyrosine binding domains) but should leave other interactions intact. We performed rapid cell lysis and immunoprecipitation procedures (a total of 60–90 min) as compared with standard protocols (e.g. several hours or overnight) to reduce dissociation of protein-protein interactions independent of tyrosine phosphorylation. Bound proteins were eluted with phenyl phosphate, separated by SDS-PAGE, subjected to in-gel trypsin digestion, and analyzed by LC-MS/MS (Fig. 1A). The relative abundance of a given protein was measured by averaging the intensities of all peptides identified for the protein at each activation time point. Three independent biological replicas (SILAC1, -2, and -3) were performed for which we obtained two data sets at TCR stimulation times 0, 0.5, 2, 5, and 10 min and one data set at TCR stimulation times 0, 2, 5, 10, and 20 min. To ensure a high confidence level of protein identification, we determined “confidence level thresholds” by using a target-decoy statistical model (9, 26, 27). Of 1240 proteins initially identified in three biological replicas, 758 were retained based on a cumulative Mascot score of ≥60 for at least two peptides (Fig. 1B, supplemental Table S1 and Fig. S1, B and C). Protein quantitation at different time points was carried out using MSQuant (9) and manually validated. A conservative threshold of 1.6-fold increase upon TCR stimulation in protein abundance above basal level in anti-Tyr(P) immunoisolation was considered significant (for details, see supplemental text and Fig. S1_A_), which classified 141 proteins as activated (Fig. 1B). Finally, manual validation taking into account protein identification in the biological replicas and two activation time series as well as the quality of the MS (for quantitation) and MS2 spectra (for peptide sequencing) distinguished 77 proteins as activated with very high confidence (Fig. 1B, supplemental Table S2; mean S.D. of protein quantitation, 11.5%, see supplemental Fig. S1_D_). In these selected proteins we identified 35 tyrosine phosphorylation sites, 3 of which have to our knowledge not been previously reported (supplemental Table S3; MS/MS spectra are shown in supplemental Fig. S6). Inspection of the final protein list using phosphoELM (protein phosphorylation data base) revealed that 31 proteins (39%) are known to be tyrosine-phosphorylated, whereas the remaining 48 (61%) might be new targets of tyrosine kinases or interaction partners of tyrosine-phosphorylated proteins. As expected, proteins that increased as a consequence of TCR-induced tyrosine phosphorylation contained typical signaling signatures such as domains of guanine nucleotide exchange (Rho GTPase guanine nucleotide exchange factor, Rho GTPase-activating protein) and kinase activities or domains required for protein-protein (Src homology 2 and 3) and protein-phospholipid interactions (pleckstrin homology, C2) (Fig. 1C). Interestingly, we observed in addition an increase in the abundance of RNA-associated proteins (Fig. 1C). Cross-referencing the protein list with the signaling pathways data base Kegg showed a prevalence of proteins previously implicated in TCR signaling and other pathways sharing components with the TCR signaling pathway (Fig. 1D). Many of the proteins identified were known elements of the TCR signaling machinery (supplemental Table S2), underscoring the high specificity and sensitivity of our SILAC analysis. This together with the stringent cut-off applied suggested that previously unnoticed proteins that increased in the immunoisolates upon stimulation could be novel components of the TCR signalosome and/or be under its control.
FIGURE 1.
Quantitative analysis of TCR-dependent protein tyrosine phosphorylation. A, experimental workflow is shown. Jurkat cells were metabolically labeled with three combinations of arginine and lysine containing light (R0, K0) or heavy (R6, K4 or R10, K8) isotopes until complete incorporation (as described under “Experimental Procedures”). Equal numbers of cells for each labeling condition were divided into two series (1 and 2), activated with anti-CD3 Ab at 37 °C for the indicated times, and immediately lysed in ice-cold lysis buffer. Post-nuclear lysates of each activation series were mixed in a 1:1:1 ratio and subjected to Tyr(P) (pY) immunoprecipitation for 60–90 min at 4 °C using a mixture of three anti-Tyr(P) Abs coupled to beads (4G10, Tyr(P)-99, Tyr(P)-20). Beads were washed and eluted with phenyl phosphate, and eluates were separated by SDS-PAGE. 10 slices per gel lane were cut and digested with trypsin overnight, and the resulting tryptic peptides were analyzed by LC-MS/MS. After protein identification and quantitation, relative protein abundance in each activation series were bridged and normalized using quantitation of the common time point (0.5 min), thus, resulting in a continuous activation profile over 5 time points. B, 758 proteins were confidently identified from three independent SILAC experiments. 141 proteins that showed a significant increase (≥1.6-fold) above basal levels in anti-Tyr(P) immunoisolates after TCR stimulation were selected, of which 77 were retained after further manual validation. Numbers within bars indicate tyrosine-phosphorylated proteins identified in each set. C, over-represented domains within identified protein sequences from the three SILAC experiments; most domains are involved in signaling events. ArfGAP, GTPase activating proteins toward Arf; CH, calponin homology domain; RRM, RNA recognition motif; C2, calcium-dependent phospholipid binding domain; RhoGAP, Rho GTPase activating protein domain; RhoGEF, guanine nucleotide exchange factor for Rho GTPases domain; PH, Pleckstrin homology domain; SH2, Src homology domain 2; SH3, Src homology domain 3. D, analysis of activated signaling pathways by cross-correlation with the signaling pathways data base Kegg is shown.
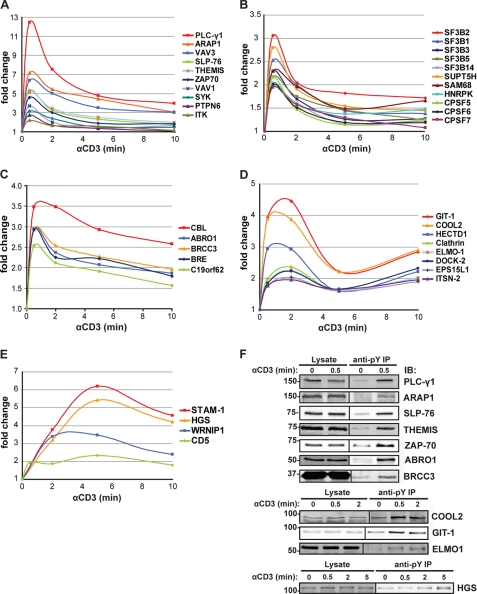
TCR-induced Functionally Distinct Waves of Tyrosine-phosphorylated Proteins
Temporal analysis of anti-Tyr(P) immunoisolates upon TCR stimulation revealed the unfolding of phosphorylation kinetics for individual proteins and their interaction partners. We clustered those with significant abundance changes (supplemental Table S2) into three main distinct kinetic profiles. The earliest (peaking at ∼0.5 min, Fig. 2, A–C) included the majority of proteins followed by an intermediate (∼0.5–2 min, Fig. 2D) and late cluster (∼2–10 min, Fig. 2E), indicating that TCR-induced multiprotein complex formation due to tyrosine phosphorylation of cytoplasmic substrates proceeded by waves until relatively late times after stimulation. The tyrosine phosphorylation-induced immunoisolation of representative proteins from each wave was confirmed by immunoblot (Fig. 2F). Thirty-five percent of proteins displaying significant changes under these stimulatory conditions were new in the context of TCR signaling (supplemental Table S2). As expected, the earliest wave corresponded to signal transducers of the TCR-proximal signaling complex such as ZAP-70, LAT, SLP-76, PLC-γ1, ITK, Vav1, and tyrosine-protein phosphatase non-receptor type 6 (PTPN6) (Fig. 2A). Some components of the TCR signalosome confidently identified in our SILAC analysis (supplemental Table S1), including LCK, ADAP, and Grb2, were not considered activated, because according to the criteria used here for significant abundance change (≥1.6-fold), they did not show consistent change above or below basal phosphorylation levels (e.g. LCK phosphorylation was considered constant, supplemental Fig. S2). Consistent with these data, recent studies in our laboratory have established that LCK is constitutively active in unstimulated Jurkat cells and human CD4+ T cells and that its global phosphorylation remains unchanged upon TCR ligation (28). We also identified three Tyr(P) sites that, to our knowledge, have not yet been reported, including a highly conserved tyrosine (Tyr-45) of LAT, Tyr-771 of ADAP, and Tyr-222 of Gads (supplemental Table S3). Importantly, we detected in the earliest signaling cluster a formerly unidentified protein, C6orf190 (human protein Q8N1K5) which we further characterized in this work (see below). Its mouse ortholog, named Themis, was recently identified and studied functionally in vivo after gene ablation (21–25).
FIGURE 2.
Kinetics of Tyr(P)-dependent signaling complexes after TCR stimulation. A, rapid, transient kinetics of known TCR-proximal signaling proteins involved in signal initiation are shown. The newly identified protein THEMIS falls in the same cluster. B, rapid, transient kinetics of nucleic acid-associated proteins (splicing factors (SF3Bs), transcription elongation factor (SUPT5H), cleavage and polyadenylation specificity factors (CPSFs)) are shown. C, rapid kinetics of CBL and components of the BRCA1 complex are shown. D, a representative group of proteins isolated at an intermediate stage after TCR activation is shown. Most proteins in this group are involved in cytoskeleton rearrangement and trafficking. E, a group of proteins isolated at the later stages after TCR activation, including proteins involved in endosomal sorting such as STAM1 and HGS is shown. F, validation of tyrosine phosphorylation by immunoblot (IB) is shown. Anti-Tyr(P) IPs of proteins isolated at early (_PLC-γ_1, ARAP1, SLP-76, THEMIS, ZAP-70, ABRO1, BRCC3), intermediate (COOL2, GIT1, ELMO-1), and later (HGS) time points after TCR activation are shown.
Similarity of phosphorylation kinetics in different proteins (or complexes) may indicate physical proximity and perhaps participation in a common signaling pathway and/or function (29, 30). Such an example is illustrated in Fig. 2C, in which the kinetics of BRE, BRCC3, and ABRO1, three components of the BRCA1 complex involved in DNA damage repair and ubiquitination, have a kinetics profile closely overlapping with that of C19orf62. Indeed, the latter is a formerly uncharacterized protein that has only recently been identified as a new component of this complex and was named MERIT40 (31, 32). Surprisingly, we also identified nucleic acid-associated proteins peaking at the earliest time point (Fig. 2B), including various proteins involved in mRNA splicing/processing and translational regulation (supplemental Table S2). Among these proteins were several splicing factor 3B subunits (SF3B), cleavage and polyadenylation specificity factors (CPSF), and small nuclear ribonucleoproteins. With the exception of Sam68, these RNA-binding proteins have not been previously reported to be tyrosine-phosphorylated upon TCR stimulation (18, 33–35).
The second protein cluster was characterized by a broader peak between 0.5 and 2 min after TCR activation and biphasic kinetics (Fig. 2D). This group predominantly includes proteins involved in cytoskeleton rearrangement and protein trafficking, two key events that regulate formation of the immune synapse (36). The third cluster contained proteins such as signal-transducing adaptor molecule 1 (STAM1), HGS, Werner helicase interacting protein 1 (WRNIP1), and CD5, whose phosphorylation is markedly sustained (Fig. 2E). Overall, our analysis reveals discrete waves of signaling events after TCR triggering that are characterized by sets of proteins with “common” functions. Notably, TCR stimulation initially leads to sharp and intense tyrosine phosphorylation signals with rapid down-regulation but propagates into broader peaks (longer-lasting phosphorylation) as the signal diverges into various signaling pathways at later time points.
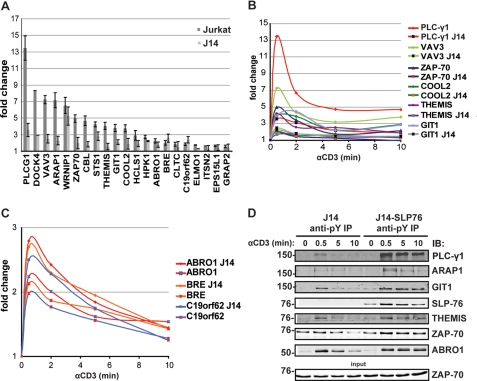
The LAT·SLP-76 Signalosome Controls Most but Not All TCR Signaling Pathways
Because the LAT·SLP-76 scaffold is a central nucleation platform on which several proteins assemble to direct most TCR-induced signaling pathways (1), we wanted to quantitatively assess the dependence of Tyr(P) kinetics on this key signaling node. SLP-76 is indispensable in tyrosine phosphorylation and activation of PLC-γ1 and the Ras pathway (37) as well as actin cytoskeletal remodeling and cell adhesion (1). Consistently, the abundance of several signaling proteins was greatly diminished in the Tyr(P) immunoisolates of TCR-stimulated SLP-76-deficient (J14) Jurkat cells as compared with SLP-76-sufficient cells with PLC-γ1 and Vav family proteins being most strongly affected (Fig. 3, A, B, and D). Immunoisolation of proteins whose precise functions in T cell activation are unknown, such as Rac/Rho/Cdc2/Arf regulators dedicator of cytokinesis 4 (DOCK4), COOL2 (ARHGEF6/PIXα), and its interacting partner GIT1 and ARAP1, which is implicated in receptor endocytosis (38), were also substantially reduced in SLP-76-deficient cells. The presence of the newly identified protein THEMIS in Tyr(P)-mediated signaling complexes was also found to be dependent on SLP-76. However, implication in TCR signaling of other proteins, such as BRCA1 complex components ABRO1, brain and reproductive organ-expressed protein BRCA1/BRCA2-containing complex subunit 45 (BRE), and C19orf62/MERIT40, ELMO-1, Intersectin-2 (ISTN2), and epidermal growth factor receptor substrate 15-like1 (EPS15L1), seemed less dependent on or independent of SLP-76 (Fig. 3, A and C). As shown in Fig. 3D, for some proteins we validated the degree of dependence on SLP-76 by immunoblotting. Interestingly and in line with previous reports (39, 40), lower levels of phospho-ZAP-70 were captured in immunoisolates from SLP-76-deficient cells, and immunoblots reproducibly showed a weak dependence on SLP-76 (Fig. 3D). The above data suggested that the involvement of a large cohort of the signaling components activated by TCR engagement, including a number of previously uncharacterized proteins, were strongly dependent on the assembly of the SLP-76·LAT scaffold. However, other proteins, such as PLC-γ1, GIT1, THEMIS, and ABRO1, were only immunoisolated at the earliest stimulation time point (0.5 min) in the absence of the SLP·76/LAT complex, whereas reconstitution of the J14 cells with SLP-76 allowed for a more sustained phosphorylation of these proteins (Fig. 3D).
FIGURE 3.
Dependence of global TCR signaling on the SLP-76·LAT signalosome. A, shown is the abundance of SLP-76-dependent and -independent proteins commonly identified from SILAC Tyr(P) IP in wild-type and SLP-76-deficient (J14) Jurkat cells at 30 s after TCR activation. B, complete anti-Tyr(P) isolation kinetics of SLP-76-dependent proteins from the same experiment as in A is shown. C, exemplary SLP-76-independent anti-Tyr(P) isolation kinetics of components of the BRCA1 complex from the same experiment as in A (note different Y axis scale) are shown. D, validation of SLP-76 dependence of Tyr(P) (pY) IPs in J14 and J14-SLP-76-FLAG cells by immunoblot probed with specific protein antibodies is shown.
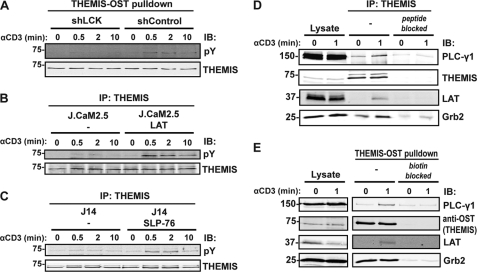
THEMIS Is a Component of the TCR-proximal Signaling Machinery
THEMIS, one of the new proteins identified in this study, is expressed exclusively in CD8+ and CD4+ T cells (BioGPS gene expression data base). Our recent work showed that THEMIS is directly phosphorylated on tyrosine after TCR stimulation in Jurkat cells and human T cells (21). Moreover, the close coincidence of kinetic profiles of THEMIS, LAT, and SLP-76 tyrosine phosphorylation (supplemental Fig. S3, B and C) suggested that THEMIS was implicated in TCR-proximal signaling and that it might be recruited to the plasma membrane via LAT and/or SLP-76. Consistent with this hypothesis, THEMIS tyrosine phosphorylation was found to be strongly dependent on LCK and LAT, as demonstrated using a Jurkat cell line knocked-down for LCK expression and the LAT-deficient Jurkat cells line J.CaM2.5 (37) (Fig. 4, A and B). Similar immunoblot experiments confirmed that THEMIS phosphorylation was also dependent on SLP-76 (41) (Fig. 4C), although to a lesser degree as compared with LCK and LAT. To directly test the hypothesis that THEMIS interacted with the LAT·SLP-76 complex, Jurkat cells were stimulated with anti-CD3 mAb, and THEMIS immunoisolates were probed for the presence of the LAT and SLP-76 as well as various signaling proteins that are part of the complex (Fig. 4D). We consistently detected a specific and TCR-inducible interaction of THEMIS with LAT (Fig. 4D). PLC-γ1 showed low levels of constitutive binding to THEMIS that increased substantially after stimulation (Fig. 4D). Relatively high levels of Grb2 were bound constitutively to THEMIS, and in most experiments a slight increase of Grb2 was seen after activation (Fig. 4D). Binding of THEMIS to SLP-76 was not consistently detected (data not shown), suggesting that LAT but not SLP-76 could be a binding partner of THEMIS. Similar results were obtained in a Jurkat cell line in which THEMIS was fused to a potent affinity tag (One-STrEP-Tag (OST)) and subjected to a Streptactin pulldown assay (Fig. 4E). Specific THEMIS/LAT interaction was confirmed by MS-based analysis of LAT immunoisolates in activated Jurkat cells (data not shown). These data strongly suggest that THEMIS is a novel component of the TCR signaling machinery that is recruited onto LAT via its constitutive interaction with Grb2 and may be necessary for signal diversification and regulation (2).
FIGURE 4.
Themis is a new component of the SLP-76·LAT signalosome. A, shown is a THEMIS-One STrEP tag (OST) pulldown assay using Streptactin-Sepharose after anti-CD3 stimulation in Jurkat cells transfected with non-targeting control shRNA or a shRNA construct targeting LCK (70% knockdown efficiency). IB, immunoblot; pY, Tyr(P). B, THEMIS IP from anti-CD3 stimulated LAT-deficient (J.CaM2.5) and reconstituted cells is shown. C, THEMIS IP from anti-CD3 stimulated SLP-76-deficient (J14) and reconstituted cells is shown. D, THEMIS IPs from resting or anti-CD3-stimulated Jurkat cells is shown. Specific THEMIS antibody saturated with the peptide against which the antibody was raised served as a control. E, a THEMIS-OST pulldown assay from resting or CD3-stimulated Jurkat cells stably expressing THEMIS-OST using biotin-saturated Streptactin-Sepharose as control is shown. Both sets of blots were probed with antibodies against TCR-proximal signaling proteins.
THEMIS Is Required for Effective TCR-induced ERK Activation
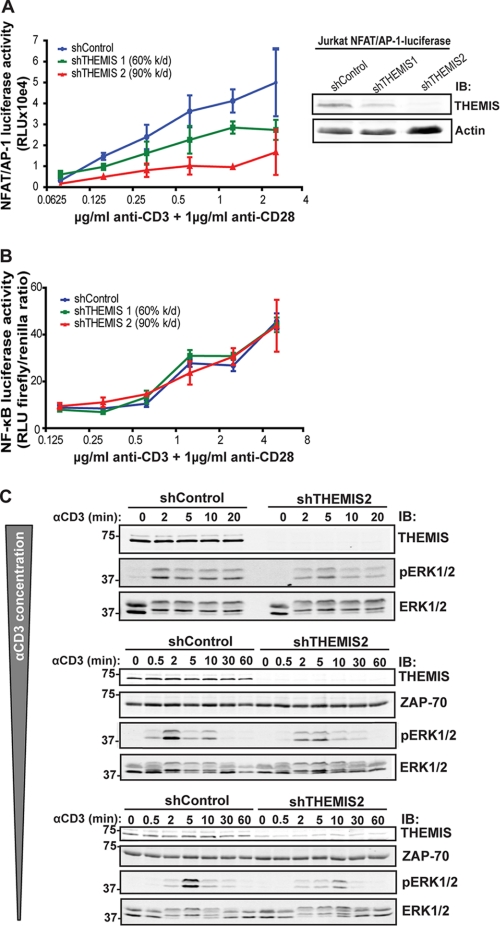
The association of THEMIS with the LAT complex suggested that it might regulate TCR-induced classic pathways, such as Ca2+/NFAT, Ras/ERK, JNK, p38, and NF-κB that control downstream gene expression (1). To test this idea, we first assessed whether IL-2 production, which depends on those pathways, was regulated by THEMIS. We used two shRNAs that reduced THEMIS expression in Jurkat cells to 40 and 10%, respectively, whereas a non-targeting shRNA and a LAT shRNA (60% knockdown) served as negative and positive controls, respectively (Fig. 5A, lower panel). Cells were either stimulated with CD3/CD28 Abs (Fig. 5A, left panel) or staphylococcal enterotoxin E-loaded Raji B cells (Fig. 5B, right panel). Reduced expression of THEMIS resulted in lower production of IL-2 as a function of THEMIS knockdown in both TCR stimulation settings, with the highest inhibition being comparable to that of the LAT control knockdown. THEMIS knockdown also significantly reduced TCR-induced IL-2 gene transcription (Fig. 5B), as assessed by using an IL-2-luciferase reporter-expressing Jurkat cell line. Importantly, re-expression of THEMIS in knock-down cells restored IL-2 reporter activity to control levels (Fig. 5B). Consistently, overexpression of THEMIS by lentiviral transduction in Jurkat cells significantly increased IL-2 gene transcription above that of cells transduced with control vector (Fig. 5B). To strengthen these data, we isolated spleen-derived CD4+CD25− T cells from Themis-deficient mice to distinguish them from CD25+ T regulatory and effector T cells and assessed their capacity to produce IL-2 after CD3/CD28 stimulation. Consistent with the data in Jurkat cells, Fig. 5C shows that IL-2 secretion was considerably affected in peripheral T cells lacking Themis. These data supported the conclusion that THEMIS plays an important role in regulating TCR-induced IL-2 gene expression. To understand which pathways were controlled by THEMIS, we assessed TCR-induced NFAT/AP-1 and NF-κB activity in THEMIS knockdown Jurkat cells (Fig. 6, A and B). THEMIS knockdown significantly reduced NFAT/AP-1-luciferase activity in NFAT/AP-1-luciferase reporter-expressing Jurkat cell lines after anti-CD3/CD28 stimulation compared with control cells (Fig. 6A), whereas no noticeable difference was observed for the NF-κB luciferase reporter (Fig. 6B). Based on these observations, we set out to determine whether TCR-induced intracellular calcium increase was affected by THEMIS expression. However, we could not detect any significant difference between knockdown and control cells (data not shown). We, therefore, deduced that the AP-1 branch needed to activate the NFAT/AP-1 reporter was affected. If this was the case, then either the Ras/ERK and/or the JNK pathway could be affected by THEMIS deficiency. To test this hypothesis we compared anti-CD3-induced ERK activation in Jurkat cells with ≥90% shRNA-mediated reduction of THEMIS expression with control cells. We found that the onset and maximal induction of ERK phosphorylation were dependent on THEMIS expression, which was especially evident at low grade stimulation using decreasing amounts of anti-CD3 Ab (Fig. 6C). However, activation of p38 and JNK was apparently unaffected (supplemental Fig. S4). Taken together, these results establish THEMIS as a positive regulator of TCR signaling affecting IL-2 gene expression, most likely through the control of ERK activation.
FIGURE 5.
THEMIS is a positive regulator of TCR-induced signaling. A, IL-2 ELISAs of shControl, shTHEMIS1, shTHEMIS2, and shLAT Jurkat cells stimulated with plate-bound anti-CD3 and soluble anti-CD28 (left panel) or staphylococcal enterotoxin E-pulsed Raji B cells (right panel) for 24 h is shown. Immunoblot (IB) analysis of the cell lines used (lower panel, shTHEMIS1 (60% knockdown), shTHEMIS2 (90% knockdown), shLAT (60% knockdown), and the non-targeting shControl). B, IL-2-luciferase assay of shControl and shTHEMIS2 (90% knockdown) is shown. IL-2-luciferase cells were transduced with either empty vector or an shRNA-resistant mutant of THEMIS (right panel) and stimulated with staphylococcal enterotoxin E-pulsed Raji B cells (left panel). C, TCR stimulation-induced IL-2 secretion in peripheral CD4+CD25− T cells from wild-type and Themis knock-out mice is shown. Conventional CD4+ cells were purified by negative selection and stimulated with plate-bound anti-CD3 and soluble anti-CD28 Ab for 48 h. IL-2 concentrations in supernatants were measured by ELISA. Shown are data from three mice of each group, p = 0.03.
FIGURE 6.
THEMIS positively modulates NFAT/AP-1 and ERK activity. A, shown is an NFAT/AP-1-luciferase assay of shControl, shTHEMIS1 (60% knockdown), and shTHEMIS2 (90% knockdown) NFAT/AP-1-luciferase Jurkat cells after stimulation with plate-bound anti-CD3 and soluble anti-CD28 (left panel) and immunoblot (IB) analysis of THEMIS expression in the cell lines used (right panel). RLU, relative light units. B, an NF-κB-luciferase assay of shControl, shTHEMIS1, and shTHEMIS2 Jurkat cells stimulated as in A (for THEMIS expression levels in cell lines used see Fig. 5A, lower panel). C, immunoblots of ERK1/2 phosphorylation kinetics in shControl and shTHEMIS2 (90% knockdown) Jurkat cells in three independent experiments using decreasing anti-CD3 concentrations (total ERK1/2 and ZAP-70 blots are shown as loading controls) are shown.
DISCUSSION
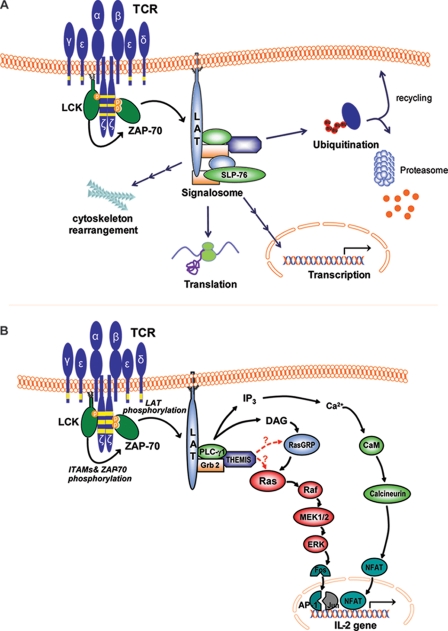
TCR ligation-induced rearrangements of cytoskeleton, protein traffic, and gene expression rely on an early burst of protein-tyrosine kinase-induced phosphorylations. Here, we have employed state of the art MS-based quantitative proteomics to improve our understanding of the spatiotemporal links between these TCR signaling events. Protein-tyrosine kinase-driven phosphorylations could be temporally resolved into at least three distinct clusters of similar phosphorylation kinetics (signaling waves). As suggested by other SILAC-based investigations (29, 30), overlapping kinetic profiles can be predictive of spatial proximity and probable engagement in a common or related cellular function. This correlation is illustrated by a subgroup in the earliest kinetics cluster that contains known TCR-induced kinase substrates, including the LAT·SLP-76 complex and its binding partners that activate major functional pathways (1). The assignment of THEMIS, a T cell-specific protein recently discovered to be implicated in T cell development (21, 22, 24), to this subgroup allowed us to correctly predict its implication in TCR-proximal signaling. Moreover, this approach allowed us to appreciate the fine details of the LAT·SLP-76 signalosome dynamics in that SLP-76 was not found to be absolutely required for initial tyrosine phosphorylation of its binding partners but to sustain it. Surprisingly, the earliest signaling wave also included several proteins involved in co- and post-transcriptional mRNA regulation, some of which shuttle between nucleus and cytoplasm in complex with mRNA (42). A second cluster contained proteins controlling actin cytoskeleton rearrangement and initiation of ubiquitination-mediated membrane protein trafficking followed by a third one comprising elements of the endosomal compartment processing proteins destined for degradation (43). Our data suggest that upon TCR stimulation protein-tyrosine kinases set the tempo for diverse cellular responses to coordinate the formation and dynamics of the immunological synapse and sustain gene expression (Fig. 7A).
FIGURE 7.
TCR-induced activation of signaling pathways observed in the present study. A, shown is a schematic representation of the cellular pathways observed in our SILAC analysis to be directly or indirectly affected by TCR stimulation-induced tyrosine phosphorylation in T cells. B, shown is a model of the possible role of THEMIS in the TCR signaling cascade. Upon TCR activation, THEMIS is phosphorylated on tyrosine and recruited to the plasma membrane where it interacts with PLC-γ1 and LAT, presumably via Grb2. THEMIS exerts a positive regulatory effect on ERK activation, presumably via a direct or indirect effect on Ras. IP3, inositol 1,4,5-trisphosphate.
TCR-driven early gene expression is controlled by a number of key transcription factors (e.g. NFAT, NF-κB, Myc, Fos/Jun) whose activation is dependent on calcium elevation and activation of small GTPases and PKCs (1). However, we found that factors involved in mRNA regulation were among the earliest direct or indirect targets of tyrosine kinases after TCR ligation. These included the transcription elongation factor SUPT5H, SF3B subunits 1, 2, 3, 5, and 14, CPSF subunits 5 (NUDT21), 6, and 7, heteronuclear ribonucleoprotein particle K (hnRNPK) which is implicated in translation activation (44, 45), and the mitotic Src kinase substrate Sam68, a hnRNPK-homology (KH) domain family member involved in alternative splicing (46–48). Some components of these complexes, including SF3B14 (35), CPSF5/NUDT21 (35), Sam68 (34), and hnRNPK (18), have been found to be tyrosine-phosphorylated in T cells. Tyrosine phosphorylation of hnRNPK appears to be crucial for translation activation (49), whereas for Sam68 it may be important for its role as a scaffold (47). These data suggest that TCR-induced early gene expression relies not only on rapid and coordinated activation of several transcription factors but also on concomitant regulation of mRNAs processing and translation. This observation warrants future investigations on the impact of these poorly understood controls of protein expression on sustained signaling at the immune synapse.
Our study also provided clues as to the time when other, not well understood TCR-induced processes, such as changes in cytoskeleton dynamics and intracellular trafficking of membrane proteins, begin to take place. We found the adaptor ELMO-1 and its binding partner DOCK2 (50), both of which contain Tyr(P) sites (phosphoELM data base), to peak at ∼2 min and, thus, become protein-tyrosine kinase targets after the formation of the LAT·SLP-76 complex. Tyrosine phosphorylation of ELMO-1 (51) contributes to the efficiency of Rac2 activation by DOCK2 (52, 53), which is required for the formation of actin-rich lamellipodia protrusions in lymphocyte migration (54), reminiscent of those formed by T cells at the immunological synapse (36). This suggests a spatiotemporal protein-tyrosine kinase regulation of membrane ruffling at the immunological synapse to optimize TCR engagement with the pMHC (36). GIT-1 and COOL2 were also present in the kinetics cluster at ∼2 min, the reported time at which they are recruited to the immunological synapse for PAK1 and sustained PLC-γ1 activation (55, 56), which may play a role in actin cytoskeleton dynamics at this stage of activation. GIT-1 and COOL2 are TCR-inducibly phosphorylated on tyrosine (supplemental Table S2 (18)) and were both dependent on SLP-76 (Fig. 4A, B and D). However, recruitment of GIT-1, COOL2, and PAK1 to the immunological synapse was previously found to be independent of SLP-76 (56), suggesting that their phosphorylation but not plasma membrane recruitment may depend on the LAT·SLP-76 signalosome.
The same cluster also included components of clathrin-coated vesicles, including the clathrin heavy polypeptide, EPS15L-1 (also called EPS-15R), and tyrosine-phosphorylated intersectin-2 (ITSN-2) (Fig. 2D, supplemental Table S2), which forms a complex with Eps15 that itself interacts with EPS15L-1 (57). HGS (also called Hrs) and its binding partner STAM-1, both of which are phosphorylated on tyrosine in response to receptor ligation (58), were isolated only at 5 min after TCR stimulation and persisted thereafter (Fig. 2E). STAM-1 (together with STAM-2) was previously shown to be required for normal T cell development and survival (59). The Hrs/STAM complex forms part of the ESCRT-0 complex that allows ubiquitinated transmembrane receptors (and their binding partners) present in clathrin-coated vesicles to travel from early endosomes to the ESCRT-I complex and the multivesicular body for subsequent lysosomal degradation (43). The functional meaning of tyrosine phosphorylation of components of the endocytotic and ESCRT-0 machinery minutes after TCR stimulation and the early presence of HECTD1 (Fig. 2D), an uncharacterized member of the HECT (homologous to E6-AP carboxyl-terminal)-domain-containing E3 ubiquitin ligase family that controls target fate (60), remain to be understood. In summary, our SILAC-based data faithfully recapitulate the sequence of endocytosis, trafficking, and degradation steps likely modulating signal strength and/or duration and represent a valuable approach to dissect the temporal mechanics of these events with higher precision.
In our investigation, we identified C6orf190, now called THEMIS, a human T cell-specific 74-kDa protein rapidly phosphorylated on tyrosine upon TCR ligation. Surprisingly, C6orf190 has gone undetected by conventional biochemical approaches and large scale proteomics analyses in T cells in recent years, suggesting that its abundance and/or phosphorylation stoichiometry may be low. We and others have recently shown that the mouse orthologue, Themis, is a critical component of the T cell development program (21–25). Themis-deficient mice have a block in thymic-positive selection, but it is unclear whether Themis plays a role in mature T cells. Although similar phenotypes have been observed in mice deficient in proximal TCR signaling components (61–63), there is disagreement as to the precise role of Themis, including the suggestion that it is not directly implicated in TCR signaling (22). Among the five independent studies of Themis-deficient mice, only ours identified subtle but reproducible TCR signaling defects (21–25). However, Themis−/− double-positive thymocytes showed decreased expression of activation markers upon TCR stimulation (21, 24) and displayed defective expression of transcription factors selectively required for CD4 lineage development (24), suggesting a defect in TCR signal strength (64). Whereas we previously observed partial but reproducible defects in both calcium flux and ERK phosphorylation in Themis−/− double-positive thymocytes (21), potential for re-wiring of cell signaling circuits due to in vivo “adaptation” to the systemic gene ablation might lead to erroneous conclusions. Therefore, the in vivo results need to be confirmed in an in vitro model. Our SILAC-based kinetics data combined with genetic, protein-protein interaction and signaling pathway analyses allowed definitive consolidation of the notion that THEMIS regulates TCR-proximal signaling and is unlikely to govern an unknown metabolic pathway, as suggested by another study (22). Indeed, THEMIS tyrosine phosphorylation was found to occur within 30 s of TCR engagement (21) and was strictly dependent on LAT, LCK, and SLP-76. Consistent with the assumption that shared kinetics clustering is predictive of protein-protein association (29, 30), we found that THEMIS associated with LAT upon TCR ligation. In agreement with previous reports (22, 24, 25), we showed that THEMIS interacts constitutively with Grb-2. One possibility is that the THEMIS-Grb2 complex binds inducibly to one of the three potential Grb-2 binding sites on LAT, and our preliminary data suggest this to be the case. Importantly, we found that THEMIS interacted with PLC-γ1 in TCR-stimulated Jurkat cells, further supporting our previous observations in Themis−/− double-positive thymocytes (21). What then could be the role of THEMIS as a regulator of TCR-proximal signaling? In line with studies in Themis−/− thymocytes (21, 22, 24, 25), we did not observe a decrease in TCR-inducible tyrosine-phosphorylation of signaling proteins in THEMIS knockdown cells (supplemental Fig. S5). On the other hand, our data clearly indicate that THEMIS controls IL-2 gene expression via an effect on AP-1, consistent with a partial but selective control on ERK activity observed here in THEMIS knockdown Jurkat cells and in our previous work in Themis−/− mice (21). Other pathways, notably NF-κB, p38 and JNK, were apparently unaffected by THEMIS deficiency. Experiments on THEMIS knockdown Jurkat cells did not show a reproducible defect in TCR-induced calcium flux (data not shown). We cannot easily reconcile these data with previous observations in Themis−/− double-positive thymocytes, but differences in cell type and/or mode of TCR stimulation might explain this apparent discrepancy. TCR signaling defects in the absence of THEMIS appear to be relatively subtle and may be missed by applying excessive TCR stimulation. Indeed, the defects in ERK signaling after THEMIS knockdown were distinctively and reproducibly observed here only under conditions of low-grade TCR triggering. The absence of a distinct defect on calcium suggests that THEMIS exerts most of its control on the ERK “arm” of IL-2 gene activation. Low grade TCR stimulation selectively activates Ras by RasGRP1 via PLC- γ1-produced diacylglycerol instead of recruiting son of seven less (SOS) proteins (65). One possibility is that THEMIS amplifies either diacylglycerol production or PLC-γ1 activity directly and by a threshold effect acts on the ERK pathway rather than on calcium. These findings suggest that THEMIS might drive T cell development by functioning as a selective amplifier to fine-tune the Ras pathway (Fig. 7B). In line with previous reports indicating that CD4 lineage commitment depends on stronger and more sustained ERK activation than CD8 lineage commitment (66–68), Themis deficiency affects the CD4 lineage more than the CD8 lineage (21, 22, 24, 25). Although further studies will be necessary to elucidate the molecular function of THEMIS and specifically its role in ERK activation, our study provides a strong indication for the role of THEMIS in the TCR signalosome.
In conclusion, our study demonstrates the power of quantitative proteomics in temporally resolving signals spreading from the immunological synapse by monitoring continuous profiles of activation with multiple time points during TCR stimulation. Moreover, it reveals the crucial role of protein-tyrosine kinases in regulating temporal succession of cellular processes such as actin cytoskeleton rearrangement, intracellular trafficking of membrane proteins, and activation of the mRNA processing machinery well beyond the TCR-proximal-signalosome (Fig. 7A). In the future, further functional and mechanistic dissection could be achieved by increasing the density of time points during TCR stimulation as well as MS sensitivity and bioinformatics robustness, especially if coupled with genetic or pharmacological perturbations. Importantly, these studies may lead to identification of unsuspected missing links of the TCR-proximal signaling machinery as demonstrated here for THEMIS and its implication in the activation of the Ras/ERK pathway (Fig. 7B).
Supplementary Material
Supplemental Data
Acknowledgments
We thank M. Tremblay for NFAT/AP-1-luciferase cells, H. Stockinger for IL-2 luciferase cells, V. Cerundolo, T. Mustelin, and D. Trono for plasmids, P. Randazzo for anti-ARAP1 antibody, B. Thomas for help with the mass-spectrometry analysis, and all members of the T cell signaling laboratory for helpful discussions.
*
This work was supported by Wellcome Trust Grant GR076558MA and EU-FP7 “Sybilla” Grant 201106 (to O. A.).
6
The abbreviations used are:
TCR
T cell antigen receptor
Ab
antibody
ABRO1
Abraxas brother protein 1
AP-1
activator protein 1
GAP
GTPase activating protein
ARAP1
Arf-GAP with Rho-GAP domain, ANK repeat, and pleckstrin homology domain-containing protein 1
GEF
guanine nucleotide exchange factor
ARHGEF
Rho GEF
BRCA1
breast cancer type 1 susceptibility protein
BRCC3
BRCA1/BRCA2-containing complex subunit 36
BRE
brain and reproductive organ-expressed protein BRCA1/BRCA2-containing complex subunit 45
DOCK
Dedicator of cytokinesis
ELMO-1
engulfment and cell motility 11
EPS15L1
epidermal growth factor receptor substrate 15-like1
HGS
hepatocyte growth factor-regulated tyrosine kinase substrate
GIT1
Arf GTPase-activating protein
Grb2
growth factor receptor-bound protein 2
ISTN2
Intersectin-2
IP
immunoprecipitation
LCK
lymphocyte-specific kinase
NFAT
nuclear factor of activated T cells
NF-κB
nuclear factor κB
SILAC
stable isotope labeling by amino acids in cell culture
STAM1
signal-transducing adaptor molecule 1
THEMIS
thymocyte-expressed molecule involved in selection
ADAP
adhesion and degranulation promoting adapter protein
SF3B
splicing factor 3B
CPSF
cleavage and polyadenylation specificity factors
OST
One-STrEP-Tag
hnRNPK
heteronuclear ribonucleoprotein particle K
LAT
activation of T cells
SLP-76
SH2 domain-containing lymphocyte protein of 76 kDa
PLC
phospholipase Cγ1
ZAP70
ζ chain-associated protein kinase of 70 kDa.
REFERENCES
- 1.Smith-Garvin J. E., Koretzky G. A., Jordan M. S. (2009) Annu. Rev. Immunol. 27, 591–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Acuto O., Di Bartolo V., Michel F. (2008) Nat. Rev. Immunol. 8, 699–712 [DOI] [PubMed] [Google Scholar]
- 3.Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., Mann M. (2006) Cell 127, 635–648 [DOI] [PubMed] [Google Scholar]
- 4.Hu Q., Noll R. J., Li H., Makarov A., Hardman M., Graham Cooks R. (2005) J. Mass Spectrom. 40, 430–443 [DOI] [PubMed] [Google Scholar]
- 5.Makarov A. (2000) Anal. Chem. 72, 1156–1162 [DOI] [PubMed] [Google Scholar]
- 6.Blagoev B., Kratchmarova I., Ong S. E., Nielsen M., Foster L. J., Mann M. (2003) Nat. Biotechnol. 21, 315–318 [DOI] [PubMed] [Google Scholar]
- 7.Gao J., Opiteck G. J., Friedrichs M. S., Dongre A. R., Hefta S. A. (2003) J. Proteome Res. 2, 643–649 [DOI] [PubMed] [Google Scholar]
- 8.Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. (2002) Mol. Cell Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 9.Schulze W. X., Mann M. (2004) J. Biol. Chem. 279, 10756–10764 [DOI] [PubMed] [Google Scholar]
- 10.Gerber S. A., Rush J., Stemman O., Kirschner M. W., Gygi S. P. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 6940–6945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ross P. L., Huang Y. N., Marchese J. N., Williamson B., Parker K., Hattan S., Khainovski N., Pillai S., Dey S., Daniels S., Purkayastha S., Juhasz P., Martin S., Bartlet-Jones M., He F., Jacobson A., Pappin D. J. (2004) Mol. Cell. Proteomics 3, 1154–1169 [DOI] [PubMed] [Google Scholar]
- 12.Cox J., Mann M. (2008) Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 13.Keller A., Eng J., Zhang N., Li X. J., Aebersold R. (2005) Mol. Syst. Biol. 1, 2005.0017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trost M., English L., Lemieux S., Courcelles M., Desjardins M., Thibault P. (2009) Immunity 30, 143–154 [DOI] [PubMed] [Google Scholar]
- 15.Kim J. E., White F. M. (2006) J. Immunol. 176, 2833–2843 [DOI] [PubMed] [Google Scholar]
- 16.Matsumoto M., Oyamada K., Takahashi H., Sato T., Hatakeyama S., Nakayama K. I. (2009) Proteomics 9, 3549–3563 [DOI] [PubMed] [Google Scholar]
- 17.Salomon A. R., Ficarro S. B., Brill L. M., Brinker A., Phung Q. T., Ericson C., Sauer K., Brock A., Horn D. M., Schultz P. G., Peters E. C. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 443–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mayya V., Lundgren D. H., Hwang S. I., Rezaul K., Wu L., Eng J. K., Rodionov V., Han D. K. (2009) Sci. Signal. 2, ra46. [DOI] [PubMed] [Google Scholar]
- 19.Nguyen V., Cao L., Lin J. T., Hung N., Ritz A., Yu K., Jianu R., Ulin S. P., Raphael B. J., Laidlaw D. H., Brossay L., Salomon A. R. (2009) Mol. Cell. Proteomics 8, 2418–2431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olsen J. V., Vermeulen M., Santamaria A., Kumar C., Miller M. L., Jensen L. J., Gnad F., Cox J., Jensen T. S., Nigg E. A., Brunak S., Mann M. (2010) Sci. Signal. 3, ra3. [DOI] [PubMed] [Google Scholar]
- 21.Fu G., Vallée S., Rybakin V., McGuire M. V., Ampudia J., Brockmeyer C., Salek M., Fallen P. R., Hoerter J. A., Munshi A., Huang Y. H., Hu J., Fox H. S., Sauer K., Acuto O., Gascoigne N. R. (2009) Nat. Immunol. 10, 848–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson A. L., Aravind L., Shulzhenko N., Morgun A., Choi S. Y., Crockford T. L., Lambe T., Domaschenz H., Kucharska E. M., Zheng L., Vinuesa C. G., Lenardo M. J., Goodnow C. C., Cornall R. J., Schwartz R. H. (2009) Nat. Immunol. 10, 831–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kakugawa K., Yasuda T., Miura I., Kobayashi A., Fukiage H., Satoh R., Matsuda M., Koseki H., Wakana S., Kawamoto H., Yoshida H. (2009) Mol. Cell. Biol. 29, 5128–5135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lesourne R., Uehara S., Lee J., Song K. D., Li L., Pinkhasov J., Zhang Y., Weng N. P., Wildt K. F., Wang L., Bosselut R., Love P. E. (2009) Nat. Immunol. 10, 840–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patrick M. S., Oda H., Hayakawa K., Sato Y., Eshima K., Kirikae T., Iemura S., Shirai M., Abe T., Natsume T., Sasazuki T., Suzuki H. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 16345–16350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elias J. E., Gibbons F. D., King O. D., Roth F. P., Gygi S. P. (2004) Nat. Biotechnol. 22, 214–219 [DOI] [PubMed] [Google Scholar]
- 27.Elias J. E., Haas W., Faherty B. K., Gygi S. P. (2005) Nat. Methods 2, 667–675 [DOI] [PubMed] [Google Scholar]
- 28.Nika K., Soldani C., Salek M., Paster W., Gray A., Etzensperger R., Fugger L., Polzella P., Cerundolo V., Dushek O., Höfer T., Viola A., Acuto O. (2010) Immunity 32, 766–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blagoev B., Ong S. E., Kratchmarova I., Mann M. (2004) Nat. Biotechnol. 22, 1139–1145 [DOI] [PubMed] [Google Scholar]
- 30.Krüger M., Kratchmarova I., Blagoev B., Tseng Y. H., Kahn C. R., Mann M. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 2451–2456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feng L., Huang J., Chen J. (2009) Genes Dev. 23, 719–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shao G., Patterson-Fortin J., Messick T. E., Feng D., Shanbhag N., Wang Y., Greenberg R. A. (2009) Genes Dev. 23, 740–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brill L. M., Salomon A. R., Ficarro S. B., Mukherji M., Stettler-Gill M., Peters E. C. (2004) Anal. Chem. 76, 2763–2772 [DOI] [PubMed] [Google Scholar]
- 34.Fusaki N., Iwamatsu A., Iwashima M., Fujisawa J. (1997) J. Biol. Chem. 272, 6214–6219 [DOI] [PubMed] [Google Scholar]
- 35.Rush J., Moritz A., Lee K. A., Guo A., Goss V. L., Spek E. J., Zhang H., Zha X. M., Polakiewicz R. D., Comb M. J. (2005) Nat. Biotechnol. 23, 94–101 [DOI] [PubMed] [Google Scholar]
- 36.Dustin M. L. (2009) Immunity 30, 482–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yablonski D., Kuhne M. R., Kadlecek T., Weiss A. (1998) Science 281, 413–416 [DOI] [PubMed] [Google Scholar]
- 38.Yoon H. Y., Lee J. S., Randazzo P. A. (2008) Traffic 9, 2236–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu H., Purbhoo M. A., Davis D. M., Rudd C. E. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 10166–10171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Houtman J. C., Houghtling R. A., Barda-Saad M., Toda Y., Samelson L. E. (2005) J. Immunol. 175, 2449–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Finco T. S., Kadlecek T., Zhang W., Samelson L. E., Weiss A. (1998) Immunity 9, 617–626 [DOI] [PubMed] [Google Scholar]
- 42.Giorgi C., Moore M. J. (2007) Semin. Cell Dev. Biol. 18, 186–193 [DOI] [PubMed] [Google Scholar]
- 43.Hurley J. H., Emr S. D. (2006) Annu. Rev. Biophys. Biomol. Struct. 35, 277–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Naarmann I. S., Harnisch C., Flach N., Kremmer E., Kühn H., Ostareck D. H., Ostareck-Lederer A. (2008) J. Biol. Chem. 283, 18461–18472 [DOI] [PubMed] [Google Scholar]
- 45.Ostareck D. H., Ostareck-Lederer A., Wilm M., Thiele B. J., Mann M., Hentze M. W. (1997) Cell 89, 597–606 [DOI] [PubMed] [Google Scholar]
- 46.Busà R., Geremia R., Sette C. (2010) Nucleic Acids Res 38, 3005–3018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lukong K. E., Richard S. (2003) Biochim Biophys Acta 1653, 73–86 [DOI] [PubMed] [Google Scholar]
- 48.Paronetto M. P., Cappellari M., Busà R., Pedrotti S., Vitali R., Comstock C., Hyslop T., Knudsen K. E., Sette C. (2010) Cancer Res. 70, 229–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ostareck-Lederer A., Ostareck D. H., Cans C., Neubauer G., Bomsztyk K., Superti-Furga G., Hentze M. W. (2002) Mol. Cell. Biol. 22, 4535–4543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sanui T., Inayoshi A., Noda M., Iwata E., Stein J. V., Sasazuki T., Fukui Y. (2003) Blood 102, 2948–2950 [DOI] [PubMed] [Google Scholar]
- 51.Yokoyama N., deBakker C. D., Zappacosta F., Huddleston M. J., Annan R. S., Ravichandran K. S., Miller W. T. (2005) Biochemistry 44, 8841–8849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brugnera E., Haney L., Grimsley C., Lu M., Walk S. F., Tosello-Trampont A. C., Macara I. G., Madhani H., Fink G. R., Ravichandran K. S. (2002) Nat. Cell Biol 4, 574–582 [DOI] [PubMed] [Google Scholar]
- 53.Côté J. F., Vuori K. (2007) Trends Cell Biol. 17, 383–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fukui Y., Hashimoto O., Sanui T., Oono T., Koga H., Abe M., Inayoshi A., Noda M., Oike M., Shirai T., Sasazuki T. (2001) Nature 412, 826–831 [DOI] [PubMed] [Google Scholar]
- 55.Ku G. M., Yablonski D., Manser E., Lim L., Weiss A. (2001) EMBO J. 20, 457–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Phee H., Abraham R. T., Weiss A. (2005) Nat. Immunol. 6, 608–617 [DOI] [PubMed] [Google Scholar]
- 57.van Bergen En Henegouwen P. M. (2009) Cell Commun. Signal. 7, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Komada M., Kitamura N. (2005) J. Biochem. 137, 1–8 [DOI] [PubMed] [Google Scholar]
- 59.Yamada M., Ishii N., Asao H., Murata K., Kanazawa C., Sasaki H., Sugamura K. (2002) Mol. Cell. Biol. 22, 8648–8658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kerscher O., Felberbaum R., Hochstrasser M. (2006) Annu. Rev. Cell Dev. Biol. 22, 159–180 [DOI] [PubMed] [Google Scholar]
- 61.Lucas J. A., Felices M., Evans J. W., Berg L. J. (2007) J. Immunol. 179, 7561–7567 [DOI] [PubMed] [Google Scholar]
- 62.Schaeffer E. M., Broussard C., Debnath J., Anderson S., McVicar D. W., Schwartzberg P. L. (2000) J. Exp. Med. 192, 987–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Turner M., Mee P. J., Walters A. E., Quinn M. E., Mellor A. L., Zamoyska R., Tybulewicz V. L. (1997) Immunity 7, 451–460 [DOI] [PubMed] [Google Scholar]
- 64.Singer A., Bosselut R., Harvey Cantor L. G. a. F. W. A. (2004) Adv. Immunol. 83, 91–131 [DOI] [PubMed] [Google Scholar]
- 65.Priatel J. J., Teh S. J., Dower N. A., Stone J. C., Teh H. S. (2002) Immunity 17, 617–627 [DOI] [PubMed] [Google Scholar]
- 66.Adachi S., Iwata M. (2002) Cell. Immunol. 215, 45–53 [DOI] [PubMed] [Google Scholar]
- 67.Kaye J. (2000) Immunol. Res. 21, 71–81 [DOI] [PubMed] [Google Scholar]
- 68.Wilkinson B., Kaye J. (2001) Cell. Immunol. 211, 86–95 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data