Bivalent recognition of nucleosomes by the tandem PHD fingers of the CHD4 ATPase is required for CHD4-mediated repression (original) (raw)
Abstract
CHD4 is a catalytic subunit of the NuRD (nucleosome remodeling and deacetylase) complex essential in transcriptional regulation, chromatin assembly and DNA damage repair. CHD4 contains tandem plant homeodomain (PHD) fingers connected by a short linker, the biological function of which remains unclear. Here we explore the combinatorial action of the CHD4 PHD1/2 fingers and detail the molecular basis for their association with chromatin. We found that PHD1/2 targets nucleosomes in a multivalent manner, concomitantly engaging two histone H3 tails. This robust synergistic interaction displaces HP1_γ_ from pericentric sites, inducing changes in chromatin structure and leading to the dispersion of the heterochromatic mark H3K9me3. We demonstrate that recognition of the histone H3 tails by the PHD fingers is required for repressive activity of the CHD4/NuRD complex. Together, our data elucidate the molecular mechanism of multivalent association of the PHD fingers with chromatin and reveal their critical role in the regulation of CHD4 functions.
Keywords: gene repression, epigenetics, histone, posttranslational modifications
Chromodomain helicase DNA-binding protein 4 (CHD4) is an ATP dependent chromatin remodeler and a major subunit of the repressive NuRD complex (1–4). The CHD4/NuRD complex plays pivotal roles in transcriptional regulation and reorganization and maintenance of chromatin structure (5, 6). It targets pericentromeric heterochromatin and has recently been implicated in DNA damage repair, being rapidly recruited to DNA double strand breaks (7, 8). Other components of the complex include a second catalytic subunit HDAC1/2 (histone deacetylase) and the nonenzymatic proteins MBD2/3 (methyl-CpG-binding domain), RbAp46/48 (retinoblastoma-associated), MTA1/2/3 (metastasis-associated) and p66α/β (Fig. 1A). The exact composition of NuRD can vary, reflecting alterations in the activity and localization of the complex.
Fig. 1.
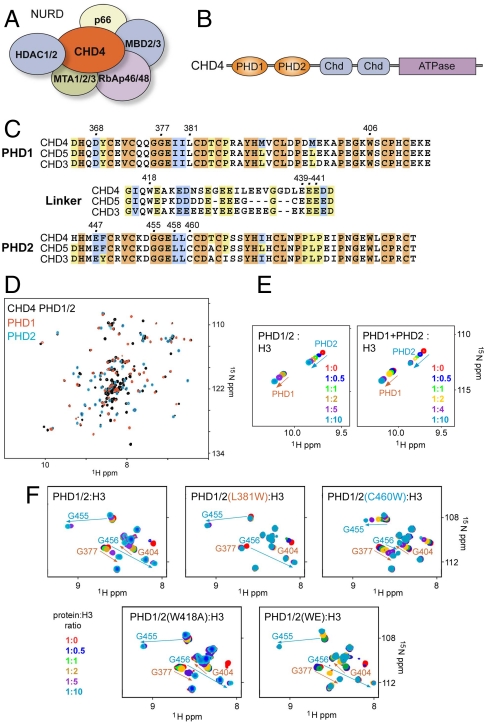
The tandem PHD fingers of CHD4 possess individual histone-binding activities. (A) The CHD4/NuRD complex. (B) CHD4 contains two N-terminal PHD fingers (orange), two chromodomains (blue) and a C-terminal ATPase module (purple). (C) Alignment of the CHD3-CHD5 PHD1/2 sequences. Residues that are strongly, moderately, and weakly conserved in PHD1 and PHD2 fingers are highlighted in orange, blue, and yellow, respectively. (D) An overlay of the 1H,15N HSQC spectra of unbound PHD1/2 (black) and the individual PHD1 (red) and PHD2 (blue) fingers. (E) An overlay of 1H,15N HSQC spectra of the tandem PHD1/2 construct (Left) and PHD1 and PHD2 separately (Right) as increasing amounts of the histone H3 tail peptide (1–12) is titrated in. Spectra are color coded according to the molar ratio of protein:peptide. Resonances of the PHD1 domain are labeled in brown and those for the PHD2 domain are labeled in blue. (F) Overlays of 1H,15N HSQC spectra of PHD1/2 (Top Left) and mutants PHD1/2(L381W) (Top Middle), PHD1/2(C460W) (Top Right), PHD1/2(W481A) (Bottom Left), and PHD1/2 (W481A/E439A/E440A/E441A) (Bottom Right) as the histone H3 tail peptide (1–12) is titrated in.
CHD4 (also known as Mi-2β) is a large multimodular protein. It belongs to the family of nine CHD proteins and, similar to its closest family members CHD3 (or Mi-2α) and CHD5, contains tandem plant homeodomain (PHD) fingers, two chromodomains, and a catalytic ATPase module (Fig. 1 B and C). The SNF2-like ATPase domain mediates nucleosome mobility, providing the energy necessary for histone displacement and sliding (9). Unlike other chromodomains that bind methylated histone marks enriched in heterochromatic regions (10–13), the CHD4 chromodomains exhibit a preference for DNA (14). The individual PHD fingers of CHD4 (PHD1 and PHD2) have been shown to possess histone-binding activity (15, 16); however, the target of the tandem PHD1/2 module and the functional consequences of its association with chromatin remain unclear.
PHD fingers comprise one of the largest families of epigenetic effectors capable of recognizing or “reading” posttranslationally modified or unmodified histone tails, consequently recruiting various nuclear complexes to chromatin (reviewed in refs. 17 and 18). PHD fingers bind primarily to histone H3 trimethylated at Lys4 (H3K4me3) or unmodified H3K4. The biological outcome of these interactions is highly context-dependent and is determined by a number of factors including the composition of the complex, adjacent effector domains present in the same protein, and neighboring posttranslational histone modifications. An adjacent effector domain can be another PHD finger, a distinct histone-binding domain such as bromodomain, chromodomain, Tudor, or a catalytic histone-associating module. The combinatorial action of linked effectors often gives rise to binding mechanisms that are fundamentally different from those seen for the individual modules. Simultaneous interactions of adjacent domains modulate the affinity and specificity of their host proteins or protein complexes for specific chromatin states and thus play a significant role in epigenetic mechanisms. Despite the widespread phenomena of the multivalent engagement of epigenetic readers, characterization of the structure and binding properties of multidomain constructs poses unique challenges. To date few examples have been reported (19, 20), and we have only begun understanding the complexity of crosstalk within polyeffector systems that defines a specific biological outcome.
In this study, we elucidate the biological function and binding mechanism of the tandem CHD4 PHD1/2 fingers connected by the native linker. Our data reveal that each PHD module of PHD1/2 possesses individual histone-binding activity. The domain organization positions PHD1/2 to concomitantly bind two histone H3 tails, directing CHD4 to chromatin in a multivalent manner. We employ a combination of NMR, mutagenesis, tryptophan fluorescence, and gel shift assays to define this unique mechanism and in vivo immunofluorescence, immunoprecipitation, shRNA knockdown, and flow cytometry analyses to establish the significance of the PHD1/2 fingers for chromatin localization and transcriptional regulation by CHD4.
Results and Discussion
The Tandem PHD Fingers of CHD4 Possess Individual Histone-Binding Activities.
The CHD4 ATPase contains an N-terminal tandem of PHD fingers (PHD1/2) linked by a stretch of 30 amino acids. To establish whether the linked PHD fingers function together, we investigated their structures and abilities to bind histones by NMR (Fig. 1 and Figs. S1 and S2). To examine the structural interrelation of the domains, the 1H, 15N HSQC spectra of PHD1/2 and the individual PHD1 and PHD2 fingers in the free state were collected and compared. An overlay of the spectra showed that the majority of cross peaks of the globular PHD modules have similar positions, indicating no significant change in the fold of these domains in the tandem construct (Fig. 1D). Additional cross peaks, which belong to the linker region, were clustered primarily in the middle of the PHD1/2 spectrum, implying that the linker is largely unstructured. A comparable linewidth (at half height) measured in the 1H,15N HSQC spectra suggested that the PHD fingers in the tandem module are rotating nearly independently of each other. The lack of resonance perturbations in the spectrum of 15N-labeled PHD1 when unlabeled PHD2 was added stepwise confirmed that the two domains do not physically interact (Fig. S2_A_). Thus, in the absence of ligands, the two domains of PHD1/2 are flexibly linked and structurally independent.
Histone-binding analysis reveals that each domain in PHD1/2 has distinct binding activity toward the N-terminal tail of H3, with PHD2 exhibiting a higher affinity than PHD1. We characterized the binding of PHD1/2 to histone tail peptides by NMR titration experiments (Fig. 1 E and F). 1H, 15N HSQC spectra of uniformly 15N-labeled PHD1/2 were collected while titrating in a peptide corresponding to the first 12 residues of the H3 tail. Addition of increasing amounts of the peptide induced substantial chemical shift changes in both of the linked modules. The pattern of chemical shift changes in PHD1/2 was similar to that observed in the separated PHD1 and PHD2 fingers mixed together upon addition of the H3 peptide (Fig. 1E). This suggests that each PHD finger in the tandem PHD1/2 module binds an individual histone tail. The pattern of resonance perturbations also indicated two distinct binding events in PHD1/2, with one set of peaks shifting significantly upon the first addition of peptide and reaching saturation at a 1∶5 protein:peptide molar ratio and another set only beginning to shift at 1∶2. Careful examination reveals that cross peaks associated with the first pattern of changes belong to the PHD2 finger and peaks associated with the second pattern belong to PHD1 (Fig. 1E). Thus, when linked, the PHD2 finger has a higher affinity for the H3 peptide than does the PHD1 finger.
The recognition of individual histone tails by each domain in PHD1/2 was supported by mutational analysis. Point mutants of PHD1/2 were generated in which histone binding of either PHD1 or PHD2 was compromised. Titration experiments using PHD1/2-L381W (a PHD1 mutant) and the unmodified H3 peptide demonstrated that the interaction of PHD1 with the peptide was abolished, whereas the interaction of PHD2 was unchanged (Fig. 1F). Similarly, mutation of the PHD2 finger (PHD1/2-C460W) led to significantly decreased binding of PHD2 to the H3 tail, whereas the interaction of PHD1 was not affected (Fig. 1F). To determine how the H3 tail peptide is recognized, we performed NMR titration experiments using the unmodified H3 peptide in which Ala1 is acetylated (H3A1ac) (Fig. S2_B_). The absence of chemical shift changes in PHD1/2 upon addition of H3A1ac implied that binding of both PHD fingers is abrogated. We concluded that each PHD finger in the tandem module requires the N-terminal Ala1 residue for binding.
Together these results reveal a unique mode of histone binding in which the two fingers of the tandem module are structurally independent and exhibit distinct binding activity, each recognizing an individual histone H3 tail with PHD2 playing a dominant role. This mechanism contrasts with the mechanisms for association of the double PHD finger of DPF3 and the double chromodomain of CHD1 with a single histone tail (21, 22). The PHD fingers of DPF3 bind sequentially to a long stretch of the tail, with one finger recognizing the N terminus of H3 and another interacting with the central region of the tail acetylated at lysine 14, whereas the chromodomains of CHD1 form a single binding pocket for the H3 tail methylated at lysine 4. Thus, CHD4 PHD1/2 is an example in which two effector domains within the same protein are seen to have duplicate histone-binding function.
CHD4 PHD1/2 Associates Bivalently with a Single Nucleosome.
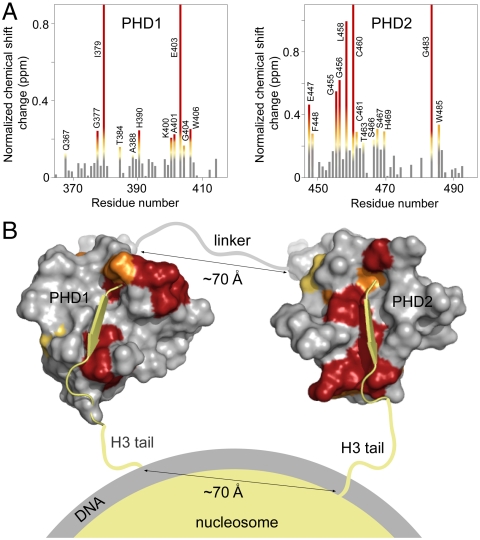
Our data demonstrate that PHD1/2 binds two histone H3 tails, suggesting that PHD1/2 associates with nucleosomes in a multivalent manner. Using NMR data and modeling we show that the interdomain orientation of PHD1/2 positions it to bivalently target a single nucleosome, concomitantly recognizing both of the histone H3 tails. The histone-binding sites of PHD1/2 were determined by analysis of chemical shift changes observed in the 1H,15N HSQC spectra of PHD1/2 upon titration of the H3 peptide. Those residues that were perturbed the most (greater than the average shift plus one-quarter standard deviation) were assumed to be directly or indirectly involved in binding to H3 (Fig. 2A). Mapping these changes onto the structures of the individual PHD fingers (2L5U.pdb and 2L75.pdb) reveals well-defined binding pockets that are similar to those previously reported for the individual PHD fingers (Fig. 2B). Association of PHD1/2 with the two peptides was modeled using NMR data and the structures of unbound PHD1 and of PHD2 in complex with a histone peptide (16, 23). PHD1 and PHD2 were oriented in order for the C-terminal end of PHD1 to align with the N-terminal end of PHD2. Further, NMR data suggest that the 30-residue linker between the two domains is largely unstructured, therefore the distance between the PHD fingers would be just over 70 Å when the linker is fully extended. This interdomain configuration could allow for binding to two histone H3 tails of a single nucleosome, which protrude from the core at a distance of 70 Å in a parallel orientation (Fig. 2B). In agreement, few chemical shift perturbations were observed in the linker region, suggesting that it is not involved in the interaction. Furthermore, mutation of the conserved Trp residue and the conserved acidic (EEE) patch in the linker did not affect the binding (Fig. 1F).
Fig. 2.
The interdomain organization of PHD1/2 results in a perfect fit for binding to two histone H3 tails in a single nucleosome. (A) The histograms show normalized 1H, 15N chemical shift changes in backbone amides of the PHD1 (Left) and PHD2 (Right) domains of PHD1/2 upon addition of the H3 peptide as a function of residue. Shifts greater than the average plus one-quarter (yellow), one-half (orange) and one (red) standard deviation are highlighted and the residues labeled. These changes are mapped onto the surface of the unbound PHD1 and H3K9me3-bound PHD2 structures in B. (B) A model of the association of CHD4 PHD1/2 with the nucleosome. The PHD1-H3 peptide complex, the linker and orientation with respect to the nucleosome were modeled.
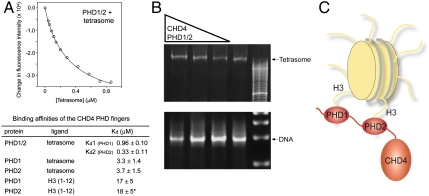
We next examined the ability of CHD4 PHD1/2 to associate with nucleosomes using reconstituted tetrasomes (the H3/H4 tetramer wrapped by 80 bp of DNA). Our data reveal that PHD1/2 can bind to a single tetrasome in a bivalent manner (Fig. 3). To evaluate the strength of the association with the tetrasomal particle, we measured binding affinities by tryptophan fluorescence. Analysis of the binding curves of PHD1/2 required a two site binding model, indicating that both PHD fingers are involved in the interaction with the tetrasome (Fig. 3A). This fitting yielded two Kd values of 0.33 μM and 0.96 μM, which in keeping with the NMR data correspond to PHD2 and PHD1, respectively. As expected, binding of the separate PHD fingers was well described by a single site model (Kds = 3–4 μM, Fig. 3A). The three- and 11-fold increase in affinity of the linked PHD fingers as compared to binding affinities of the individual domains is likely entropically driven, with both the PHD fingers and histone tails being prealigned for the interaction.
Fig. 3.
PHD1/2 associates bivalently with a single nucleosome. (A) A representative tryptophan fluorescence curve and the two-site fit for PHD1/2 as the tetrasome particle is titrated in. The table shows K d values of the linked PHD1/2 fingers and individual PHD1 and PHD2 for the tetrasome and the unmodified H3 tail peptides, residues 1–12 (averaged over three experiments). (*) Taken from ref. 15. (B) The tetrasome particle free and in the presence of increasing amounts of PHD1/2 was run on a nondenaturing acrylamide gel and detected by ethidium bromide staining. The tetrasome bands (Top) and the free DNA bands (Bottom) are shown. (C) A model of the concomitant interaction of both PHD modules with the two histone H3 tails.
Binding of the CHD4 PHD1/2 fingers to a single tetrasome was substantiated by gel shift assays. PHD1/2 was first incubated with the tetrasome in 0.2∶1, 1∶1 and 5∶1 molar ratios, and the reaction mixtures were run on a nondenaturing acrylamide gel and stained with ethidium bromide for detection. A shift of the tetrasome band (approximately 102 kDa) upon formation of the complex with PHD1/2 (approximately 119 kDa) indicated that PHD1/2 binds to a single tetrasome but does not crosslink a pair of tetrasomes of the overall size of 221 kDa (Fig. 3B). No shift was observed in the case of free DNA, suggesting that PHD1/2 does not interact with the tetrasomal DNA. Together, the NMR, tryptophan fluorescence, and gel shift assays demonstrate that PHD1/2 can associate bivalently with a single nucleosome.
PHD1/2 Association with H3 Is Modulated by Histone PTMs.
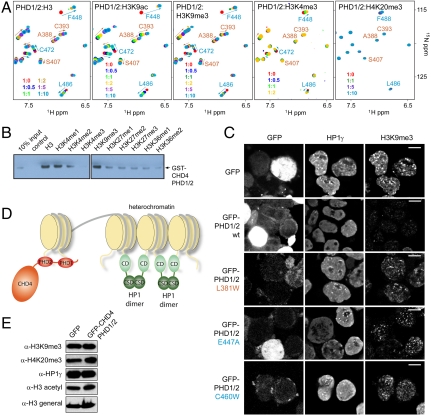
Histones undergo a number of posttranslational modifications (PTMs), particularly in their N-terminal tails. We found that methylation or acetylation of Lys9 strengthens binding of both domains in PHD1/2 to the histone H3 tail, whereas methylation of Lys4 significantly weakens it. The association of the tandem construct with several modified histone H3 tail peptides was tested by NMR and pull-down assays (Fig. 4 A and B). 1H, 15N HSQC spectra of PHD1/2 were recorded in the presence of increasing amounts of H3K9ac, H3K9me3, H3K4me3, or H4K20me3 and recognition of methylation marks on H3K4, H3K9, H3K27, and H3K36 was examined by Western blot analysis. As compared to the unmodified H3 peptide, titration of H3K9ac or H3K9me3 into 15N-labeled PHD1/2 showed that both PHD fingers reach saturation at lower concentration of the peptide, indicating that binding is facilitated by an increase in the hydrophobicity of Lys9 (Fig. 4A). However, the linked PHD fingers retained distinct binding modes, with PHD2 prevailing over PHD1. In contrast, titration of H3K4me3 diminished binding of both modules. Moreover, the more methyl groups Lys4 contained, the weaker this interaction became (Fig. 4B). A lack of chemical shift perturbations in either domain implied that PHD1/2 does not recognize H4K20me3 and little to no association with methylated Lys27 or Lys36 was observed in pull-down assays.
Fig. 4.
Histone modifications modulate the interaction of PHD1/2. (A) Overlays of 1H, 15N HSQC spectra of PHD1/2 as peptides corresponding to unmodified H3, H3K9ac, H3K9me3, H3K4me3 or H4K20me3 (from left to right) are titrated in. Spectra are color coded according to the protein:peptide molar ratio. (B) Pull-down assays of GST-PHD1/2 with different singly modified histone tail peptides. (C) CHD4 PHD1/2 disrupts pericentric heterochromatin. Representative images of immunofluorescence performed in sorted GFP and GFP-CHD4 PHD1/2 (wild type and the indicated mutants) cells. Cells were spotted onto slides and stained for H3K9me3 and HP1γ. Scale bar, 10 μm. (D) A model of how intranucleosomal binding of CHD4 PHD1/2 can modulate the internucleosomal interaction of HP1γ. (E) Western blot analysis of H3K9me3, H4K20me3, HP1γ, and total H3 acetylation 48 hr after transfection with either GFP or GFP-CHD4 PHD1/2. Total histone H3 (H3 general) was used as a loading control.
CHD4 PHD1/2 Disrupts Pericentric Heterochromatin.
NuRD has been shown to localize to heterochromatin and display a CHD4-dependent activity in heterochromatin maintenance and assembly. To determine if the PHD fingers of CHD4 are necessary for localization of CHD4 to pericentric heterochromatin, we generated a GFP-fusion of PHD1/2 (GFP-CHD4-PHD1/2) (Fig. 4). HEK293T cells were transfected to express the fusion protein or the GFP tag alone. GFP+ cells were sorted 48 h post transfection and the subcellular localization of GFP was examined (Fig. 4C). As a control for the integrity of pericentric heterochromatin regions, we assessed the localization pattern of H3K9me3 and HP1γ. As expected, in cells transfected with GFP alone we observed the normal focal accumulation of H3K9me3 and HP1γ. However, in cells transfected with GFP-CHD4-PHD1/2, we observed diffuse nuclear localization of the fusion protein and loss of the normal focal accumulation of the pericentric heterochromatin markers. Such a disruption of heterochromatin markers by the PHD1/2 fingers of CHD4 could result from decreased levels of these markers or their redistribution within the nucleus. To differentiate between these possibilities, we performed Western blot analysis and examined global levels of H3K9me3 and HP1γ (Fig. 4E). H3 acetylation and H4K20me3 were measured as controls. The absence of any changes in these markers 48 h after transfection suggested that the PHD1/2 fingers of CHD4 specifically disrupt pericentric heterochromatin structure, possibly through displacement of HP1γ.
Schizosaccharomyces pombe HP1 displays a 0.12 μM affinity for the H3K9me3-containing nucleosomes (24). We found that the linked PHD fingers of CHD4 bind to the unmodified H3 tails of the tetrasome with the Kds of 0.33 and 0.96 μM, and these affinities are expected to increase significantly upon methylation of Lys9 (a 10-fold increase is seen with H3 peptides alone). These data support a model wherein the tandem PHD1/2 fingers of CHD4 compete with chromodomain (CD) of HP1γ for H3K9me3-enriched nucleosomes, resulting in displacement of HP1γ (Fig. 4D). In contrast to CHD4 PHD1/2, HP1γ associates with two spatially proximal nucleosomes through binding of its CD to H3K9me3 and dimerization via its chromoshadow domain (CSD) (24), providing a mechanism for the formation and spreading of heterochromatin. The antagonistic action of CHD4 PHD1/2 would lead to the disruption of pericentric heterochromatin assembly and the subsequent dispersion of H3K9me3 and HP1γ observed by light microscopy (Fig. 4C). To test this idea, we generated the GFP-fusion mutants of CHD4 PHD1/2, in which histone binding of either PHD1 or PHD2 was compromised, and examined the impact of expression of these mutants on pericentric heterochromatin. As shown in Fig. 4C, the L381W mutant (PHD1 is impaired) or the E447A and C460W mutants (PHD2 is impaired) were unable to disrupt the normal focal accumulation of H3K9me3 and HP1γ, therefore supporting the notion that the concomitant engagement of both PHD fingers of CHD4 is required to elicit this effect. Lastly, we note that further studies are necessary to more fully explore the interplay of PHD1/2 and HP1γ in the context of full length CHD4 and in the context of intact NuRD complex.
PHD1/2 Histone-Binding Activity Is Crucial for Repressive Functions of CHD4/NuRD CRCs in Vivo.
The role of the PHD1/2-histone interactions in transcriptional regulation by CHD4/NuRD was examined by assessing the ability of wild-type and mutated CHD4 to repress transcription of the mb-1 (Cd79a) gene in B cells. Display of membrane-bound IgM (mIgM) on B cells requires transcription of the mb-1 gene, which encodes the Ig-α subunit of the B cell receptor (25). The display of mIgM on μM.2 murine plasmacytoma cells is proportional to mb-1 transcript abundance (26). In μM.2 cells, mb-1 transcription is activated approximately 80-fold by the Early B cell factor 1 (EBF1) and Paired box protein 5 (Pax5) transcription factors but is restrained by CHD4/NuRD CRCs (27). In this context, depletion of endogenous CHD4 by shRNA increases activation of mb-1 promoters significantly by enhancing chromatin accessibility and demethylation of mb-1 promoter CpGs (27).
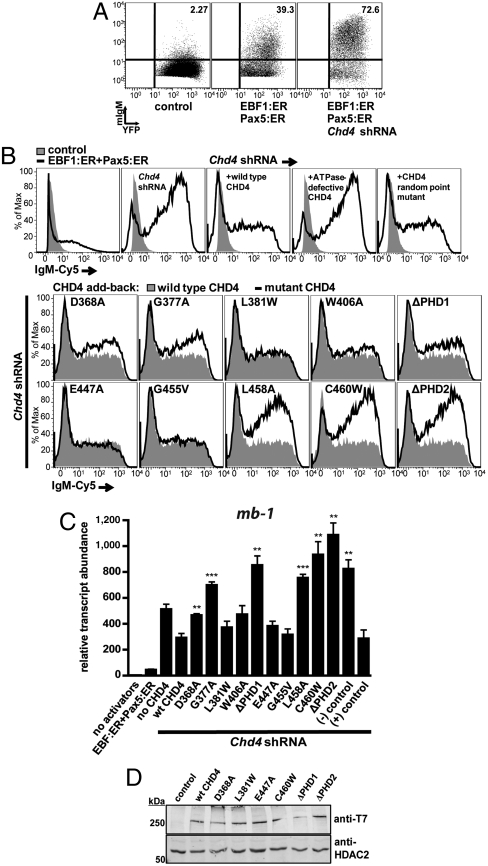
In μM.2 cells expressing estrogen-dependent versions of EBF1 and Pax5 (EBF1:ER, Pax5:ER) and 3′ UTR-specific Chd4 shRNAs, highly efficient display of mIgM was detected due to activation of endogenous mb-1 transcription (Fig. 5A, Right, and Fig. 5B, Top, Second from Left). In this context, expression of exogenous wild-type CHD4 attenuated mb-1 transcription as measured by a significant reduction in mIgM (Fig. 5B, Top, Third from Left). To demonstrate that enzymatically active CHD4 is required for this activity, we generated a point mutation (K757R) that inactivates the ATPase domain of CHD4 (28). This mutation is predicted to block nucleosome mobilization by CHD4. We also generated a mutation (R1034A) with the potential to interfere with DNA-binding function as predicted based on the crystal structure of SWI2/SNF2 (29). As expected, the K757R mutation abrogated reduction of IgM display by CHD4 in the presence of activated transcription factors (Fig. 5B, Top, Fourth from Left). However, the R1034A mutant retained wild type activity. This mutant demonstrates that mutation of CHD4 does not intrinsically result in loss of repressive activity (Fig. 5B, Top Right).
Fig. 5.
PHD fingers of CHD4 are necessary for transcriptional repressive functions in vivo. (A) Increased display of membrane-bound IgM (mIgM) in response to activated transcription factors (EBF1:ER, Pax5:ER) and depletion of CHD4·μM.2 plasmacytoma cells do not express mIgM in the absence of transcriptional activators (Left). When activated by 4-hydroxytamoxifen (4-OHT), the transcription factors EBF1 (as EBF1:ER) and Pax5 (as Pax5:ER) synergistically activate endogenous Cd79a genes, which enables the assembly of mIgM on the B cell surface at low levels (Middle). shRNA-mediated knockdown of CHD4 greatly increases Cd79a transcription (shown in C) and the display of mIgM in response to EBF1 and Pax5 (Right). Empty retroviruses and luciferase shRNA were used as controls. (B) Effect of mutations in PHD1 and PHD2 in CHD4 on mIgM expression in μM.2 cells. Expression of wild-type CHD4 restores repressive functions when endogenous CHD4 is depleted by 3′UTR-specific shRNA. (Top Row, First Panel) Background mIgM in the presence of EBF1:ER and Pax5:ER. Depletion of CHD4 greatly enhances mIgM (Top Row, Second Panel). Add-back of wild type CHD4 (Top Row, Third Panel) or mutant CHD4 (Top Row, Fourth and Fifth Panels and Middle and Bottom Rows). (C) Relative abundance of mb-1 transcripts from sorted cells in B. CHD4 R1034A (positive control) did not reduce CHD4 activity, while CHD4 K757R (negative control) is inactive. All conditions have activated EBF1:ER and Pax5:ER except control (no activators). Asterisks indicate statistical significance relative to wild type: **= p < 0.001, ***= p < 0.0001. (D) Western blot of CHD4 mutants. Whole cell extracts were prepared from μM.2 cells transduced with CHD4 or control (empty vector) retroviruses and analyzed by blotting with anti-T7 epitope antibodies. HDAC2 served as a nuclear protein loading control.
The add-back assay was used to assess the importance of recognition of histone H3 tails by the PHD1/2 fingers for CHD4 transcriptional repressive function in vivo. We introduced mutations in the PHD1 and PHD2 fingers of the full length protein that have been shown to significantly reduce binding of the individual PHD modules to histone H3 peptides (15, 16). Additionally, PHD finger deletion mutants (ΔD365-C411, ΔPHD1 and ΔD443-C490, ΔPHD2) of CHD4 were generated. Of the five PHD1 mutants tested, D368A, G377A and ΔPHD1 attenuated repressive activity of CHD4, while the L381W and W406A mutants did not result in significant effects (Fig. 5B, Second Row). However, expression of the PHD2 finger mutants L458A, C460W, and ΔPHD2 completely abrogated CHD4 repressive function, as evidenced by increased IgM display (Fig. 5B, Third Row). Statistical analysis of both mIgM mean fluorescence intensities (MFI) and mb-1 transcript abundance confirmed that PHD1 finger mutants D368A, G377A, and ΔPHD1 and PHD2 finger mutants L458A, C460W, and ΔPHD2 exhibit significantly reduced activity compared to wild-type CHD4 (Fig. 5C and Fig. S4). As controls, Western blotting detected the expression of (3X)T7-tagged wild-type and mutant CHD4 in transduced μM.2 cells (Fig. 5D). These results support two significant conclusions: (i) histone tail binding by CHD4 PHD fingers is necessary for transcriptional repression by the CHD4/NuRD complex in B cells, and (ii) although PHD1 and PHD2 each contribute to the activity of CHD4, PHD2 plays a more important role than PHD1, which is in agreement with measured binding affinities.
Concluding Remarks
In this study, we determined the biological function and a unique binding mechanism of the tandem PHD1/2 fingers of the CHD4 ATPase. We found that the PHD1/2 fingers target nucleosomes in a bivalent manner, concomitantly recognizing two histone H3 tails with high affinity. This interaction is further modulated by histone modifications, with methylation or acetylation of Lys9 enhancing binding, and methylation of Lys4 or acetylation of Ala1 abolishing it. Our functional data reveal that histone binding by the CHD4 PHD1/2 fingers is required for transcriptional repressive activity of the CHD4/NuRD complex and support the mechanism of multivalent engagement.
The interdomain organization of CHD4 PHD1/2 allows for intranucleosomal interactions. Although the biochemical data do not preclude the ability of PHD1/2 to engage adjacent nucleosomes in trans, disruption of heterochromatin structure upon displacement of HP1γ suggests that the CHD4 PHD fingers may not be as effective as HP1γ at internucleosomal bridging. Thus, the crosstalk between linked effector domains may be essential not only for recognition of a set of posttranslational histone modifications but also for distinguishing a particular chromatin landscape as defined by internucleosomal spacing. It is also of note that rather than defining the binding interaction, histone modifications appear to fine-tune the affinity of CHD4 for nucleosomes in a functionally relevant way. The ability to recognize unmodified histone H3 tails is critical in CHD4 function in transcriptional regulation, whereas the additional affinity imparted by Lys9 methylation is important in heterochromatin targeting.
Materials and Methods
Protein Purification, PCR Mutagenesis, NMR Spectroscopy, Fluorescence Spectroscopy, Gel Shift Assays, Cell Culture and Western Analysis, Immunofluorescence, Pull-Down Assays, RNA Isolation, and qPCR.
Tetrasome Reconstitution.
The tetrasome was reconstituted as previously reported (30). Briefly, histones H3 and H4 (with the modifications C110A and T71C) were expressed in Escherichia. coli Rosetta2(DE3) pLysS cells grown in 2XTY media. The histones were solubilized, purified over ion exchange columns, and lyophilized. An 80-bp segment corresponding to a portion of the Widom 601 sequence was ordered from Integrated DNA Technologies. The single-stranded DNA was purified by ethanol precipitation before annealing. The histones were refolded into tetramer form into 10 mM Tris pH 7.5, 1 mM EDTA, 5 mM β-mercaptoethanol and 2M NaCl following the protocol in ref. 31 and further purified over a sephacryl S-100 column (without EDTA). The purified tetramer was then mixed at a 1∶1 molar ratio with the DNA at approximately 0.1 mg/mL and reconstituted into tetrasome form using a continuous salt-gradient dialysis method, starting in 300 mL of 10 mM Tris pH 7.5, 0.5 mM TCEP and 2M NaCl and adding 10 mM Tris pH 7.5 and 0.5 mM TCEP at 1.5 mL/ min to reach a final NaCl concentration of 150 mM.
Cell Lines, Transfection, Retroviral Infection, and Flow Cytometry.
The μM.2 and μM.2 EBF:ER stably transfected cell lines were cultured as described previously (27). Generation of retroviruses and infection of cells were performed as described (26) with the following exceptions: For generation of CHD4 wild type and mutant retroviruses, 67.2 μg retroviral plasmid DNA and 56 μL Lipofectamine™ 2000 (Invitrogen) were used to transfect 60–80% confluent 100-mm dishes of ΦNX cells. Resulting retroviral supernatants were concentrated using Retro-Concentin™ (System Biosciences). Precipitated viral pellets were resuspended in μM.2 growth media to 1/100 of the original volume of supernatant. All viral transductions were performed in 100-mm dishes. Eight ml of retroviral supernatant was used for transductions of shRNA and control retroviruses, while the equivalent of 24 mL retroviral supernatant (concentrated retrovirus) was used for transductions of CHD4 wild type and mutant retroviruses. Eight mL of cells (6.25 × 105 cells/mL) and 18.8 μg/mL polybrene (Sigma-Aldrich) was used for all transductions. ER fusion proteins (EBF1:ER and Pax5:ER) were induced using 0.5 μM 4-OHT (Sigma-Aldrich) 48 hrs after retroviral transduction. Flow cytometry was performed 5 d after retroviral transduction.
For flow cytometry, APC-eFluor® 780- or Pacific Blue-conjugated anti-CD90.1 was obtained from eBioscience and Cy5-conjugated anti-IgM from Jackson Research Laboratories. Staining of cells was performed as previously described and detected using a CyAn™ flow cytometer (Dako). Live cells were gated mCherry fluorescent protein+ (Chd4 shRNA), CD90.1-PB+ (exogenous Chd4) and YFP+ (Pax5:ER). Data was analyzed using FloJo™ software.
Supplementary Material
Supporting Information
Acknowledgments.
We thank F. Davrazou, J. Scorgie, and B. Hirsch for help with the experiments and discussions; G. Blobel for the CHD4 cDNA plasmid; and K. Luger for the histone H3 and H4 plasmids. This research is supported by grants from the National Institutes of Health (NIH) (GM096863 and CA113472) and the Cancer League of Colorado to T.G.K. and NIH Grants R01 AI054661 and AI081878 to J.H. This work is also supported in part by the Intramural Research Program of the National Institute of Environmental Health Sciences, NIH (Z01ES101765 to P.W.). J.R. was supported by a generous grant from the Rocky Mountain Chapter of the Arthritis Foundation and NIH Postdoctoral Training Grant T32 AI07405. C.A.M. is an NIH National Research Service Award postdoctoral fellow (NHLBI, F32HL096399).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Tong JK, Hassig CA, Schnitzler GR, Kingston RE, Schreiber SL. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature. 1998;395:917–921. doi: 10.1038/27699. [DOI] [PubMed] [Google Scholar]
- 2.Wade PA, Jones PL, Vermaak D, Wolffe AP. A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr Biol. 1998;8:843–846. doi: 10.1016/s0960-9822(98)70328-8. [DOI] [PubMed] [Google Scholar]
- 3.Xue Y, et al. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell. 1998;2:851–861. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Y, LeRoy G, Seelig HP, Lane WS, Reinberg D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell. 1998;95:279–289. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]
- 5.Denslow SA, Wade PA. The human Mi-2/NuRD complex and gene regulation. Oncogene. 2007;26:5433–5438. doi: 10.1038/sj.onc.1210611. [DOI] [PubMed] [Google Scholar]
- 6.Wang HB, Zhang Y. Mi2, an auto-antigen for dermatomyositis, is an ATP-dependent nucleosome remodeling factor. Nucleic Acids Res. 2001;29:2517–2521. doi: 10.1093/nar/29.12.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smeenk G, et al. The NuRD chromatin-remodeling complex regulates signaling and repair of DNA damage. J Cell Biol. 2010;190:741–749. doi: 10.1083/jcb.201001048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Larsen DH, et al. The chromatin-remodeling factor CHD4 coordinates signaling and repair after DNA damage. J Cell Biol. 2010;190:731–740. doi: 10.1083/jcb.200912135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hall JA, Georgel PT. CHD proteins: A diverse family with strong ties. Biochem Cell Biol. 2007;85:463–476. doi: 10.1139/O07-063. [DOI] [PubMed] [Google Scholar]
- 10.Jacobs SA, Khorasanizadeh S. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science. 2002;295:2080–2083. doi: 10.1126/science.1069473. [DOI] [PubMed] [Google Scholar]
- 11.Nielsen PR, et al. Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature. 2002;416:103–107. doi: 10.1038/nature722. [DOI] [PubMed] [Google Scholar]
- 12.Fischle W, et al. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 2003;17:1870–1881. doi: 10.1101/gad.1110503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Min J, Zhang Y, Xu RM. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev. 2003;17:1823–1828. doi: 10.1101/gad.269603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bouazoune K, et al. The dMi-2 chromodomains are DNA binding modules important for ATP-dependent nucleosome mobilization. EMBO J. 2002;21:2430–2440. doi: 10.1093/emboj/21.10.2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Musselman CA, et al. Binding of the CHD4 PHD2 finger to histone H3 is modulated by covalent modifications. Biochem J. 2009;423:179–187. doi: 10.1042/BJ20090870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mansfield RE, et al. Plant homeodomain (PHD) fingers of CHD4 are histone H3-binding modules with preference for unmodified H3K4 and methylated H3K9. J Biol Chem. 2011;286:11779–11791. doi: 10.1074/jbc.M110.208207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Musselman CA, Kutateladze TG. PHD fingers: Epigenetic effectors and potential drug targets. Mol Interv. 2009;9:314–323. doi: 10.1124/mi.9.6.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Musselman CA, Kutateladze TG. Handpicking epigenetic marks with PHD fingers. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr613. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Z, Patel DJ. Combinatorial readout of dual histone modifications by paired chromatin-associated modules. J Biol Chem. 2011;286:18363–18368. doi: 10.1074/jbc.R111.219139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruthenburg AJ, et al. Recognition of a mononucleosomal histone modification pattern by BPTF via multivalent interactions. Cell. 2011;145:692–706. doi: 10.1016/j.cell.2011.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeng L, et al. Mechanism and regulation of acetylated histone binding by the tandem PHD finger of DPF3b. Nature. 2010;466:258–262. doi: 10.1038/nature09139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flanagan JF, et al. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature. 2005;438:1181–1185. doi: 10.1038/nature04290. [DOI] [PubMed] [Google Scholar]
- 23.Kwan AHY, et al. Engineering a protein scaffold from a PHD finger. Structure. 2003;11:803–813. doi: 10.1016/s0969-2126(03)00122-9. [DOI] [PubMed] [Google Scholar]
- 24.Canzio D, et al. Chromodomain-mediated oligomerization of HP1 suggests a nucleosome-bridging mechanism for heterochromatin assembly. Mol Cell. 2011;41:67–81. doi: 10.1016/j.molcel.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hombach J, Tsubata T, Leclercq L, Stappert H, Reth M. Molecular components of the B-cell antigen receptor complex of the IgM class. Nature. 1990;343:760–762. doi: 10.1038/343760a0. [DOI] [PubMed] [Google Scholar]
- 26.Maier H, et al. Requirements for selective recruitment of Ets proteins and activation of mb-1/Ig-alpha gene transcription by Pax-5 (BSAP) Nucleic Acids Res. 2003;31:5483–5489. doi: 10.1093/nar/gkg785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao H, et al. Opposing effects of SWI/SNF and Mi-2/NuRD chromatin remodeling complexes on epigenetic reprogramming by EBF and Pax5. Proc Natl Acad Sci USA. 2009;106:11258–11263. doi: 10.1073/pnas.0809485106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Linder B, et al. CHD4/Mi-2beta activity is required for the positioning of the mesoderm/neuroectoderm boundary in Xenopus. Genes Dev. 2007;21:973–983. doi: 10.1101/gad.409507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Durr H, Korner C, Muller M, Hickmann V, Hopfner KP. X-ray structures of the Sulfolobus solfataricus SWI2/SNF2 ATPase core and its complex with DNA. Cell. 2005;121:363–373. doi: 10.1016/j.cell.2005.03.026. [DOI] [PubMed] [Google Scholar]
- 30.Donham DC, 2nd, Scorgie JK, Churchill ME. The activity of the histone chaperone yeast Asf1 in the assembly and disassembly of histone H3/H4-DNA complexes. Nucleic Acids Res. 2011;39:5449–5458. doi: 10.1093/nar/gkr097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dyer PN, et al. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 2004;375:23–44. doi: 10.1016/s0076-6879(03)75002-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information