methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles (original) (raw)
Abstract
DNA methylation is a chemical modification of cytosine bases that is pivotal for gene regulation, cellular specification and cancer development. Here, we describe an R package, methylKit, that rapidly analyzes genome-wide cytosine epigenetic profiles from high-throughput methylation and hydroxymethylation sequencing experiments. methylKit includes functions for clustering, sample quality visualization, differential methylation analysis and annotation features, thus automating and simplifying many of the steps for discerning statistically significant bases or regions of DNA methylation. Finally, we demonstrate methylKit on breast cancer data, in which we find statistically significant regions of differential methylation and stratify tumor subtypes. methylKit is available at http://code.google.com/p/methylkit.
Rationale
DNA methylation is a critical epigenetic modification that guides development, cellular differentiation and the manifestation of some cancers [1,2]. Specifically, cytosine methylation is a widespread modification in the genome, and it most often occurs in CpG dinucleotides, although non-CpG cytosines are also methylated in certain tissues such as embryonic stem cells [3]. DNA methylation is one of the many epigenetic control mechanisms associated with gene regulation. Specifically, cytosine methylation can directly hinder binding of transcription factors and methylated bases can also be bound by methyl-binding-domain proteins that recruit chromatin-remodeling factors [4,5]. In addition, aberrant DNA methylation patterns have been observed in many human malignancies and can also be used to define the severity of leukemia subtypes [6]. In malignant tissues, DNA is either hypo-methylated or hyper-methylated compared to the normal tissue. The location of hyper- and hypo-methylated sites gives distinct signatures within many diseases [7]. Often, hypomethylation is associated with gene activation and hypermethylation is associated with gene repression, although there are many exceptions to this trend [7]. DNA methylation is also involved in genomic imprinting, where the methylation state of a gene is inherited from the parents, but de novo methylation also can occur in the early stages of development [8,9].
A common technique for measuring DNA methylation is bisulfite sequencing, which has the advantage of providing single-base, quantitative cytosine methylation levels. In this technique, DNA is treated with sodium bisulfite, which deaminates cytosine residues to uracil, but leaves 5-methylcytosine residues unaffected. Single-base resolution, %methylation levels are then calculated by counting the ratio of C/(C+T) at each base. There are multiple techniques that leverage high-throughput bisulfite sequencing such as: reduced representation bisulfite sequencing (RRBS)[10] and its variants [11], whole-genome shotgun bisulfite sequencing (BS-seq) [12], methylC-Seq [13], and target capture bisulfite sequencing [14]. In addition, 5-hydroxymethylcytosine (5hmC) levels can be measured through a modification of bisulfite sequencing techniques [15].
Yet, as bisulfite sequencing techniques have expanded, there are few computational tools available to analyze the data. Moreover, there is a need for an end-to-end analysis package with comprehensive features and ease of use. To address this, we have created methylKit, a multi-threaded R package that can rapidly analyze and characterize data from many methylation experiments at once. methylKit can read DNA methylation information from a text file and also from alignment files (for example, SAM files) and carry out operations such as differential methylation analysis, sample clustering and annotation, and visualization of DNA methylation events (See Figure 1 for a diagram of possible operations). methylKit has open-source code and is available at [16] and as Additional file 1 (see also Additional file 2 for the user guide and Additional file 3 for the package documentation ). Our data framework is also extensible to emerging methods in quantization of other base modifications, such as 5hmC [14], or sites discovered through single molecule sequencing [17,18]. For clarity, we describe only examples with DNA methylation data.
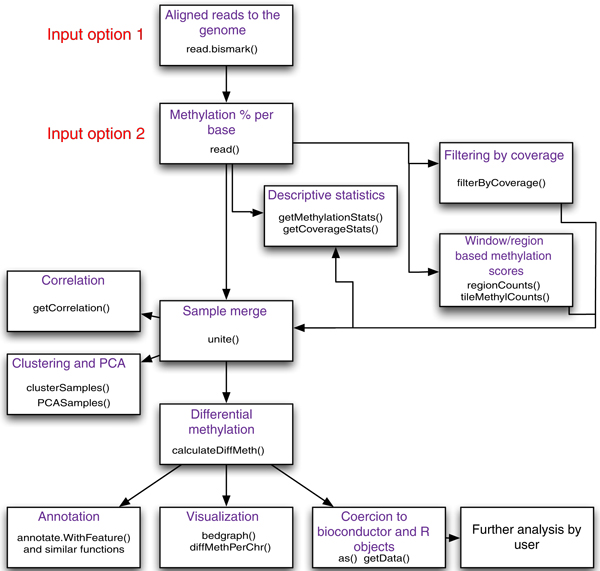
Figure 1.
Flowchart of possible operations by methylKit. A summary of the most important_methylKit_ features is shown in a flow chart. It depicts the main features of _methylKit_and the sequential relationship between them. The functions that could be used for those features are also printed in the boxes.
Flexible data integration and regional analysis
High-throughput bisulfite sequencing experiments typically yield millions of reads with reduced complexity due to cytosine conversion, and there are several different aligners suited for mapping these reads to the genome (see Frith et al. [19] and Krueger et al. [20] for a review and comparison between aligners). Since methylKit only requires a methylation score per base for all analyses, it is a modular package that can be applied independent of any aligner. Currently, there are two ways that information can be supplied to_methylKit_:: 1) methylKit can read per base methylation scores from a text file (see Table 1 for an example of such a file); and, 2) methylKit can read SAM format [21] alignments files obtained from Bismark aligner [22]. If a SAM file is supplied, methylkit first processes the alignment file to get %methylation scores and then reads that information into memory.
Table 1.
Sample text file that can be read by methylKit.
| chrBase | chr | base | strand | coverage | freqC | freqT |
|---|---|---|---|---|---|---|
| chr21.9764539 | chr21 | 9764539 | R | 12 | 25 | 75 |
| chr21.9764513 | chr21 | 9764513 | R | 12 | 0 | 100 |
| chr21.9820622 | chr21 | 9820622 | F | 13 | 0 | 100 |
| chr21.9837545 | chr21 | 9837545 | F | 11 | 0 | 100 |
| chr21.9849022 | chr21 | 9849022 | F | 124 | 72.58 | 27.42 |
| chr21.9853326 | chr21 | 9853326 | F | 17 | 70.59 | 29.41 |
Most bisulfite experiments have a set of test and control samples or samples across multiple conditions, and methylKit can read and store (in memory) methylation data simultaneously for N-experiments, limited only by memory of the node or computer. The default setting of the processing algorithm requires that there be least 10 reads covering a base and each of the bases covering the genomic base position have at least 20 PHRED quality score. Also, since DNA methylation can occur in CpG, CHG and CHH contexts (H = A, T, or C) [3], users of methylKit have the option to provide methylation information for all these contexts: CpG, CHG and CHH from SAM files.
Summarizing DNA methylation information over pre-defined regions or tiling windows
Although base-pair resolution DNA methylation information is obtained through most bisulfite sequencing experiments, it might be desirable to summarize methylation information over tiling windows or over a set of predefined regions (promoters, CpG islands, introns, and so on). For example, Smith et al. [9] investigated methylation profiles with RRBS experiments on gametes and zygote and summarized methylation information on 100bp tiles across the genome. Their analysis revealed a unique set of differentially methylated regions maintained in early embryo. Using tiling windows or predefined regions, such as promoters or CpG islands, is desirable when there is not enough coverage, when bases in close proximity will have similar methylation profiles, or where methylation properties of a region as a whole determines its function. In accordance with these potential analytic foci, methylKit provides functionality to do either analysis on tiling windows across the genome or predefined regions of the genome. After reading the base pair methylation information, users can summarize the methylation information on pre-defined regions they select or on tiling windows covering the genome (parameter for tiles are user provided). Then, subsequent analyses, such as clustering or differential methylation analysis, can be carried out with the same functions that are used for base pair resolution analysis.
Example methylation data set: breast cancer cell lines
We demonstrated the capabilities of methylKit using an example data set from seven breast cancer cell lines from Sun et al. [23]. Four of the cell lines express estrogen receptor-alpha (MCF7, T47D, BT474, ZR75-1), and from here on are referred to as ER+. The other three cell lines (BT20, MDA-MB-231, MDA-MB-468) do not express estrogen receptor-alpha, and from here on are referred to as ER-. It has been previously shown that ER+ and ER- tumor samples have divergent gene expression profiles and that those profiles are associated with disease outcome [24,25]. Methylation profiles of these cell lines were measured using reduced RRBS [10]. The R objects contained the methylation information for breast cancer cell lines and functions that produce plots and other results that are shown in the remainder of this manuscript are in Additional file 4.
Whole methylome characterization: descriptive statistics, sample correlation and clustering
Descriptive statistics on DNA methylation profiles
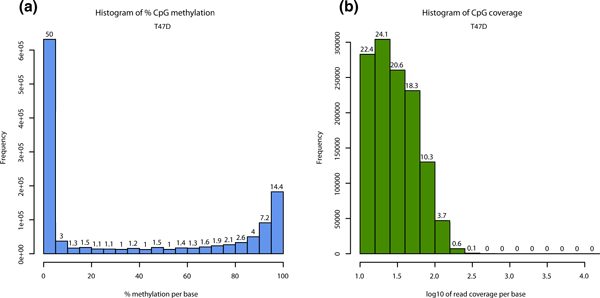
Read coverage per base and % methylation per base are the basic information contained in the_methylKit_ data structures. methylKit has functions for easy visualization of such information (Figure 2a and 2b for % methylation and read coverage distributions, respectively - for code see Additional file 4). In normal cells, % methylation will have a bimodal distribution, which denotes that the majority of bases have either high or low methylation. The read coverage distribution is also an important metric that will help reveal if experiments suffer from PCR duplication bias (clonal reads). If such bias occurs, some reads will be asymmetrically amplified and this will impair accurate determination of % methylation scores for those regions. If there is a high degree of PCR duplication bias, read coverage distribution will have a secondary peak on the right side. To correct for this issue,methylKit has the option to filter bases with very high read coverage.
Figure 2.
Descriptive statistics per sample. (a) Histogram of %methylation per cytosine for ER+ T47D sample. Most of the bases have either high or low methylation. (b) Histogram of read coverage per cytosine for ER+ T47D sample. ER+, estrogen receptor-alpha expressing.
Measuring and visualizing similarity between samples
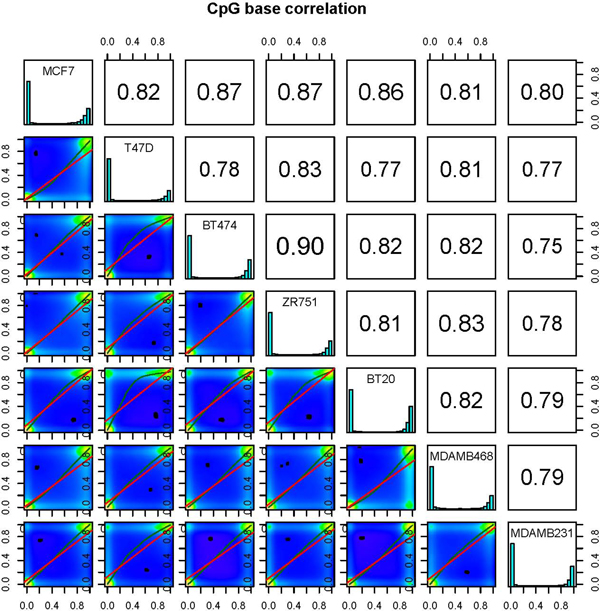
We have also included methods to assess sample similarity. Users can calculate pairwise correlation coefficients (Pearson, Kendall or Spearman) between the %methylation profiles across all samples. However, to ensure comparable statistics, a new data structure is formed before these calculations, wherein only cytosines covered in all samples are stored. Subsequently, pairwise correlations are calculated, to produce a correlation matrix. This matrix allows the user to easily compare correlation coefficients between pairs of samples and can also be used to perform hierarchical clustering using 1- correlation distance. methylKit can also further visualize similarities between all pairs of samples by creating scatterplots of the %methylation scores (Figure 3). These functions are essential for detecting sample outliers or for functional clustering of samples based on their molecular signatures.
Figure 3.
Scatter plots for sample pairs. Scatter plots of %methylation values for each pair in seven breast cancer cell lines. Numbers on upper right corner denote pair-wise Pearson's correlation scores. The histograms on the diagonal are %methylation histograms similar to Figure 2a for each sample.
Hierarchical clustering of samples
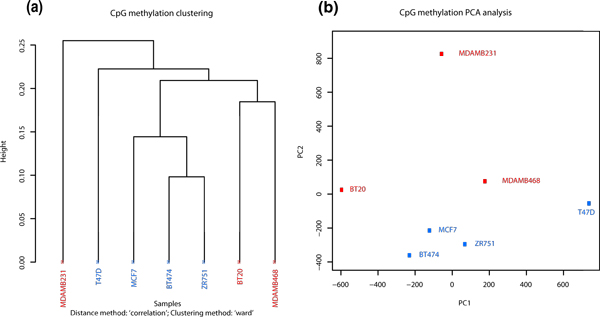
methylKit can also be used to cluster samples hierarchically in a variety of ways. The user can specify the distance metric between samples ('1 - correlation' 'Euclidean', 'maximum', 'manhattan', 'canberra', 'binary' or 'minkowski') as well as the agglomeration method to be used in the hierarchical clustering algorithm (for example, 'Ward's method', or 'single/complete linkage', and so on). Results can either be returned as a dendrogram object or a plot. Dendrogram plots will be color coded based on user defined groupings of samples. For example, we found that most ER+ and ER- samples clustered together except MDMB231 (Figure 4a). Moreover, the user may be interested in employing other more model-intensive clustering algorithms to their data. Users can easily obtain the %methylation data from methylKit object and perform their own analysis with the multitude of R-packages already available for clustering. An example of such a procedure (k-means clustering) is shown in Additional file 4.
Figure 4.
Sample clustering. (a) Hierarchical clustering of seven breast cancer methylation profiles using 1-Pearson's correlation distance. (b) Principal Component Analysis (PCA) of seven breast cancer methylation profiles, plot shows principal component 1 and principal component 2 for each sample. Samples closer to each other in principal component space are similar in their methylation profiles.
Principal component analysis of samples
methylKit can be used to perform Principal Component Analysis (PCA) on the samples' %-methylation profiles (see for example [26]). PCA can reduce the high dimensionality of a data set by transforming the large number of regions to a few principal components. The principal components are ordered so that the first few retain most of the variation present in the original data and are often used to emphasize grouping structure in the data. For example, a plot of the first two or three principal components could potentially reveal a biologically meaningful clustering of the samples. Before the PCA is performed, a new data matrix is formed, containing the samples and only those cytosines that are covered in all samples. After PCA, methylKit then returns to the user a 'prcomp' object, which can be used to extract and plot the principal components. We found that in the breast cancer data set, PCA reveals a similar clustering to the hierarchical clustering where MDMB231 is an outlier.
Differential methylation calculation
Parallelized methods for detecting significant methylation changes
Differential methylation patterns have been previously described in malignancies [27-29] and can be used to differentiate cancer and normal cells [30]. In addition, normal human tissues harbor unique DNA methylation profiles [7]. Differential DNA methylation is usually calculated by comparing methylation levels between multiple conditions, which can reveal important locations of divergent changes between a test and a control set. We have designed methylKit to implement two main methods for determining differential methylation across all regions: logistic regression and Fisher's exact test. However, the data frames in methylKit can easily be used with other statistical tests and an example is shown in Additional file 4 (using a moderated t-test, although we maintain that most natural tests for this kind of data are Fisher's exact and logistic regression based tests). For our example data set we compared ER+ to ER- samples, with our 'control group' being the ER- set.
Method #1: logistic regression
In logistic regression, information from each sample is specified (the number of methylated Cs and number of unmethylated Cs at a given region), and a logistic regression test will be applied to compare fraction of methylated Cs across the test and the control groups. More specifically, at a given base/region we model the methylation proportion Pi, for sample i= 1,...,n (where n is the number of biological samples) through the logistic regression model:
| log(Pi/(1 - Pi)) = β0+β1*Ti | (1) |
|---|
where Ti denotes the treatment indicator for sample i, Ti = 1 if sample i is in the treatment group and Ti = 0 if sample i is in control group. The parameter β0 denotes the log odds of the control group and β1 the log oddsratio between the treatment and control group. Therefore, independent tests for all the bases/regions of interest are against the null hypothesis H0: β1= 0. If the null hypothesis is rejected it implies that the logodds (and hence the methylation proportions) are different between the treatment and the control group and the base/region would subsequently be classified as a differentially methylated cytosine (DMC) or region (DMR). However, if the null hypothesis is not rejected it implies no statistically significant difference in methylation between the two groups. One important consideration in logistic regression is the sample size and in many biological experiments the number of biological samples in each group can be quite small. However, it is important to keep in mind that the relevant sample sizes in logistic regression are not merely the number of biological samples but rather the total read coverages summed over all samples in each group separately. For our example dataset, we used bases with at least 10 reads coverage for each biological sample and we advise (at least) the same for other users to improve power to detect DMCs/DMRs.
In addition, we have designed methylKit such that the logistic regression framework can be generalized to handle more than two experimental groups or data types. In such a case, the inclusion of additional treatment indicators is analogous to multiple regression when there are categorical variables with multiple groups. Additional covariates can be incorporated into model (1) by adding to the right side of the model:
α1*Covariate1,i+...+αK*CovariateK,i
where Covariate1,i, ..., CovariateK,i denote K measured covariates (continuous or categorical) for sample i = 1,...,n and α1,..., αkdenote the corresponding parameters.
Method #2: Fisher's exact test
The Fisher's exact test compares the fraction of methylated Cs in test and control samples in the absence of replicates. The main advantage of logistic regression over Fisher's exact test is that it allows for the inclusion of sample specific covariates (continuous or categorical) and the ability to adjust for confounding variables. In practice, the number of samples per group will determine which of the two methods will be used (logistic regression or Fisher's exact test). If there are multiple samples per group, methylKit will employ the logistic regression test. Otherwise, when there is one sample per group, Fisher's exact test will be used.
Following the differential methylation test and calculation of _P_-values, _methylKit_will use the sliding linear model (SLIM) method to correct _P_-values to q-values [31], which corrects for the problem of multiple hypothesis testing [32,33]. However, we also implemented the standard false discovery rate (FDR)-based method (Benjamini-Hochberg) as an option for _P_-value correction, which is faster but more conservative. Finally, methylKit can use multi-threading so that differential methylation calculations can be parallelized over multiple cores and be completed faster.
Extraction and visualization of differential methylation events
We have designed methylKit to allow a user to specify the parameters that define the DMCs/DMRs based on: q-value, %methylation difference, and type of differential methylation (hypo-/hyper-). By default, it will extract bases/regions with a q-value <0.01 and %methylation difference >25%. These defaults can easily be changed when calling _get.methylDiff()function. In addition, users can specify if they want hyper-methylated bases/regions (bases/regions with higher methylation compared to control samples) or hypo-methylated bases/regions (bases/regions with lower methylation compared to control samples). In the literature, hyper- or hypo-methylated DMCs/DMRs are usually defined relative to a control group. In our examples, and in_methylKit in general, a control group is defined when creating the objects through supplied treatment vector, and hyper-/hypomethylation definitions are based on that control group.
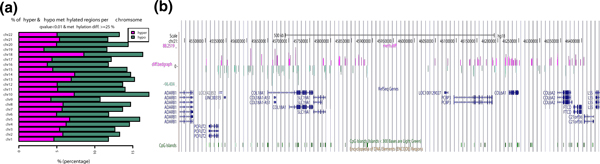
Furthermore, DMCs/DMRs can be visualized as horizontal barplots showing percentage of hyper- and hypo-methylated bases/regions out of covered cytosines over all chromosomes (Figure 5a). We observed higher levels of hypomethylation than hypermethylation in the breast cancer cell lines, which indicates that ER+ cells have lower levels of methylation. Since another common way to visualize differential methylation events is with a genome browser, methylKit can output bedgraph tracks (Figure 5b) for use with the UCSC Genome Browser or Integrated Genome Viewer.
Figure 5.
Visualizing differential methylation events. (a) Horizontal bar plots show the number of hyper- and hypomethylation events per chromosome, as a percent of the sites with the minimum coverage and differential. By default this is a 25% change in methylation and all samples with 10X coverage. (b) Example of bedgraph file uploaded to UCSC browser. The bedraph file is for differentially methylated CpGs with at least a 25% difference and q-value <0.01. Hyper- and hypo-methylated bases are color coded. The bar heights correspond to % methylation difference between ER+ and ER- sets. ER+, estrogen receptor-alpha expressing; ER-, estrogen receptor-alpha non-expressing. UCSC, University of California Santa Cruz.
Annotating differential methylation events
Annotation with gene models and CpG islands
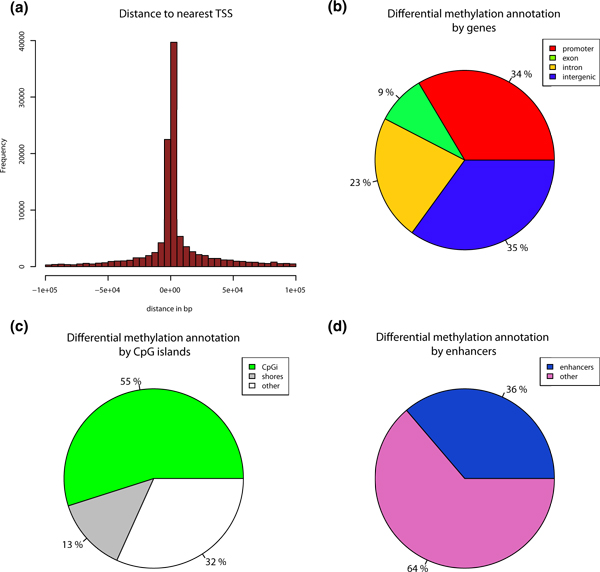
To discern the biological impact of differential methylation events, each event must be put into its genomic context for subsequent analysis. Indeed, Hansen et al. [34] showed that most variable regions in terms of methylation in the human genome are CpG island shores, rather than CpG islands themselves. Thus, it is interesting to know the location of differential methylation events with regard to CpG islands, their shores, and also the proximity to the nearest transcription start site (TSS) and gene components. Accordingly, methylKit can annotate differential methylation events with regard to the nearest TSS (Figure 6a) and it also can annotate regions based on their overlap with CpG islands/shores and regions within genes (Figures 6b and 6c are output from methylKit).
Figure 6.
Annotation of differentially methylated CpGs. (a) Distance to TSS for differentially methylated CpGs are plotted from ER+ versus ER- analysis. (b) Pie chart showing percentages of differentially methylated CpGs on promoters, exons, introns and intergenic regions. (c) Pie chart showing percentages of differentially methylated CpGs on CpG islands, CpG island shores (defined as 2kb flanks of CpG islands) and other regions outside of shores and CpG islands. (d) Pie chart showing percentages of differentially methylated CpGs on enhancers and other regions. ER+, estrogen receptor-alpha expressing; ER-, estrogen receptor-alpha non-expressing, TSS, transcription start site.
Annotation with custom regions
As with most genome-wide assays, the regions of interest for DNA methylation analysis may be quite numerous. For example, several reports show that Alu elements are aberrantly methylated in cancers [35,36] and enhancers are also differentially methylated [37,38]. Since users may need to focus on specific genomic regions and require customized annotation for capturing differential DNA methylation events, methylKit can annotate differential methylation events using user-supplied regions. As an example, we identified differentially methylated bases of ER+ and ER- cells that overlap with ENCODE enhancer regions [39], and we found a large proportion of differentially methylated CpGs overlapping with the enhancer marks, and then plotted them with methylKit (Figure 6d).
Analyzing 5-hydroxymethylcytosine data with methylKit
5-Hydroxymethylcytosine is a base modification associated with pluropotency, hematopoiesis and certain brain tissues (reviewed in [40]). It is possible to measure base-pair resolution 5hmC levels using variations of traditional bisulfite sequencing. Recently, Yu et al. [41] and Booth et al. [15] published similar methods for detecting 5hmC levels in base-pair resolution. Both methods require measuring 5hmC and 5mC levels simultaneously and use 5hmC levels as a substrate to deduce real 5mC levels, since traditional bisulfite sequencing cannot distinguish between the two [42]. However, both the 5hmC and 5mC data generated by these protocols are bisulfite sequencing based, and the alignments and text files of 5hmC levels can be used directly in_methylKit_. Furthermore, methylKit has an adjust.methylC() function to adjust 5mC levels based on 5hmC levels as described in Booth et al. [15].
Customizing analysis with convenience functions
methylKit is dependent on Bioconductor [43] packages such as GenomicRanges and its objects are coercible to_GenomicRanges_ objects and regular R data structures such as data frames via provided convenience functions. That means users can integrate methylKit objects to other Bioconductor and R packages and customize the analysis according to their needs or extend the analysis further by using other packages available in R.
Conclusions
Methods for detecting methylation across the genome are widely used in research laboratories, and they are also a substantial component of the National Institutes of Health's (NIH's) EpiGenome roadmap and upcoming projects such as BLUEPRINT [44]. Thus, tools and techniques that enable researchers to process and utilize genome-wide methylation data in an easy and fast manner will be of critical utility.
Here, we show a large set of tools and cross-sample analysis algorithms built into_methylKit_, our open-source, multi-threaded R package that can be used for any base-level dataset of DNA methylation or base modifications, including 5hmC. We demonstrate its utility with breast cancer RRBS samples, provide test data sets, and also provide extensive documentation with the release.
Abbreviations
5hmC: 5-hydroxymethylcytosine; 5mC: 5-methylcytosine; bp: base pair; BS-seq,:bisulfite sequencing; DMC: differentially methylated cytosine; DMR: differentially methylated region; ER: estrogen receptor alpha; FDR: false discovery rate; PCA: principal component analysis; PCR: polymerase chain reaction; RRBS: reduced representation bisulfite sequencing; SLIM: sliding linear model; TSS: transcription start site.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
AA designed methylKit, developed the first codebase, and added most features. MK designed the logistic regression based statistical test for methylKit and worked on statistical modeling and initial clustering features. SL wrote some of the features in _methylKit_and prepared plots for the manuscript. MEF, FGB and AM tested the code and provided initial data for development of methylKit. CEM supervised the work, tested code, and coordinated test data for validation. All authors have read and approved the manuscript for publication.
Supplementary Material
Additional file 1
methylKit v0.5.3. This version of methylKit is included for archival purposes only. Please download the most recent version from [16].
Additional file 2
methylKit User Guide. A vignette file to accompany the methylKit software package; the most recent software and vignette can be downloaded at [16].
Additional file 3
methylKit documentation. Documentation for functions and classes in the methylKit software package; the most recent software and documentation can be downloaded at [16].
Additional file 4
R script for example analysis. The file contains R commands that are needed to do analysis and to produce graphs used in this manuscript. The file contains both the commands and detailed comments on how those commands can be used. An up to date version of this script will be consistently maintained at [16].
Contributor Information
Altuna Akalin, Email: ala2027@med.cornell.edu.
Matthias Kormaksson, Email: mk375@cornell.edu.
Sheng Li, Email: shl2018@med.cornell.edu.
Francine E Garrett-Bakelman, Email: frg9015@nyp.org.
Maria E Figueroa, Email: marfigue@med.umich.edu.
Ari Melnick, Email: amm2014@med.cornell.edu.
Christopher E Mason, Email: chm2042@med.cornell.edu.
Acknowledgements
We wish to acknowledge the invaluable contribution of the WCMC Epigenomics Core Facility. MEF is supported by the Leukemia & Lymphoma Society Special Fellow Award and a Doris Duke Clinical Scientist Development Award. FGB is supported by a Sass Foundation Judah Folkman Fellowship. AM is supported by an LLS SCOR grant (7132-08) and a Burroughs Wellcome Clinical Translational Scientist Award. AM and CEM are supported by a Starr Cancer Consortium grant (I4-A442). CEM is supported by the National Institutes of Health (I4-A411, I4-A442, and 1R01NS076465-01).
References
- Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–2210. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo Q-M, Edsall L, Antosiewicz-Bourget J, Stewart R, Ruotti V, Millar AH, Thomson JA, Ren B, Ecker JR. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird AP, Wolffe AP. Methylation-induced repression--belts, braces, and chromatin. Cell. 1999;99:451–454. doi: 10.1016/S0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol Cell Biol. 1998;18:6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF, Tallman MS, Sun Z, Wolniak K, Peeters JK, Liu W, Choe SE, Fantin VR, Paietta E, Löwenberg B, Licht JD, Godley LA, Delwel R, Valk PJM, Thompson CB, Levine RL, Melnick A. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez AF, Assenov Y, Martin-Subero JI, Balint B, Siebert R, Taniguchi H, Yamamoto H, Hidalgo M, Tan A-C, Galm O, Ferrer I, Sanchez-Cespedes M, Villanueva A, Carmona J, Sanchez-Mut JV, Berdasco M, Moreno V, Capella G, Monk D, Ballestar E, Ropero S, Martinez R, Sanchez-Carbayo M, Prosper F, Agirre X, Fraga MF, Graña O, Perez-Jurado L, Mora J, Puig S. et al. A DNA methylation fingerprint of 1628 human samples. Genome Res. 2012;22:407–419. doi: 10.1101/gr.119867.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E, Beard C. Role for DNA methylation in genomic imprinting. Nature. 1993;366:362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- Smith ZD, Chan MM, Mikkelsen TS, Gu H, Gnirke A, Regev A, Meissner A. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature. 2012;484:339–344. doi: 10.1038/nature10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, Gnirke A, Jaenisch R, Lander ES. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akalin A, Garrett-Bakelman FE, Kormaksson M, Busuttil J, Zhang L, Khrebtukova I, Milne TA, Huang Y, Biswas D, Hess JL, Allis D, Roeder RG, Valk PJM, Lo B, Paietta E, Tallman MS, Schroth GP, Mason CE, Melnick A, Figueroa ME. Base-pair resolution DNA methylation sequencing reveals profoundly divergent epigenetic landscapes in acute myeloid leukemia. PLoS Genet. 2012;8:e1002781. doi: 10.1371/journal.pgen.1002781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, O'Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball MP, Li JB, Gao Y, Lee J-H, LeProust EM, Park I-H, Xie B, Daley GQ, Church GM. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat Biotechnol. 2009;27:361–368. doi: 10.1038/nbt.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth MJ, Branco MR, Ficz G, Oxley D, Krueger F, Reik W, Balasubramanian S. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science. 2012;336:934–937. doi: 10.1126/science.1220671. [DOI] [PubMed] [Google Scholar]
- methylKit. http://code.google.com/p/methylkit
- Flusberg BA, Webster DR, Lee JH, Travers KJ, Olivares EC, Clark TA, Korlach J, Turner SW. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat Methods. 2010;7:461–465. doi: 10.1038/nmeth.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherf GM, Lieberman KR, Rashid H, Lam CE, Karplus K, Akeson M. Automated forward and reverse ratcheting of DNA in a nanopore at 5-Å precision. Nat Biotechnol. 2012;30:344–348. doi: 10.1038/nbt.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frith MC, Mori R, Asai K. A mostly traditional approach improves alignment of bisulfite-converted DNA. Nucleic Acids Res. 2012;40:e100. doi: 10.1093/nar/gks275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F, Kreck B, Franke A, Andrews SR. DNA methylome analysis using short bisulfite sequencing data. Nat Methods. 2012;9:145–151. doi: 10.1038/nmeth.1828. [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Asmann YW, Kalari KR, Bot B, Eckel-Passow JE, Baker TR, Carr JM, Khrebtukova I, Luo S, Zhang L, Schroth GP, Perez EA, Thompson EA. Integrated analysis of gene expression, CpG island methylation, and gene copy number in breast cancer cells by deep sequencing. PloS One. 2011;6:e17490. doi: 10.1371/journal.pone.0017490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van 't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AAM, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, Schreiber GJ, Kerkhoven RM, Roberts C, Linsley PS, Bernards R, Friend SH. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- Sotiriou C, Neo S-Y, McShane LM, Korn EL, Long PM, Jazaeri A, Martiat P, Fox SB, Harris AL, Liu ET. Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc Natl Acad Sci USA. 2003;100:10393–10398. doi: 10.1073/pnas.1732912100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joliffe I. Principal Component Analysis. 2. New York, USA, Springer; 2002. [Google Scholar]
- Esteller M, Corn PG, Baylin SB, Herman JG. A gene hypermethylation profile of human cancer. Cancer Res. 2001;61:3225–3229. [PubMed] [Google Scholar]
- Baylin SB, Herman JG. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends Genet. 2000;16:168–174. doi: 10.1016/S0168-9525(99)01971-X. [DOI] [PubMed] [Google Scholar]
- Costello JF, Frühwald MC, Smiraglia DJ, Rush LJ, Robertson GP, Gao X, Wright FA, Feramisco JD, Peltomäki P, Lang JC, Schuller DE, Yu L, Bloomfield CD, Caligiuri MA, Yates A, Nishikawa R, Su Huang H, Petrelli NJ, Zhang X, O'Dorisio MS, Held WA, Cavenee WK, Plass C. Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nat Genet. 2000;24:132–138. doi: 10.1038/72785. [DOI] [PubMed] [Google Scholar]
- Doi A, Park I-H, Wen B, Murakami P, Aryee MJ, Irizarry R, Herb B, Ladd-Acosta C, Rho J, Loewer S, Miller J, Schlaeger T, Daley GQ, Feinberg AP. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat Genet. 2009;41:1350–1353. doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H-Q, Tuominen LK, Tsai C-J. SLIM: a sliding linear model for estimating the proportion of true null hypotheses in datasets with dependence structures. Bioinformatics. 2011;27:225–231. doi: 10.1093/bioinformatics/btq650. [DOI] [PubMed] [Google Scholar]
- Storey J. A direct approach to false discovery rates. J R Stat Soc Series B Stat Methodol. 2002;64:479–498. doi: 10.1111/1467-9868.00346. [DOI] [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KD, Timp W, Bravo HC, Sabunciyan S, Langmead B, McDonald OG, Wen B, Wu H, Liu Y, Diep D, Briem E, Zhang K, Irizarry R a, Feinberg AP. Increased methylation variation in epigenetic domains across cancer types. Nat Genet. 2011;43:768–775. doi: 10.1038/ng.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M. DNA hypomethylation in cancer cells. Epigenomics. 2009;1:239–259. doi: 10.2217/epi.09.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez J, Vives L, Jordà M, Morales C, Muñoz M, Vendrell E, Peinado MA. Genome-wide tracking of unmethylated DNA Alu repeats in normal and cancer cells. Nucleic Acids Res. 2008;36:770–784. doi: 10.1093/nar/gkm1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Schöler A, Wirbelauer C, Oakeley EJ, Gaidatzis D, Tiwari VK, Schübeler D. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480:490–495. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- Wiench M, John S, Baek S, Johnson TA, Sung M-H, Escobar T, Simmons CA, Pearce KH, Biddie SC, Sabo PJ, Thurman RE, Stamatoyannopoulos JA, Hager GL. DNA methylation status predicts cell type-specific enhancer activity. EMBO J. 2011;30:3028–3039. doi: 10.1038/emboj.2011.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, Ku M, Durham T, Kellis M, Bernstein BE. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branco MR, Ficz G, Reik W. Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nat Rev Genet. 2011;13:7–13. doi: 10.1038/nrg3080. [DOI] [PubMed] [Google Scholar]
- Yu M, Hon GC, Szulwach KE, Song C-X, Zhang L, Kim A, Li X, Dai Q, Shen Y, Park B, Min J-H, Jin P, Ren B, He C. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149:1368–1380. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Pastor WA, Shen Y, Tahiliani M, Liu DR, Rao A. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PloS One. 2010;5:e8888. doi: 10.1371/journal.pone.0008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JYH, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:r80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams D, Altucci L, Antonarakis SE, Ballesteros J, Beck S, Bird A, Bock C, Boehm B, Campo E, Caricasole A, Dahl F, Dermitzakis ET, Enver T, Esteller M, Estivill X, Ferguson-Smith A, Fitzgibbon J, Flicek P, Giehl C, Graf T, Grosveld F, Guigo R, Gut I, Helin K, Jarvius J, Küppers R, Lehrach H, Lengauer T, Lernmark Å, Leslie D. et al. BLUEPRINT to decode the epigenetic signature written in blood. Nat Biotechnol. 2012;30:224–226. doi: 10.1038/nbt.2153. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1
methylKit v0.5.3. This version of methylKit is included for archival purposes only. Please download the most recent version from [16].
Additional file 2
methylKit User Guide. A vignette file to accompany the methylKit software package; the most recent software and vignette can be downloaded at [16].
Additional file 3
methylKit documentation. Documentation for functions and classes in the methylKit software package; the most recent software and documentation can be downloaded at [16].
Additional file 4
R script for example analysis. The file contains R commands that are needed to do analysis and to produce graphs used in this manuscript. The file contains both the commands and detailed comments on how those commands can be used. An up to date version of this script will be consistently maintained at [16].