Quantitative Epigenetics Through Epigenomic Perturbation of Isogenic Lines (original) (raw)
Abstract
Interindividual differences in chromatin states at a locus (epialleles) can result in gene expression changes that are sometimes transmitted across generations. In this way, they can contribute to heritable phenotypic variation in natural and experimental populations independent of DNA sequence. Recent molecular evidence shows that epialleles often display high levels of transgenerational instability. This property gives rise to a dynamic dimension in phenotypic inheritance. To be able to incorporate these non-Mendelian features into quantitative genetic models, it is necessary to study the induction and the transgenerational behavior of epialleles in controlled settings. Here we outline a general experimental approach for achieving this using crosses of epigenomically perturbed isogenic lines in mammalian and plant species. We develop a theoretical description of such crosses and model the relationship between epiallelic instability, recombination, parent-of-origin effects, as well as transgressive segregation and their joint impact on phenotypic variation across generations. In the limiting case of fully stable epialleles our approach reduces to the classical theory of experimental line crosses and thus illustrates a fundamental continuity between genetic and epigenetic inheritance. We consider data from a panel of Arabidopsis epigenetic recombinant inbred lines and explore estimates of the number of quantitative trait loci for plant height that resulted from a manipulation of DNA methylation levels in one of the two isogenic founder strains.
SYSTEMATIC or stochastic changes in chromatin states, such as gains or losses of DNA or histone methylation, are sometimes transmitted across generations with significant phenotypic effects (Richards 2006). Since different chromatin variants (epialleles) can exist in the same sequence background (i.e., on the same sequence allele), they can produce a dimension of functional variation at the population level that cannot be captured by an analysis based on DNA sequence alone (Johannes et al. 2008). How much of this epigenetic variation is routinely missed in linkage or association mapping studies is an open question (Johannes et al. 2008; Maher 2008; Manolio et al. 2009; Eichler et al. 2010), but preliminary estimates in plants suggest that it can account for up to 30% of the variation in commonly studied phenotypes such as height and flowering time (Johannes et al. 2009).
Unlike DNA sequence alleles, epialleles can exhibit a high degree of instability across generations (Rakyan et al. 2002; Mathieu et al. 2007). Because of these dynamic properties the quantitative implications of epigenetic inheritance in the context of human health, evolution, and agriculture have remained largely speculative (Johannes et al. 2008; Richards 2008; Bossdorf et al. 2008; Petronis 2010; Biemont 2010). To overcome this limitation, it is necessary to obtain a basic inventory of the transgenerational behavior of epialleles in both mammals and plants and to formally incorporate these properties into our current models of quantitative inheritance in natural and experimental populations (Johannes et al. 2008). The aim of this article is to outline both experiment and theory to achieve this.
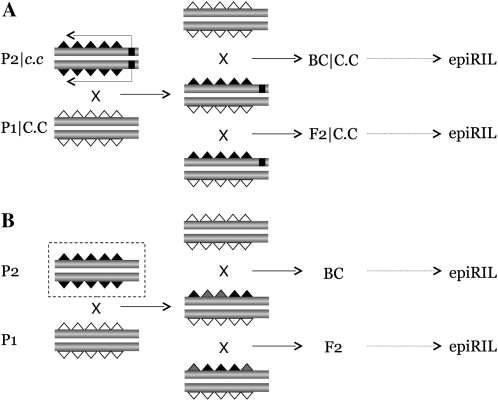
A powerful experimental approach for studying the induction and propagation of epigenetic variation is through crosses of epigenomically perturbed isogenic strains. In the model plant Arabidopsis two groups have recently implemented such an approach by constructing so-called epigenetic recombinant inbred lines (epiRILs) (Johannes et al. 2009; Reinders et al. 2009). These populations were derived from crosses between individuals with virtually identical DNA sequences but drastically divergent epigenomic profiles. In both cases, the cross was initiated from a wild-type (wt) plant and a plant carrying a single loss-of-function mutation in ddm1 (Johannes et al. 2009) or met1 (Reinders et al. 2009), two genes involved in DNA methylation control. As a result, mutant plants exhibit significant global changes in DNA methylation (Vongs et al. 1993; Cokus et al. 2008; Lister et al. 2008; Reinders et al. 2009). Although mobilization of transposable elements also occurs in these epiRILs, they nonetheless provide a unique opportunity to study the transgenerational behavior of induced epigenetic variation against a nearly invariant DNA sequence background (Figure 1A). The experimental setup for constructing such populations represents a general strategy. Similar approaches could be considered in mammals and/or through the use of environmental triggers to initiate the epigenomic changes in the parental generation (Figure 1B).
Figure 1.—
Construction of epigenetic recombinant inbred lines (epiRIL): (A) Induction of epigenomic perturbation by means of a mutation in genes involved in chromatin control, followed by selfing (plants) or sibling mating (mammals) of conditional intercross (_F_2 | C.C) or backcross (BC | C.C) progeny. It is assumed that the parental strains have nearly identical DNA sequences (shaded horizontal bars) but different epigenomic profiles (white, gray, and black triangles). The exceptions are rare de novo sequence changes (not shown for simplicity) resulting from compromised chromatin states in the mutant parent (_P_2 | c.c). (B) Same crossing scheme as above but using environmental manipulations in place of mutations to invoke the initial perturbation.
Molecular profiling of the two Arabidopsis epiRIL populations has shown that only a fraction of induced epialleles remain stable in subsequent generations, the rest being subject to dynamic modifications (Johannes et al. 2009; Reinders et al. 2009). Two basic patterns are beginning to emerge. The first pattern indicates that a subset of epialleles undergo rapid and stochastic fluctuations over a wide spectrum of chromatin states, many of which are outside of the parental range (Reinders et al. 2009). While such alterations can be causative of phenotypes within a given generation, they probably do not contribute to phenotypic inheritance (Slatkin 2009) and can therefore be regarded as noise in the underlying heritable substrate. The second, and more important pattern, is a systematic and gradual reversion of mutant epiallelic states to those of the wt over the course of several generations. This process represents an intrinsic rescue system that is invoked to restore proper genome function and integrity. Loci that meet this pattern tend to correspond to sequences that are continuously targeted by the RNA-directed DNA methylation (RdDM) machinery (Johannes et al. 2009; Teixeira et al. 2009; Teixera and Colot 2010).
Epiallelic instabilities, as described above, create a complex source of heritable variation. These properties pose challenges to the way we have to approach quantitative inheritance in the epiRIL or similar populations. The key task is to simultaneously account for two processes: The first involves the meiotic transmission of maternal and paternal DNA sequence haplotypes according to Mendelian laws. The second is a dynamic process that governs continuous changes in the chromatin states (epialleles) harbored by these haplotypes and leads to non-Mendelian patterns of inheritance. Here we develop the necessary theoretical foundation to quantify these processes using epiRILs (Figure 1A) as a model system. We find that epiallelic reversion, recombination, parent-of-origin effects, and transgressive segregation are key parameters in these populations: Their joint effects can produce complex and highly dynamic inheritance patterns that cannot be predicted from strictly Mendelian models. In the limiting case of fully stable epialleles our model reduces to the classical theory of experimental line crosses and thus illustrates a fundamental continuity between genetic and epigenetic inheritance. In what follows we present the first comprehensive attempt to quantify epigenetic inheritance in model organisms.
THEORY
Conceptual basis:
Consider a locus, L, extensively involved in genome-wide chromatin control. Genotype C.C at this locus corresponds to proper chromatin maintenance, whereas the mutant genotype c.c induces global chromatin changes (e.g., modifications of DNA or histone methylation). We start with two inbred parents, a wild-type parent, _P_1 | C.C, and a mutant parent, _P_2 | c.c. By design, the two parents have identical DNA sequences (except at locus L and inevitably at a small number of other loci; Mirouze et al. 2009; Tsukahara et al. 2009), but drastically divergent chromatin profiles (Figure 1A).
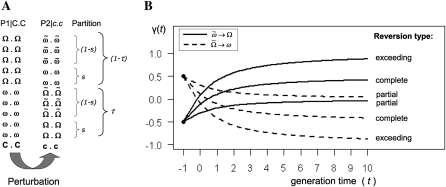
As a result of the epigenomic perturbation induced by the mutation, the two parents will differ in their chromatin states at N loci determining a quantitative trait y. Suppose that in the _P_1 | C.C parent N(1 − τ) of these loci have stable epigenotype Ω.Ω and _N_τ of the loci have stable epigenotype ω.ω (Figure 2A). Here epiallele Ω corresponds to a phenotypically increasing and ω to a decreasing chromatin state. Relative to _P_1 | C.C, the mutant parent, _P_2 | c.c, will have undergone the following possible epiallelic changes at the N loci: Ω → ω, Ω→ω˜, ω → Ω, and ω→Ω˜, where the tilde (∼) signifies an unstable epiallelic state (Figure 2A).
Figure 2.—
Epigenomic structure of the parental strains and epiallelic reversion: (A) As a result of the perturbation the wt (_P_1|C.C) and the mutant (_P_2|c.c) parents will differ in their diploid chromatin states (epigenotypes) at N loci. The mutant (_P_2|c.c) parent will have a proportion τ of phenotypically increasing epigenotypes (Ω.Ω and Ω˜.Ω˜) as well as a proportion (1 − τ) of decreasing epigenotypes (ω.ω and ω˜.ω˜), with s and (1 − s) of these being either stable or unstable, respectively (see text). (B) Unstable epialleles (Ω˜,ω˜) induced in the mutant parent have the capacity to revert to wt states over time according to some function, γ(t). This reversion can be perfect or imperfect. We quantify epialleles in the parental generation by setting Ω=Ω˜=12 and ω=ω˜=−12. Starting at t = 0 (the _F_2|C.C or BC|C.C generation) the state values of reversible epialleles begin to change.
We suppose that a proportion s of the newly induced epialleles remain stable in subsequent generations (Ω and ω) (Figure 2A). This is even the case when proper chromatin maintenance function is restored. Since stable epialleles behave like DNA sequence changes at the population level, we make no formal distinction between them. Only direct molecular profiling and sequencing of the two parents and their cross-derivatives will make it possible to uncover the physical basis of such stable induced alterations.
Apart from stable epialleles, we also assume a proportion (1 − s) of newly induced epialleles (Ω˜ and ω˜) with the capacity to revert to approximate wild-type states over generations. We quantify this physical process through a function γ(t), which describes the progressive changes of epiallelic states in continuous time (Figure 2B). Specifically, Ω˜ and ω˜ correspond to sequences that are targeted by the RdDM machinery (Johannes et al. 2009; Reinders et al. 2009; Teixeira et al. 2009; Teixera and Colot 2010) or possibly other correction mechanism. For the cross design shown in Figure 1A, this reversion begins after the C.C genotype has been reintroduced at L, that is, in the conditional F2 (_F_2|C.C) or backcross (BC|C.C) populations (Figure 2, A and B). Upon further propagation of individual lines from these conditional populations by means of selfing (plants) or sibling mating (mammals), progressive epiallelic reversion continues through recurrent maintenance action at each generation. Including the initial perturbation, the different epiallelic fates outlined above can be summarized schematically as follows:
Ω↗↘ω˜→reversion γ(t)Ωω→stable inheritanceω;ω↗↘Ω˜→reversion γ(t)ωΩ→stable inheritanceΩ.
Our goal is to model this process for any generation of inbreeding. This allows us to draw direct connections between the basic properties of epialleles and their impact on heritable variation at the population level.
Transgenerational epigenetic dynamics:
We quantify the epigenotype at locus j at time t using the coding introduced in Table 1.
TABLE 1.
Coding of epigenotypes at a locus
| Value | |||
|---|---|---|---|
| Epigenotype | Additivity | Mutant dominance | wt dominance |
| Ω.Ω | +1 | +1 | +1 |
| ω.ω | −1 | −1 | −1 |
| Ω.ω | 0 | +1 (if Ω is the mutant) | −1 (if Ω is the mutant) |
| −1 (if ω is the mutant) | +1 (if ω is the mutant) | ||
| Ω˜.Ω˜ | 2γ(t) | 2γ(t) | 2γ(t) |
| ω˜.ω˜ | −2γ(t) | −2γ(t) | −2γ(t) |
| Ω˜.ω | 1/2 + γ(t) | 2γ(t) | −1 |
| Ω.ω˜ | 1/2 − γ(t) | −2γ(t) | +1 |
Parental generation:
Since all individuals are assumed homozygous in the parental generation, the total phenotypic variance can be expressed as
| σP2(y)=σε2+[Nδ(2τ−1)]2, | (1) |
|---|
where σε2 is the pooled within-line (environmental) variance, and δ is henceforth defined as the average contribution of a single locus to the between-parental phenotypic mean difference D (Serebrovsky 1928):
This expression assumes that δ is equivalent over all N causative loci; that is, δ1 = δ2 = … = δ_N_ = δ.
Note in Equation 1 that when the transgression parameter τ = 0.5 we have that σP2(y)=σε2. In this limiting case the total phenotypic variance in the parental generation is purely environmental, despite drastic functional divergence between the parents. This situation provides the condition for a maximum gain in transgressive variance in subsequent generations. It follows from the fact that each parent is fixed for both phenotypically decreasing and increasing states so that recombination events can produce offspring with more extreme epigenotypes (Riesenberg et al. 1999). We therefore refer to parameter τ as a measure of “transgression potential” in the parental generation, which can become “realized,” in some sense, in subsequent generations. This phenomenon is discussed in detail below.
F1 and base-population:
Crossing _P_1|C.C × _P_2|c.c yields the _F_1 generation. We assume that epiallelic states induced in the _P_2|c.c parent remain stable (Teixeira et al. 2009), but this assumption can be easily relaxed if necessary. As a consequence, the phenotypic variance may or may not (Richards 2009) be equivalent to the environmental variance. As shown in Figure 1A the _F_1|C.c are used to derive the base population for advanced inbreeding generations either through a backcross (_F_1|C.c × _P_1|C.C) or through an intercross (_F_1|C.c × _F_1|C.c). From these crosses, only the C.C progeny (BC|C.C or _F_2|C.C) are selected to initiate the inbreeding process through selfing or sibling mating. This permits a detailed study of the time-dependent behavior of parental epialleles independent of the recurrent action of the c.c genotype. For simplicity we ignore the introgression of wt epigenotypes surrounding locus L as a result of this selection procedure.
Advanced inbreeding generations:
At any time point t of inbreeding, the variance in trait y is the sum of an epigenetic component, σ2(η,t), and an environmental (error) component, σ2(ε):
| σ2(y,t)=σ2(η,t)+σ2(ε). | (3) |
|---|
We assume that epigenotypes are uncorrelated with the error term ε, and that the error terms are uncorrelated across generation times. With equal effect sizes across the N causative loci, the epigenetic variance can be approximated as
| σ2(η,t)≃δ2N{σ2(ηj|t)+(N−1)σ(ηj,ηk|t,r−)}, | (4) | | ------------------------------------------ | --- |
where σ2(η_j_ | t) is the variance at a single locus j at time t and σ(ηj,ηk|t,r−) is the covariance between any two loci j and k separated by an average pairwise recombination fraction r− (Franklin 1970). It can be shown (see Appendix A) that Equation 4 has the explicit form
| σ2(η,t)≃Nδ24{q1[4γ(γ+1)(s−1)−3s−1]+(N−1)q2[1+s+2γ(1−s)]2(1−2τ)2}, | (5) |
|---|
where we put γ = γ(t) to lighten the notation. The parameters _q_1 and _q_2 depend on the mode of action (additivity or dominance), the type of inbreeding scheme (selfing or sibling mating), and the base population (backcross or F2-intercross) used to initiate the inbreeding processes. Table 2 provides a summary of the specific form of _q_1 and _q_2 in each of these cases.
TABLE 2.
Parameter values _q_1 and _q_2
| Base | Effect | Parameter values |
|---|---|---|
| Selfing | ||
| F2 | Add. | q1=12t+1−1 |
| q2=2r−−12r−+1((1−2r−)t+12t+1−1) | ||
| F2 | Dom. | q1=1−22+2t22t+2 |
| q2=12t+1((1+2r−(r−−1))t(1−2r−2+4r−3)1+2r−−(1−2r−)t+21+2r−)+1−2r−1+2r−−12t+2 | ||
| BC | Add. | q1=12t+1−34 |
| q2=2r−−12r−+1((1−2r−)t(1−r−)2t+1−34) | ||
| BC | Dom. | q1=1+2t+1ξ−3×22t22t+2,ξ=1→ wt dominant,ξ=−1→ mutant dominant |
| q2=−12t+1((1−4r−2)ξ2r−+1+(r−−1)[(1+2r−(r−−1))t−(1−2r−)t+11+2r−])−342r−−12r−+1−12t+2,ξ=1→wt dominant,ξ=−1→mutant dominant | ||
| Sib.mat | ||
| F2 | Add. | q1=−1−14(1−54)t(35−1)+14(1+54)t(35+1) |
| F2 | Dom. | q1=15((−1)t+121+2t−123+4t[(35−7)(1−5)2t−(35+7)(1+5)2t]−5) |
| BC | Add. | q1=−34−14(1−54)t(25−1)+14(1+54)t(25+1) |
| BC | Dom. | q1=15((−1)t23+2t+122+2t[ξ(5−25)(1−5)t+ξ(5+25)(1+5)t+122+2t((9−45)(1−5)2t+(9+45)(1+5)2t)]−154),ξ=1→wt dominant,ξ=−1→mutant dominant |
An important observation is that dominance (complete dominance in our case) appears as a time-dependent phenomenon, owing not only to the progressive depletion of heterozygote epigenotypes, but also to the reversion of mutant epialleles to wt states. It is therefore necessary to distinguish two types of dominance effects, one being attributable to epialleles inherited from the mutant parent (_P_2|c.c) and the other being due to epialleles deriving from the wt parent (_P_2|C.C) (see Tables 1 and 2). As we show, this distinction has an effect on the epigenetic variation when inbreeding is carried forward from a backcross base population, as a result of the initial asymmetry of the epigenotype frequencies. In the case of a F2 base population, on the other hand, the epigenetic variance component is equivalent under the two dominance scenarios (what differs is the phenotypic mean of the population).
Estimation of the number of induced quantitative trait loci (QTL):
As an extension of previous biometrical approaches (Castle 1921; Serebrovsky 1928; Lande 1981; Zeng 1992), Equation 5 can be used directly to obtain conservative estimates of the number of QTL (N) resulting from the initial epigenomic perturbation in the parental generation. To achieve this, we substitute δ from Equation 2 into the equation for the epigenetic variance (Equation 5), and solve for N at any generation t of inbreeding to obtain
| N=D2{q1[1−4γ(1+γ)(s−1)+3s]+q2[1−2γ(s−1)+s]2(1−2τ)2}(1−2τ)2{D2q2[1−2γ(s−1)+s]2−16σ2(η,t)}. | (6) |
|---|
It is perhaps interesting to note that if we consider the restrictive case of a F2 base population with additivity, s = 1 (fully stable epialleles), τ = 0 (no transgression), r−=0.5 (linkage equilibrium among N loci), and t = 0, Equation 6 reduces to the well-known Castle–Wright estimator (Castle 1921).
The most important result of this manuscript is Equation 5. It formalizes the relationship between epiallelic reversion (via γ(t) and s), recombination (via r−), transgression (via τ) and parent-of-origin effects (by keeping track of epiallelic origins). This relationship jointly determines the epigenetic variation in the population during inbreeding. In the following section we examine more closely the complex and highly dynamic patterns of heritable variation that can arise from it.
RESULTS
Classical theory of experimental line crosses typically assumes no transgression (τ = 0) and the transmission of fully stable parental alleles (s = 1). Sometimes, it is further assumed that all loci are in linkage equilibrium (r−=0.5). These constraints represent special cases of the theory developed here. We treat these scenarios as a reference against which to compare the rich spectrum of epigenetic inheritance in epigenomically perturbed line crosses. The phenotypic variance at any time point is simultaneously determined by all of the parameters specified in Equation 5. For clarity we assess their influences systematically by varying them one at the time. Several key population-level phenomena are considered for the case of selfing starting from a backcross base population. The case of selfing from a F2 base population can be found in the supporting information. Throughout, we fix the average recombination rate (r−) at 0.44, a value that is based on the Arabidopsis genetic map (Lynch and Walsh 1998).
Inheritance of unstable alleles:
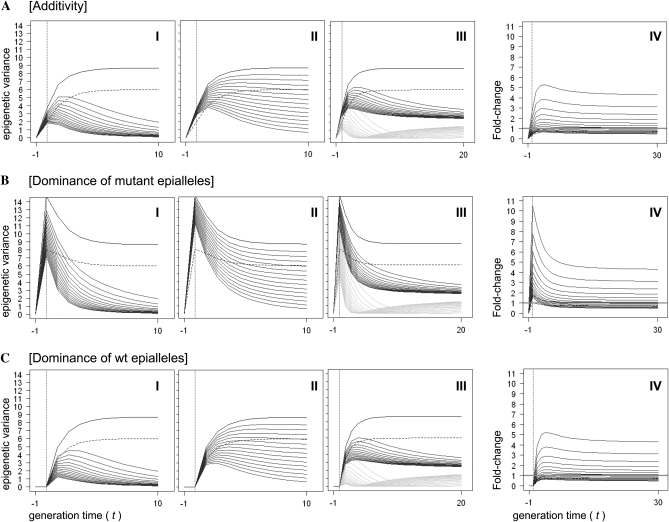
We first consider the case of complete epiallelic instability (s = 0) and no transgression (τ = 0). In this case all induced mutant epialleles are effectively reverted to the wt state over time. The specific form of the reversion function that governs this process is currently unknown, but should be of substantial interest in future empirical studies (Johannes et al. 2008). Although it is reasonable to assume that reversion is locus specific, for the purpose of providing an average description of the system it suffices to consider an average (between-locus) reversion function, γ(t). We tentatively posit the form γ(t) = 1/2−2/πarctan(β(t+1)), where β is the rate parameter (Figure 2B). We vary β so that we can examine the full range from slow to fast reversion. In contrast to stable Mendelian inheritance (Figure 3, A–C (I), black solid and dashed line), the reversion function has the effect of eroding heritable variation over time so that as t →∞ the heritable variation in the population is progressively lost (Figure 3, A–C (I), dark gray solid lines).
Figure 3.—
Transgenerational dynamics of epigenetic variation. We show the dynamic behavior of epigenetic variation in the case of additivity (A), complete dominance of mutant epialleles (B), and complete dominance of wt epialleles (C). Throughout we show the Mendelian inheritance case with linkage (r− = 0.44, s = 1 and τ = 0, solid black line) and with no linkage (r− = 0.5, s = 1 and τ = 0, dashed black line). For each mode of action we consider four key phenomena: inheritance of unstable epialleles (A–C, I): s = 0, τ = 0, and variable reversion function with rate parameter β evaluated at equal increments over the range 0.1 (top line) to 0.5 (bottom line); _x_-axis plots generation time and _y_-axis gives the epigenetic variance, σ2(η, t). Mixed inheritance of stable and unstable epialleles (A–C, II): τ = 0, fixed reversion function with β = 0.2, and variable s evaluated at equal increments over the range 0 (bottom line) to 0.9 (top line); _x_-axis and _y_-axis defined as above. Effects of imperfect epigenomic resetting (A–C, III): Parameter settings were chosen as in (A–C, I) but reversion was assumed to be imperfect with transitions to epiallelic states that are twice the initial wt state (dark gray solid lines) or intermediate between wt and mutant (light gray solid lines); _x_-axis and _y_-axis defined as above. Realized transgression potentials (A–C, IV): s = 0.5, fixed reversion function with β = 0.2, and variable τ evaluated in equal increments over the range 0 (bottom line) to 0.35 (top line); the _x_-axis plots generation time and the _y_-axis gives the fold change in epigenetic variance relative to the between-parental variance, σ2(η,t)[σP2(y)−σε2]−1.
Mixed inheritance of stable and unstable epialleles:
Complete instability of epialleles is an extreme case. It is more likely that a proportion of the induced epialleles remain stable and produce Mendelian inheritance patterns (Johannes et al. 2009; Reinders et al. 2009). Indeed, preliminary molecular profiling of the _ddm_1-derived epiRILs suggests that only up to one-half of the tested epialleles are reversible with the remaining loci being stable for at least eight generations of inbreeding (Johannes et al. 2009; Teixeira et al. 2009). Moreover, gross perturbations of the epigenome can lead to de novo sequence variation through the insertion of remobilized transposable elements or other structural abnormalities. These induced alterations in the parental generation also contribute to the fraction of stable segregating variation. The precise proportions depend on the particular perturbation, organism, and experimental setup and needs to be determined on a case-by-case basis.
We explore the effect of different proportions assuming a fixed reversion function and no transgression, τ = 0. Figure 3, A–C (II), illustrates the effect of this type of mixed inheritance with the phenotypic variance being decreased over generation time due to reversion but converging to a stable value as t → ∞. The final variance represents the stable heritable substrate that can be gleaned from the initial perturbation. It should be clear that as the proportion of stable epialleles in the genome approaches unity (s → 1), our model converges to the familiar case of Mendelian inheritance, which is the basis of the classical theory of experimental line crosses.
Effects of imperfect epigenomic resetting:
The reversion of mutant epialleles to the wt state can be an imperfect process (Figure 2B). It is possible that epialleles converge to values outside of the parental range (Reinders et al. 2009). For example, the remethylation of _hypo_methylated mutant epialleles could produce _hyper_methylated states in subsequent generations relative to the wt (Reinders et al. 2009). We consider this case by letting all unstable epialleles revert to state values that are twice that of the wt parent. This implies that the reversion of mutant states must first pass through wt states before they reach their final stable state. This process leaves an interesting signature at the population level: It leads to an initial depletion of epigenetic variance, before there is an unexpected gain in heritable variance at later generations (Figure 3, A–C (III) light gray solid lines).
A different pattern occurs in situations where epiallelic reversion converges to intermediate parental values (0.5 of wt values). In this case, heritable variation is never completely lost (Figure 3, A–C (III), dark gray solid lines). Note that this latter pattern mimics the situation observed for the case of mixed inheritance (previous section). Distinguishing these two possibilities empirically therefore depends on detailed knowledge of the average reversion function, γ(t), and the proportion of stable epialleles, s, operating in a given population.
Realized transgression potentials:
It has been shown that transgressive segregation is widespread in experimental line crosses (Riesenberg et al. 1999). A major reason for this is that the parents have often not undergone divergent selection prior to crossing and are therefore each fixed for both increasing and decreasing genotypes. In QTL experiments this is reflected in the detection of QTL with opposite signs relative to the parental means.
In perturbation-derived populations as the ones described here, transgressive segregation is likely to be a key aspect of quantitative inheritance. The removal of methylation, for instance, can lead to active or inactive chromatin states in the mutant parent, which can translate into increasing or decreasing phenotypic values, depending on whether the underlying sequences are involved in inhibitory or facilitating functions in the networks that connect (epi)genotype with phenotype (Figure 2A).
We explore the effect of different transgression potentials (τ) for a fixed reversion function and s = 0.5. We vary τ between 0 and 0.35 (τ = 0.5 being the maximum). The effect for large τ is dramatic. Under additivity, and assuming τ = 0.35, we find that heritable variation increases in the order of fivefold relative to the between parental variance (Figure 3A, IV). This effect is further exaggerated when mutant epialleles act dominantly. In this case we observe a nearly 11-fold increase in heritable variation at early generations (Figure 3B, IV) followed by a gradual decrease due to the progressive loss of hererozygote epigenotypes.
Our treatment of transgression represents the first theoretical approach to highlight the importance of transgressive segregation in generating phenotypic innovation in experimental line crosses.
Application:
A recent analysis of the _ddm_1-derived Arabidopsis epiRILs reported large heritable variation for plant height. This work has shown that perturbations of plant methylomes are sufficient to induce lasting phenotypic consequences in commonly studied complex traits. An important question concerns the specific features of the heritable architecture that has been set up in the epiRILs, such as its physical basis (e.g., sequence vs. methylation based) or the number and sizes of induced QTL. Definitive answers to this question require genome-wide epigenetic profiling techniques (e.g., ChIP-chip, ChIP-seq, or BS-seq) in combination with QTL mapping methods as well as resequencing of each line (Johannes et al. 2009). While such efforts are underway, we here consider using Equation 6 as the basis for deriving the first estimates of the number of QTL (and their average effects sizes) underlying plant height in this population.
Since phenotypic data from only two time points is available (parental generation and generation eight of inbreeding), it is necessary to make informed assumptions about several of the unknown parameters specified in Equation 6. These assumptions rely heavily on previous molecular or phenotypic observations of this population (Johannes et al. 2009; Teixeira et al. 2009) and are explicitly listed below:
- σ2(η, t) = 11.2: estimated from a random-effects model.
- D = 9.92: difference in phenotypic means (in centimeters) between the _ddm_1 and wt parents.
- s = 0.5: based on an analysis of a random subset of loci.
- r−=0.44: average recombination fraction for the Arabidopsis genome (Lynch and Walsh 1998).
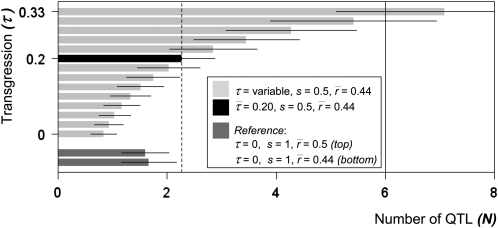
Values for the transgression potential, τ, remain elusive because this quantity cannot be directly measured using molecular techniques. Keeping this limitation in mind, we show various estimates for the number of QTL (N) for different values of τ (Figure 4). They range from as low as 1 (for τ = 0) to as high as 6 (for τ = 0.31). A unique estimate of N can be obtained by fixing τ at its theoretical average (see Appendix B). In this case we find that N−=2.26 (95% CI = ± 0.31) (Figure 4, black bar). This (conservative) estimate suggests the induction of a polygenic heritable architecture, with each QTL explaining ∼14% of the phenotypic variance in plant height. Given these considerably large effect sizes, the underlying causative loci should be mappable in future integrative QTL studies, even with relatively small sample sizes.
Figure 4.—
Estimates of the number of QTL. Plotted are estimates of number of QTL (N) against the transgression potential parameter τ. Parameters were set according to the shade coding shown in the figure. Confidence intervals (95%) were calculated using 1000 nonparametric bootstrap samples (Appendix B). The black solid vertical line marks the expected maximum detectable N in a backcross-derived RIL, which is approximately 6 (see Appendix B).
DISCUSSION
Epigenetic modifications, such as DNA methylation, are not only widely conserved across species (Zemach et al. 2010; Feng et al. 2010), but also show substantial interindividual variation within populations (Vaughn et al. 2007; Zhang et al. 2008; Kaminsky et al. 2009). The possibility that this type of epigenetic variation can influence phenotypes independent of DNA sequence variation poses major challenges to our current understanding of complex trait inheritance.
The first problem is that sequence-based mapping approaches (i.e., linkage or association mapping) may be insufficient to fully capture the heritable architecture of complex traits (Johannes et al. 2008). A recent study by Kong et al. (2009), for example, illustrates that knowledge of the epigenetic status of sequence alleles (in this case, the parent-of-origin of the alleles) is often necessary to establish significant associations with phenotypes. Although the authors found that these effects were sex specific, it raises the need to routinely include both levels of variation (DNA sequence or chromatin state) in the analysis.
The second and partially related problem is that the potential temporal instability of epigenetic variation can produce a level of phenotypic dynamics at the population-level that cannot be predicted from strictly Mendelian models of inheritance. Here we have outlined an experimental and theoretical approach to begin to address this issue. The approach relies on the use of a perturbation strategy to induce epigenetic variation in isogenic lines, followed by a transgenerational assessment of derivative populations. This provides an ideal platform for studying the temporal properties of epialleles and permits a theoretical description of these properties in connection with the inheritance of complex traits observed in such populations.
Extensions using environmental triggers:
Two experimental examples of this approach have recently been implemented in the model plant Arabidopsis using loss-of-function mutations in ddm1 and met1, two genes involved in genome-wide methylation maintenance, to initiate the perturbation. While this continues to be a valuable resource, future experiments should attempt to produce similar populations using environmental manipulations in place of mutations (Figure 1B). Bossdorf et al. (2010), for instance, demonstrated that treatment of Arabidopsis ecotypes with demethylation agents is sufficient to invoke phenotypically relevant methylation changes. Similarly, Verhoeven et al. (2010) used environmental stressors, such as chemical induction of herbivore and pathogen defenses, and noted heritable DNA methylation changes in asexual dandelion. These types of interventions may be sufficient to induce and propagate epigenetic variation in the parents and their cross derivatives. A demonstration of this should have important implications for evolutionary theory, which traditionally draws a clear divider between the environment and the heritable material. There is certainly need for modeling approaches that incorporate environmentally induced epigenetic changes into evolutionary theory. More general attempts to assess the role of epigenetic inheritance in the context of selection and adaptation have been undertaken (Csaba 1998; Pál and Miklós 1999; Bonduriansky and Day 2009). The inclusion of epiallelic reversion, as we have formalized it here, would be an appealing extension. It is tempting to speculate that such reversion processes have evolved to facilitate short-term adaptation of populations to rapid environmental changes. These issues are outside of the scope of this work.
Considerations for mammalian populations:
Studies of epigenetic inheritance have been primarily pursued in plant species, and much less is known about the transgenerational behavior of epialleles in mammalian populations. The dominant paradigm dictates that the epigenome is completely reset during early mammalian development (Reik 2007; Feng et al. 2010), which implies that induced epigenetic effects are not carried to subsequent generations. Formally, this suggests a reversion process that follows a step function, dropping to wt levels at t = −1 (first-generation progeny of two parents). However, several well-documented single-locus examples of epiallelic inheritance exist in mammalian systems (Rakyan et al. 2003; Blewitt et al. 2006; Daxinger and Whitelaw 2010). These findings are probably no exception as more recent genome-wide surveys of mouse gametes show clear instances of transmitted parental DNA methylation profiles (Borgel et al. 2010), suggesting that epigenetic inheritance may be much more widespread in mammalian populations than previously acknowledged.
By help of transgenerational phenotypic data, concrete hypotheses about the extent of epigenomic resetting can be tested on statistical grounds using the experimental framework outlined in this article. This can be achieved by considering alternative reversion functions in a fit to the data. Such a proposal is akin to the approach recently developed by Tal et al. (2010), which permits hypothesis tests about effect of epigenomic resetting events on the covariance between relatives.
We argue that the construction of mammalian crosses between isogenic parents with perturbed and unperturbed epigenomes will be critically important to begin to extrapolate results to humans. The theory outlined in this article, particularly the results for sibling mating, is entirely compatible with an analysis of experimental mammalian populations (e.g., mice or rats). However, the initial construction of such crosses using perturbations may be more complicated than in plants, given that mutations in genes controlling DNA methylation tend to be lethal (Li et al. 1992). Instead, one could consider other mutants, partial knockdowns, or any suitable environmental manipulation strategy. Another complication is to distinguish instances of maternal or paternal imprinting. One solution would be to set up reciprocal crosses to delineate these effects (i.e., perturbation of progenitor mother vs. father).
Conclusion:
Our theory attempts to connect recent observations of the dynamic properties of epialleles (i.e., DNA methylation variants) to a long tradition of quantitative genetics. We have shown that in the case of fully stable epialleles, genetic and epigenetic inheritance are formally indistinguishable. This illustrates that there is actually no dichotomy between these two modes of inheritance. Rather, they should be viewed as different points on a continuum that ranges from stable to unstable inheritance. We therefore hope that our work will help to bridge the gap between the fields of genetics and epigenetics.
APPENDIX A
This appendix shows the derivation of the epigenetic variance in the population, Equation 5. For the details on the values of the parameters presented, we refer the reader to the supporting information. The epigenetic variance can be written as
σ2(η,t(≃δ2N{σ2(ηj|t(+(N−1(σ(ηj,ηk|t,r−(},
where σ2(η_j_|t) is the variance at a single locus j at time t and σ(ηj,ηk|t,r−) is the covariance between any two loci j and k separated by an average pairwise recombination fraction r− (Franklin 1970).
Variance at a single locus:
The 12 different epigenotypes at locus j resulting from the initial cross (Figure 2A) can be classified into four different classes of three elements each. Denote these four classes by _s_1,…, _s_4 with corresponding probability weights _W_1,…, _W_4 (supporting material in File S1, 1.1). Since each locus can belong to any one of these classes, its variance can be written as the sum of the within-class variances weighted by their probabilities:
| σ2(ηj,t)=∑m=14Wmσ2(ηj|sm,t)=∑m=14Wm(∑i=14K(i)(t)ηsm(i)(t)2−(∑i=14K(i)(t)ηsm(i)(t))2), | (7) |
|---|
where K(t) is a vector of expected single-locus epigenotype probabilities and η(t) is a vector of expected epigenotypes at locus j at time t (supporting material in File S1, 1.2). The probability vector K(t) is obtained following a Markov chain approach. Below we specify the cases for selfing and sibling mating.
Selfing:
K(t=0) is a three-dimensional vector with the probabilities of each single-locus epigenotype, determined by the type of base population (F2 or BC) (supporting material in File S1, 1.3.1). The transition matrix Tˆ for each class is a 3 × 3 matrix of transition probabilities (supporting material in File S1, 1.3.2). Using a Markov chain approach we calculate the frequency of each epigenotype at time t using K(t)=K(0)Tˆt. Using Equation 7 the variance at a single locus is given by
| σ2(ηj,t)=14{q1[4γ(t)(γ(t)+1)(s−1)−3s−1]}. | (8) |
|---|
The value of the parameter _q_1 differs for the type of base population considered (F2 or BC) and for the type of epigenotypic effect considered (additivity or dominance); see Table A1.
TABLE A1.
| Base | Effect | _q_1 |
|---|---|---|
| F2 | Add. | 12t+1−1 |
| F2 | Dom. | 1−22+2t22t+2 |
| BC | Add. | 12t+1−34 |
| BC | Dom. | 1+2t+1ξ−3×22t22t+2, ξ = 1 → wt dominant, ξ = −1 → mutant dominant |
Sibling mating:
Consider, instead of the probabilities of each single-locus, the probabilities of their mating types (Bulmer 1985). There are 16 possible mating types in each class, which can be reduced to 6 considering the following basic symmetries (supporting material in File S1, 1.4.1): Ω.ω˜=ω˜.Ω, and Ω.ω˜ × Ω.Ω=Ω.Ω × Ω.ω˜. The transition matrix Tˆ is the collection of the probabilities of going from one mating type to another in one generation of sibling mating; it has dimension 6 × 6 (supporting material in File S1, 1.4.3) (for a detailed description on how to construct such a matrix, see Bulmer 1985, Chap. 3). The initial probabilities of each mating class, Q(0), is a 6-dimensional vector given by Q(l)(0) = K(i)(0)K(j)(0), where i and j are the single-locus epigenotypes involved in mating type l. At any generation t, Q(t)=Q(0)Tˆt. The probabilities for the 4 different single-locus epigenotypes are given by K(i)(t)=∑l=16pilQ(l)(t), where pil is the proportion of single-locus i involved in mating type l (supporting material in File S1, 1.4.2). Using Equation 7 the variance at a single locus is given by
| σ2(ηj,t)=14{q1[4γ(t)(γ(t)+1)(s−1)−3s−1]}. | (9) |
|---|
The value of the parameter _q_1 depends on the case considered (Table A2).
TABLE A2.
| Base | Effect | _q_1 |
|---|---|---|
| F2 | Add. | −1−14(1−54)t(35−1)+14(1+54)t(35+1) |
| F2 | Dom. | 15((−1)t+121+2t−123+4t[(35−7)(1−5)2t−(35+7)(1+5)2t]−5) |
| BC | Add. | −34−14(1−54)t(25−1)+14(1+54)t(25+1) |
| BC | Dom. | 15((−1)t23+2t−122+2t[ξ(5−25)(1−5)t+ξ(5+25)(1+5)t+122+2t((9−45)(1−5)2t+(9+45)(1+5)2t)]−154),ξ=−1→wt dominant,ξ=−1→mutant dominant |
Covariance between loci:
To calculate the covariance term between loci j and k separated by an average recombination factor r− we use a similar classification for the possible 160 different two-locus epigenotypes resulting from the initial cross (Figure 2A). They can be assigned to 16 different classes of 10 pairs each, _d_1,…, _d_16, with probability Vi(ηj, ηk) (supporting material in File S1, 2.1). Since each two-locus epigenotype can belong to any one of these 16 classes we can write the covariance as:
| σ(ηj,ηk|t,r−(≃∑m=116Vm(ηj,ηk(σ(ηj,ηk|dm,t,r−(=∑m=116Vm(ηj,ηk([∑i=110R(i)(t(ηdm,1(i)(t(ηdm,2(i)(t(−(∑i=110R(i)(t(ηdm,1(i)(t()(∑i=110R(i)(t(ηdm,2(i)(t()], | (10) | | ----------------------------------------------------------------------------------------------------------------------------------------------------------------- | ---- |
where R(t) is a 10-dimensional vector of two-locus epigenotype probabilities (i.e., parental haplotypes). Here, ηdm,1 and ηdm,2 are the two different expected epigenotypes in a given class dm (supporting material in File S1, 2.2). The probability vector R(t) is obtained following a Markov chain approach; below we specify the cases for selfing and sibling mating. In the simplified case of purely Mendelian inheritance (s = 1), no transgression (τ = 0), and t = ∞, the derivation of R(t∞) has received considerable attention as a problem in its own right (Haldane and Waddington 1931; Wright 1933; Kimura 1963; Broman 2005).
Selfing:
For each of the 16 classes R(t=0) is calculated depending on the type of base population considered (F2 or BC) (supporting material in File S1, 2.3.1). The transition probability matrix Tˆ is a 10 × 10 matrix of transition probabilities of each two-locus epigenotype crossed with itself (supporting material in File S1, 2.3.2), and R(t)=R(0)Tˆt. Using Equation 10 we obtain
| σ(ηj,ηk|t,r−)≃14{q2[1+s+2γ(1−s)]2(1−2τ)2}. | (11) |
|---|
The analytical value of the parameter _q_2 is shown in Table A3.
TABLE A3.
| Base | Effect | _q_2 |
|---|---|---|
| F2 | Add. | 2r−−12r−+1((1−2r−)t+12t+1−1) |
| F2 | Dom. | 12t+1((1+2r−(r−−1))t(1−2r−2+4r−3)1+2r−−(1−2r−)t+21+2r−)+1−2r−1+2r−−12t+2 |
| BC | Add. | 2r−−12r−+1((1−2r−)t(1−r−)2t+1−34) |
| BC | Dom. | −12t+1(ξ(1−4r−2)2r−+1+(r−−1)[(1+2r−(r−−1))t−(1−2r−)t+11+2r−])−342r−−12r−+1−12t+2,ξ=1→wt dominant,ξ=−1→mutant dominant |
Sibling mating:
Consider the 55 different mating types between the 10 two-locus epigenotypes in each class (supporting material in File S1, 2.4.1) (Bulmer 1985). Taking into account the symmetries mentioned above we can reduce them to 22 different mating types for the F2- generated population and to 34 for the BC one. In the same way as for the single-locus case, Q(0) are the initial probabilities of each mating class, Q(t)=Q(0)Tˆt, and the probabilities for the 10 two-locus epigenotypes, R(t), can be extracted from the mating type probabilities Q(t) (supporting material in File S1, 2.4.2).
Unfortunately, the transition matrix Tˆ cannot be diagonalized symbolically because its dimension is too large (see Figure S2). For this reason we cannot obtain an analytical expression (as a function of the parameter r−) for the probabilities R(t). However, we can fix r− to a numerical value before calculating the power Tˆt, and thus obtain the exact probability values at any time t. Moreover, it is possible to write symbolically the probability vector as R(t)={R(1)(t),R(2)(t),…,R(10)(t)} and, using Equation 10 and taking into account ∑i=110R(i)=1, we can write the covariance for sibling mating in the form
| σ(ηj,ηk|t,r−)≃14{q2[1+s+2γ(1−s)]2(1−2τ)2}, | (12) |
|---|
where the constant _q_2 is calculated exactly for any value of r− for a F2- or a BC-based population.
Epigenetic variance:
Finally, combining Equations 8 and 9 with Equations 11 and 12, and multiplying by the number of loci and their mean phenotypic effect, _N_δ2, yields Equation 5 in the main text.
APPENDIX B
In this appendix we show how we estimate the number of QTL in the _ddm_1-derived epiRILs. Consider the equation for N in the main text (Equation 6) at t = ∞ (i.e., fully inbred), under the assumption of perfect resetting (γ(t =∞) = −12):
N=3D2s(s(1−2τ)2+(2r¯+1)/(2r¯−1))(3D2s2+16σ2(η,t)(2r¯+1)/(2r¯−1))(1−2τ)2=f(Ψ,t),
where Ψ is a vector of all the parameters specified on the right-hand side of the equation. Substituting the values for D, σ2(η, t), s, and r− provided in the main text yields one equation and two unknowns (N and τ). With heritability data from only one generation, it is not possible to find unique solutions, and at least one additional generation of phenotypic measurements is required.
Obtaining N−:
In the absence of such additional data one strategy is to calculate an average N by integrating over the theoretical range of τ,
where u = τ|(N = _N_max) is the upper integration limit. To find a value for u we solve Equation 5 for τ and evaluate it at the expected maximum number of QTL, _N_max, which can be detected given the particular mating scheme.
Obtaining the expected value for _N_max:
In a population of RILs, let Rj denote the probability of a recombinant type over the entire length of the _j_th chromosome, Rj=∑i∈ℛR(i)(t=∞), where R(i)(t) are the components of R(t) (Appendix A) at fixation and ℛ is the ensemble of recombinant two-locus epigenotypes. Using our Markov chain approach for selfing, the value of Rj is given by
| Rj|F2=2rj(1+2rj),Rj|BC=3rj(2+4rj), | (13) | | ------------------------------------- | ---- |
where rj is the recombination fraction at meiosis between the beginning and the end of chromosome j. Note that the result for Rj|_F_2 is consistent with Haldane and Waddington (1931). Given a known genetic map, rj can be calculated using any map function, as long as its inverse is available (Liu 1998). The probability of a recombinant type implies at least one recombination breakpoint in the interval, thus generating two potential QTL segments flanking the breakpoint. Assuming s = 1 (all epialleles are stable), the expected maximum number of QTL occurs in a situation where each generated segment is occupied by a QTL. This expectation can be approximated by
where C is the total number of chromosomes, and the ratio on the right-hand side is the odds ratio of a recombination vs. no recombination breakpoint on chromosome j. As a rule of thumb, it is safe to say that E(_N_max|F_2)≃2_C and E(Nmax|BC)≃65C. These latter expressions assume linkage equilibrium between the beginning and end of chromosome j, that is rj = 0.5.
Bootstrap standard errors:
We obtain standard errors for N− using a nonparametric bootstrap approach. To achieve this we take the following steps:
- Recalculate D on the basis of a random sample of size n from each of the two parental phenotypic vectors.
- Draw a random, stratified bootstrap sample from the epiRILs phenotypic vector, and approximate the epigenetic variance, σ2(η, t), using a random intercepts model: yi,j = β0 + bizi,j + ε_i_,j, where β0 is a common fixed intercept, bi is the random intercept of the i_th line, zi,j is an index variable, and ε_i, j is the error. We assume that bi ∼ N(0, σ2(η, t = ∞)), ε_i_, j ∼ N(0, σ2_i_), and Cov(ε_i_, j, ε_i_, _j_′) = 0.
- Use the estimates for 1 and 2, and determine the upper integration limit (u) for τ.
- Determine N− by calculating N−=∫0uf(Ψ,t)dτ/∫0udτ.
- Repeat steps 1–4 a large number of times. The standard deviation of the resulting bootstrap distribution is an approximation for the standard error of N−.
Note that these sampling errors will be slightly underestimated, because the values for s and γ(t) are assumed known from molecular analysis, and the variation in _N_max is also neglected for simplicity.
Acknowledgments
We thank several anonymous reviewers for detailed suggestions. We also thank Ritsert C. Jansen and Vincent Colot for helpful comments on earlier versions of this manuscript. F. Johannes was supported by a Horizon Breakthrough grant (Netherlands Organisation for Scientific Research (NOW)), and M. Colomé-Tatché acknowledges support from the Centre for Quantum Engineering and Space-Time Research (QUEST).
LITERATURE CITED
- Biemont C., 2010. Inbreeding effects in the epigenetic era. Nat. Rev. Genet. 11: 234. [DOI] [PubMed] [Google Scholar]
- Blewitt M., Vickaryous N., Paldi A., Koseki H., Whitelaw E., 2006. Dynamic reprogramming of DNA methylation at an epigenetically sensitive allele in mice. PLoS Genet. 2: e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonduriansky R., Day T., 2009. Nongenetic inheritance and its evolutionary implications. Annu. Rev. Ecol. Evol. Syst. 40: 103–125 [Google Scholar]
- Borgel J., Guibert S., Li Y., Chiba H., Schübeler D., et al. , 2010. Targets and dynamics of promoter DNA methylation during early mouse development. Nat. Genet. 42: 1093–1100 [DOI] [PubMed] [Google Scholar]
- Bossdorf O., Richards C., Pigliucci M., 2008. Epigenetics for ecologists. Ecol. Lett. 11: 106–115 [DOI] [PubMed] [Google Scholar]
- Bossdorf O., Arcuri D., Richards C., Pigliucci M., 2010. Experimental alteration of DNA methylation affects the phenotypic plasticity of ecologically relevant traits in Arabidopsis thaliana. Evol. Ecol. 24: 541–553 [Google Scholar]
- Broman K., 2005. The genomes of recombinant inbred lines. Genetics 169: 1133–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulmer M. G., 1985. The Mathematical Theory of Quantitative Genetics. Clarendon Press, Oxford [Google Scholar]
- Castle W., 1921. An improved method of estimating the number of genetic factors concerned in cases of blending inheritance. Science 54: 223. [DOI] [PubMed] [Google Scholar]
- Cokus S., Feng S., Zhang X., Chen Z., Merriman B., et al. , 2008. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452: 215–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csaba P., 1998. Plasticity, memory and the adaptive landscape of the genotype. Proc. R. Soc. Lond. B 265: 1319–1323 [Google Scholar]
- Daxinger L., Whitelaw E., 2010. Transgenerational epigenetic inheritance: more questions than answers. Genome Res. 20: 1623–1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler E., Flint J., Gibson G., Kong A., Leal S., et al. , 2010. Missing heritability and strategies for finding the underlying causes of complex disease. Nat. Rev. Genet. 11: 446–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., Cokus S., Zhang X., Chen P.-Y., Bostick M., et al. , 2010. Conservation and divergence of methylation patterning in plants and animals. Proc. Nat. Acad. Sci. USA 107: 8689–8694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin I., 1970. Average recombination frequencies. Genetics 66: 709–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldane J., Waddington C., 1931. Inbreeding and linkage. Genetics 16: 357–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannes F., Colot V., Jansen R., 2008. Epigenome dynamics: a quantitative genetics perspective. Nat. Rev. Genet. 9: 883–890 [DOI] [PubMed] [Google Scholar]
- Johannes F., Porcher E., Teixeira F., Saliba-Colombani V., Simon M., et al. , 2009. Assessing the impact of transgenerational epigenetic variation on complex traits. PLoS Genet. 5: e1000530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminsky Z., Tang T., Wang S.-C., Ptak C., Oh G., et al. , 2009. DNA methylation profiles in monozygotic and dizygotic twins. Nat. Genet. 41: 240–245 [DOI] [PubMed] [Google Scholar]
- Kimura M., 1963. A probability method for treating inbreeding systems, especially with linked genes. Biometrics 19: 1–17 [Google Scholar]
- Kong A., Steinthorsdottir V., Masson G., Thorleifsson G., Sulem P., et al. , 2009. Parental origin of sequence variants associated with complex diseases. Nature 462: 868–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande R., 1981. The minimum number of genes contributing to quantitative variation between and within populations. Genetics 99: 541–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E., Bestor T. H., Jaenisch R., 1992. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69: 915–926 [DOI] [PubMed] [Google Scholar]
- Lister R., O'Malley R., Tonti-Filippini J., Gregory B., Berry C., et al. , 2008. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133(3): 523–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B. H., 1998. Statistical Genomics. CRC Press, Boca Raton, FL [Google Scholar]
- Lynch M., Walsh B., 1998. Genetics and the Analysis of Quantitative Traits. Sinauer Associates, Sunderland, MA [Google Scholar]
- Maher B., 2008. Personal genomes: the case of the missing heritability. Nature 456: 18–21 [DOI] [PubMed] [Google Scholar]
- Manolio T., Collins F., Cox N., Goldstein D., Hindorff L., et al. , 2009. Finding the missing heritability of complex diseases. Nature 461: 747–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu O., Reinders J., Caikovski M., Smathajitt C., Paszkowski J., 2007. Transgenerational stability of the Arabidopsis epigenome is coordinated by CG methylation. Cell 130(5): 851–862 [DOI] [PubMed] [Google Scholar]
- Mirouze M., Reinders J., Bucher E., Nishimura T., Schneeberger K., et al. , 2009. Selective epigenetic control of retrotransposition in Arabidopsis. Nature 461: 427–430 [DOI] [PubMed] [Google Scholar]
- Pál C., Miklós I., 1999. Epigenetic inheritance, genetic assimilation and speciation. J. Theor. Biol. 200: 19–37 [DOI] [PubMed] [Google Scholar]
- Petronis A., 2010. Epigenetics as a unifying principle in the aetiology of complex traits and diseases. Nature 465: 721–727 [DOI] [PubMed] [Google Scholar]
- Rakyan V., Blewitt M., Druker R., Preis J., Whitelaw E., 2002. Metastable epialleles in mammals. Trends Genet. 18(7): 348–351 [DOI] [PubMed] [Google Scholar]
- Rakyan V., Chong S., Champ M., Cuthbert P., Morgan H., et al. , 2003. Transgenerational inheritance of epigenetic states at the murine Axin(Fu) allele occurs after maternal and paternal transmission. Proc. Natl. Acad. Sci. USA 100: 2538–2543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik W., 2007. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature 447: 425–432 [DOI] [PubMed] [Google Scholar]
- Reinders J., Wulff-Brande B., Mirouze M., Mari-Ordonez A., Dapp M., et al. , 2009. Compromised stability of DNA methylation and transposon immobilization in mosaic Arabidopsis epigenomes. Genes Dev. 23: 939–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards E., 2006. Inherited epigenetic variation–revisiting soft inheritance. Nat. Rev. Genet. 7: 395–401 [DOI] [PubMed] [Google Scholar]
- Richards E., 2008. Population epigenetics. Curr. Opin. Genet. Dev. 18(2): 221–226 [DOI] [PubMed] [Google Scholar]
- Richards E., 2009. Quantitative epigenetics: DNA sequence variation need not apply. Genes Dev. 23: 1601–1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riesenberg L., Archer M., Wayne R., 1999. Transgressive segregation, adaptation and speciation. Heredity 83: 363–372 [DOI] [PubMed] [Google Scholar]
- Serebrovsky A., 1928. An analysis of the inheritance of quantitative transgressive characters. Z. Indukt. Abstammungs. Verbungsl. 48: 229–243 [Google Scholar]
- Slatkin M., 2009. Epigenetic inheritance and the missing heritability problem. Genetics 182: 845–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tal O., Kisdi E., Jablonka E., 2010. Epigenetic contribution to covariance between relatives. Genetics 184: 1037–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixera F., Colot V., 2010. Repeat elements and the Arabidopsis DNA methylation landscape. Heredity 105: 14–23 [DOI] [PubMed] [Google Scholar]
- Teixeira F., Heredia F., Sarazin A., Roudier F., Boccara M., et al. , 2009. A role for RNAi in the selective correction of DNA methylation defects. Science 323: 1600–1604 [DOI] [PubMed] [Google Scholar]
- Tsukahara S., Kobayashi A., Kawabe A., Mathieu O., Miura A., et al. , 2009. Bursts of retrotransposition reproduced in Arabidopsis. Nature 461: 423–426 [DOI] [PubMed] [Google Scholar]
- Vaughn M., Tanurdzic M., Lippman Z., Jiang H., Carrasquillo R., et al. , 2007. Epigenetic natural variation in Arabidopsis thaliana. PLoS Biol. 5: e174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhoeven K., Jansen J., van Dijk P., Biere A., 2010. Stress-induced DNA methylation changes and their heritability in asexual dandelion. New Phytol. 185: 10.1111/j.1469–8137.2009.03121.x [DOI] [PubMed] [Google Scholar]
- Vongs A., Kakutani T., Martienssen R., Richards E., 1993. Arabidopsis thaliana DNA methylation mutants. Science 260: 1926–1928 [DOI] [PubMed] [Google Scholar]
- Wright S., 1933. Inbreeding and homozygosis. Proc. Natl. Acad. Sci. USA 19: 411–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A., McDaniel I., Silva P., Zilberman D., 2010. Genome-wide evolutionary analysis of eurkaryotic DNA methylation. Science 328: 916–919 [DOI] [PubMed] [Google Scholar]
- Zeng Z.-B., 1992. Correcting the bias of Wright’s estimates of the number of genes affecting a quantitative character: a further improvement. Genetics 131: 987–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Shiu S., Cal A., Borevitz J., 2008. Global analysis of genetic and epigenetic and transcriptional polymorphisms in Arabidopsis thaliana using whole genome tiling arrays. PLoS Genet. 4: e1000032. [DOI] [PMC free article] [PubMed] [Google Scholar]