Suppression of Survival Signalling Pathways by the Phosphatase PHLPP (original) (raw)
. Author manuscript; available in PMC: 2014 Jan 1.
Summary
The recently discovered PH (pleckstrin homology) domain Leucine rich repeat Protein Phosphatase (PHLPP) family is emerging as a central component in suppressing cell survival pathways. Originally discovered in a rational search for a phosphatase that directly dephosphorylates and inactivates Akt, PHLPP is now known to potently suppress cell survival both by inhibiting proliferative pathways and by promoting apoptotic pathways. In the first instance, PHLPP directly dephosphorylates a conserved regulatory site (termed the hydrophobic motif) on Akt, protein kinase C (PKC), and S6 kinase, thereby terminating signalling by these pro-survival kinases. In the second instance, PHLPP dephosphorylates and thus activates the pro-apoptotic kinase Mst1, thereby promoting apoptosis. PHLPP is deleted in a large number of cancers and the genetic deletion of one isozyme in a PTEN (phosphatase and tensin homolog located on chromosome 1) +/− prostate cancer model results in increased tumourigenesis, underscoring the role of PHLPP as a tumour suppressor. This review summarises the targets and cellular actions of PHLPP, with emphasis on its role as a tumour suppressor in the oncogenic PI3K (phosphoinositide 3-kinase)/Akt signalling cascade.
Keywords: PHLPP, Akt, PKC, PI3K, mTOR, PTEN, phosphatase, prostate cancer, chronic lymphocytic leukaemia
Introduction
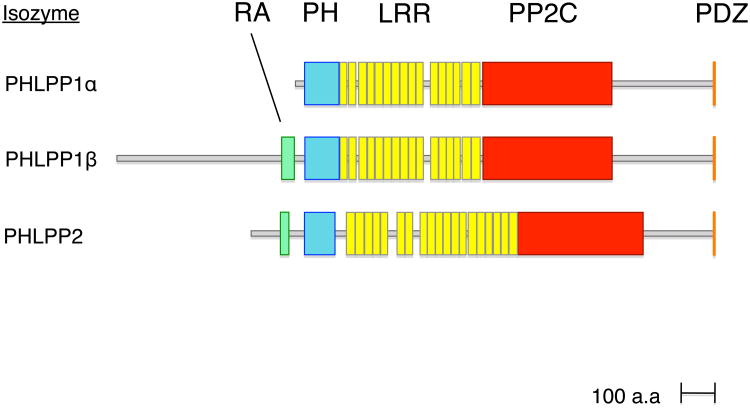
The PHLPP family of serine/threonine phosphatases contains three isozymes (Figure 1): the alternatively-spliced PHLPP1β (also known as suprachiasmatic nucleus oscillatory protein, SCOP [1]) and PHLPP1α [2], and PHLPP2 [3]. PHLPP1α and PHLPP1β differ only in the first exon (see [4] for diagram), resulting in an approximately 50 kDa N-terminal extension to PHLPP1β.
Figure 1.
Domain structure of the human PHLPP isozymes. All PHLPP family members contain a pleckstrin homology (PH) domain, a series of leucine-rich repeats (LRR), a PP2C phosphatase domain, and a C-terminal PDZ ligand. In addition, PHLPP1β and PHLPP2 contain a putative Ras association (RA) domain near their N-termini.
PHLPP1 and PHLPP2 share a common architecture (Figure 1), including a phosphatase domain, which is 58% conserved between PHLPP1 and PHLPP2 [5]. Sequence analysis reveals that the phosphatase domain belongs to the PP2C branch of the PPM family, whose members rely on Mg2+ or Mn2+ for activity and are insensitive to common phosphatase inhibitors such as okadaic acid [2, 6]. Conserved regulatory domains in PHLPP include a pleckstrin homology (PH) domain, a leucine rich repeat segment (LRR), and a C-terminal PDZ(post synaptic density protein (PSD95), Drosophila disc large tumor suppressor (Dlg1), and zonula occludens-1 protein (zo-1)) ligand. [7]. PHLPP1β and PHLPP2 also contain a putative Ras association (RA) domain near their N-termini, although the function of these RA domains is as yet unverified. The PH domain has a relatively low affinity for phosphoinositides [8], as it contains only the middle arginine of the R-X-R-S-F motif required for phosphoinositide binding [5]. The full-length protein has been reported not to bind phosphatidylinotisol 3,4-bisphosphate or phosphatidylinotisol 3,4,5-trisphosphate (PIP3) in vitro [9]; however, whether phosphoinositide binding occurs in cells remains to be established. The PH domain is, however, important for protein interactions and is essential for the regulation of one PHLPP substrate, PKC [10]. The series of leucine-rich repeats have been reported to regulate signalling through the extracellular signal-regulated kinase (ERK) pathway [7]. Finally, a PDZ ligand is present at the C-terminus of PHLPP1 (DTPL) and PHLPP2 (DTAL); the PDZ scaffold NHERF (Na+/H+ exchanger regulatory factor) has recently been reported to bind both sequences [11]. PHLPP is conserved in eukaryotes. Interestingly, the yeast homologue CYR1 contains an adenylate cyclase domain near the C-terminus but no PDZ ligand [6].
The PHLPP family was identified in a rational, systematic search for genes predicted to encode a phosphatase domain linked to a PH domain [2], criteria hypothesized to be important for a phosphatase that would dephosphorylate the lipid second messenger kinases Akt (itself controlled by a PH domain) and protein kinase C (also controlled by membrane-targeting modules). The mRNA of PHLPP1β had previously been identified in a screen for transcripts whose levels oscillate in a circadian fashion in the rat suprachiasmatic nucleus and had thus been termed SCOP [1]. Biochemical and cellular studies validated PHLPP1 and PHLPP2 as functional phosphatases that dephosphorylate and inactivate Akt at its hydrophobic motif site, serine 473 [2, 3]. Since their identification, the PHLPP isozymes have been shown to be widely expressed in human and mouse tissues, with particularly high expression in brain (Table 1) [1, 5, 12]; both PHLPP proteins appear be localised at the membrane and in the cytosol and nucleus of multiple cell types [5].
Table 1.
PHLPP levels in cancer and normal tissue.
| Sample Type | PHLPP expression | Detection | Reference |
|---|---|---|---|
| Human Cancer | |||
| Breast | PHLPP1 mRNA is 2.0 fold lower in invasive ductal breast carcinoma | Array | [74] |
| Breast | PHLPP1 mRNA is 2.3 fold lower in ductal breast carcinoma | Array | [75] |
| CLL | PHLPP1 mRNA is 12.4 fold lower in CLL | Array | [76] |
| CLL | PHLPP1 mRNA is 5.5 fold lower in CLL | Array | [77] |
| CLL with 13q14 deletion | PHLPP1 mRNA is absent in >50% of cases with 13q14 deletion | qPCR | [78] |
| Colon | PHLPP2 mRNA is 5 fold lower in colon cancer | Array | [79] |
| Colon | PHLPP2 mRNA is 6.1-fold lower in rectal adenoma | Array | [79] |
| Colon | PHLPP2 mRNA is 3.5-3.9 fold lower in several types of colon adenoma | Array | [80] |
| Colon | PHLPP2 mRNA is 2 fold lower in colorectal adenoma | Array | [81] |
| Colon | PHLPP1 and PHLPP2 protein expressions are lost or reduced in 78 and 86% of tumour tissue samples, respectively | IHC | [22] |
| Oesophageal | PHLPP1 mRNA is 5.0 fold lower in oesophageal adenocarcinoma | Array | [82] |
| Oesophageal | PHLPP2 mRNA is 3.8 fold lower in oesophageal adenocarcinoma | Array | [82] |
| GBM | PHLPP1 mRNA is 2.1 fold lower in GBM | Array | [83] |
| Liver, Pancreas, Stomach | PHLPP1 protein expression is significantly decreased in liver, pancreas, and stomach tumour samples | IHC | [32] |
| Mantle Cell Lymphoma | PHLPP1 mRNA is 4.3 fold lower in mantle cell lymphoma | Array | [76] |
| Melanoma | PHLPP2 mRNA is 2.3 fold lower in benign melanocytic skin novus | Array | [84] |
| Pancreatic ductal adenocarcinoma | Low PHLPP1 but not PHLPP2 protein expression is negatively correlated with survival | IHC | [25] |
| Prostate | PHLPP1 mRNA is significantly reduced in high Gleason Score tumours | qPCR | [47] |
| Prostate | PHLPP1 and PHLPP2 mRNAs are absent in 30% and 50% of prostate tumours, respectively | Array | [26] |
| Prostate | PHLPP1 protein expression is 4 fold lower in PTEN-null, AR-null prostate cancer compared to benign lesions | IHC | [60] |
| Normal tissue | |||
| Various rat tissues | PHLPP1β protein is expressed in testis, hypothalamus, hippocampus, cortex, olfactory bulb, and cerebellum | WB | [1] |
| Various human tissues | PHLPP1 mRNA is strongly expressed in brain, heart, skeletal muscle, kidney, liver, small intestine, placenta, lung | Northern blot | [23] |
| Mouse T cells | PHLPP1 and 2 mRNAs are increased by 7 and two fold, respectively, in Tregs compared to Tconv | RT-PCR | [35] |
| Mouse hippocampus | PHLPP1 and 2 protein levels are 30-50% lower in CA1 compared to CA3 | WB | [12] |
| Rat SCN | PHLPP1β mRNA is higher at clock time 0 than clock time 12 | RT-PCR | [1] |
| Mouse striatum | PHLPP1 protein levels are ∼50% lower in mouse models of Huntington's disease | WB | [34] |
| Human putamen | PHLPP1 protein levels are ∼80% lower in Huntington's patients | WB | [34] |
| Rat liver | PHLPP2 mRNA levels are 23.4 fold higher in rats given lanthanum nitrate | Array | [85] |
| Human skeletal muscle | PHLPP1 protein is increased ∼two fold in obese versus non-obese subjects | WB | [21] |
| Human white adipose tissue | PHLPP1 protein is increased ∼two fold in obese versus non-obese subjects | WB | [21] |
Molecular, cellular, and physiological functions of PHLPP
Targets of PHLPP
PHLPP was originally identified as the phosphatase for a conserved C-terminal phosphorylation site first identified on PKC [13, 14] and S6 kinase [15] that is conserved in many members of the AGC family, including Akt [16]. This segment is flanked by hydrophobic residues and is thus referred to as the hydrophobic motif [16].
Akt
The PHLPP isozymes bind to the oncogenic serine/threonine kinase Akt and dephosphorylate it under serum-starved, agonist-stimulated, and normal conditions [2, 3]. Upon recruitment to the plasma membrane, Akt is activated by sequential phosphorylations on its activation loop (at threonine 308 in Akt1) by PDK-1 and hydrophobic motif (serine 473) by an mTORC2-facilitated mechanism. Akt phosphorylated at T308 alone is only about 10% as active as the fully phosphorylated form but retains activity towards a group of substrates [17]. Phosphorylation of S473 stabilizes the active conformation of the kinase, allowing for full activation and phosphorylation of all its known substrates [18-20]. Thus, dephosphorylation of S473 by PHLPP is critical for regulation of Akt activity. PHLPP1 and PHLPP2 display a remarkable preference for S473 over T308 in vivo [2, 3]. Accordingly, PHLPP expression decreases Akt activity in vitro and the phosphorylation of numerous Akt substrates in cells, while depletion of either or both PHLPP isozymes results in increased Akt substrate phosphorylation [2, 3, 21-24]. Further supporting the high degree of substrate specificity, knockdown of PHLPP1 versus PHLPP2 affects different subsets of Akt substrates: PHLPP1 knockdown results in increased phosphorylation of GSK (glycogen synthase kinase) 3α and β, MDM2 (murine double minute 2), and TSC2 (tuberous sclerosis complex 2)/tuberin, whereas PHLPP2 knockdown increases the phosphorylation of GSK3β, FoxO (Forkhead Box O) 1, p27, and TSC2 [3]. This selectivity arises because, in certain cell lines, PHLPP1 binds to and dephosphorylates Akt2 and Akt3 but not Akt1, while PHLPP2 binds and regulates Akt1 and Akt3 but not Akt2 [3, 25]. However, it should be noted that this selectivity has only been observed in lung and pancreatic cancer cell lines in culture; in the prostate, which expresses primarily Akt2 and 3 [20], deletion of PHLPP1 still results in an increase in Akt phosphorylation [26]. The mechanisms behind this observed specificity are unknown but could involve differential scaffolding of the isozymes: the PDZ ligand of PHLPP1 is necessary for its regulation of Akt [2], and the PDZ ligands of the two isozymes differ slightly. Indeed, the PDZ ligand of PHLPP2 appears to bind many more recombinant PDZ domains on a PDZ domain array than that of PHLPP1 (Kunkel, M.T., Garcia, E.L., Hall, R.A., and Newton, A.C., unpublished data).
Protein kinase C
Both PHLPP1 and PHLPP2 dephosphorylate the hydrophobic motif of PKC (serine 657 in PKCα) in vitro and in cells [10]. Dephosphorylation of this site in PKC does not acutely impair its activity; rather, it shunts the protein to the detergent-insoluble pellet where it is further dephosphorylated at okadaic acid-sensitive sites (PDK-1 site and turn motif), ubiquitinated, and degraded by a proteosome-dependent mechanism [27]. Thus, knockdown of PHLPP results in increased steady-state levels of PKC. Indeed, PKC levels correlate inversely with PHLPP levels in both cancer and normal cell lines [10]. PHLPP1 binds PKC via the C1 domain of the kinase and the PP2C and PH domains of the phosphatase; notably, the PH domain of PHLPP is required for PHLPP to dephosphorylate PKC in cells, suggesting that this module may play a scaffolding role [10].
S6 kinase
PHLPP has recently been shown to directly dephosphorylate the hydrophobic motif of Akt's close relative S6K1 in vitro; Liu and colleagues elegantly demonstrated that PHLPP knockdown or deletion results in increased phosphorylation of S6K1 and its downstream substrate S6 even when Akt is not activated, or when S6K1 phosphorylation is uncoupled from Akt upon treatment with rapamycin [28]. Interestingly, PHLPP-mediated repression of S6K1 leads to increased insulin-stimulated signalling via loss of feedback inhibition of insulin receptor substrate 1 (IRS-1; see Figure 2), with the apparently paradoxical result that PHLPP knockdown leads to decreased Akt phosphorylation at both the hydrophobic motif and activation loop in the colon cancer cells used in this study.
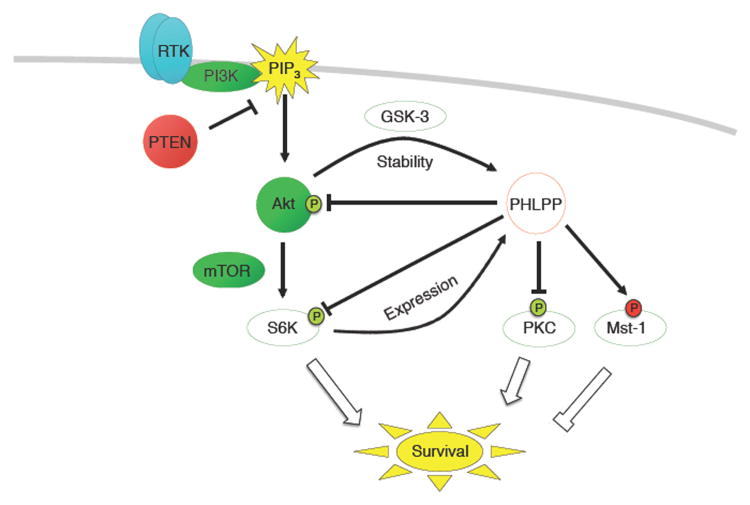
Figure 2.
PHLPP suppresses the PI3K/Akt signalling pathway. Upon activation of growth factor receptors, PI3K is recruited by insulin receptor substrate-1 (IRS-1) and other proteins to the receptors, where it is activated by phosphorylation, resulting in production of the second messenger PIP3 at the plasma membrane. Upon this stimulus, Akt translocates to the plasma membrane, where it is activated by phosphorylation at the activation loop (T308) and hydrophobic motif (S473). Fully phosphorylated, active Akt phosphorylates a range of downstream substrates, including TSC2, which indirectly regulates mTOR, and GSK3β. Several phosphatases (bright red) act to restrain PI3K/Akt pathway activation, including the lipid phosphatase PTEN, which dephosphorylates PIP3, removing the upstream signal for Akt activation, and the protein phosphatases PP2A and PHLPP, which dephosphorylate Akt at T308 (the activation loop) and S473 (the hydrophobic motif), respectively. Note that PHLPP also dephosphorylates S6K1 at its hydrophobic motif, repressing protein synthesis and activating the S6K/IRS-1 feedback loop. PHLPP expression is tightly regulated downstream of Akt, resulting in negative feedback via two mechanisms. First, mTORC1 activation increases PHLPP1 and PHLPP2 protein translation via activation of S6K and inhibition of 4EBP1; second, GSK3β-mediated phosphorylation of PHLPP1 results in increased PHLPP degradation mediated by the E3 ligase β-TrCP. Thus, repression of GSK3β by Akt prevents PHLPP degradation and increases PHLPP protein levels.
The ERK signalling pathway
There is some evidence that PHLPP plays a role in regulating the activation of ERK, although its direct target in this context is unknown. Overexpression of PHLPP1 represses phosphorylation of ERK induced by multiple agonists, including phorbol esters, cytokines, CpG oligonucleotides, and CD40 [7, 24], and siRNA-mediated depletion of PHLPP1 increases the levels of phospho-ERK in neural and B-cell models [12, 24, 29]. Interestingly, the LRR region of the yeast PHLPP ortholog CYR1 binds Ras, allowing Ras-dependent activation of the associated adenylate cyclase [30]. This observation led Shimizu et al. [7] to propose that the LRR of PHLPP1β 1) competes with nucleotides for binding to Ras and 2) is necessary for PHLPP1β to repress phorbol ester-stimulated ERK phosphorylation. However, it is unclear if the regulation of ERK by PHLPP in this context results from dampening of PKC activation or from direct effects on Ras, and further investigation into possible mechanisms is warranted.
Mst1
PHLPP has recently been shown to catalyze an activating dephosphorylation of the pro-apoptotic kinase Mst1 (the human homologue of Hippo), thus providing another mechanism to suppress cell survival. Specifically, Pardee and coworkers reported that in certain gastrointestinal cancer cells, overexpression of PHLPP significantly increased apoptosis in the absence of significant decreases in the phosphorylation of its known substrates, Akt, ERK, or PKC, leading to a search for cell type-specific downstream partners of PHLPP. A mass spectrometry-based screen for PHLPP1-interacting proteins identified Mst1, which was then shown to be a direct substrate for PHLPP1 and PHLPP2. In certain pancreatic cancer cells, PHLPP dephosphorylates an inhibitory residue (threonine 387) on Mst1, which, interestingly, is phosphorylated by Akt [31]. PHLPP-mediated dephosphorylation results in activation of the kinase and increased downstream signalling through p38 and c-Jun N-terminal kinase (JNK), ultimately leading to apoptosis and growth arrest [32]. Interestingly, another study showed that PHLPP1 and 2 do act to dephosphorylate Akt2 and 1, respectively, in a different pancreatic cancer cell line [25], indicating that even within a given cancer type, PHLPP can modulate different targets depending on the specific cell line. These data highlight the cell type-dependent actions of PHLPP and reveal the multiple ways in which PHLPP acts to oppose survival signalling.
Cellular and physiological functions of PHLPP
By both 1] terminating signalling by two survival kinases, Akt and PKC, and 2] activating signalling by a pro-apoptotic kinase, Mst1, PHLPP strongly opposes cell growth and survival; this has been shown in numerous cellular and xenograft models [2, 3, 22, 24, 25, 32]. In addition, PHLPP1 and PHLPP2 decrease protein synthesis and cell size by a mechanism dependent on downregulation of S6K [28]. PHLPP1 and PHLPP2 also appear to repress cellular migration, and both isozymes are less highly expressed in metastatic breast cancer lines compared to primary breast cancer or normal breast lines [32, 33], suggesting that PHLPP may play a role in cancer cell motility.
In normal physiology, PHLPP1 depletion or deletion is cardioprotective: by increasing Akt activation, PHLPP1 knockdown blocks doxorubicin-induced apoptosis in neonatal rat ventricular myocytes, and ischemic injury is reduced in PHLPP1 knockout mouse hearts [23]. Similarly, PHLPP1 depletion may be neuroprotective, as it has been shown to increase Akt signalling in hippocampal and striatal neurons [12, 34]. The effects of PHLPP1 on Akt are also important for regulatory T cells (Tregs), which require suppression of Akt signalling for their development and function and which have elevated expression of PHLPP1 and PHLPP2 compared to conventional T cells. Deletion of PHLPP1 inhibits the development of conventional T cells into Tregs positive for the marker protein Foxp3, and Tregs from PHLPP1 knockout mice are less able to suppress the proliferation of conventional T cells [35].
Given the high level of PHLPP expression in the brain, it is not surprising that PHLPP has been postulated to play roles in several neural functions, namely long-term memory and the regulation of circadian rhythms. Inducible overexpression of PHLPP1β in the murine forebrain blocks the formation of long-term novel object memory [29], while deletion of PHLPP1 interferes with light-induced resetting of the circadian clock [36]. Interestingly, injection of a PKC inhibitor into the suprachiasmatic nucleus has been shown to affect the circadian response to a light pulse [37], raising the possibility that the regulation of PKC by PHLPP is essential for the proper function of the circadian clock.
PHLPP in cancer
Several lines of data support a role for PHLPP as a tumour suppressor in vivo. First, as previously mentioned, PHLPP overexpression in glioblastoma (GBM), breast, colon, and pancreatic cancer cell lines decreases colony formation and growth in vitro and in xenograft models [2, 22, 25, 32, 38]. Second, PHLPP expression is lost or decreased in a variety of human cancers (as detailed in the regulation section below). Furthermore, the PHLPP1 knockout mouse displays an increased incidence of prostate intraepithelial neoplasia, which eventually progresses to frank carcinoma when combined with partial loss of PTEN (phosphatase and tensin homolog located on chromosome 10) [26]. Interestingly, it appears that PHLPP1 loss cooperates with PTEN loss and p53 mutation to promote prostate carcinogenesis. PHLPP1-/-, PTEN+/- mouse prostate cells initially undergo senescence; however, upon spontaneous inactivation of p53, this genetic combination results in increased proliferation in the prostate, carcinoma formation, and increased mortality. Moreover, PHLPP1-/, PTEN+/-, p53-mutant mouse embryonic fibroblasts are dependent upon Akt activity for their increased abilities to proliferate, survive, and form colonies in soft agar, suggesting that in this context, at least, PHLPP blocks carcinogenesis through its actions on Akt. The idea that PHLPP1 and PTEN downregulation cooperate to increase carcinogenesis is supported by studies in humans: Kaplan-Meier analysis of relapse after radical prostatectomy revealed that a “PTEN/PHLPP1 low” status predicts relapse better than Gleason scoring [26]. Finally, though few variations in the PHLPP protein sequences have been described, a single nucleotide polymorphism in PHLPP2 (L1016S, located in the phosphatase domain) has been shown to result in reduced inhibition of basal Akt phosphorylation. This polymorphism has relevance for human cancer, as grade 3 but not grade 2 breast tumours often display loss of heterozygosity at this locus, resulting in preferential loss of expression of only the more active leucine allele [39].
Regulation of PHLPP
Downregulation of PHLPP in cancer
The genetic locus for PHLPP1, which lies at 18q21.33, undergoes loss of heterozygosity in a high percentage of colon cancers [40, 41]; similarly, the PHLPP2 locus at 16q22.3-16q23.1, which contains a fragile site, is susceptible to loss of heterozygosity in breast and ovarian cancers, Wilms' tumours, and prostate and hepatocellular carcinomas [42-46].
Consistent with a role for PHLPP in the progression of prostate cancer, several studies have shown that PHLPP levels are decreased in this disease. Chen et al. [26] discovered that PHLPP1 and PHLPP2 protein expression is lost in 30% and 45%, respectively, of human prostate cancer samples, while Hellwinkel et al. [47] found that PHLPP1 mRNA levels were frequently downregulated in high-grade but not low-grade prostate cancer samples, supporting the idea that PHLPP1 loss participates with other genetic abnormalities to promote prostate cancer progression. In fact, loss of both PHLPP1 and PHLPP2 is associated with PTEN and p53 loss in metastatic but not primary prostate cancer, which suggests that p53 loss is a late event that cooperates with PHLPP loss in metastasis [26]. The effects of PHLPP1 deletion on prostate carcinogenesis may be explained by the strong dependence of prostate cancer on PI3K/Akt signalling; a survey of 218 prostate tumours found that the PI3K pathway is altered in almost half of the primary tumours and all metastatic samples. (Notably, this same study found downregulation of PHLPP1 or PHLPP2 in 11% of primary tumours and 37% of metastases [48].)
In addition to prostate cancer, PHLPP levels are decreased in a number of other malignancies (summarised in Table 1), particularly chronic lymphocytic leukaemia (CLL), GBM, and colon cancer. However, the factors that regulate PHLPP expression in normal and cancer cells are incompletely characterised and deserve further attention.
Regulation of PHLPP expression
Transcriptional regulation of the PHLPP1 and PHLPP2 genes is still very much a black box, although there are several hints at possible regulators. SMAD3, a downstream effector of transforming growth factor-β, binds the PHLPP1 promoter, and this binding has been shown to correlate with increased PHLPP1 expression during the development of regulatory T cells [35]. Interestingly, the SMAD3 binding partner SMAD4 acts as a tumour suppressor in PTEN-null prostate cancer and is often co-deleted with PHLPP1, with which it shares a genetic locus [26, 49]. Expression of Huntington's disease-associated alleles of huntingtin decreases the mRNA levels of PHLPP1, perhaps via nuclear transcription factor Y (NF-Y), which has putative binding sites located in the PHLPP1 promoter [34]. PHLPP1 mRNA expression is also repressed by the microRNA miR-190, which is upregulated by exposure to the carcinogen arsenic and which is overexpressed in many cancers, including CLL [50]. There may also be a connection between Akt signalling and PHLPP1 mRNA expression, though the mechanism is unclear; one study found a slight elevation in PHLPP1 but not PHLPP2 mRNA in primary myotubes from type 2 diabetics, which displayed decreased Akt activation in response to insulin. Conversely, Warfel et al. [51] examined microarray data from the NCI-60 panel of cancer cell lines and found decreased levels of PHLPP1 mRNA in GBM lines compared to astrocytoma lines, correlating with increased Akt activation in GBM. However, effects on PHLPP1 protein synthesis and degradation downstream of Akt often complicate the interpretation of Akt-dependent changes in PHLPP protein levels, which may involve transcriptional, translational, and/or protein-degradation mechanisms.
The post-translational regulation of PHLPP is better understood than its transcriptional regulation. To date, two mechanisms controlling the steady-state levels of PHLPP protein have been described. Both act downstream of Akt, implying the existence of at least two negative feedback loops involving PHLPP and Akt (see Figure 2). First, the rate of PHLPP translation is controlled by mTOR (mammalian target of rapamycin): treating cells with the mTOR inhibitor rapamycin or genetically interfering with the mTORC1 complex decreases PHLPP1 and PHLPP2 protein levels without affecting protein degradation or mRNA expression, suggesting that mTOR activation downstream of Akt results in increased translation of PHLPP1 and PHLPP2 [38]. Consistent with this, PHLPP1 depletion in 3T3 fibroblasts and the subsequent increase in Akt signalling increase PHLPP2 protein but not mRNA levels in an mTOR-dependent fashion [26]. Second, the rate of degradation of PHLPP is controlled by Akt activity, whose inhibitory phosphorylation of GSK3β results in stabilization of PHLPP1 protein [52]. Specifically, casein kinase- and GSK3β-mediated phosphorylation of PHLPP1 at threonine 851 and serine 847, respectively, contributes to recognition of PHLPP1 by the E3 ligase β-TrCP (β-transducin repeats-containing protein) and subsequent ubiquitin-mediated degradation. Notably, this latter feedback loop is frequently lost in GBM (both in GBM cell lines and primary GBM neurospheres from human tumours); in these samples, β-TrCP is sequestered in the nucleus, away from its substrate PHLPP1, and the rate of PHLPP1 degradation is insensitive to Akt activation [51]. Either or both of these Akt-dependent mechanisms may explain the increase in PHLPP1 protein levels seen under conditions of Akt activation, including insulin stimulation of hepatoma cells [21], treatment of rat ventricular myocytes with leukaemia inhibitory factor, and transgenic overexpression of insulin-like growth factor 1 in the mouse heart [23]. It should be noted that β-TrCP-dependent degradation of PHLPP1 is not the only mechanism by which PHLPP1 is degraded: the calcium-dependent protease calpain has also been shown to degrade PHLPP1β in vitro and in vivo, and inhibition of calpain in the hippocampus blocks long-term novel object memory formation, which is opposed by PHLPP1β expression (see “cellular and physiological functions of PHLPP”, above) [29]. Also, the deubiquitinase UCH-L1 (ubiquitin carboxy terminal-hydrolase L1), which acts as a tumour promoter in CLL, decreases PHLPP1β protein but not mRNA expression in a manner that depends upon its deubiquitinase activity [53].
Regulation of PHLPP by Protein Scaffolds
The key role of protein-protein interactions in achieving spatial and temporal specificity of signalling by enzymes has come to the forefront of the signal transduction field in recent years [54]. PHLPP is no exception: a number of protein scaffolds for PHLPP have been proposed to be essential for the regulation of PHLPP targets. First, NHERF1, which binds PHLPP1, PHLPP2, and PTEN via its two PDZ domains, has been reported to localise PHLPP1 to the membrane in GBM cells, an interaction that allows the phosphatase to exert anti-proliferative effects [11]. Interestingly, samples from patients with high-grade GBM often display concomitant reductions in PHLPP1, PTEN, and NHERF protein levels, and upon PTEN depletion in LN229 cells, PHLPP1 becomes better able to reduce Akt activation, likely because of increased scaffold availability. These data again highlight the cooperation between PHLPP1 loss and PTEN loss and suggest that in the absence of PTEN, PHLPP1 provides an additional layer of negative regulation of the PI3K pathway [11]. The scaffold Scribble has been proposed to play a similar role in colon cancer cells, where it coordinates PHLPP1 (via Scribble's LRR region) and Akt at the membrane, allowing PHLPP1 to dephosphorylate S473 [55]. Finally, the putative tumour suppressor FKBP51 (FK506 binding protein 51) has been reported to scaffold PHLPP to Akt in pancreatic cancer cells [56]. FKBP51 is essential for PHLPP to dephosphorylate Akt and exert its apoptotic effects in pancreatic cancer cells; accordingly, overexpression of FKBP51 results in decreases in S473 phosphorylation of Akt and cell survival that are dependent on the presence of PHLPP [57]. Other studies suggest that this interaction may play a role in suppressing Akt signalling in lung and prostate cancers. In non-small cell lung cancer cells, repression of insulin-stimulated Akt phosphorylation downstream of the P2 × 7 purinergic receptor requires both PHLPP and the activity of FKBP51, which scaffolds Akt in these cells and may recruit PHLPP, PTEN, and calcineurin to oppose Akt phosphorylation [58]. In models of prostate cancer, castration or androgen receptor inhibition decreases PHLPP1 protein levels in a manner that depends upon FKBP51 depletion, suggesting that FKBP51 not only scaffolds PHLPP but also plays a role in maintaining its stability [59, 60]. In human prostate cancer tissue samples, PHLPP1 protein levels are correlated with the levels of PTEN, FKBP51, and the androgen receptor, raising the possibility that the androgen and PI3K pathways may interact via inhibition of PHLPP [60].
PHLPP has also been shown to interact with several other signalling factors, though the effects of these interactions are mainly unknown. PHLPP2 binds to adenylate cyclase in rat cardiomyocytes, and this interaction appears to contribute to forskolin-stimulated downregulation of Akt [61]. Interestingly, this interaction recapitulates the proximity between a PP2C phosphatase domain linked and adenylate cyclase seen in yeast, where these two domains are part of the same protein, CYR1. In addition, a recent proteomic screen of the human deubiquitinases revealed ubiquitin-specific peptidases (USPs) 12 and 46 as binding partners for both PHLPP1 and PHLPP2 [62]. These and other interactions remain to be further characterised and may offer hints to mechanisms of PHLPP regulation or the discovery of new PHLPP targets.
Phosphatase activity of PHLPP
Little is yet known about how the intrinsic catalytic activity of PHLPP is controlled in cells. However, a recent screen for specific inhibitors of PHLPP revealed insights into the structure of the phosphatase domain. Homology modelling of the structure of the PP2C domain of PHLPP2 onto the known structure of PP2Cα [6], combined with knowledge about the structure of several validated PHLPP2 inhibitors, allowed Sierecki et al. to construct a set of models for the active site, all of which included one, two, or three manganese ions, and to determine key residues for the catalytic activity, which include aspartate 806, glutamate 989, and aspartate 1024 [63]. Virtual screening using the best of these theoretical catalytic domain structures allowed the identification of numerous new predicted inhibitors, a few of which were experimentally tested and shown to inhibit PHLPP activity in vitro and in vivo. As expected, these inhibitors increase phosphorylation of Akt and its downstream substrates and attenuate etoposide-induced apoptosis in cell culture [63].
Endogenous regulators of PHLPP activity per se remain to be identified. Both isozymes have an abundance of predicted phosphorylation sites, and phosphorylation is a likely candidate to control the catalytic activity of the enzyme. Indeed, it has been suggested that factors downstream of the insulin receptor may modulate PHLPP activity [64], and serum stimulation or overexpression of Akt1, 2, or 3 was reported to decrease the activity of tagged, exogenously expressed PHLPP [9]. Further study of the regulation of PHLPP activity in vivo in the coming years is likely to unveil mechanisms that control the catalytic activity of PHLPP.
Phosphatases in the PI3K pathway
The PI3K/Akt signalling cascade (see Figure 2) is a well-studied, complex pathway that promotes cell growth and survival. Attesting to the importance of precise control of Akt signalling for cellular homeostasis, signalling through this pathway is often increased in primary and metastatic tumours [48, 65], and its activation, combined with that of other factors such as Ras [66], is crucial for driving the majority of human cancers [67]. In fact, several inhibitors of the pathway targeted at PI3K, Akt, and/or mTOR are currently in clinical trials as cancer therapeutics [65]. Though much attention has been focussed on the mutation of factors that positively affect this pathway (e.g., growth factor receptors and PI3K), negative regulation is equally important and often involves dephosphorylation, which opposes signalling through this pathway via several mechanisms. These include 1) downregulation of PI3K signalling via removal of its product PIP3 by the lipid phosphatases PTEN [68] and SHIP (which dephosphorylates the 5′ position to yield PI(3,4)P2) [69], 2) dephosphorylation of Akt at its activation loop and hydrophobic motif by PP2A and PHLPP, respectively [2, 3, 70], and 3) desphosphorylation/activation of the Akt substrate and Hippo homologue Mst1, which acts to increase apoptosis and limit organ size [31, 71]. The importance of proper phosphatase regulation of this pathway is highlighted by several recent studies showing that depletion of both PHLPP and PTEN is necessary for full activation of Akt in GBM [11] and predicts metastasis and relapse in prostate cancer [26].
This exquisitely tuned pathway is subject to several layers of negative feedback regulation (Figure 2). One well-characterised loop involves S6K-mediated inhibitory phosphorylation of IRS-1, which dampens activation of PI3K in the face of excess mTOR activation [72]. Also, Yu et al. [73] recently showed that mTORC1 phosphorylates and stabilizes the adaptor protein Grb10, which negatively regulates signalling through growth factor receptors. Recent studies (described in more detail above) have demonstrated the existence of three more feedback loops, these ones involving PHLPP. First, mTOR activation positively regulates the translation of PHLPP1 and PHLPP2, thereby resulting in inactivation of Akt by PHLPP-mediated dephosphorylation [38]. Second, increased Akt activation results in phosphorylation and inhibition of GSK3β, resulting in decreased phosphorylation of PHLPP1 at serine 847. PHLPP1 that is not phosphorylated at serine 847 cannot be recognized by its E3 ligase β-TrCP and is therefore stabilized, leading to increased levels of PHLPP and decreased activation of Akt [51, 52]. Notably, this feedback loop is lost in GBM because of sequestration of β-TrCP in the nucleus, away from its substrate PHLPP1 [51]. Finally, PHLPP also dephosphorylates the activation loop of S6K1, thus activating PI3K/Akt signalling through upregulation of IRS-1 [28].
Perspective
Repression of signalling by protein phosphatases, originally conceived as a promiscuous, general “off” signal, has increasingly been recognized as an important layer of specific and dynamic regulation in mammalian cells. The PHLPP phosphatases, which dephosphorylate the oncogenic kinase Akt (among other targets), provide good examples of specificity of signalling. First, they are highly selective for the hydrophobic motif of Akt over the activation loop; second, they display a remarkable isozyme specificity, with PHLPP1 binding and dephosphorylating Akt2 and 3 and PHLPP2 binding and dephosphorylating Akt1 and 3. Their localization and binding to Akt also appear to be tightly regulated by the scaffolding proteins NHERF1, Scribble, and FKBP51 in a cell type-dependent manner, and the requirement for various protein-protein interaction domains for binding to these factors and to PHLPP substrates highlights the additional layer of regulation provided by such non-catalytic domains. Finally, the existence of several feedback loops involving upregulation of PHLPP protein levels downstream of Akt activation suggest that precise regulation of this phosphatase is essential for the modulation of signalling through the PI3K/Akt pathway.
The rise in studies concerning PHLPP since its characterisation in 2005 is beginning to unveil the targets and upstream regulators of this phosphatase. With recent evidence demonstrating a tumour suppressor role of PHLPP, future studies of the regulation of this enzyme in normal and cancer cells is likely to provide major insights into this enzyme as a therapeutic target and/or biomarker in human cancer.
Acknowledgments
This work was supported by NIH GM43154 and NIH P01 DK54441 (to A.C.N.). A.O. and M.N. were supported in part by the University of California at San Diego Graduate Training Program in Cellular and Molecular Pharmacology through NIGMS, National Institutes of Health Institutional Training Grant T32 GM007752.
Abbreviations
PHLPP
PH domain leucine rich repeat protein phosphatase
PH
pleckstrin homology
PKC
protein kinase C
PTEN
phosphatase and tensin homolog located on chromosome 10
PI3K
phosphoinositide 3-kinase
SCOP
suprachiasmatic nucleus oscillatory protein
LRR
leucine rich repeat
PDZ
post synaptic density protein (PSD95), Drosophila disc large tumor suppressor (DLG), and zonula occludens-1 protein (zo-1)
RA
Ras association
PIP3
phosphatidylinotisol 3,4,5-trisphosphate
ERK
extracellular signal-regulated kinase
NHERF
Na+/H+ exchanger regulatory factor
IRS-1
insulin receptor substrate 1
Tregs
regulatory T cells
GBM
glioblastoma
CLL
chronic lymphocytic leukaemia
mTOR
mammalian target of rapamycin
β-TrCP
β-transducin repeats-containing protein
FKBP51
FK506 binding protein 51
IHC
immunohistochemistry
WB
Western blot
Tconv
conventional T cells
SCN
suprachiasmatic nucleus
References
- 1.Shimizu K, Okada M, Takano A, Nagai K. SCOP, a novel gene product expressed in a circadian manner in rat suprachiasmatic nucleus. FEBS Lett. 1999;458:363–9. doi: 10.1016/s0014-5793(99)01190-4. [DOI] [PubMed] [Google Scholar]
- 2.Gao T, Furnari F, Newton AC. PHLPP: a phosphatase that directly dephosphorylates Akt, promotes apoptosis, and suppresses tumor growth. Mol Cell. 2005;18:13–24. doi: 10.1016/j.molcel.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 3.Brognard J, Sierecki E, Gao T, Newton AC. PHLPP and a second isoform, PHLPP2, differentially attenuate the amplitude of Akt signaling by regulating distinct Akt isoforms. Mol Cell. 2007;25:917–31. doi: 10.1016/j.molcel.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 4.O'Neill A, Newton AC. PHLPP1 (PH domain leucine-rich repeat protein phosphatase 1) Atlas Genet Cytogenet Oncol Haematol. 2009 URL: http://AtlasGeneticsOncology.org/Genes/PHLPP1ID44544ch18q21.html.
- 5.Brognard J, Newton AC. PHLiPPing the switch on Akt and protein kinase C signaling. Trends Endocrinol Metab. 2008;19:223–30. doi: 10.1016/j.tem.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barford D, Das AK, Egloff MP. The structure and mechanism of protein phosphatases: insights into catalysis and regulation. Annu Rev Biophys Biomol Struct. 1998;27:133–64. doi: 10.1146/annurev.biophys.27.1.133. [DOI] [PubMed] [Google Scholar]
- 7.Shimizu K, Okada M, Nagai K, Fukada Y. Suprachiasmatic nucleus circadian oscillatory protein, a novel binding partner of K-Ras in the membrane rafts, negatively regulates MAPK pathway. J Biol Chem. 2003;278:14920–5. doi: 10.1074/jbc.M213214200. [DOI] [PubMed] [Google Scholar]
- 8.Park WS, Heo WD, Whalen JH, O'Rourke NA, Bryan HM, Meyer T, Teruel MN. Comprehensive identification of PIP3-regulated PH domains from C. elegans to H. sapiens by model prediction and live imaging. Mol Cell. 2008;30:381–92. doi: 10.1016/j.molcel.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kanan Y, Matsumoto H, Song H, Sokolov M, Anderson RE, Rajala RV. Serine/threonine kinase akt activation regulates the activity of retinal serine/threonine phosphatases, PHLPP and PHLPPL. J Neurochem. 2010;113:477–88. doi: 10.1111/j.1471-4159.2010.06609.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao T, Brognard J, Newton AC. The phosphatase PHLPP controls the cellular levels of protein kinase C. J Biol Chem. 2008;283:6300–11. doi: 10.1074/jbc.M707319200. [DOI] [PubMed] [Google Scholar]
- 11.Molina JR, Agarwal NK, Morales FC, Hayashi Y, Aldape KD, Cote G, Georgescu MM. PTEN, NHERF1 and PHLPP form a tumor suppressor network that is disabled in glioblastoma. Oncogene. 2011 doi: 10.1038/onc.2011.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jackson TC, Verrier JD, Semple-Rowland S, Kumar A, Foster TC. PHLPP1 splice variants differentially regulate AKT and PKCalpha signaling in hippocampal neurons: characterization of PHLPP proteins in the adult hippocampus. J Neurochem. 2010;115:941–55. doi: 10.1111/j.1471-4159.2010.06984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keranen LM, Dutil EM, Newton AC. Protein kinase C is regulated in vivo by three functionally distinct phosphorylations. Current biology: CB. 1995;5:1394–1403. doi: 10.1016/s0960-9822(95)00277-6. [DOI] [PubMed] [Google Scholar]
- 14.Tsutakawa SE, Medzihradszky KF, Flint AJ, Burlingame AL, Koshland DE., Jr Determination of in vivo phosphorylation sites in protein kinase C. J Biol Chem. 1995;270:26807–12. doi: 10.1074/jbc.270.45.26807. [DOI] [PubMed] [Google Scholar]
- 15.Pearson RB, Dennis PB, Han JW, Williamson NA, Kozma SC, Wettenhall RE, Thomas G. The principal target of rapamycin-induced p70s6k inactivation is a novel phosphorylation site within a conserved hydrophobic domain. The EMBO journal. 1995;14:5279–87. doi: 10.1002/j.1460-2075.1995.tb00212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Newton AC. Regulation of the ABC kinases by phosphorylation: protein kinase C as a paradigm. The Biochemical journal. 2003;370:361–71. doi: 10.1042/BJ20021626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fayard E, Tintignac LA, Baudry A, Hemmings BA. Protein kinase B/Akt at a glance. J Cell Sci. 2005;118:5675–8. doi: 10.1242/jcs.02724. [DOI] [PubMed] [Google Scholar]
- 18.Guertin DA, Stevens DM, Thoreen CC, Burds AA, Kalaany NY, Moffat J, Brown M, Fitzgerald KJ, Sabatini DM. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev Cell. 2006;11:859–71. doi: 10.1016/j.devcel.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 19.Jacinto E, Facchinetti V, Liu D, Soto N, Wei S, Jung SY, Huang Q, Qin J, Su B. SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell. 2006;127:125–37. doi: 10.1016/j.cell.2006.08.033. [DOI] [PubMed] [Google Scholar]
- 20.Yang ZZ, Tschopp O, Hemmings-Mieszczak M, Feng J, Brodbeck D, Perentes E, Hemmings BA. Protein kinase B alpha/Akt1 regulates placental development and fetal growth. J Biol Chem. 2003;278:32124–31. doi: 10.1074/jbc.M302847200. [DOI] [PubMed] [Google Scholar]
- 21.Andreozzi F, Procopio C, Greco A, Mannino GC, Miele C, Raciti GA, Iadicicco C, Beguinot F, Pontiroli AE, Hribal ML, Folli F, Sesti G. Increased levels of the Akt-specific phosphatase PH domain leucine-rich repeat protein phosphatase (PHLPP)-1 in obese participants are associated with insulin resistance. Diabetologia. 2011;54:1879–87. doi: 10.1007/s00125-011-2116-6. [DOI] [PubMed] [Google Scholar]
- 22.Liu J, Weiss HL, Rychahou P, Jackson LN, Evers BM, Gao T. Loss of PHLPP expression in colon cancer: role in proliferation and tumorigenesis. Oncogene. 2009;28:994–1004. doi: 10.1038/onc.2008.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miyamoto S, Purcell NH, Smith JM, Gao T, Whittaker R, Huang K, Castillo R, Glembotski CC, Sussman MA, Newton AC, Brown JH. PHLPP-1 negatively regulates Akt activity and survival in the heart. Circ Res. 2010;107:476–84. doi: 10.1161/CIRCRESAHA.109.215020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suljagic M, Laurenti L, Tarnani M, Alam M, Malek SN, Efremov DG. Reduced expression of the tumor suppressor PHLPP1 enhances the antiapoptotic B-cell receptor signal in chronic lymphocytic leukemia B-cells. Leukemia. 2010;24:2063–71. doi: 10.1038/leu.2010.201. [DOI] [PubMed] [Google Scholar]
- 25.Nitsche C, Edderkaoui M, Moore RM, Eibl G, Kasahara N, Treger J, Grippo PJ, Mayerle J, Lerch MM, Gukovskaya AS. The Phosphatase PHLPP1 Regulates Akt2, Promotes Pancreatic Cancer Cell Death, and Inhibits Tumor Formation. Gastroenterology. 2011 doi: 10.1053/j.gastro.2011.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen M, Pratt CP, Zeeman ME, Schultz N, Taylor BS, O'Neill A, Castillo-Martin M, Nowak DG, Naguib A, Grace DM, Murn J, Navin N, Atwal GS, Sander C, Gerald WL, Cordon-Cardo C, Newton AC, Carver BS, Trotman LC. Identification of PHLPP1 as a Tumor Suppressor Reveals the Role of Feedback Activation in PTEN-Mutant Prostate Cancer Progression. Cancer Cell. 2011;20:173–86. doi: 10.1016/j.ccr.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gysin S, Imber R. Phorbol-ester-activated protein kinase C-alpha lacking phosphorylation at Ser657 is down-regulated by a mechanism involving dephosphorylation. Eur J Biochem. 1997;249:156–60. doi: 10.1111/j.1432-1033.1997.t01-2-00156.x. [DOI] [PubMed] [Google Scholar]
- 28.Liu J, Stevens PD, Li X, Schmidt MD, Gao T. PHLPP-mediated dephosphorylation of S6K1 inhibits protein translation and cell growth. Mol Cell Biol. 2011 doi: 10.1128/MCB.05799-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shimizu K, Phan T, Mansuy IM, Storm DR. Proteolytic degradation of SCOP in the hippocampus contributes to activation of MAP kinase and memory. Cell. 2007;128:1219–29. doi: 10.1016/j.cell.2006.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Field J, Xu HP, Michaeli T, Ballester R, Sass P, Wigler M, Colicelli J. Mutations of the adenylyl cyclase gene that block RAS function in Saccharomyces cerevisiae. Science. 1990;247:464–7. doi: 10.1126/science.2405488. [DOI] [PubMed] [Google Scholar]
- 31.Jang SW, Yang SJ, Srinivasan S, Ye K. Akt phosphorylates MstI and prevents its proteolytic activation, blocking FOXO3 phosphorylation and nuclear translocation. J Biol Chem. 2007;282:30836–44. doi: 10.1074/jbc.M704542200. [DOI] [PubMed] [Google Scholar]
- 32.Qiao M, Wang Y, Xu X, Lu J, Dong Y, Tao W, Stein J, Stein GS, Iglehart JD, Shi Q, Pardee AB. Mst1 is an interacting protein that mediates PHLPPs' induced apoptosis. Mol Cell. 2010;38:512–23. doi: 10.1016/j.molcel.2010.03.017. [DOI] [PubMed] [Google Scholar]
- 33.Qiao M, Iglehart JD, Pardee AB. Metastatic potential of 21T human breast cancer cells depends on Akt/protein kinase B activation. Cancer Res. 2007;67:5293–9. doi: 10.1158/0008-5472.CAN-07-0877. [DOI] [PubMed] [Google Scholar]
- 34.Saavedra A, Garcia-Martinez JM, Xifro X, Giralt A, Torres-Peraza JF, Canals JM, Diaz-Hernandez M, Lucas JJ, Alberch J, Perez-Navarro E. PH domain leucine-rich repeat protein phosphatase 1 contributes to maintain the activation of the PI3K/Akt pro-survival pathway in Huntington's disease striatum. Cell Death Differ. 2009;17:324–35. doi: 10.1038/cdd.2009.127. [DOI] [PubMed] [Google Scholar]
- 35.Patterson SJ, Han JM, Garcia R, Assi K, Gao T, O'Neill A, Newton AC, Levings MK. Cutting edge: PHLPP regulates the development, function, and molecular signaling pathways of regulatory T cells. J Immunol. 2011;186:5533–7. doi: 10.4049/jimmunol.1002126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Masubuchi S, Gao T, O'Neill A, Eckel-Mahan K, Newton AC, Sassone-Corsi P. Protein phosphatase PHLPP1 controls the light-induced resetting of the circadian clock. Proc Natl Acad Sci U S A. 2010;107:1642–7. doi: 10.1073/pnas.0910292107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee B, Almad A, Butcher GQ, Obrietan K. Protein kinase C modulates the phase-delaying effects of light in the mammalian circadian clock. Eur J Neurosci. 2007;26:451–62. doi: 10.1111/j.1460-9568.2007.05664.x. [DOI] [PubMed] [Google Scholar]
- 38.Liu J, Stevens PD, Gao T. mTOR-dependent regulation of PHLPP expression controls the rapamycin sensitivity in cancer cells. J Biol Chem. 2010;286:6510–20. doi: 10.1074/jbc.M110.183087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brognard J, Niederst M, Reyes G, Warfel N, Newton AC. Common polymorphism in the phosphatase PHLPP2 results in reduced regulation of Akt and protein kinase C. J Biol Chem. 2009;284:15215–23. doi: 10.1074/jbc.M901468200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goel A, Arnold CN, Niedzwiecki D, Chang DK, Ricciardiello L, Carethers JM, Dowell JM, Wasserman L, Compton C, Mayer RJ, Bertagnolli MM, Boland CR. Characterization of sporadic colon cancer by patterns of genomic instability. Cancer Res. 2003;63:1608–14. [PubMed] [Google Scholar]
- 41.Johnson-Pais TL, Nellissery MJ, Ammerman DG, Pathmanathan D, Bhatia P, Buller CL, Leach RJ, Hansen MF. Determination of a minimal region of loss of heterozygosity on chromosome 18q21.33 in osteosarcoma. Int J Cancer. 2003;105:285–8. doi: 10.1002/ijc.11070. [DOI] [PubMed] [Google Scholar]
- 42.Patael-Karasik Y, Daniely M, Gotlieb WH, Ben-Baruch G, Schiby J, Barakai G, Goldman B, Aviram A, Friedman E. Comparative genomic hybridization in inherited and sporadic ovarian tumors in Israel. Cancer Genet Cytogenet. 2000;121:26–32. doi: 10.1016/s0165-4608(00)00224-7. [DOI] [PubMed] [Google Scholar]
- 43.Rakha EA, Green AR, Powe DG, Roylance R, Ellis IO. Chromosome 16 tumor-suppressor genes in breast cancer. Genes Chromosomes Cancer. 2006;45:527–35. doi: 10.1002/gcc.20318. [DOI] [PubMed] [Google Scholar]
- 44.Safford SD, Goyeau D, Freemerman AJ, Bentley R, Everett ML, Grundy PE, Skinner MA. Fine mapping of Wilms' tumors with 16q loss of heterozygosity localizes the putative tumor suppressor gene to a region of 6.7 megabases. Ann Surg Oncol. 2003;10:136–43. doi: 10.1245/aso.2003.03.038. [DOI] [PubMed] [Google Scholar]
- 45.Torring N, Borre M, Sorensen KD, Andersen CL, Wiuf C, Orntoft TF. Genome-wide analysis of allelic imbalance in prostate cancer using the Affymetrix 50K SNP mapping array. Br J Cancer. 2007;96:499–506. doi: 10.1038/sj.bjc.6603476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsuda H, Zhang WD, Shimosato Y, Yokota J, Terada M, Sugimura T, Miyamura T, Hirohashi S. Allele loss on chromosome 16 associated with progression of human hepatocellular carcinoma. Proc Natl Acad Sci U S A. 1990;87:6791–4. doi: 10.1073/pnas.87.17.6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hellwinkel OJ, Rogmann JP, Asong LE, Luebke AM, Eichelberg C, Ahyai S, Isbarn H, Graefen M, Huland H, Schlomm T. A comprehensive analysis of transcript signatures of the phosphatidylinositol-3 kinase/protein kinase B signal-transduction pathway in prostate cancer. BJU Int. 2008;101:1454–60. doi: 10.1111/j.1464-410X.2008.07540.x. [DOI] [PubMed] [Google Scholar]
- 48.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva B, Antipin Y, Mitsiades N, Landers T, Dolgalev I, Major JE, Wilson M, Socci ND, Lash AE, Heguy A, Eastham JA, Scher HI, Reuter VE, Scardino PT, Sander C, Sawyers CL, Gerald WL. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ding Z, Wu CJ, Chu GC, Xiao Y, Ho D, Zhang J, Perry SR, Labrot ES, Wu X, Lis R, Hoshida Y, Hiller D, Hu B, Jiang S, Zheng H, Stegh AH, Scott KL, Signoretti S, Bardeesy N, Wang YA, Hill DE, Golub TR, Stampfer MJ, Wong WH, Loda M, Mucci L, Chin L, DePinho RA. SMAD4-dependent barrier constrains prostate cancer growth and metastatic progression. Nature. 2011;470:269–73. doi: 10.1038/nature09677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Beezhold K, Liu J, Kan H, Meighan T, Castranova V, Shi X, Chen F. miR-190-mediated downregulation of PHLPP contributes to arsenic-induced Akt activation and carcinogenesis. Toxicol Sci. 2011 doi: 10.1093/toxsci/kfr188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Warfel NA, Niederst M, Stevens MW, Brennan PM, Frame MC, Newton AC. Mislocalization of the E3 ligase, beta-transducin repeat-containing protein 1 (beta-TrCP1), in the pleckstrin homology domain leucine-rich repeat protein phosphatase 1 (PHLPP1) and Akt. J Biol Chem. 2011;286:19777–88. doi: 10.1074/jbc.M111.237081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li X, Liu J, Gao T. beta-TrCP-mediated ubiquitination and degradation of PHLPP1 are negatively regulated by Akt. Mol Cell Biol. 2009;29:6192–205. doi: 10.1128/MCB.00681-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hussain S, Foreman O, Perkins SL, Witzig TE, Miles RR, van Deursen J, Galardy PJ. The de-ubiquitinase UCH-L1 is an oncogene that drives the development of lymphoma in vivo by deregulating PHLPP1 and Akt signaling. Leukemia. 2010;24:1641–55. doi: 10.1038/leu.2010.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pawson CT, Scott JD. Signal integration through blending, bolstering and bifurcating of intracellular information. Nature structural & molecular biology. 2010;17:653–8. doi: 10.1038/nsmb.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li X, Yang H, Liu J, Schmidt MD, Gao T. Scribble-mediated membrane targeting of PHLPP1 is required for its negative regulation of Akt. EMBO Rep. 2011;12:818–24. doi: 10.1038/embor.2011.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Teti A, Colucci S, Grano M, Argentino L, Zambonin Zallone A. Protein kinase C affects microfilaments, bone resorption, and [Ca2+]o sensing in cultured osteoclasts. The American journal of physiology. 1992;263:C130–9. doi: 10.1152/ajpcell.1992.263.1.C130. [DOI] [PubMed] [Google Scholar]
- 57.Pei H, Li L, Fridley BL, Jenkins GD, Kalari KR, Lingle W, Petersen G, Lou Z, Wang L. FKBP51 affects cancer cell response to chemotherapy by negatively regulating Akt. Cancer Cell. 2009;16:259–66. doi: 10.1016/j.ccr.2009.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mistafa O, Ghalali A, Kadekar S, Hogberg J, Stenius U. Purinergic receptor-mediated rapid depletion of nuclear phosphorylated Akt depends on pleckstrin homology domain leucine-rich repeat phosphatase, calcineurin, protein phosphatase 2A, and PTEN phosphatases. J Biol Chem. 2010;285:27900–10. doi: 10.1074/jbc.M110.117093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Carver BS, Chapinski C, Wongvipat J, Hieronymus H, Chen Y, Chandarlapaty S, Arora VK, Le C, Koutcher J, Scher H, Scardino PT, Rosen N, Sawyers CL. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell. 2011;19:575–86. doi: 10.1016/j.ccr.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mulholland DJ, Tran LM, Li Y, Cai H, Morim A, Wang S, Plaisier S, Garraway IP, Huang J, Graeber TG, Wu H. Cell autonomous role of PTEN in regulating castration-resistant prostate cancer growth. Cancer Cell. 2011;19:792–804. doi: 10.1016/j.ccr.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gao MH, Miyanohara A, Feramisco JR, Tang T. Activation of PH-domain leucine-rich protein phosphatase 2 (PHLPP2) by agonist stimulation in cardiac myocytes expressing adenylyl cyclase type 6. Biochem Biophys Res Commun. 2009;384:193–8. doi: 10.1016/j.bbrc.2009.04.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sierecki E, Sinko W, McCammon JA, Newton AC. Discovery of small molecule inhibitors of the PH domain leucine-rich repeat protein phosphatase (PHLPP) by chemical and virtual screening. J Med Chem. 2010;53:6899–911. doi: 10.1021/jm100331d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang M, Riedel H. Insulin receptor kinase-independent signaling via tyrosine phosphorylation of phosphatase PHLPP1. J Cell Biochem. 2009;107:65–75. doi: 10.1002/jcb.22095. [DOI] [PubMed] [Google Scholar]
- 65.Courtney KD, Corcoran RB, Engelman JA. The PI3K pathway as drug target in human cancer. J Clin Oncol. 2010;28:1075–83. doi: 10.1200/JCO.2009.25.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wong KK, Engelman JA, Cantley LC. Targeting the PI3K signaling pathway in cancer. Curr Opin Genet Dev. 2010;20:87–90. doi: 10.1016/j.gde.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brognard J, Hunter T. Protein kinase signaling networks in cancer. Curr Opin Genet Dev. 2011;21:4–11. doi: 10.1016/j.gde.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Maehama T, Dixon JE. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem. 1998;273:13375–8. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- 69.Ooms LM, Horan KA, Rahman P, Seaton G, Gurung R, Kethesparan DS, Mitchell CA. The role of the inositol polyphosphate 5-phosphatases in cellular function and human disease. The Biochemical journal. 2009;419:29–49. doi: 10.1042/BJ20081673. [DOI] [PubMed] [Google Scholar]
- 70.Beaulieu JM, Sotnikova TD, Marion S, Lefkowitz RJ, Gainetdinov RR, Caron MG. An Akt/beta-arrestin 2/PP2A signaling complex mediates dopaminergic neurotransmission and behavior. Cell. 2005;122:261–73. doi: 10.1016/j.cell.2005.05.012. [DOI] [PubMed] [Google Scholar]
- 71.Zhao B, Tumaneng K, Guan KL. The Hippo pathway in organ size control, tissue regeneration and stem cell self-renewal. Nature cell biology. 2011;13:877–83. doi: 10.1038/ncb2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–84. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 73.Yu Y, Yoon SO, Poulogiannis G, Yang Q, Ma XM, Villen J, Kubica N, Hoffman GR, Cantley LC, Gygi SP, Blenis J. Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science. 2011;332:1322–1326. doi: 10.1126/science.1199484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Karnoub AE, Dash AB, Vo AP, Sullivan A, Brooks MW, Bell GW, Richardson AL, Polyak K, Tubo R, Weinberg RA. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature. 2007;449:557–63. doi: 10.1038/nature06188. [DOI] [PubMed] [Google Scholar]
- 75.Richardson AL, Wang ZC, De Nicolo A, Lu X, Brown M, Miron A, Liao X, Iglehart JD, Livingston DM, Ganesan S. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell. 2006;9:121–32. doi: 10.1016/j.ccr.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 76.Basso K, Margolin AA, Stolovitzky G, Klein U, Dalla-Favera R, Califano A. Reverse engineering of regulatory networks in human B cells. Nat Genet. 2005;37:382–90. doi: 10.1038/ng1532. [DOI] [PubMed] [Google Scholar]
- 77.Haslinger C, Schweifer N, Stilgenbauer S, Dohner H, Lichter P, Kraut N, Stratowa C, Abseher R. Microarray gene expression profiling of B-cell chronic lymphocytic leukemia subgroups defined by genomic aberrations and VH mutation status. J Clin Oncol. 2004;22:3937–49. doi: 10.1200/JCO.2004.12.133. [DOI] [PubMed] [Google Scholar]
- 78.Ouillette P, Erba H, Kujawski L, Kaminski M, Shedden K, Malek SN. Integrated genomic profiling of chronic lymphocytic leukemia identifies subtypes of deletion 13q14. Cancer Res. 2008;68:1012–21. doi: 10.1158/0008-5472.CAN-07-3105. [DOI] [PubMed] [Google Scholar]
- 79.Sabates-Bellver J, Van der Flier LG, de Palo M, Cattaneo E, Maake C, Rehrauer H, Laczko E, Kurowski MA, Bujnicki JM, Menigatti M, Luz J, Ranalli TV, Gomes V, Pastorelli A, Faggiani R, Anti M, Jiricny J, Clevers H, Marra G. Transcriptome profile of human colorectal adenomas. Mol Cancer Res. 2007;5:1263–75. doi: 10.1158/1541-7786.MCR-07-0267. [DOI] [PubMed] [Google Scholar]
- 80.Kaiser S, Park YK, Franklin JL, Halberg RB, Yu M, Jessen WJ, Freudenberg J, Chen X, Haigis K, Jegga AG, Kong S, Sakthivel B, Xu H, Reichling T, Azhar M, Boivin GP, Roberts RB, Bissahoyo AC, Gonzales F, Bloom GC, Eschrich S, Carter SL, Aronow JE, Kleimeyer J, Kleimeyer M, Ramaswamy V, Settle SH, Boone B, Levy S, Graff JM, Doetschman T, Groden J, Dove WF, Threadgill DW, Yeatman TJ, Coffey RJ, Jr, Aronow BJ. Transcriptional recapitulation and subversion of embryonic colon development by mouse colon tumor models and human colon cancer. Genome Biol. 2007;8:R131. doi: 10.1186/gb-2007-8-7-r131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gaspar C, Cardoso J, Franken P, Molenaar L, Morreau H, Moslein G, Sampson J, Boer JM, de Menezes RX, Fodde R. Cross-species comparison of human and mouse intestinal polyps reveals conserved mechanisms in adenomatous polyposis coli (APC)-driven tumorigenesis. Am J Pathol. 2008;172:1363–80. doi: 10.2353/ajpath.2008.070851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hao Y, Triadafilopoulos G, Sahbaie P, Young HS, Omary MB, Lowe AW. Gene expression profiling reveals stromal genes expressed in common between Barrett's esophagus and adenocarcinoma. Gastroenterology. 2006;131:925–33. doi: 10.1053/j.gastro.2006.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bredel M, Bredel C, Juric D, Harsh GR, Vogel H, Recht LD, Sikic BI. High-resolution genome-wide mapping of genetic alterations in human glial brain tumors. Cancer Res. 2005;65:4088–96. doi: 10.1158/0008-5472.CAN-04-4229. [DOI] [PubMed] [Google Scholar]
- 84.Talantov D, Mazumder A, Yu JX, Briggs T, Jiang Y, Backus J, Atkins D, Wang Y. Novel genes associated with malignant melanoma but not benign melanocytic lesions. Clin Cancer Res. 2005;11:7234–42. doi: 10.1158/1078-0432.CCR-05-0683. [DOI] [PubMed] [Google Scholar]
- 85.Zhao H, Hao WD, Xu HE, Shang LQ, Lu YY. Gene expression profiles of hepatocytes treated with La(NO3)3 of rare earth in rats. World J Gastroenterol. 2004;10:1625–9. doi: 10.3748/wjg.v10.i11.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]