An Archaeal Immune System Can Detect Multiple Protospacer Adjacent Motifs (PAMs) to Target Invader DNA (original) (raw)
Background: CRISPR/Cas systems allow archaea and bacteria to resist invasion by foreign nucleic acids.
Results: The CRISPR/Cas system in Haloferax recognized six different PAM sequences that could trigger a defense response.
Conclusion: The PAM sequence specificity of the defense response in type I CRISPR systems is more relaxed than previously thought.
Significance: The PAM sequence requirements for interference and adaptation appear to differ markedly.
Keywords: Archaea, Microbiology, RNA, RNA Metabolism, RNA Processing, CRISPR/Cas, Haloferax volcanii, PAM
Abstract
The clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated (Cas) system provides adaptive and heritable immunity against foreign genetic elements in most archaea and many bacteria. Although this system is widespread and diverse with many subtypes, only a few species have been investigated to elucidate the precise mechanisms for the defense of viruses or plasmids. Approximately 90% of all sequenced archaea encode CRISPR/Cas systems, but their molecular details have so far only been examined in three archaeal species: Sulfolobus solfataricus, Sulfolobus islandicus, and Pyrococcus furiosus. Here, we analyzed the CRISPR/Cas system of Haloferax volcanii using a plasmid-based invader assay. Haloferax encodes a type I-B CRISPR/Cas system with eight Cas proteins and three CRISPR loci for which the identity of protospacer adjacent motifs (PAMs) was unknown until now. We identified six different PAM sequences that are required upstream of the protospacer to permit target DNA recognition. This is only the second archaeon for which PAM sequences have been determined, and the first CRISPR group with such a high number of PAM sequences. Cells could survive the plasmid challenge if their CRISPR/Cas system was altered or defective, e.g. by deletion of the cas gene cassette. Experimental PAM data were supplemented with bioinformatics data on Haloferax and Haloquadratum.
Introduction
To ensure their survival and persistence in the environment, prokaryotes have developed several defense strategies against invading genetic elements, such as viruses and plasmids. Innate defense mechanisms have been known for years and include restriction modification systems, the alteration of virus receptors on the cell surface, and the secretion of extracellular polymers that prevent virus attachment (1). Recently, an invader-specific and adaptive defense system was discovered (2) and named after its typical arrangement of sequence repeats, i.e. clustered regularly interspaced short palindromic repeats (CRISPR).2 The repeats generally occur nearby a group of protein-coding genes named CRISPR-associated (cas) genes (3). Recently, CRISPR/Cas systems were classified into three major types and several subtypes based on Cas protein sequences (4).
CRISPR/Cas-mediated immunity is achieved via three phases: adaptation, expression, and interference. In the first stage, a piece of the invader DNA is integrated as new spacer into the 5′-end of the CRISPR locus. Transcription of the CRISPR gene in the expression stage produces a long primary CRISPR RNA (pre-crRNA) that is processed by Cas proteins to generate mature crRNA species. In type III systems, crRNAs are matured with the help of the endogenous RNase III, the Cas9 protein, and a short RNA, which is termed tracrRNA (5). In the interference phase, the invading nucleic acid is recognized by the respective crRNA (displayed on the surface of Cas proteins) and silenced. For a detailed description of all three steps, see recent reviews (6–12).
An essential factor for a successful interference and presumably also for the adaptation stage is the dual recognition of both the protospacer sequence and the nearby protospacer adjacent motif (PAM) which is found only in the natural target. This dual recognition mechanism prevents autoimmunity at the spacer encoded by the chromosomal CRISPR gene (13). PAM sequences appear to be conserved and show a distinct relationship to the CRISPR repeat sequences, which also show significant conservation, providing a means of classification into CRISPR groups (14, 15). (We are using the following terms here: “CRISPR/Cas type” as classified by Makarova et al. (4) that describes the whole immune system with CRISPR RNAs and the type-specific and subtype-specific Cas proteins and “CRISPR repeat clusters” or “CRISPR group” as defined by Kunin et al. (14) and Mojica et al. (15) that describes the classification of CRISPR groups by their repeats.) PAM sequence requirements and position vary between CRISPR/Cas types; for example, in type I systems, the PAM sequences are found directly upstream of the protospacer, whereas in CRISPR/Cas type II systems, they are located immediately downstream of the protospacer sequence (4). Up to now, a requirement for PAM sequences by CRISPR/Cas type III systems has not been reported (4). In the adaptation phase, PAM sequences probably play a crucial role in the selection of protospacers from the invading nucleic acid, but details of the recognition mechanism remain unclear (4, 15).
The importance of PAM sequences in the interference stage of CRISPR/Cas type I systems was recently reported by Semenova et al. (16) using Escherichia coli and by Gudbergsdottir et al. (17) using Sulfolobus islandicus. Mutation of the PAM sequence resulted in escape from interference in both organisms, showing that a correct PAM sequence is essential for target recognition by the CRISPR/Cas type I system (16, 17). In contrast, the CRISPR/Cas type III system of Staphylococcus epidermidis did not require any PAM sequence for interference, suggesting that type III systems in general do not require a PAM sequence (13).
PAM sequences are easily determined if spacer sequences of CRISPR loci can be matched to known virus or plasmid sequences (15). However, it is often difficult to find matching sequences to spacers because of the limited sequence information of prokaryotic viruses and plasmids, but the current rapid expansion in whole genome, metagenomic, and metaviromic sequence studies is beginning to provide useful data even in extreme environments, such as hypersaline waters (18).
Here, we provide the first insights into the function and specific roles of the CRISPR/Cas system of the halophilic euryarchaeon Haloferax volcanii. This organism possesses three CRISPR loci, one on the main chromosome and two closely spaced loci on the minichromosome pHV4 (Fig. 1). Between the two pHV4-encoded loci is the only Cas protein gene cassette comprising proteins belonging to CRISPR/Cas type I-B. To identify functional PAM sequences for Haloferax, we used a systematic approach using a plasmid-based invader assay.
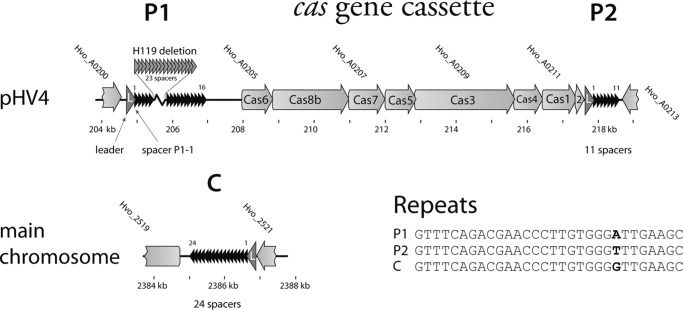
FIGURE 1.
The CRISPR/Cas system of H. volcanii. Two CRISPR genes (P1 and P2) are encoded on minichromosome pHV4 flanking the cas gene cluster. The third CRISPR gene is encoded on the chromosome (C). The cas gene cluster codes for the Cas proteins Cas1, Cas2, Cas3, Cas4, Cas5, Cas6, Cas7, and Cas8b. Sequencing of the CRISPR P1 locus of the Haloferax strain used in this study (H119) showed that part of this locus was deleted. 23 repeats and 23 spacers are missing in the P1 locus in strain H119. In Haloferax strain DS2, the P1 locus contains 40 repeats and 39 spacers. The repeat sequences of the three CRISPR RNAs are identical except for one nucleotide (bottom right).
EXPERIMENTAL PROCEDURES
Strains
H. volcanii strains H26 (Δ_pyrE2_) and H119 (19) were grown aerobically at 45 °C in Hv-YPC (yeast extract, peptone, and casamino acids) medium or in Hv-Ca medium (20). E. coli strains DH5α (Invitrogen) and GM121 (21) were grown aerobically at 37 °C in 2YT medium (22).
Construction of Plasmids
In the initial screen to identify PAM sequences, di- and trinucleotide combinations were introduced upstream and downstream of spacer1 of CRISPR locus P1 (P1-1) of H. volcanii using an overlap extension reaction with Pfu polymerase (Fermentas) and different sets of oligonucleotides (supplemental Table 1). Once the upstream location of the PAM sequence had been established, all PAM candidate sequences were introduced only upstream of the spacer sequence in the subsequent cloning reactions. Reaction products were cloned into the EcoRV-digested vector pTA409 (23), and the resulting plasmids were sequenced. Plasmids were passaged through E. coli GM121 cells (to avoid methylation) and then introduced into Haloferax cells using the Polyethylene glycol method (19, 24).
Transformation of H. volcanii
Plasmids containing spacer P1-1 and different test PAM sequences were introduced into H. volcanii strain H26 (Δ_pyrE2_). Transformants were selected on Hv-Ca plates without uracil. As a positive control, strain H26 was transformed with plasmid pTA409, which is the vector without any inserts. Transformation rates were calculated as the number of colonies obtained by transformation of 1 μg of plasmid DNA (cfu/μg of DNA). Each transformation reaction was conducted at least twice using independent preparations of plasmid. To confirm the identification of a functional PAM sequence, H. volcanii cells were transformed at least three times with the plasmid-PAM construct using at least two different plasmid preparations. As observed in the similar study by Gudbergsdottir et al. (17), transformation rates cannot be estimated very accurately; therefore, PAM sequences that led to an at least 100-fold reduction in transformation rates in this plasmid assay are defined as a functional PAM sequence for H. volcanii.
Isolation of Escape Mutants
After transformation of H26 with pTA409-PAM9 (PAM ACT), 30 clones that survived the plasmid challenge were selected for further analysis. Twenty-six clones (escape mutants 1–26) were from one transformation experiment, and four clones (escape mutants 27–30) were from another transformation experiment.
Northern Blot Hybridization
Total RNA was isolated from exponentially growing H. volcanii H119 cells as described (25). To analyze expression of CRISPR RNAs, cells were grown at different temperatures (30, 45, and 48.5 °C) and different salt concentrations (15, 18, and 23%). Cells were harvested at exponential phase (OD650 = 0.5) and at stationary phase (OD650 = 1.1–1.4), and RNA was extracted. Then 10 μg samples were separated on 8% denaturing gels and subsequently transferred to nylon membranes (Hybond-N+, GE Healthcare). DNA oligonucleotide probes complementary to the repeat sequence (RepeatP1) or to different spacer sequences (Spacer1P1, Spacer1P2, and Spacer1C) were used as probes for hybridization (for primer sequences, see supplemental Table 1).
Southern Blot Hybridization
Southern blot analysis was carried out as described in Sambrook and Russel (26) with the following modifications. Genomic DNA was isolated from Haloferax strains as described (19) and digested using SmaI. Ten micrograms of digested DNA were separated on a 0.7% agarose gel and transferred to a nylon membrane (Hybond-N+, GE Healthcare). The hybridization probe Cas1do was generated by PCR using genomic Haloferax DNA as template and DIG-dUTP (dNTP-labeling mixture of DIG DNA Labeling kit, Roche Applied Science) and primers Cas2.1/Cas2.2, yielding an amplimer of 460 bp. A 5′ and 3′ DIG-labeled oligonucleotide (Spacer1.3) was used to detect spacer1 of the CRISPR locus P1 in H. volcanii as well as to detect spacer1 encoded on the invader plasmid in plasmid transformants. After hybridization, DIG-labeled probes were detected using the DIG Luminescent Detection kit according to the manufacturer's protocol (Roche Applied Science).
PCR and Sequence Analysis of H. volcanii Wild-type Strains and Transformants
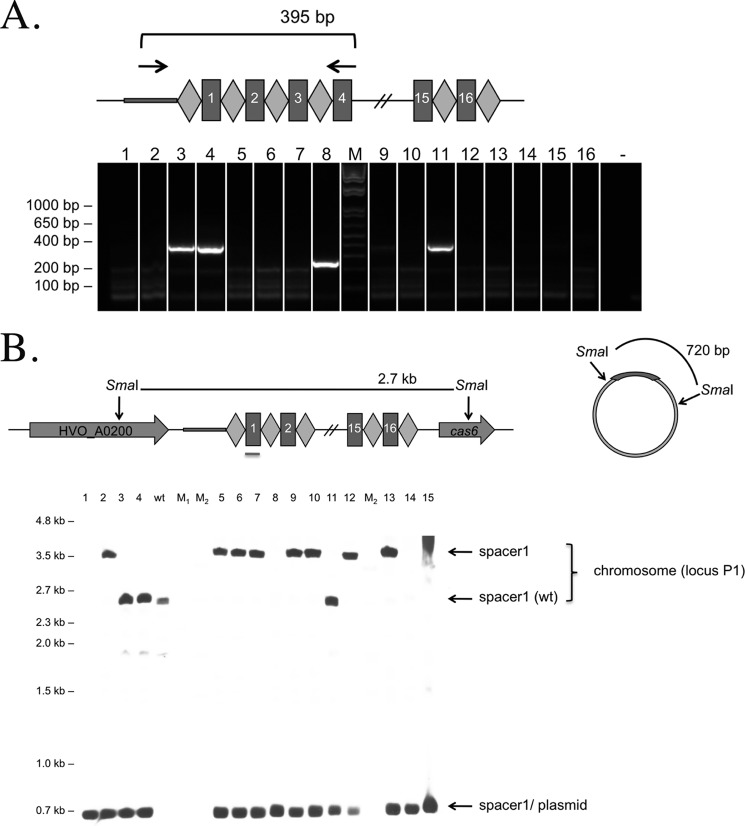
PCR analysis of CRISPR loci was performed with GoTaq polymerase (Promega) using genomic DNA from H. volcanii strains H26 and H119 and primers C1 and C2 for locus C, P1.1 and P1.2 for locus P1 and P2.1, and P2.2 for locus P2. The resulting PCR fragments were cloned and sequenced. PCR for analysis of the cas gene cluster was conducted with GoTaq polymerase (Promega) using oligos Cas3/Cas2.1 on genomic DNA from Haloferax wild-type strain and transformants as template. This yielded a 4554-bp product from wild-type strain DNA that spanned the cas genes cas3, cas4, and cas1. For the investigation of a deletion of spacer1 in the CRISPR locus P1, oligos P1.3 and P1.4 were used, yielding a product of 395 bp in the wild type. The presence of spacer1 in the invader plasmid was investigated by amplifying the plasmid insert with primers US1 and RS1, yielding a product of about 300 bp.
Two escape mutants (25 and 26), which deleted the spacer sequence from the plasmid and did not yield any product upon PCR with primers US1 and RS1, were investigated with a second PCR using primers ColE1 and pyrE2. Here, a PCR product for only one mutant was obtained (escape mutant 25).
PCR products of spacer sequences from the genomic CRISPR locus and PCR products of PAM and spacer sequences from the plasmid were sequenced to detect potential point mutations. From escape mutant strains (escape mutants 3, 4, 17, 24, 27, and 29; Table 3) for which no deletions in the cas gene cluster or the spacer could be detected, the cas gene cluster was amplified using several primers (primers Cas1.1–Cas8.6; supplemental Table 1), and the resulting PCR products were sequenced.
TABLE 3.
Escape mutants utilize different mechanisms for escaping the CRISPR/Cas-mediated defense
The mechanisms for evasion of the CRISPR/Cas-mediated defense were analyzed for 30 escape mutants using PCR, sequencing, and Southern blot analyses. If the presence of spacer1 (on the chromosome or the plasmid) or cas genes could be shown, this is indicated with “+”; otherwise a “−” is indicated. In column 1, labeled “Escape mutant,” the 30 mutants are listed. In column 2, “Spacer1 (PCR),” the results for the PCR analysis of the presence of spacer1 in CRISPR locus P1 are listed. In column 3, “Spacer1 (Southern),” the results for the Southern analysis of the presence of spacer1 in CRISPR locus P1 are listed. Mutants showing signals of wild-type size (2.7 kb) are indicated with (WT). In column 4, “Protospacer1 (Southern),” the results for the Southern analysis of the presence of spacer1 in the plasmid are listed. In column 5, “cas genes (PCR),” the results for the PCR analysis of the presence of the cas genes are listed. In column 6, “cas genes (sequencing),” the cas gene clusters of mutants 3, 4, 17, 24, 27, and 29 were sequenced. For mutant 27, no mutations were found in the cas gene cluster. In column 7, “cas genes (Southern),” the results for the Southern analysis of the presence of the cas genes are listed. Major changes determined in the respective deletion mutant are shown in bold.
| Escape mutant | Spacer1 (PCR) | Spacer1 (Southern) | Protospacer1 (Southern) | cas genes (PCR) | cas genes (sequencing) | cas genes (Southern) |
|---|---|---|---|---|---|---|
| 1 | − | − | + | + | NDa | + |
| 2 | − | + | + | − | ND | − |
| 3 | + | + (WT) | + | + | Mutation in cas8b | + |
| 4 | + | + (WT) | + | + | Mutation in cas3 | + |
| 5 | − | + | + | − | ND | − |
| 6 | − | + | + | − | ND | − |
| 7 | − | + | + | − | ND | − |
| 8 | − | − | + | + | ND | + |
| 9 | + | + | + | − | ND | − |
| 10 | − | + | + | − | ND | − |
| 11 | + | + (WT) | +b | + | ND | + |
| 12 | − | + | + | − | ND | − |
| 13 | − | + | + | − | ND | − |
| 14 | − | − | + | + | ND | + |
| 15 | − | + | + | − | ND | − |
| 16 | − | + | + | − | ND | − |
| 17 | + | + (WT) | + | + | Mutation in cas8b | + |
| 18 | − | + | + | − | ND | − |
| 19 | − | + | + | − | ND | − |
| 20 | + | + | + | − | ND | − |
| 21 | + | + | + | − | ND | − |
| 22 | − | + | + | − | ND | − |
| 23 | − | + | + | − | ND | − |
| 24 | + | + (WT) | + | + | Mutation in cas3 | + |
| 25 | + | + (WT) | − | + | ND | + |
| 26 | + | + (WT) | − | + | ND | + |
| 27c | + | + | + | ND | No mutations | + |
| 28 | + | + | + | ND | ND | − |
| 29 | + | + | + | ND | Mutation in cas8b | + |
| 30 | − | ND | + | ND | ND | − |
RESULTS
The Haloferax CRISPR Genes Are Transcribed and Processed
The Haloferax CRISPR/Cas system belongs to the subtype I-B and consists of three CRISPR loci and a cas gene cassette that encodes eight Cas proteins (Fig. 1). The repeat sequences of all three CRISPR RNAs are 30 nucleotides long and differ by a single nucleotide (Fig. 1). They belong to the CRISPR group 9 (unfolded archaeal repeats) of the classification described by Kunin et al. (14) and Mojica et al. (15). The leader sequences of all three CRISPR loci are also highly similar, suggesting that the CRISPR loci might be derived from a common ancestral locus.
PCR amplification of all three CRISPR loci in Haloferax strains H119 and H26 confirmed the expected sizes of the C and P2 loci but revealed that the P1 locus was 1.5 kb shorter than expected from the genome sequence of the DS2 strain reported previously (27). Sequence analysis showed that this locus in the H119 strain was shortened by 23 repeat and 23 spacer sequences in comparison with the DS2 strain (Fig. 1), but no other changes were observed. In summary, the CRISPR loci of H. volcanii strain H119 encode 24 spacers (locus C), 16 spacers (locus P1), and 11 spacers (locus P2), respectively.
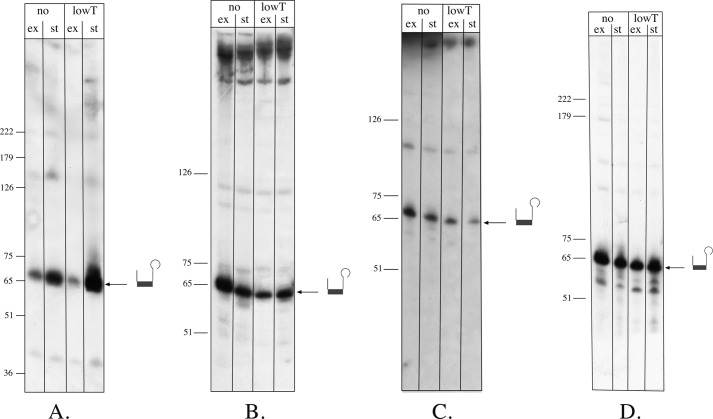
Expression of the three Haloferax CRISPR genes was analyzed by Northern blot hybridization (Fig. 2). All three CRISPR transcripts were detectable and could be seen to be processed into small crRNAs under all conditions monitored, i.e. different temperatures, salt concentrations, and growth phases (see “Experimental Procedures”) (Fig. 2 and data not shown).
FIGURE 2.
Expression of CRISPR genes. RNA from Haloferax cultures grown under different conditions was isolated, separated on 8% denaturing polyacrylamide gels, transferred to membranes, and then hybridized to either a P1 repeat probe (A) or the first spacer of each CRISPR locus (B–D). The P1 repeat probe binds to the repeats of all three CRISPR loci (only one mismatch to the repeat sequences of P2 and C; see Fig. 1). The other probes (spacer1 from P1 (B), spacer1 from locus P2 (C), and spacer1 from locus C (D)) are specific to their respective loci. Lanes no, standard conditions; lanes lowT, lower temperature (30 °C); lanes ex, exponential phase; lanes st, stationary phase. The sizes of a DNA marker are shown on the left in nucleotides. The crRNA is shown schematically on the right.
Matches to Spacers in the Haloferax Genome and Other Genomes
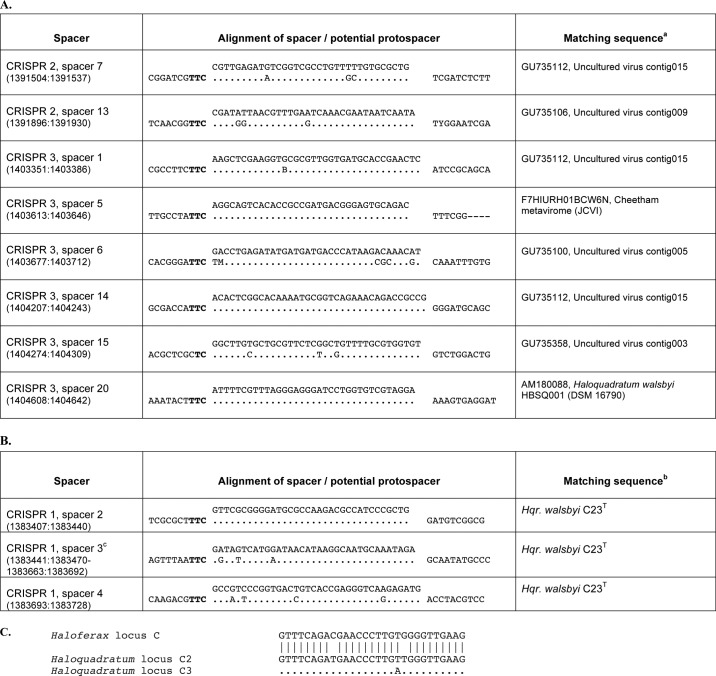
The 74 spacer sequences of the three CRISPR loci of H. volcanii DS2 were compared with sequences in GenBankTM (BLASTN) as well as to environmental sequences at the J. Craig Venter Institute.3 Only two significant matches were detected (Table 1). Spacer C-14 was highly similar to a sequence about 800 kb distant on the same chromosome within HVO_0372. Although the overall similarity is 76%, so precluding autoimmune targeting by the CRISPR/Cas defense, the alignment shows that the two sequences are identical but for a contiguous 9-nucleotide region located near the 3′-end. HVO_0372 encodes a protein with no known homologues or conserved domains. It occurs within a likely phage/plasmid integrant, bordered at one end by a tRNA gene (HVO_0361) and at the other by an XerC/D-like integrase (HVO_0385) and a nearby partial tRNA copy. Except for one or two cases, such as a phage type integrase (HVO_0379), most of the intervening ORFs are obscure in origin and function. A foreign origin of the chromosomal region from HVO_0361 to HVO_0385 (330–341 kb) is also supported by tetranucleotide frequency analysis (TETRA) (28). This showed a distinct change in tetramer composition in this region compared with the average for the entire chromosome (data not shown).
TABLE 1.
Haloferax (Hfx.) spacer sequence matches to genomic and metagenomic sequences
All 74 spacers encoded in the three CRISPR loci of strain DS2 were compared with sequences in GenBank and with a metagenomic data set (GS84) collected by the J. Craig Venter Institute. Here, the two highest scoring matches are shown. In each alignment, the spacer sequence is shown above the matching sequence with dots representing identical bases. Spacer sequences are oriented so that the leader sequence of the CRISPR is upstream (to the left). Potential PAM sequence motifs in the retrieved sequence are shown in bold.
Spacer P1-2 matched closely (88%) to an environmental sequence recovered from a salt lake in Australia (Lake Tyrrell). The alignment shows a perfect match in the 5′-part of the sequence. The matching sequence occurs within an ORF predicted to encode a ParBc domain protein (COG1475), which could function in plasmid partition. As can be seen in the alignments of Table 1, the trinucleotide motifs upstream of the protospacer sequences are CAC and TTC (Table 1). With only two matches to Haloferax spacers, it was not possible to identify potential PAM sequences in the flanking regions. Additional data were required, and this was achieved by a systematic, experimental approach.
The Haloferax CRISPR/Cas System Can Recognize Several PAMs
To determine the PAM sequence required for the CRISPR/Cas system in Haloferax, we used a plasmid-based invader assay because it has been shown previously that plasmids bearing protospacer sequences with a functional PAM efficiently trigger CRISPR/Cas-mediated defense in cells carrying the corresponding spacer sequence (17, 29). To that end, DNA fragments were generated containing spacer1 of CRISPR locus P1 (spacer P1-1) flanked by dinucleotide combinations (supplemental Table 1: pTA409-PAM4–14 and pTA409-PAM16–17; Fig. 3) and three known trinucleotide PAM sequences (15) (supplemental Table 1: pTA409-PAM1–3). These fragments were subsequently cloned into the Haloferax vector pTA409, yielding a total of 16 plasmids with different candidate PAM motifs upstream as well as downstream of the spacer1 sequence (supplemental Table 1: pTA409-PAM1–14 and pTA409-PAM16–17; Fig. 3).
FIGURE 3.
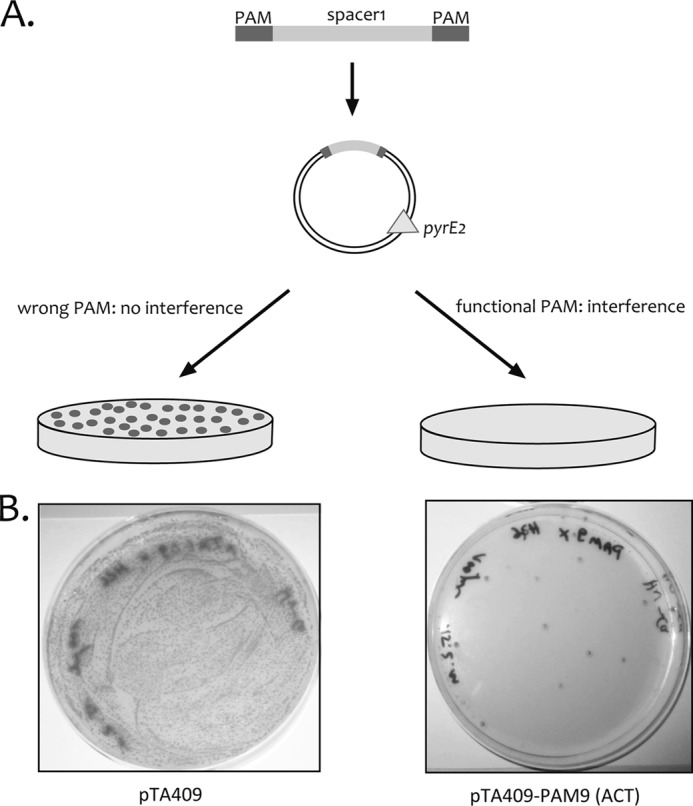
A plasmid-based invader assay for the identification of PAM sequences. A, the spacer sequence P1-1 was cloned adjacent to potential PAM sequences into the vector pTA409. For initial screening, the potential PAM sequence was cloned upstream as well as downstream of the spacer sequence. The second screening was done with constructs containing the PAM sequences only upstream of the spacer sequence. Selection for transformants was achieved by growth without uracil, which is only possible with the pyrE2 selection marker encoded on the vector. Cells can grow if the invader plasmid is retained, such as when it does not trigger a CRISPR/Cas interference response. If a nucleotide combination is active as a PAM, the plasmid will be recognized by the CRISPR/Cas system and degraded, so precluding growth of transformants on selective medium. B, transformation of Haloferax cells with invader plasmids carrying the correct PAM sequence results in failure to grow for almost all cells (right plate, pTA409-PAM9 (ACT)), whereas transformation with the vector (without PAM and P1-1) (left plate, pTA409; only a dilution of the transformed cells has been plated) results in a lawn of cells.
Plasmid constructs were introduced into Haloferax H26 cells, which cannot grow without uracil as the strain lacks the pyrE2 gene. Transformants were selected by uracil prototrophy conferred by the pyrE2 gene on the plasmid (Fig. 3). Plasmid elimination via the CRISPR/Cas defense of the host cell should lead to reduced transformation rates. Of the 16 plasmids tested with different sequences adjacent to the spacer1 sequence, two showed very low transformation rates (at least a 100-fold reduction in transformation) compared with the vector pTA409 (Table 2). The two plasmids that triggered the defense reaction contained the sequences TTC and CT as PAM sequences. TTC was predicted as a PAM for CRISPR/Cas systems belonging to the CRISPR repeat cluster 3 (15), whereas to our knowledge, the motif CT has not been reported for any CRISPR/Cas system in the literature.
TABLE 2.
Transformation rates of PAM constructs
Each construct was tested at least three times, and transformation rates were averaged (see “Experimental Procedures”). The transformation rates of the functional PAM constructs are listed; transformation rates for the non-functional PAM constructs were the same as the transformation rates of the control vector pTA409.
| Plasmid | Transformation rate |
|---|---|
| cfu/μ_g DNA_−1 | |
| pTA409 | 23,809 |
| pTA409-PAM3 (TTC) | 40 |
| pTA409-PAM9 (ACT) | 5 |
| pTA409-PAM25 (TAA) | 180 |
| pTA409-PAM26 (TAT) | 15 |
| pTA409-PAM27 (TAG) | 5 |
| pTA409-PAM54 (CAC) | 105 |
The PAM Sequence Must Be Located Upstream of the Protospacer
The initial PAM plasmids contained the potential PAM sequences both upstream as well as downstream of the spacer P1-1 sequence in each construct. To determine whether the PAM sequence has to be located 5′ or 3′ to the spacer P1-1 sequence, each of the two identified PAM sequences was cloned either upstream or downstream of spacer P1-1. Challenge of H. volcanii cells with these plasmids showed that only constructs in which the PAM sequence was upstream of the spacer P1-1 sequence displayed drastically reduced transformation rates. If the PAM sequences were located downstream of the spacer P1-1 sequence, then transformation rates were equal to those of the control vector pTA409.
The H. volcanii PAMs Are Three Nucleotides Long
With one dinucleotide (CT) and one trinucleotide (TTC) being successful in our assay, we wanted to know whether CT indeed functions as a dinucleotide or acts as an ACT trinucleotide (with the first nucleotide A originating from the pTA409 vector). Therefore, 46 different trinucleotide combinations were tested using the same plasmid-based invader assay (this time, motifs were cloned only upstream of spacer1). The previously identified dinucleotide motif CT turned out to be a trinucleotide with ACT because GCT, CCT, and TCT did not work as PAMs. Furthermore, of the 46 new plasmid constructs, four additional PAM sequences were identified: TAA, TAT, TAG, and CAC. Plasmids with these sequences showed an at least 100-fold drop in transformation rates compared with the vector pTA409 (Table 2). Thus, altogether, we could identify six PAM sequences for Haloferax: TTC, ACT, TAA, TAT, TAG, and CAC.
Cells Survive by Generating Deletions in CRISPR/Cas Genes
Upon transformation of Haloferax with the invader plasmid carrying a functional PAM sequence, we observed a few transformants that had survived the plasmid challenge. The integrity of the CRISPR/Cas genes of these “escape mutants” was examined to understand how they managed to survive. Possible escape mechanisms would be (i) the mutation or deletion of Cas protein genes involved in the defense reaction, (ii) mutation or deletion of the spacer P1-1 sequence in the CRISPR locus (P1), and (iii) mutation or deletion of the PAM and/or spacer P1-1 sequence on the invader plasmid. After introduction of the invader plasmid pTA409-PAM9 (with the PAM sequence ACT) into H. volcanii, 30 transformants (escape mutants) were selected for further analysis. PCR and Southern blot hybridization showed that 18 of the mutants (60%) had lost the complete cas gene cluster (Fig. 4, Fig. 5, and Table 3). The region abutting the deletion in these _cas_− mutants was amplified using PCR, and sequence analysis showed that for 12 mutants the deletion was achieved by recombination between repeat 3 of the upstream CRISPR locus (P1) and repeat 3 of the downstream CRISPR locus (P2); in two cases, the recombination had occurred between repeat 4 (P1) and repeat 10 (P2); and in one mutant, it was between repeat 13 (P1) and repeat 9 (P2). One other mutant was generated by recombination in the leader regions of locus P1 and P2, thereby deleting in addition to the cas gene cluster the complete P1 locus (including spacer1). The 12 mutants that were generated by recombinations at the same sites most likely were generated independently because after transformation cells do not have time to divide. Another possibility is that the recombination occurred in a Haloferax cell prior to transformation.
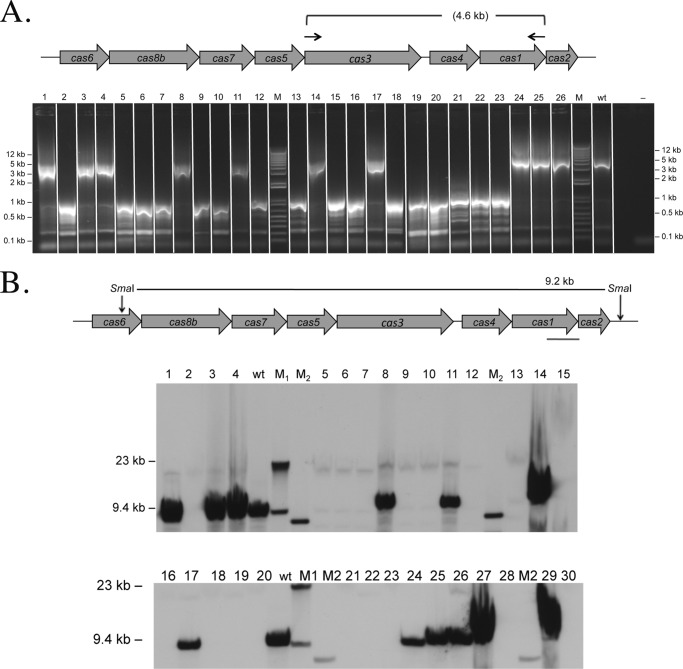
FIGURE 4.
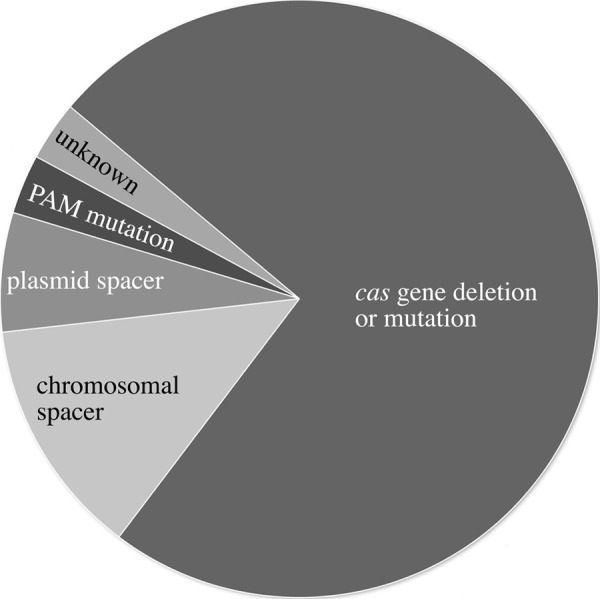
Analysis of CRISPR/Cas escape mutants. Thirty individual colonies of escape mutants that grew on selective medium after challenge with invader plasmid pTA409-PAM9 (ACT) were analyzed using PCR and Southern blot analyses and grouped into five categories according to the lesions identified. Most surviving colonies (77%) had deletions or mutations of the cas gene cluster. Additional ways of escape were the deletion of the spacer P1-1 from the CRISPR locus (10%) or the plasmid (7%). For one escape mutant (3%), the mutations leading to survival could not be identified. One mutant (3%) had a mutation in the PAM sequence, rendering it non-functional. One mutant had a deletion that covered the cas gene cluster and CRISPR locus P1 (including spacer1); this mutant is only shown under cas gene deletion.
FIGURE 5.
Analyses of escape mutants for cas gene cluster. A, PCR analyses. Twenty-six escape mutants were analyzed by PCR on chromosomal DNA for the presence of the cas genes cas3–cas1. The cas gene locus is shown schematically at the top. Primers hybridized as indicated with arrows in the cas3 gene and cas1 gene. For 10 escape mutants, a full-length PCR product is detected (lanes 1, 3, 4, 8, 11, 14, 17, and 24–26); for the other escape mutants, a shorter PCR product of approximately 1 kb is visible. Lanes 1–26, PCR products with chromosomal DNA from escape mutants; lane M, DNA size marker (sizes are given at both sides in kilobase pairs); lane wt, DNA from H119; lane −, control reaction without DNA. B, Southern blot of escape mutants. The cas gene locus is shown schematically at the top. Chromosomal DNA of escape mutants was digested with SmaI, separated on an agarose gel, and blotted. A DIG-labeled probe was used for hybridization as indicated with the red line below the cas1 gene in the scheme. If no deletion in the locus occurred, a 9.2-kb fragment should be detected. The wild-type cas gene locus fragment is present in escape mutants 1, 3, 4, 8, 11, 14, 17, 24–27, and 29, confirming the PCR results (see A and Table 3). The probe used does not pick up any signal for escape mutants 2, 5–7, 9, 10, 12, 13, 15, 16, 18–20, 21–23, 28, and 30. Lanes 1–30, DNA from wild-type and escape mutants; lane wt, DNA from H119; lanes M1 and M2, DNA size marker (sizes are given at the left in kilobase pairs).
PCR and Southern blot analyses showed that most escape mutants still contained the spacer P1-1 in the genomic CRISPR locus (Figs. 4, 5, and 7). Only four of the 30 mutants were found to have lost the spacer P1-1 sequence in the CRISPR locus P1, one of which was already described above because it had a deletion in the cas gene cluster as well as the whole P1 locus. The second mutant had deletions in spacers 1–8, the third had a deletion in spacer1 and spacer2, and the fourth had a deletion in spacers 1–11. In Southern blots, the expected wild-type fragment for the P1 locus of about 2.7 kb was observed for seven mutants, and these also yielded a wild-type sized PCR product for the spacer P1-1 region (Fig. 6). The other mutants showed a larger fragment in the Southern blot with a probe against spacer P1-1. The larger fragment in these isolates is a result of deletion of the cas gene cassette, so generating a SmaI fragment of a different size.
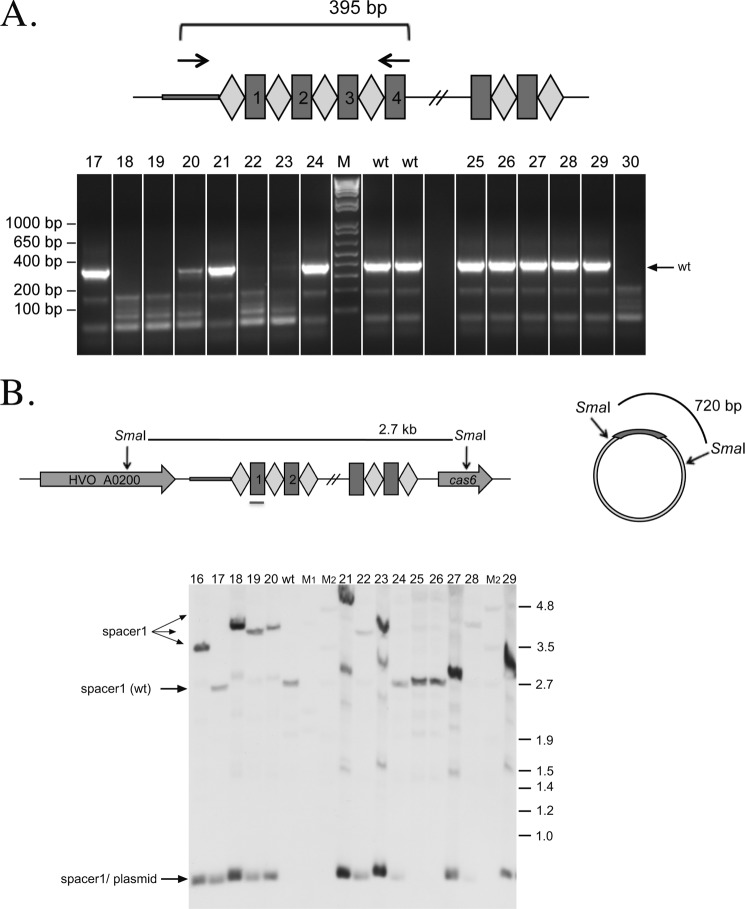
FIGURE 7.
Analyses of escape mutants 16–30 for the presence of spacer P1-1. A, PCR analysis. Fourteen escape mutants were analyzed by PCR on chromosomal DNA for the presence of spacer P1-1. The CRISPR locus P1 is shown schematically at the top. Repeats are shown as diamonds, and spacers are shown as rectangles. Primers used are indicated with arrows in the leader region and spacer4 of the CRISPR locus P1. If P1-1 is not deleted, the expected PCR product would have a size of 395 bp (arrow wt). For nine escape mutants, a full-length PCR product was detected (lanes 17, 20, 21, and 24–29). For all other mutants, no product was generated, suggesting that there is a longer deletion in this region that prevents binding of one or both PCR primers. Lanes 16–30, PCR products with chromosomal DNA from escape mutants; lane M, DNA size marker (sizes are given at the left of the gel in base pairs); lane wt, control PCR with wild-type DNA. B, Southern blot of escape mutants. The CRISPR locus P1 and flanking regions as well as the invader plasmid carrying the P1-1 and PAM sequences are shown schematically at the top. Repeats are shown as diamonds, and spacers are shown as rectangles. Chromosomal DNA and plasmids of escape mutants were digested with SmaI, separated on an agarose gel, and blotted. A DIG-labeled oligonucleotide complementary to spacer P1-1 was used for hybridization as indicated with the red line below spacer1 in the scheme. Using this probe, the P1-1 sequence could be detected on the chromosome as well as on the plasmid (if present). If no deletion in the CRISPR locus occurred, a 2.7-kb fragment should be detected (arrow wt), and if the P1-1 was not removed from the plasmid, a 720-bp fragment should be visible. The wild-type CRISPR locus fragment of 2.7 kb is present in escape mutants 17, 24–27, and 29, confirming the PCR results (see A). A fragment longer than the wild-type fragment is detected in mutants 18–23, 28, and 30. In these mutants, a rearrangement of this locus has occurred as observed later upon analysis of the neighboring cas gene cluster (Figs. 5 and 6). Some mutants show the presence of spacer1 in the Southern blot (escape mutants 18, 19, 22, 23, and 30) (data not shown) with fragments larger than the expected wild-type fragment of 2.7 kb, but they do not give a signal in the PCR (see A and Table 3). This is due to the rearrangement in this locus that deletes the complete cas gene cluster along with parts of the adjoining CRISPR loci. In these cases, spacer4 from CRISPR locus P1 was also deleted; therefore, the PCR primer cannot bind, and the PCR could not work, but spacer1 is still present. Except for mutants 25 and 26, all escape mutants analyzed here still contain the P1-1 spacer on the plasmid because in these lanes the 700-bp fragment is visible. Lanes 16–30, plasmid and chromosomal DNA from escape mutants; lane wt, DNA from H119; lanes M1 and M2, DNA size marker (sizes are given at the left in kilobase pairs).
FIGURE 6.
A, PCR analysis of escape mutants for the presence of spacer P1-1. Sixteen escape mutants were analyzed by PCR on chromosomal DNA for the presence of spacer P1-1. The CRISPR locus P1 is shown schematically at the top. Repeats are shown as diamonds, and spacers are shown as rectangles. PCR primers bind as indicated by arrows in the leader region and spacer4 of the CRISPR locus P1. If P1-1 is present, the expected PCR product would have a size of 395 bp. Three escape mutants generated a full-length PCR product (lanes 3, 4, and 11), whereas one escape mutant showed a shorter PCR product of approximately 250 bp (lane 8). Sequence analysis showed that this mutant had a deletion of two repeats and two spacers (spacer1 and spacer2). For all other mutants, no product was generated, suggesting that there is a longer deletion in this region that prevents binding of one or both PCR primers. Lanes 1–16, PCR products with chromosomal DNA from escape mutants; lane M, DNA size marker (sizes are given at the left of the gel in base pairs); lane −, control PCR without addition of template DNA. B, Southern blot of escape mutants. The CRISPR locus P1 and flanking regions as well as the invader plasmid carrying the P1-1 and PAM sequences are shown schematically at the top. Repeats are shown as diamonds, and spacers are shown as rectangles. Total DNA (genomic DNA and plasmids (pTA409-PAM9)) was isolated from escape mutants, digested with SmaI, separated on an agarose gel, blotted onto a membrane, and hybridized to a DIG-labeled oligonucleotide complementary to spacer P1-1 (indicated by the gray line below spacer1 in the diagram). Using this probe, the P1-1 sequence can be detected on the chromosome as well as on the plasmid pTA409-PAM9. If no deletion in the CRISPR locus occurred, a 2.7-kb fragment should be detected. If the P1-1 spacer was not removed from the plasmid, a 720-bp fragment should be visible. The wild-type CRISPR locus fragment is present in escape mutants 3, 4, and 11, confirming the PCR results (see A). The probe did not pick up any signal for escape mutants 1, 8, and 14. In escape mutant 8, at least spacer1 is deleted, but the leader and spacer4 are, according to the PCR product, still present (see A). Mutants 2, 5–7, 9, 10, 12, 13, and 15 show a fragment of about 3.5 kb, which is larger than the wild-type band, suggesting a major change in this region. This was confirmed later as all these mutants had the complete cas gene cluster and parts of the adjoining CRISPR loci deleted (Fig. 5). All escape mutants analyzed here still contain the P1-1 spacer on the plasmid because in all lanes the 700-bp fragment is visible. Lanes 1–15, plasmid and chromosomal DNA from escape mutants; lane wt, DNA from H119; lanes M1 and M2, DNA size marker (sizes are given at the left in kilobase pairs).
The same hybridization showed that except for two escape mutants the spacer P1-1 sequence was still present on the “invader” plasmid (Figs. 4, 5, and 7). The two mutants that had lost the plasmid-borne P1-1 spacer were investigated further with PCR and sequence analysis. For one mutant, we were not able to amplify a PCR product from the plasmid, whereas the other mutant had a deletion covering the multiple cloning site (including the inserted PAM and spacer sequences).
Finally, seven mutants remained that were shown to have retained spacer P1-1 on both the chromosome and the plasmid as well as a complete cas gene cluster. We sequenced the relevant parts of the plasmid (the insert containing the PAM and spacer1 sequence) and the chromosomal spacer1 sequences of these mutants, which revealed that one mutant had a point mutation in the PAM sequence (ACT → GCT), confirming nicely that a correct PAM is essential for interference. For the remaining six mutants, we amplified the cas gene cluster and sequenced it. Three of these mutants had mutations in the cas8b gene, and two had mutations in the cas3 gene, rendering them non-functional. One mutant remained for which we could not find any changes in the known CRISPR/Cas genes. We assume that this mutant has mutations in other genes that are required for the immune system but not yet known to be involved in the system.
PAM Sequences Are Conserved in Haloarchaea
The CRISPR spacers of H. volcanii DS2 showed very few matches to known sequences, probably reflecting the age of isolation of the DS2 strain (i.e. 1974). Although genomic and metagenomic data have recently become available from the Dead Sea, it is not likely to include viruses and plasmids that were common in those waters 38 years ago. However, there are many other haloarchaea that carry type I-B CRISPR/Cas systems and for which recent metagenomic data are also available. These would be expected to provide more frequent sequence matches and broader insights into the PAM sequences used by this CRISPR/Cas subtype. One such example is Haloquadratum walsbyi, a species that has two strains that have been completely sequenced (30, 31) and for which environmental DNA sequences are available from both of the crystallizer ponds from which these strains were isolated (i.e. autochthonous DNA). As shown in Table 4, A and B, matches of spacer sequences to autochthonous DNA were more frequent than found for Haloferax, and the matching environmental sequences were often within or nearby ORFs related to known or predicted halovirus/phage genes or to chromosomal loci that have been previously found to be common targets of CRISPR/Cas systems (e.g. cas genes) (32). The sequence TTC, which was also identified experimentally in this study, was commonly observed in the matching environmental DNA sequences just upstream of the spacer-contig alignment (Table 4).
TABLE 4.
PAM sequences in Haloquadratum
A, spacers of the three CRISPR loci of H. walsbyi C23T were compared with sequences in GenBank and with environmental sequences of the Cheetham saltern crystallizer in Geelong, Australia (from the J. Craig Venter Institute (JCVI) (J. M. Hoffman, D. Fadrosh, M. Lewis, and S. J. Williamson, unpublished data) and Ref. 48). In each alignment, the spacer sequence is shown above the matching sequence with dots representing identical bases. Spacer sequences are oriented so that the leader sequence of the CRISPR is upstream (to the left). Potential PAM sequence motifs in the retrieved sequence are shown in bold. B = C or G or T and M = A or C; these ambiguities are present in the sequences deposited in GenBank. B, spacers of H. (Hqr.) walsbyi HBSQ001 as described in Dyall-Smith et al. (31) are aligned with closely matching sequences found in the C23T strain of H. walsbyi. The alignments and potential PAM sequences are presented as described for A. C, repeat sequence of H. volcanii CRISPR locus C (top line) aligned to the repeats of CRISPR loci C2 and C3 of H. walsbyi (lines 2 and 3). Dots in the locus C3 repeat sequence indicate bases identical to the repeats of CRISPR locus C2.
DISCUSSION
An Active Defense System
Using an invader plasmid carrying a Haloferax CRISPR spacer sequence, it was possible to mimic cell invasion by foreign DNA and trigger a sequence-specific defense. This resulted in dramatically reduced rates of transformation, most likely effected by Cas-mediated cleavage of the plasmid DNA (33). These results were consistent with the observed CRISPR RNA expression data, and together, they demonstrated that both the expression and defense phases of the CRISPR/Cas system in Haloferax were active. As observed here, only a very few Haloferax cells grew on medium without uracil when transformed with the invader plasmid carrying a functional PAM sequence. Targeting of the introduced spacer by the CRISPR/Cas system would degrade the plasmid, including the pyrE2 gene, rendering the cell unable to grow on uracil-free medium. Thus, in our experimental system, interference was most likely directed at the plasmid DNA.
The CRISPR/Cas system of H. volcanii has remained remarkably intact given that this organism was isolated over 30 years ago. Although the sequenced strain, DS2, was purchased directly from a culture collection (ATCC 29605) and probably underwent few laboratory passages from submission to sequencing (27), the H119 strain examined in the present study has had a long history of laboratory culture (19). It is a Δ_pyrE2_ Δ_trpA_ Δ_leuB_ mutant described in 2004 (19) and was developed from strain DS70, a derivative of strain DS2 (National Collection of Industrial, Food and Marine Bacteria, 2012) which has been cured of plasmid pHV2 (created in 1996 and described in 2001) (34). The P1 CRISPR locus of the H119 strain was found to have suffered an internal deletion of 23 repeat/spacer pairs compared with the ancestral DS2 strain. Genomic analyses of Halobacterium salinarum revealed similar kinds of changes where passage of the same original strain in two different laboratories accumulated a number of differences, mainly indels (35).
All three H. volcanii CRISPR loci were found to be constitutively transcribed, and the primary transcripts were processed to crRNAs. There is only one cas gene cluster in H. volcanii on minichromosome pHV4, so processing of the chromosomal CRISPR transcript (from locus C) was presumably facilitated in trans by the pHV4-encoded Cas proteins. Plasmid carriage of the main functional CRISPR/Cas system in Haloferax is consistent with other studies showing that these defense systems are often found on mobile genetic elements, suggesting frequent lateral transfer (6, 36, 37). The constitutive nature of CRISPR RNA expression in Haloferax differs from the situation in E. coli where expression is usually repressed by a histone-like nucleoid structuring protein, H-NS (38).
Six Different PAM Sequences Can Trigger the Defense Reaction against DNA
To identify functional PAM sequences in Haloferax, the invader plasmid system was used to systematically test (proto)spacer adjacent sequence motifs. Six PAM sequences were identified that when present upstream of the spacer sequence activated the CRISPR/Cas response: ACT, TAA, TAT, TAG, TTC, and CAC. This is the highest number of PAMs for a single CRISPR repeat group identified so far. Two of these PAM sequences (TTC and CAC) were also found upstream of the protospacer sequence by our in silico analysis. TTC as an upstream PAM sequence was also identified in Streptococcus mutans (39) and in Xanthomonas and other organisms belonging to the CRISPR repeat cluster 3 (15). In E. coli (CRISPR/Cas subtype I-E), AWG was identified as an upstream PAM (15, 16). The only other archaeal PAM sequences identified so far, CC and CT, are from the CRISPR/Cas type I system in Sulfolobus and are also located upstream of the protospacer (17, 40).
Our system investigates PAM requirements during the defense reaction. The ability of multiple sequence motifs to activate this response indicates that the system is quite flexible, allowing protection from a number of sequence variants within the natural population of foreign DNA elements. This would make sense considering that viruses mutate to escape the CRISPR/Cas system, and tolerating more PAM sequences would counteract mutations made by the virus in that motif. However, the data collected with the in silico analysis of Haloquadratum regarding protospacer selection for inclusion into CRISPR loci indicate that the adaptation step is more restrictive. It appears that in Haloquadratum there is a strong bias toward using protospacers containing only a limited repertoire of PAM sequences with the most common being TTC. Because H. volcanii and H. walsbyi belong to the same CRISPR system (type I-B) and have very similar repeat sequences, they might have similar PAM requirements. Thus, taking the in silico data for Haloquadratum and the experimental data for Haloferax together, the PAM sequence requirements for interference and for adaptation appear to differ, and relaxed PAM sequence requirements for interference may be advantageous because they hamper invader escape by simple mutations without compromising autoimmunity protection.
Recognition of several different PAM sequences on invading plasmids was also observed in Streptococcus thermophilus (41). Here, the authors discussed that the selective pressure for plasmids compared with bacteriophages is lower, allowing a more degenerate PAM for plasmids than for bacteriophages (41).
In Staphylococcus epidermis, the requirement for the sequence upstream of the protospacer is that it must differ from the repeat sequence, which is upstream of the spacer in the CRISPR locus (i.e. the last eight nucleotides of the repeat sequence) (13). This is an even more relaxed sequence requirement for interference.
The general properties of the CRISPR/Cas system in H. volcanii fit well with those of other type I systems, which typically have short (2–5-nucleotide) PAM sequences located upstream of the protospacer sequence and target DNA. Furthermore, results presented clearly show that a “correct” PAM sequence is essential for a successful defense reaction as has been shown for Sulfolobus and E. coli (16, 17).
Conservation of CRISPR/Cas Types in Haloarchaea
Comparison of the Haloferax CRISPR system with the haloarchaeal organism H. walsbyi showed that both contain a CRISPR/Cas system of type I-B and that the repeat sequences are very similar to each other (Table 4C). In silico analysis of the Haloquadratum spacers with genome databases and metavirome data revealed several matches, allowing the identification of the PAM sequence TTC, which is identical to one of the six PAM sequences found experimentally for Haloferax in this study. This conservation confirms earlier observations that PAM sequences are connected to the CRISPR/Cas type and to the CRISPR repeat cluster (42).
Escape through Deletion of the cas Gene Cluster
The plasmid-based invader assay used in this study to mimic foreign DNA differs from a natural invasion (e.g. by a virus) in that successful degradation of the plasmid leads not to survival but to the inability to grow on the selective medium used to detect plasmid uptake. However, the clear difference observed in transformation rates of invader plasmid constructs that differed by only 1–3 nucleotides was persuasive evidence of the important role of the PAM sequence in target recognition. Another useful feature of the experimental design was the ability to recover stable transformants that had taken up an invader plasmid carrying a functional PAM. As expected, the majority of these were mutants that had survived (or escaped selection) because of lesions in their CRISPR/Cas system, including many that had lost the entire cas gene cluster (Fig. 4). In almost all cases, deletion of the cas genes was achieved by recombination between repeats of the upstream and downstream CRISPR loci. Here, more than 60% of the mutants were generated by recombination at the same site. Five mutants had mutations in the cas3 or cas8b gene, which rendered them non-functional. Several escape mutants succeeded in deleting the P1-1 spacer on pHV4; one even deleted the entire P1 CRISPR locus. Removal of spacer sequences by recombination between repeats seems to be a common reaction because internal deletions in CRISPR loci have been observed in several instances (43–45), including the Haloferax strain used in this study. Such events may help regulate CRISPR length or allow survival upon successful invasion (2, 17, 40, 46, 47). In addition, these deletions are reminiscent of repeat-associated deletions that have been detected upon comparison of Haloquadratum strains (31).
Two mutants removed the protospacer from the invader plasmid. Interestingly, one mutant survived because of a point mutation in the PAM sequence. One mutant did not mutate or delete the PAM sequence or the spacer sequences (neither in the plasmid nor on the chromosome) nor did it delete or mutate the cas genes. This mutant may carry a deletion or mutation in another gene important for the defense reaction.
In contrast to the clear preference to survive by deletion of the cas gene cluster observed in this study, a previous study in Sulfolobus showed that in this organism escape mutants used a variety of deletions to escape death by the immune system. A wide range of deletion sizes was observed with very few being identical (17). This difference might be because we used the first spacer of the CRISPR locus in our invader plasmid, leaving only the first repeat or the leader sequence for recombination. In addition, in Haloferax, the cas gene cluster is flanked by CRISPR genes encoded in the same orientation, allowing deletion of the cas gene cluster by recombination between repeats from the flanking CRISPR gene clusters.
Supplementary Material
Supplemental Data
Acknowledgments
We thank Elli Bruckbauer for expert technical assistance. Furthermore, we are grateful to members of the Deutsche Forschungsgemeinschaft FOR1680 for helpful discussions. M. D. S. thanks Jeff Hoffman, Doug Fadrosh, Matt Lewis, and Shannon Williamson of the J. Craig Venter Institute (who acknowledge funding from the United States Department of Energy, Office of Science, and Office of Biological and Environmental Research) for providing environmental sequence data from Lake Tyrrell and Cheetham saltern.
*
This work was supported by the Deutsche Forschungsgemeinschaft in the frame of Research Group 1680, “Unravelling the prokaryotic immune system.”
3
J. M. Hoffman, D. Fadrosh, M. Lewis, and S. J. Williamson, unpublished data.
2
The abbreviations used are:
CRISPR
clustered regularly interspaced short palindromic repeat
Cas
CRISPR-associated
PAM
protospacer adjacent motif
crRNA
CRISPR RNA
DIG
digoxigenin.
REFERENCES
- 1.Labrie S. J., Samson J. E., Moineau S. (2010) Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 8, 317–327 [DOI] [PubMed] [Google Scholar]
- 2.Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D. A., Horvath P. (2007) CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712 [DOI] [PubMed] [Google Scholar]
- 3.Jansen R., Embden J. D., Gaastra W., Schouls L. M. (2002) Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Microbiol. 43, 1565–1575 [DOI] [PubMed] [Google Scholar]
- 4.Makarova K. S., Haft D. H., Barrangou R., Brouns S. J., Charpentier E., Horvath P., Moineau S., Mojica F. J., Wolf Y. I., Yakunin A. F., van der Oost J., Koonin E. V. (2011) Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 9, 467–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deltcheva E., Chylinski K., Sharma C. M., Gonzales K., Chao Y., Pirzada Z. A., Eckert M. R., Vogel J., Charpentier E. (2011) CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471, 602–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deveau H., Garneau J. E., Moineau S. (2010) CRISPR/Cas system and its role in phage-bacteria interactions. Annu. Rev. Microbiol. 64, 475–493 [DOI] [PubMed] [Google Scholar]
- 7.Garrett R. A., Vestergaard G., Shah S. A. (2011) Archaeal CRISPR-based immune systems: exchangeable functional modules. Trends Microbiol. 19, 549–556 [DOI] [PubMed] [Google Scholar]
- 8.Horvath P., Barrangou R. (2010) CRISPR/Cas, the immune system of bacteria and archaea. Science 327, 167–170 [DOI] [PubMed] [Google Scholar]
- 9.Karginov F. V., Hannon G. J. (2010) The CRISPR system: small RNA-guided defense in bacteria and archaea. Mol. Cell 37, 7–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marchfelder A., Fischer S., Brendel J., Stoll B., Maier L. K., Jäger D., Prasse D., Schmitz R., Randau L. (2012) Small RNAs for defence and regulation in archaea. Extremophiles doi: 10.1007/S00792-012-0469-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marraffini L. A., Sontheimer E. J. (2010) CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat. Rev. Genet. 11, 181–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Terns M. P., Terns R. M. (2011) CRISPR-based adaptive immune systems. Curr. Opin. Microbiol. 14, 321–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marraffini L. A., Sontheimer E. J. (2010) Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature 463, 568–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kunin V., Sorek R., Hugenholtz P. (2007) Evolutionary conservation of sequence and secondary structures in CRISPR repeats. Genome Biol. 8, R61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mojica F. J., Díez-Villaseñor C., García-Martínez J., Almendros C. (2009) Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 155, 733–740 [DOI] [PubMed] [Google Scholar]
- 16.Semenova E., Jore M. M., Datsenko K. A., Semenova A., Westra E. R., Wanner B., van der Oost J., Brouns S. J., Severinov K. (2011) Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc. Natl. Acad. Sci. U.S.A. 108, 10098–10103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gudbergsdottir S., Deng L., Chen Z., Jensen J. V., Jensen L. R., She Q., Garrett R. A. (2011) Dynamic properties of the Sulfolobus CRISPR/Cas and CRISPR/Cmr systems when challenged with vector-borne viral and plasmid genes and protospacers. Mol. Microbiol. 79, 35–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Santos F., Yarza P., Parro V., Briones C., Antón J. (2010) The metavirome of a hypersaline environment. Environ. Microbiol. 12, 2965–2976 [DOI] [PubMed] [Google Scholar]
- 19.Allers T., Ngo H. P., Mevarech M., Lloyd R. G. (2004) Development of additional selectable markers for the halophilic archaeon Haloferax volcanii based on the leuB and trpA genes. Appl. Environ. Microbiol. 70, 943–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dyall-Smith M. L. (2008) The Halohandbook, Spinger-Verlag, Wien, Austria [Google Scholar]
- 21.Allers T., Barak S., Liddell S., Wardell K., Mevarech M. (2010) Improved strains and plasmid vectors for conditional overexpression of His-tagged proteins in Haloferax volcanii. Appl. Environ. Microbiol. 76, 1759–1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller J. H. (1972) Experiments in Molecular Genetics, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 23.Hölzle A., Fischer S., Heyer R., Schütz S., Zacharias M., Walther P., Allers T., Marchfelder A. (2008) Maturation of the 5S rRNA 5′ end is catalyzed in vitro by the endonuclease tRNase Z in the archaeon H. volcanii. RNA 14, 928–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cline S. W., Schalkwyk L. C., Doolittle W. F. (1989) Transformation of the archaebacterium Halobacterium volcanii with genomic DNA. J. Bacteriol. 171, 4987–4991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nieuwlandt D. T., Palmer J. R., Armbruster D. T., Kuo Y.-P., Oda W., Daniels C. J. (1995) in Archaea: A Laboratory Manual (Robb F. T., Place A. R., Sowers K. R., Schreier H. J., DasSarma S., Fleischmann E. M., eds) pp. 161–162, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 26.Sambrook J., Russell D. (2001) Molecular Cloning: A Laboratory Manual, pp. 6.33–6.46, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 27.Hartman A. L., Norais C., Badger J. H., Delmas S., Haldenby S., Madupu R., Robinson J., Khouri H., Ren Q., Lowe T. M., Maupin-Furlow J., Pohlschroder M., Daniels C., Pfeiffer F., Allers T., Eisen J. A. (2010) The complete genome sequence of Haloferax volcanii DS2, a model archaeon. PLoS One 5, e9605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Teeling H., Waldmann J., Lombardot T., Bauer M., Glöckner F. O. (2004) TETRA: a web-service and a stand-alone program for the analysis and comparison of tetranucleotide usage patterns in DNA sequences. BMC Bioinformatics 5, 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manica A., Zebec Z., Teichmann D., Schleper C. (2011) In vivo activity of CRISPR-mediated virus defence in a hyperthermophilic archaeon. Mol. Microbiol. 80, 481–491 [DOI] [PubMed] [Google Scholar]
- 30.Bolhuis H., Palm P., Wende A., Falb M., Rampp M., Rodriguez-Valera F., Pfeiffer F., Oesterhelt D. (2006) The genome of the square archaeon Haloquadratum walsbyi: life at the limits of water activity. BMC Genomics 7, 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dyall-Smith M. L., Pfeiffer F., Klee K., Palm P., Gross K., Schuster S. C., Rampp M., Oesterhelt D. (2011) Haloquadratum walsbyi: limited diversity in a global pond. PLoS One 6, e20968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brodt A., Lurie-Weinberger M. N., Gophna U. (2011) CRISPR loci reveal networks of gene exchange in archaea. Biol. Direct 6, 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wiedenheft B., Sternberg S. H., Doudna J. A. (2012) RNA-guided genetic silencing systems in bacteria and archaea. Nature 482, 331–338 [DOI] [PubMed] [Google Scholar]
- 34.Wendoloski D., Ferrer C., Dyall-Smith M. L. (2001) A new simvastatin (mevinolin)-resistance marker from Haloarcula hispanica and a new Haloferax volcanii strain cured of plasmid pHV2. Microbiology 147, 959–964 [DOI] [PubMed] [Google Scholar]
- 35.Pfeiffer F., Schuster S. C., Broicher A., Falb M., Palm P., Rodewald K., Ruepp A., Soppa J., Tittor J., Oesterhelt D. (2008) Evolution in the laboratory: the genome of Halobacterium salinarum strain R1 compared to that of strain NRC-1. Genomics 91, 335–346 [DOI] [PubMed] [Google Scholar]
- 36.Georg J., Voss B., Scholz I., Mitschke J., Wilde A., Hess W. R. (2009) Evidence for a major role of antisense RNAs in cyanobacterial gene regulation. Mol. Syst. Biol. 5, 305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Greve B., Jensen S., Brügger K., Zillig W., Garrett R. A. (2004) Genomic comparison of archaeal conjugative plasmids from Sulfolobus. Archaea 1, 231–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pul U., Wurm R., Arslan Z., Geissen R., Hofmann N., Wagner R. (2010) Identification and characterization of E. coli CRISPR-cas promoters and their silencing by H-NS. Mol. Microbiol. 75, 1495–1512 [DOI] [PubMed] [Google Scholar]
- 39.van der Ploeg J. R. (2009) Analysis of CRISPR in Streptococcus mutans suggests frequent occurrence of acquired immunity against infection by M102-like bacteriophages. Microbiology 155, 1966–1976 [DOI] [PubMed] [Google Scholar]
- 40.Lillestøl R. K., Shah S. A., Brügger K., Redder P., Phan H., Christiansen J., Garrett R. A. (2009) CRISPR families of the crenarchaeal genus Sulfolobus: bidirectional transcription and dynamic properties. Mol. Microbiol. 72, 259–272 [DOI] [PubMed] [Google Scholar]
- 41.Garneau J. E., Dupuis M. È., Villion M., Romero D. A., Barrangou R., Boyaval P., Fremaux C., Horvath P., Magadán A. H., Moineau S. (2010) The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468, 67–71 [DOI] [PubMed] [Google Scholar]
- 42.Al-Attar S., Westra E. R., van der Oost J., Brouns S. J. (2011) Clustered regularly interspaced short palindromic repeats (CRISPRs): the hallmark of an ingenious antiviral defense mechanism in prokaryotes. Biol. Chem. 392, 277–289 [DOI] [PubMed] [Google Scholar]
- 43.Deveau H., Barrangou R., Garneau J. E., Labonté J., Fremaux C., Boyaval P., Romero D. A., Horvath P., Moineau S. (2008) Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J. Bacteriol. 190, 1390–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Horvath P., Coûté-Monvoisin A. C., Romero D. A., Boyaval P., Fremaux C., Barrangou R. (2009) Comparative analysis of CRISPR loci in lactic acid bacteria genomes. Int. J. Food Microbiol. 131, 62–70 [DOI] [PubMed] [Google Scholar]
- 45.Tyson G. W., Banfield J. F. (2008) Rapidly evolving CRISPRs implicated in acquired resistance of microorganisms to viruses. Environ. Microbiol. 10, 200–207 [DOI] [PubMed] [Google Scholar]
- 46.Lillestøl R. K., Redder P., Garrett R. A., Brügger K. (2006) A putative viral defence mechanism in archaeal cells. Archaea 2, 59–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pourcel C., Salvignol G., Vergnaud G. (2005) CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology 151, 653–663 [DOI] [PubMed] [Google Scholar]
- 48.Burns D. G., Camakaris H. M., Janssen P. H., Dyall-Smith M. L. (2004) Combined use of cultivation-dependent and cultivation-independent methods indicates that members of most haloarchaeal groups in an Australian crystallizer pond are cultivable. Appl. Environ. Microbiol. 70, 5258–5265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Burns D. G., Camakaris H. M., Janssen P. H., Dyall-Smith M. L. (2004) Cultivation of Walsby's square haloarchaeon. FEMS Microbiol. Lett. 238, 469–473 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data