Protein typing of circulating microvesicles allows real-time monitoring of glioblastoma therapy (original) (raw)
. Author manuscript; available in PMC: 2012 Dec 11.
Published in final edited form as: Nat Med. 2012 Nov 11;18(12):1835–1840. doi: 10.1038/nm.2994
Abstract
Glioblastomas shed large quantities of small, membrane-bound microvesicles (MVs) into the circulation. While these hold promise as potential biomarkers of therapeutic response, their identification and quantitation remain challenging. Here, we describe a highly sensitive and rapid analytical technique for profiling circulating MVs directly from blood samples of glioblastoma patients. MVs, introduced onto a dedicated microfluidic chip, are labeled with target-specific magnetic nanoparticles and detected by a miniaturized nuclear magnetic resonance system. Compared with current methods, this integrated system has a much higher detection sensitivity, and can differentiate glioblastoma multiforme (GBM) MVs from non-tumor host cell-derived MVs. We also show that circulating GBM MVs can serve as a surrogate for primary tumor mutations and a predictive metric of treatment-induced changes. This platform could provide both an earlier indicator of drug efficacy and a potential molecular stratifier for human clinical trials.
Many cancers shed materials into the peripheral circulation. These appear as circulating tumor cells (CTCs)1 and soluble proteins2,3, and are being exploited as surrogate markers of tumor staging and response to therapy. Systemic and intracranial tumors also release microvesicles (MVs)4–6 into the peripheral circulation. In particular, tumors of the central nervous system, lying behind a partially intact blood brain barrier, often do not release CTCs nor are they commonly associated with detectable soluble protein biomarkers. Large quantities of MVs, however, have been found within blood of patients with glioblastoma multiforme (GBM)3,5, and these thus offer new hope for treatment monitoring of this devastating disease.
MVs in circulation are made up of membrane-bound vesicles (50 nm – 1 μm in diameter), which differ in their cellular origin, abundance and biogenesis4. The population includes exosomes (50 – 100 nm and positive for CD63, HSP90, Flotillins) released from multivesicular endosomes, larger shed microvesicles, membrane particles, apoptotic vesicles and exosome-like vesicles originating from multivesicular bodies of other cell organelles4,7,8. Different subtypes of MVs can have overlapping size and often co-purify if separated by size only4,9. MVs contain cell surface proteins10, including EGFR and EGFRvIII6,11, as well as RNA5 and DNA12. Current analyses generally require large numbers of MVs to be concentrated and processed using time-consuming Western blotting or enzyme-linked immunosorbent assays (ELISA), making them impractical in a typical clinical setting.
Herein, we describe a highly sensitive and rapid analytical technique for profiling proteins in MVs from GBM cell cultures and from GBM patient blood samples (circulating MVs). We use both size and immunoaffinity (vesicles 50 – 150 nm and CD63-positive) to define a population of circulating MVs, which consist primarily of exosomes. MVs are labeled with target-specific magnetic nanoparticles (MNPs), and detected by a miniaturized (micro) nuclear magnetic resonance (μNMR) system13,14. A prototype μNMR system was previously used to detect whole tumor cells (>10 μm target size range)15. Adapting μNMR to MV detection, however, presented significant engineering challenges since these targets are smaller than tumor cells by 1 – 2 orders of magnitude. We thus developed a new microfluidic system and analytical technology specifically for MV detection and profiling in GBM patients, that can differentiate glioma-derived MVs from host cell-derived MVs. Employing this technology, we describe findings to evaluate the comparative protein profiles of glioma-derived MVs against those from parental GBM cells. We likewise report on the ability of this system to detect specific circulating MVs from blood of GBM patients and non-GBM control subjects, and whether circulating MVs can be used in longitudinal studies to monitor and predict response to GBM therapies.
RESULTS
Magnetic nanosensor technology for MV detection
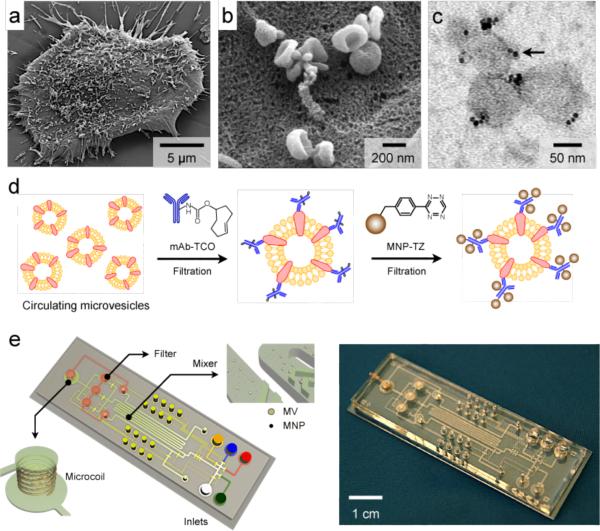
GBM cell lines in culture produced abundant MVs (Fig. 1a). MV counting based on nanoparticle tracking analysis (NTA; Supplementary Fig. 1a) reported a typical concentration of 108 – 109 MVs mL−1 in culture media. In situ scanning electron microscopy analysis of MVs on cell surface revealed that many of the MVs were saucer-shaped16, a pattern typical of exosomes (Fig. 1b). For detection by the microfluidic μNMR, we purified and labeled MVs with magnetic nanoparticles (MNPs, core diameter 7 nm) by targeting MV protein markers (Fig. 1c). Such magnetic labeling renders MV superparamagnetic, which results in faster decay of the 1H NMR signal. The decay rate (_R_2) is proportional to the MNP concentration, thus enabling the quantitation of target MV protein concentration.
Figure 1. Human glioblastoma cells produce abundant microvesicles (MVs) which can be analyzed by micro nuclear magnetic resonance (μNMR).
(a) Scanning electron microscopy image of a primary human glioblastoma cell (GBM20/3) grown in culture, releasing abundant MVs. (b) High magnification image shows that many of the MVs on the cell surface assumed typical saucer-shaped characteristics of exosomes. (c) Transmission electron microscopy image of MVs (~ 80 nm) targeted with magnetic nanoparticles (MNPs) via CD63 antibody. The samples were purified by membrane filtration to collect small MVs. The MNPs appear as black dots (indicated by an arrow). (d) Labeling procedure for extravesicular markers. The two-step BOND-2 assay configuration uses bioorthogonal amplification chemistry to maximize MNP binding onto target proteins on MVs (not drawn to scale). (e) Microfluidic system for on-chip detection of circulating MVs. The system was designed to i) allow MNP-targeting of MVs, ii) concentrate MNP-tagged MVs while removing unbound MNPs, and iii) provide in-line μNMR detection.
We used a two-step bioorthogonal approach (BOND-2; Fig. 1d) for MNP labeling. MV protein markers were first targeted with antibodies modified with _trans_-cyclooctene (TCO), and then coupled with MNPs derivatized with 1,2,4,5-tetrazine (TZ). The fast, covalent cycloaddition between TZ and TCO maximizes MNP binding and increases the magnetic signals (_R_2) by > 300% compared to that achieved by direct MNP-antibody conjugation17. Because both TZ and TCO are small (~ 200 Daltons each), this chemical conjugation does not appreciably increase the size of the antibody or the MNP. It is thus possible to use size-selective filtration to remove excess antibodies or MNPs, while retaining targeted MVs. Figure 1e shows a prototype device developed for clinical, point-of-care MV analysis. It contains three essential components: i) a chaotic mixer for reacting MVs with antibodies and MNPs, ii) a membrane filter for washing and concentrating targeted MVs, and iii) a microcoil for NMR detection (sample volume: 1 μL). MVs purified by the microfluidic device typically have a size distribution ranging 50 – 150 nm (with > 75% smaller than 120 nm in diameter), due to the cut-off sizes of membrane filtration (Supplementary Fig. 1b). To streamline the assay procedure (Supplementary Fig. 2), the fluidic flow is controlled by automated pneumatic valves (Supplementary Fig. 3).
Optimized assay for MV protein typing
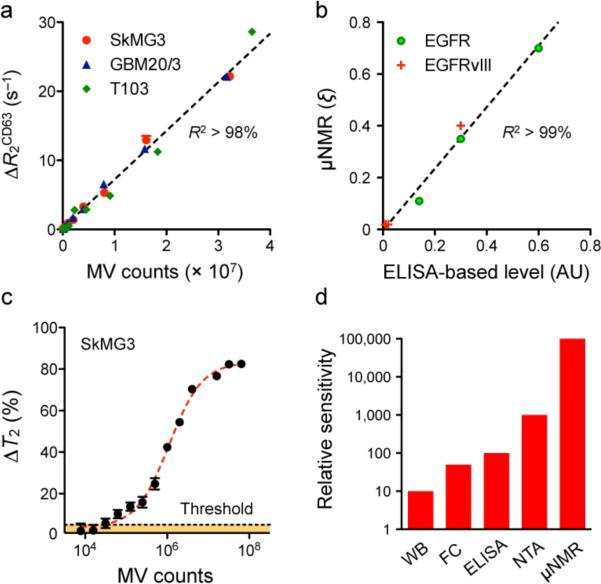
We hypothesized that in multicellular environments, CD63 expression could be used as an internal measure of total MV counts from different cell sources (Supplementary Fig. 4a)4,7. Figure 2 summarizes the results of the validation study. When MVs were analyzed for CD63 expression, the corresponding _R_2 changes (Δ_R_2CD63) were found to be linearly proportional to MV counts, which were independently confirmed by NTA (Fig. 2a). Importantly, these _R_2 changes were statistically identical even among MVs from different cell lines (P > 0.16). The μNMR measurements were highly reproducible and accurate with < 1% instrumental errors. The expression levels of other protein biomarkers were then determined by normalizing the marker-associated Δ_R_2mAb by Δ_R_2CD63 (ξmAb = Δ_R_2mAb/Δ_R_2CD63). Such normalization automatically accounts for differences in MV counts, obviating the need to quantify MVs by other measures such as NTA. The μNMR also showed excellent agreement (_R_2 > 99%) with fluorescence ELISA (Fig. 2b), and its detection sensitivity (Fig. 2c) far surpassed that of other analytical methods, being 104, 103, and 102-fold more sensitive than Western blotting, ELISA, and NTA respectively (Fig. 2d and Supplementary Fig. 4b). Since μNMR detection sensitivity depends on biomarker protein integrity, we also tested the detection level as a function of time (Supplementary Fig. 5a) and showed that signal degradation can be largely prevented by fixation (Supplementary Fig. 5b).
Figure 2. μNMR assay for MV detection.
(a) Correlation between μNMR measurements for CD63 and MV numbers in a dilution series. MV numbers were estimated by nanoparticle tracking analysis (NTA). The transverse relaxation (_R_2), as determined by μNMR, varied linearly with MV numbers (_R_2 > 98%). Importantly, the _R_2 changes (Δ_R_2CD63) were statistically identical across different cell lines (P > 0.16), which validates its use as a universal measure for quantitating MVs. Western blotting (Supplementary Fig. 4a) also revealed a consistent and higher expression of CD63 in the prepared MVs. (b) Using MVs from model cell lines, the expression levels of EGFR and EGFRvIII were measured by μNMR. The MV expression (ξ) of a target protein marker was obtained by normalizing a marker-associated _R_2 against _R_2CD63. μNMR measurements showed excellent agreement (_R_2 > 99%) with fluorescence ELISA. (c) Detection threshold of μNMR assay for MVs. With CD63-tagged MVs, the detection threshold of μNMR, as measured by the relative changes in the transverse relaxation time (_T_2 = 1/_R_2) with respect to controls, was approximately ~104 MVs. (d) Comparison of MV detection sensitivity. In a series of MV dilution assays, μNMR was considerably more sensitive than Western blotting (WB; Supplementary Fig. 4b), flow cytometry (FC), ELISA and NTA. All measurements were performed in triplicate, and the data is displayed as mean ± s.e.m.
MV molecular signature for GBM detection
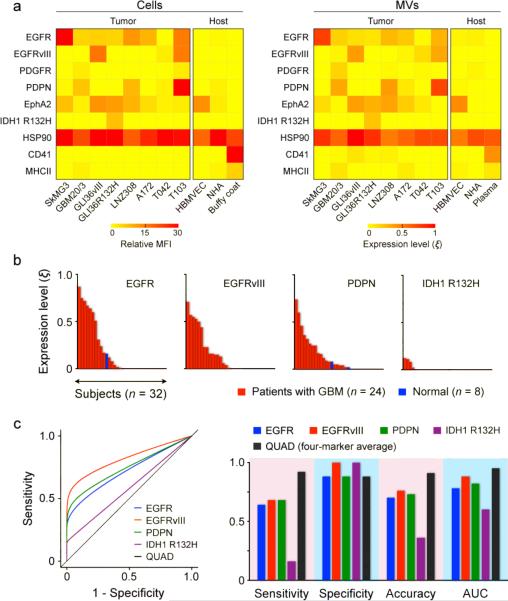
We next determined whether GBM-derived MVs show similar protein profiles to that of intact parental cells, and whether molecular markers could be used to differentiate between GBM and host cell (non-GBM) MVs. Based on prior reports3,4,18, we measured the following seven extravesicular and two intravesicular protein markers: epidermal growth factor receptor (EGFR)11, platelet-derived growth factor receptor (PDGFRα)19, podoplanin (PDPN)20, ephrin type-A receptor 2 (EphA2)21 for their elevated expressions in glioma; EGFRvIII6 and cytosolic isocitrate dehydrogenase 1 mutation (IDH1 R132H)22,23 for their specific expressions in glioma; cytosolic heat shock protein 90 (HSP90)3,4 as a positive control for MVs; CD4124 and major histocompatibility complex class II (MHCII)4 to represent host cell markers. Comparative analyses confirmed that MVs indeed reflect the protein profiles of their parental cells (Fig. 3a). GBM-derived MVs exhibit a distinct molecular signature; elevated expressions of EGFR, EGFRvIII, PDPN and IDH1 R132H together allowed effective discrimination of GBM and host cell MVs.
Figure 3. Protein typing of glioblastoma multiforme (GBM)-derived MVs from cell lines and patient samples.
(a) GBM markers (EGFR, EGFRvIII, PDGFR, PDPN, EphA2 and IDH1 R132H), positive MV control marker (HSP90) as well as host cell markers (CD41, MHCII) were profiled in both parental cells (left) and their corresponding MVs (right). A four GBM marker combination (EGFR, EGFRvIII, PDPN and IDH1 R132H) was able to distinguish GBM-derived MVs from host cell-derived MVs. HBMVEC, human brain microvascular endothelial cell; NHA, normal human astrocyte; buffy coat and plasma were isolated from whole blood donated by healthy volunteers. (b) Analysis of clinical patient samples. Waterfall plots show the expression levels of different biomarkers sorted from high (left) to low (right). Note the increased expression of EGFR and PDPN, as well as the unique expression of EGFRvIII and IDH1 R132H, in patient samples. (c) Receiver operating characteristic curves (left) were generated to compare the detection sensitivity, specificity and accuracy of each marker. Overall, the accuracy was < 76% for a single marker alone (right). When all markers were combined (QUAD), the detection accuracy considerably improved (> 90%). AUC: area under curve.
We next profiled MVs from blood samples of GBM patients (n = 24) and healthy volunteers (n = 8; Supplementary Table 1). Samples for this study had been selected in a blinded fashion but were enriched in samples positive for EGFR amplification or EGFRvIII mutation. For each patient, there was considerable heterogeneity in the expression levels of individual markers (Supplementary Fig. 6a). The waterfall plot for each marker revealed a broad spectrum of expression level (Fig. 3b). Consistent with prior report22, IDH1 R132H, albeit highly specific to GBM, showed lower prevalence in primary GBM patients. The accuracy of each marker in GBM detection, obtained from the receiver operating characteristic (ROC) curves, was < 76% (Fig. 3c and Supplementary Fig. 6b). However, by combining the results of all four markers, the accuracy increased to > 90% (Supplementary Table 2).
Efficacy of drug treatment revealed by MVs
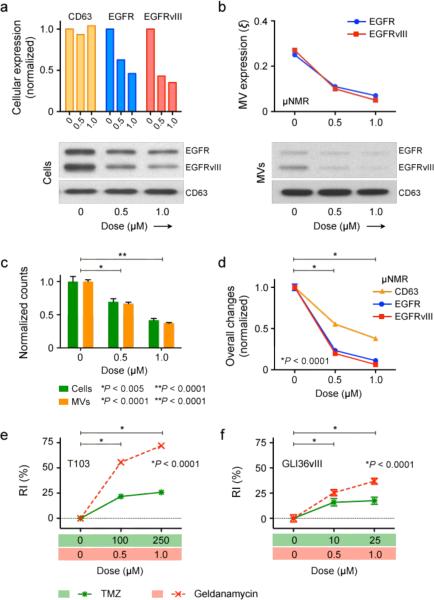
We investigated the potential use of MVs for monitoring treatment responses. Two drug regimens were selected: 1) an alkylating agent, temozolomide (TMZ), which is a first line adjuvant drug currently used with concomitant radiation therapy for the treatment of newly diagnosed GBM; and 2) a HSP90 inhibitor, geldanamycin (17AAG), whose binding to HSP90 can enhance the degradation of signaling molecules (e.g., EGFR)25–27. Treatment effects on T103 mouse GBM cells and MVs are compared in Supplementary Fig. 7 (for TMZ) and Fig. 4 (for geldanamycin). TMZ treatment did not elicit significant changes in the cellular expression of CD63, EGFR or EGFRvIII, as determined by flow cytometry and Western blotting (Supplementary Fig. 7a). Likewise, corresponding μNMR assays on MVs reported comparable expression levels for EGFR and EGFRvIII (Supplementary Fig. 7b). TMZ treatment, however, resulted in a dose-dependent decrease in both cell and MV counts (Supplementary Fig. 7c). Total MV expression of CD63, EGFR, and EGFRvIII, as measured by the μNMR, thus showed a similar dose-dependent decline, with increasing concentration of TMZ applied (Supplementary Fig. 7d).
In contrast to TMZ, geldanamycin resulted in a significant decrease in EGFR and EGFRvIII expression, but not in CD63, for both cells and MVs (Figs. 4a and b). Consequently, as MV numbers decreased in proportion to drug concentration (a change attributed to cell loss; Fig. 4c), the total MV expression of EGFR and EGFRvIII each showed a much more pronounced decline than MV-CD63 (Fig. 4d). To account for such additive effects, we defined a drug response index (RI) as RI = [(1-Δ_n_) + Σ(1-Δξ_k_)]/N, where 1-Δ_n_ and 1-Δξ_k_ are the relative changes in glioma MV numbers and MV biomarker expression levels (ξ_k_), respectively, and N is the total number of markers monitored. Conversely, the complementary of RI was defined as a tumor progression index (TPI = 1 - RI). A plot of RI captured the drug efficacies of TMZ and geldanamycin (Fig. 4e). These findings were corroborated in repeated studies with the human GBM GLI36vIII line, which showed nearly identical trends (Fig. 4f and Supplementary Fig. 8), indicating that normalized detection of MV biomarkers is highly sensitive to reveal the differences between treatment mechanisms.
Figure 4. Effects of geldanamycin treatment on T103 GBM model.
(a) On the cellular level, geldanamycin treatment did not alter the expression of CD63, but considerably reduced the amount of EGFR and EGFRvIII, as determined by flow cytometry and Western blotting. (b) Upon geldanamycin treatment, μNMR detection showed that MVs exhibit a similar decrease in EGFR and EGFRvIII profiles (ξ; normalized with respect to CD63 expression) as that observed in whole cells. (c, d) Total number of cells and MVs (c) decreased in a dose-dependent manner, upon drug treatment. However, the total EGFR and EGFRvIII levels in MVs (d) showed a steeper decline due to the combined effects of reduced MV number and decreased marker (EGFR, EGFRvIII) expression per MV. (e, f) To use MV readouts as an indicator for drug efficacy, a response index (RI) was defined, that recapitulates changes in both MV number and MV molecular expression. Compared to TMZ, the RI of geldanamycin was higher for both T103 (e) and GLI36vIII (f) cell lines due to the drug's ability to reduce both MV number as well as receptor expression. All changes with respect to untreated samples were statistically significant (P < 0.001). All analytical measurements were performed in triplicate, and the data is shown as mean ± s.e.m.
Circulating MVs predict drug efficacy in vivo
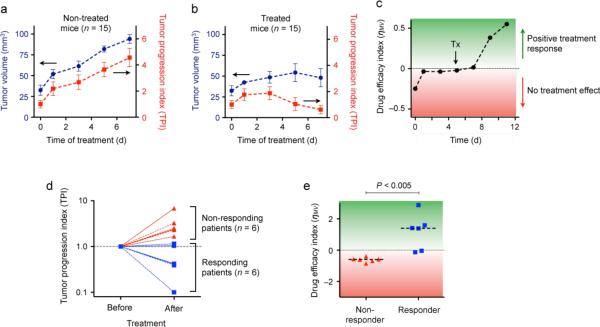
We applied the platform to monitor treatment effects in vivo. We first used a mouse model with xenografted human GBM28. Circulating MVs in cohorts of T103 tumor-bearing mice were profiled during tumor growth as well as following TMZ treatment (80 mg/kg daily). In untreated animals, the tumor progression index (TPI) increased over time and paralleled tumor volume measured by MRI (Fig. 5a). In TMZ treated animals, TPI changes occurred several days prior to reductions in tumor volume as measured by imaging (Fig. 5b). To visualize the temporal onset of therapeutic effects, we plotted an efficacy index (η_MV_), defined as the temporal change in 1/TPI. As shown in Fig. 5c, the efficacy index η_MV_ was close to zero for expanding tumors. Upon TMZ administration, however, η_MV_ rose rapidly, indicating the effectiveness of the treatment.
Figure 5. Analysis of circulating MVs in GBM mice and human patients undergoing treatment.
(a) Circulating MVs in untreated tumor-bearing animals (n = 15). The tumor progression index (TPI) is used to reflect changes in both MV number and MV molecular expression. Note the close correlation between increasing tumor volumes and TPI values over time. (b) In TMZ treated mice (n = 15), TPI values from μNMR measurements revealed response to treatment before apparent changes in tumor size. (c) Since the decline rate of TPI represents a time-sensitive indicator of treatment efficacy, we define the drug efficacy index (η_MV_) as the temporal change in TPI−1. With TMZ treatment, η_MV_ switched from negative (tumor progression) to positive (treatment response). (d, e) Clinical trial. Blood samples were collected from the same patients before and after TMZ/radiation treatment, and circulating MVs were profiled using μNMR. Both TPI (d) and η_MV_ (e) confirmed that longitudinal MV profiling can be used to predict treatment outcomes and differentiate between responders and non-responders. Dashed lines in (e) indicate the median values.
We next extended the study to clinical GBM patients whose blood samples were collected prior to and after standard-of-care TMZ/radiation treatment (Supplementary Table 3). In this longitudinal study, the tumor progression index (TPI) was evaluated upon samples became available. Responder and non-responder status was later defined by a neuro-oncologist without knowledge of the μNMR results, and based on subsequent clinical and MRI data. TPI allowed the identification and prediction of treatment outcomes, especially for non-responding cases (Fig. 5d). The corresponding efficacy index (η_MV_) also showed significant difference (P < 0.005) between responders and non-responders (Fig. 5e).
DISCUSSION
GBM is the most common primary malignancy of the central nervous system29,30. Amplification of EGFR is the most frequent genetic abnormality associated with GBM, and EGFR overexpression has been shown in up to 85% of cases29. GBM also often expresses EGFRvIII, a genomic deletion variant of EGFR that is constitutively active and highly oncogenic31,32. It is likely that the recent identification of circulating MVs containing EGFRvIII specific RNA5 and GBM associated proteins10 will not only be immediately relevant to this subset of GBM patients, but could also be expanded to other GBM-mutation evaluation. Likewise, circulating MVs may provide new avenues for cancer diagnostics and expand our understanding of cellular communication.
Evaluating circulating MVs could lead to a paradigm shift in clinical care. Phase 1 and phase 2 trials of targeted agents presently require molecular stratification of GBM tumors. In addition, there remains an urgent need to provide sequential indices of tumor molecular response to these agents. While imaging remains useful as a clinical tool, the standard RECIST (Response Evaluation Criteria In Solid Tumors) and volumetric criteria of response are insensitive therapeutic markers in patients receiving vascular-targeted agents such as Avastin. New and more sensitive imaging approaches33–36 are currently in development; however, they are not universally available, are often expensive37 and impractical for rapid sequential evaluations. As a result, there has been intense interest in finding serologic biomarkers for GBM.
Our findings show that GBM-derived circulating MVs can be rapidly detected in clinical blood samples with high sensitivity using a nanotechnology-inspired biosensor. The system combines on-chip micro-filtration and μNMR principles, to enable quantitative detection of MV numbers and protein expression. Measurements are performed on small sample volumes without the need for extensive purification or time-consuming detection techniques. By employing a bioorthogonal targeting approach to specifically target and densely pack MNPs onto MVs, the current platform has achieved a detection sensitivity that surpasses standard ELISA and flow cytometry analyses by several orders of magnitude (Fig. 2). We believe that this could be further enhanced with the use of newer magnetic nanomaterials (with higher magnetization)38, improved assay types, additional amplification steps, and new bioorthogonal approaches. Likewise, further device optimization are expected. By incorporating differential, multistep filtering system, MVs can be isolated from whole cells; multiple microcoils can be embedded for extensive parallel profiling of a larger number of MV proteins. Such system could realize a comprehensive yet portable lab-on-a-chip for MV analysis.
We further envision other clinical applications in which protein typing of circulating MVs would be useful. The above-described methodology could be extended to examine other primary tumors, particularly since many cancers secrete much greater quantities of circulating MVs than CTCs. It could also be adapted to monitor circulating MVs in a variety of inflammatory and infectious diseases, using blood samples, cerebral spinal fluid, urine, saliva or other biofluids. With its capacity for molecular diagnostics at the bedside, the developed platform could potentially redefine the current standard-of-care for patients.
Materials and Methods
Cell culture
The following human GBM cell lines were cultured in Dulbecco's modified essential medium (DMEM, Cellgro) containing 10% fetal bovine serum (FBS, Cellgro) and supplemented with penicillin-streptomycin (Cellgro): SkMG3 was provided by Dr. Timothy Chan, Memorial Sloan-Kettering Cancer Center; GBM20/3, GLI36vIII (overexpressing human EGFRvIII), and GLI36R132H (overexpressing human IDH1 R132H mutant protein) were provided by Dr. Xandra Breakefield, Massachusetts General Hospital (MGH); LNZ308 was provided by Dr. Mikael Pittet, MGH; A172 was purchased from American Type Culture Collection. Mouse model cell lines overexpressing human wild-type EGFR and human EGFRvIII (T103: WT/vIII, T042: vIII only) were generated as previously reported, and provided by Dr. Alain Charest (Tufts University). These cell lines were cultured on gelatin and in DMEM containing 20% FBS, supplemented with penicillin-streptomycin. Normal human brain microvascular endothelial cells (HBMVECs, Cell Systems) were cultured in endothelial basal medium supplemented with SingleQuots (Lonza). Normal human astrocytes (NHA, Lonza) were cultured in astrocyte basal medium supplemented with SingleQuots, as recommended by the manufacturer.
Microvesicle (MV) isolation from cell culture and quantification
Cells at passage 1–15 were cultured in MV-free medium (DMEM containing 5% depleted FBS). After 48 hours, conditioned medium from ~ 107 cells was collected and filtered through a 0.2 μm filter (Millipore), and MVs were purified by differential centrifugation as described previously5,12. MV numbers were determined by Nanosight LM10 nanoparticle characterization system (Nanosight). All nanoparticle tracking analyses (NTA) were done with identical experiment settings. For optimal results, MV concentrations were adjusted to obtain ~ 50 MVs in the field of view.
MV isolation from clinical samples
Blood samples were collected from healthy donors and GBM patients under protocols approved by the MGH Institutional Review Board. Blood was collected in vacutainer tubes (Becton Dickinson). Buffy coat was obtained via centrifugation and remaining plasma was then sterile-filtered through a 0.8 μm filter (Millipore). All clinical samples were frozen at —80 °C within two hours of collection. Thawed samples were processed for MV isolation using the same method described above. Clinical samples were processed unidentified for μNMR detection.
MV labeling and detection
Isolated MVs were resuspended in PBS and labeled with antibodies (10 μg/mL) for 45 minutes at 4 °C. The samples were then washed and membrane-filtered (Nuclepore, Whatman) to concentrate the MVs and to remove unbound antibodies. For ELISA measurements, fluorescein (FITC)-conjugated antibodies were used for targeting and scanning with fluorescence reader (Safire, Tecan). For μNMR detection with the microfluidic chip, MVs were mixed with TCO-modified antibodies. The antibody-targeted MVs and CLIO-TZ were then mixed within the microfluidic device and processed for NMR measurements (see details in Supplementary Fig. 2). For detection of intravesicular proteins, MV lysates were immuno-captured by antibody-conjugated beads (500 nm in diameter), incubated with TCO-modified detection antibody and processed for NMR measurements after coupling with CLIO-TZ in the microfluidic device. All experiments were performed with TCO-modified isotype control antibodies to determine nonspecific background binding17. MV biomarker expression profiles were normalized by their CD63 expression to account for variations in MV numbers.
μNMR measurement
We performed NMR measurements using a previously described miniaturized NMR relaxometer14. The operating magnetic field, generated by a portable permanent magnet, was 0.5 T. The _R_2 relaxation was measured on 1 μL sample volumes using Carr-Purcell-Meiboom-Gill pulse sequences with the following parameters: echo time, 4 ms; repetition time, 1 s; the number of 180° pulses per scan, 50; the number of scans, 8. All measurements were done in triplicate and data is displayed as mean ± standard error of mean.
Clinical samples
The study was approved by the MGH Institutional Review Board, and informed consent was obtained from all participants. For the profiling study, we obtained clinical blood samples prior to tumor excision from patients, assumed to harbor newly diagnosed or recurrent glioblastoma. The diagnosis was subsequently confirmed by neuropathologic examination. We subdivided glioblastoma into glioblastoma (GBM) and glioblastoma with oligodendroglial component (GBMO) according to the WHO classification of tumors of the nervous system. EGFR amplification was determined on pathologic tissues using standardized approaches. EGFRvIII mutations were evaluated on tumor tissues using techniques previously described5. Peripheral white blood cell (WBC) count was determined at the time of initial operation. MRI was performed prior to operation and evaluated for the presence of gadolinium enhancement (MRI+/−) and the maximum cross-sectional size of the tumor (area in cm2). For the longitudinal treatment response evaluation, blood samples were collected from the same patients before and after treatment. Responder and non-responder status was defined by a neuro-oncologist based on subsequent clinical and MRI data.
Supplementary Material
01
Acknowledgments
The authors thank Dr. T. Reiner for preparing TCO, Dr. N. Sergeyev for synthesizing MNPs, Dr. S. Hilderbrand for synthesizing reactive TZ, Dr. M. Pittet for LNZ308 cells, Dr. T. Chan for SkMG3 cells, and Drs. M. Liong and A. Ghazani for assay assistance, B. Marinelli for μNMR measurements, C. Min for software implementation, M. McKee for transmission electron microscopy, Dr. J. Skog for advice on NTA measurements, L. Zhu and S. Sivaraman for technical assistance and Dr. Y. Fisher-Jeffes for critically reading the manuscript. Special thanks to Drs. Carter, Castro, Carlson and clinical colleagues for many helpful discussions. This work was supported in part by: U54CA151884, R01EB010011, R01EB004626, P01CA069246, P50CA86355, U01CA141556, U24CA092782, R21CA14122; H. Shao received a B.S.-Ph.D. National Science Scholarship awarded by the Singapore Agency for Science, Technology and Research; A. Charest received an American Cancer Society Research Scholar Award 117409.
Footnotes
Author Contributions H.S., R.W. and H.L. designed the study. H.S., J.C., L.B. and H.L. performed the experiments. H.S., J.C., R.W. and H.L. analyzed the data and wrote the manuscript. A.C. generated the mouse T103 model. D.D.B. recommended GBM biomarkers and generated the EGFRvIII antibody. F.H.H. and X.O.B. coordinated the clinical study and analyzed the results.
References
- 1.Maheswaran S, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med. 2008;359:366–377. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kulasingam V, Pavlou MP, Diamandis EP. Integrating high-throughput technologies in the quest for effective biomarkers for ovarian cancer. Nat Rev Cancer. 2010;10:371–378. doi: 10.1038/nrc2831. [DOI] [PubMed] [Google Scholar]
- 3.Simpson RJ, Lim JW, Moritz RL, Mathivanan S. Exosomes: proteomic insights and diagnostic potential. Expert Rev Proteomics. 2009;6:267–283. doi: 10.1586/epr.09.17. [DOI] [PubMed] [Google Scholar]
- 4.Thery C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009;9:581–593. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- 5.Skog J, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Al-Nedawi K, et al. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat Cell Biol. 2008;10:619–624. doi: 10.1038/ncb1725. [DOI] [PubMed] [Google Scholar]
- 7.Simons M, Raposo G. Exosomes--vesicular carriers for intercellular communication. Curr Opin Cell Biol. 2009;21:575–581. doi: 10.1016/j.ceb.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 8.Lee TH, et al. Microvesicles as mediators of intercellular communication in cancer--the emerging science of cellular `debris'. Semin Immunopathol. 2011;33:455–467. doi: 10.1007/s00281-011-0250-3. [DOI] [PubMed] [Google Scholar]
- 9.Cocucci E, Racchetti G, Meldolesi J. Shedding microvesicles: artefacts no more. Trends Cell Biol. 2009;19:43–51. doi: 10.1016/j.tcb.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 10.Graner MW, et al. Proteomic and immunologic analyses of brain tumor exosomes. FASEB J. 2009;23:1541–1557. doi: 10.1096/fj.08-122184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanderson MP, et al. Generation of novel, secreted epidermal growth factor receptor (EGFR/ErbB1) isoforms via metalloprotease-dependent ectodomain shedding and exosome secretion. J Cell Biochem. 2008;103:1783–1797. doi: 10.1002/jcb.21569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balaj L, et al. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat Commun. 2011;2:180. doi: 10.1038/ncomms1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee H, Sun E, Ham D, Weissleder R. Chip-NMR biosensor for detection and molecular analysis of cells. Nat Med. 2008;14:869–874. doi: 10.1038/nm.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Issadore D, et al. Miniature magnetic resonance system for point-of-care diagnostics. Lab Chip. 2011;11:2282–2287. doi: 10.1039/c1lc20177h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haun JB, et al. Micro-NMR for rapid molecular analysis of human tumor samples. Sci Transl Med. 2011;3:71ra16. doi: 10.1126/scitranslmed.3002048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2:569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 17.Haun JB, Devaraj NK, Hilderbrand SA, Lee H, Weissleder R. Bioorthogonal chemistry amplifies nanoparticle binding and enhances the sensitivity of cell detection. Nat Nanotechnol. 2010;5:660–665. doi: 10.1038/nnano.2010.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thery C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006;Chapter 3(Unit 3.22) doi: 10.1002/0471143030.cb0322s30. [DOI] [PubMed] [Google Scholar]
- 19.Fleming TP, et al. Amplification and/or overexpression of platelet-derived growth factor receptors and epidermal growth factor receptor in human glial tumors. Cancer Res. 1992;52:4550–4553. [PubMed] [Google Scholar]
- 20.Mishima K, et al. Increased expression of podoplanin in malignant astrocytic tumors as a novel molecular marker of malignant progression. Acta Neuropathol. 2006;111:483–488. doi: 10.1007/s00401-006-0063-y. [DOI] [PubMed] [Google Scholar]
- 21.Wykosky J, Gibo DM, Stanton C, Debinski W. EphA2 as a novel molecular marker and target in glioblastoma multiforme. Mol Cancer Res. 2005;3:541–551. doi: 10.1158/1541-7786.MCR-05-0056. [DOI] [PubMed] [Google Scholar]
- 22.Parsons DW, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dang L, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Italiano JEJ, Mairuhu AT, Flaumenhaft R. Clinical relevance of microparticles from platelets and megakaryocytes. Curr Opin Hematol. 2010;17:578–584. doi: 10.1097/MOH.0b013e32833e77ee. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pedersen NM, et al. Expression of epidermal growth factor receptor or ErbB3 facilitates geldanamycin-induced down-regulation of ErbB2. Mol Cancer Res. 2009;7:275–284. doi: 10.1158/1541-7786.MCR-07-2183. [DOI] [PubMed] [Google Scholar]
- 26.Trepel J, Mollapour M, Giaccone G, Neckers L. Targeting the dynamic HSP90 complex in cancer. Nat Rev Cancer. 2010;10:537–549. doi: 10.1038/nrc2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu H, et al. The novel Hsp90 inhibitor NXD30001 induces tumor regression in a genetically engineered mouse model of glioblastoma multiforme. Mol Cancer Ther. 2010;9:2618–2626. doi: 10.1158/1535-7163.MCT-10-0248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu H, et al. Oncogenic EGFR signaling cooperates with loss of tumor suppressor gene functions in gliomagenesis. Proc Natl Acad Sci U S A. 2009;106:2712–2716. doi: 10.1073/pnas.0813314106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Verhaak RG, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pedersen MW, Meltorn M, Damstrup L, Poulsen HS. The type III epidermal growth factor receptor mutation. Biological significance and potential target for anti-cancer therapy. Ann Oncol. 2001;12:745–760. doi: 10.1023/a:1011177318162. [DOI] [PubMed] [Google Scholar]
- 32.Mellinghoff IK, et al. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N Engl J Med. 2005;353:2012–2024. doi: 10.1056/NEJMoa051918. [DOI] [PubMed] [Google Scholar]
- 33.Galban CJ, et al. The parametric response map is an imaging biomarker for early cancer treatment outcome. Nat Med. 2009;15:572–576. doi: 10.1038/nm.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Galban CJ, et al. Prospective analysis of parametric MRI biomarkers: Identification of early and distinct glioma response patterns not predicted by standard radiographic assessment. Clin Cancer Res. 2011 doi: 10.1158/1078-0432.CCR-10-2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsien C, et al. Parametric response map as an imaging biomarker to distinguish progression from pseudoprogression in high-grade glioma. J Clin Oncol. 2010;28:2293–2299. doi: 10.1200/JCO.2009.25.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Elkhaled A, et al. Magnetic Resonance of 2-Hydroxyglutarate in IDH1-Mutated Low-Grade Gliomas. Sci Transl Med. 2012;4:116ra5. doi: 10.1126/scitranslmed.3002796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wei LH, et al. Changes in tumor metabolism as readout for Mammalian target of rapamycin kinase inhibition by rapamycin in glioblastoma. Clin Cancer Res. 2008;14:3416–3426. doi: 10.1158/1078-0432.CCR-07-1824. [DOI] [PubMed] [Google Scholar]
- 38.Yoon TJ, Lee H, Shao H, Weissleder R. Highly magnetic core-shell nanoparticles with a unique magnetization mechanism. Angew Chem Int Ed Engl. 2011;50:4663–4666. doi: 10.1002/anie.201100101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01