Application of proteomics to cerebrovascular disease (original) (raw)
. Author manuscript; available in PMC: 2013 Dec 1.
Published in final edited form as: Electrophoresis. 2012 Dec;33(24):10.1002/elps.201200481. doi: 10.1002/elps.201200481
Abstract
While neurovascular diseases such as ischemic and hemorrhagic stroke are the leading causes of disability in the world, the repertoire of therapeutic interventions has remained remarkably limited. There is a dire need to develop new diagnostic, prognostic, and therapeutic options. The study of proteomics is particularly enticing for cerebrovascular diseases such as stroke, which most likely involve multiple gene interactions resulting in a wide range of clinical phenotypes. Currently, rapidly progressing neuroproteomic techniques have been employed in clinical and translational research to help identify biologically relevant pathways, to understand cerebrovascular pathophysiology, and to develop novel therapeutics and diagnostics. Future integration of proteomic with genomic, transcriptomic, and metabolomic studies will add new perspectives to better understand the complexities of neurovascular injury. Here, we review cerebrovascular proteomics research in both preclinical (animal, cell culture) and clinical (blood, urine, cerebrospinal fluid, microdialyates, tissue) studies. We will also discuss the rewards, challenges, and future directions for the application of proteomics technology to the study of various disease phenotypes. To capture the dynamic range of cerebrovascular injury and repair with a translational targeted and discovery approach, we emphasize the importance of complementing innovative proteomic technology with existing molecular biology models in preclinical studies, and the need to advance pharmacoproteomics to directly probe clinical physiology and gauge therapeutic efficacy at the bedside.
Keywords: Biomarker, Cerebrovascular disease, Cerebrospinal fluid (CSF), Mass spectrometry, Pharmaco-proteomics, Stroke
1 Introduction
Cerebrovascular disorders, such as stroke, vascular cognitive impairment, and even certain migraine subtypes, are leading causes of disability in the world. Human neurovascular disease has a wide range of phenotypes involving multiple cell types of the brain, the blood brain barrier (BBB), and interaction with the peripheral systemic circulation. Unlike cardiovascular disease, which largely involves in situ vascular thrombosis, neurovascular disease can induce focal injury with thrombotic (involving larger and small vessels), embolic (from cardiac source or periphery), hemorrhagic (intracerebral and subarachnoid), inflammatory, and traumatic mechanisms, to name only a few. Or it can result in global ischemic injury from lack of perfusion, such as from sudden cardiac arrest. While the major public health impact and the heterogeneity of neurovascular disease phenotypes prompted a multitude of preclinical and clinical investigations, the repertoire of therapeutic interventions for these devastating conditions has remained remarkably limited, and so are tools for triaging their use. For example, currently, tissue plasminogen activator (tPA), a therapeutic protease and a clot-dissolving agent, is the only FDA-approved medical treatment for acute ischemic stroke, improving clinical outcome by more than 30% at 3 months poststroke [1]. While the use of tPA is a significant step in the right direction, now more than a decade after its approval in 1996, even with evidence of benefit beyond the initial approved time-window, it is still underutilized in ischemic stroke patients at later time windows, due to fear of a tenfold increased risk of intracranial hemorrhage (ICH) as a major side effect [1–7]. So one important goal in acute stroke is to develop diagnostic and prognostic tools for better patient selection for existing high-benefit treatments with significant risks, and to develop new targets of therapy [8, 9].
Multiple studies of bedside biomarkers of known individual factors measurable by immunoassays such as ELISA have been done on this subject [10]. However, it remains challenging to gauge disease activity or overcome individual-to-individual variation and S/N ratio with single or even a panel of markers. Proteomics holds great promise to advance the field, as it has in many other specialties already [11–13]. Advances in sequencing technologies resulted in the discovery of multiple gene–gene interactions that ultimately affect protein expression. Thus, the understanding of the spectrum of proteins present in a cell or an organism after extensive transcriptional and posttranslational processing is one step closer to the ultimate clinical phenotype. While many advances have been made in terms of using differential genomic expression patterns of white blood cells—a very promising complimentary tool to clinical stroke phenotyping, it should be noted that genomic expression and protein levels do not always match in stroke [14–18]. In particular, studies have shown that on the cellular level, genetic expression may not be an accurate reflection of the protein expression during focal ischemia, which is what ultimately affects functional outcome [14]. Thus, studying proteomics is particularly enticing for neurovascular diseases such as stroke, which most likely involve multiple gene interactions resulting in a wide range of clinical phenotypes. Innovative proteomic approaches have helped to identify networks of both well-characterized and novel factors, not only improving our understanding of cerebrovascular pathologies, but also having applicability to clinical stroke care. Here, we review cerebrovascular proteomics research in both preclinical and clinical studies.
2 Application of proteomic technology to neurovascular diseases
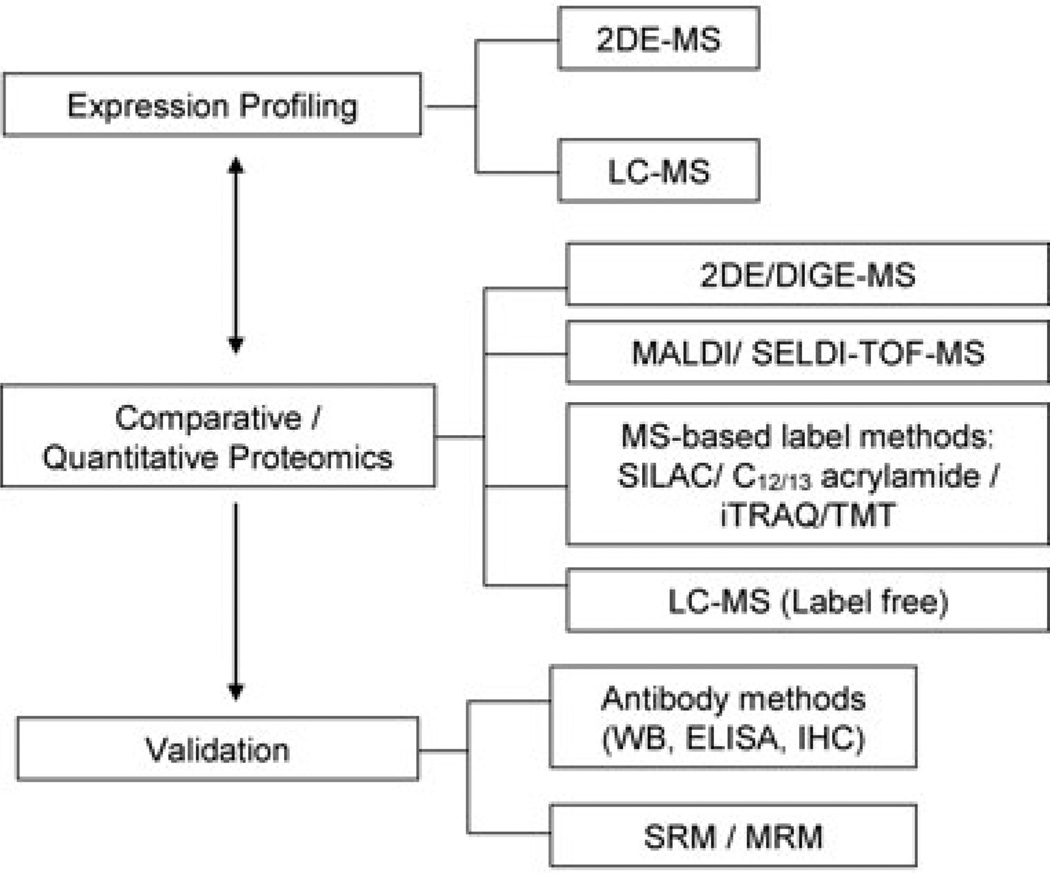
Proteomic techniques used to study stroke are broadly summarized in Fig. 1. Most initial studies were analyzed by 2D gel, LC-MS, and more recently by quantitative isotope labeled methodology and selected reaction monitoring (SRM). We searched PubMed for all literature related to cerebrovascular disease and proteomics and outlined and summarize specific studies in order of sample type, year of publication, methods, and brief highlight of results (Tables 1 and 2). MeSH terms used include “cerebrovascular disease,” “neurovascular disease,” “stroke,” and major stroke subtypes such as “ischemia”, “ischemic stroke,” “hemorrhagic stroke” in conjunction with “proteomics” or “proteome”. We understand that this is a limited portion of the existing published literature and body of knowledge, but we made every effort to include an extensive range of both preclinical (Table 1) and clinical studies (Table 2).
Figure 1.
Proteomic methodologies used to study cerebrovascular disease in publications reviewed. Methods were grouped into three categories: (i) Expression Profiling—discovery approach using shotgun MS analysis with in-gel or LC separation; (ii) Comparative/Quantitative Proteomics—semiquantitative isotope labeled and label-free technique for tissue and cell media; (iii) Validation—targeted quantitative approaches measuring specific protein level using antibody based and nonantibody-based methods. DIGE, 2D difference gel electrophoresis; SELDI-TOF, surface-enhanced laser desorption/ionization time-of-flight; SILAC, stable isotope labeling with amino acids in cell culture; C12/13 acrylamide; iTRAQ, isobaric tags for relative and absolute quantification; TMT, tandem mass tags; WB, Western blot; IHC, immunohistochemistry; SRM, selected reaction monitoring; MRM, multiple reaction monitoring.
Table 1.
Proteomics studies of neurovascular injury using animal model and cell line
| Ref./year | Authors | Samples | Methods | Findings |
|---|---|---|---|---|
| Animal model (Section 2.1) | ||||
| Hypertensive stroke-prone rat model | ||||
| [19]/2001 | Sironi et al. | Serum and urine of three hypertensive rat strains Wistar-Kyoto, spontaneously hypertensive rats, spontaneously hypertensive stroke-prone rats (SHRSP) | 2DE-MS | Urinary (transferring, hemopexin, albumin, a2-HS-glycoprotein, kallikrein-binding protein, a1-antitrypsin, Gc-globulin, transthyretin) and serum (thiostatin) protein are changed 4 wk before clinically evident stroke |
| [20]/2004 | Sironi et al. | CSF/brain tissue of SHRSP | 2DE-MS | Found larger plasma-derived protein in CSF and outlined sequence of microvascular inflammatory events leading to stroke |
| [21]/2011 | Bergerat et al. | Cerebral microvessels in stroke-prone, transgenic[hCETP]-hyperlipidemic, Dahl salt-sensitive hypertensive rats | MS/MS | Aquaporin-4 and laminin- α 1 are associated with stroke susceptibility |
| Middle cerebral artery occlusion (MCAO) model | ||||
| [22]/2004 | Dhodda et al. | Cortex of rat preconditioned with 10-min transient MCAO | 2DE-MS | Expression of HSP70, HSP27, HSP90, guanylyl cyclase, muskelin, platelet-activating factor receptor and β -actin increased at 24 h after preconditioning |
| [23]/2004 | Suzuyama et al. | CSF of rats with MCAO | SELDI-TOF-MS and 2DE-MS | Monomeric transthyretin as ischemia-specific CSF marker |
| [24]/2006 | Focking et al. | Brain extracts from MCAO mice (1 h post-MCAO thread occlusion) | Multi-Western blot | Hypoxia-inducible factor-1, phosphotyrosine, cyclin, STAT3 upregulated |
| [25]/2007 | Chen et al. | Cortex of rat 24 h post-MCAO | 2DE-MS | DRP-2, spectrin and Tmod2 upregulated compared to sham |
| [33]/2009 | Wu et al. | Combination-rat transient focal ischemia model (30 min); siRNA, PPAR-gamma P465L dominant-negative mutant | 2DE-MS | Ligand-activated PPAR-gamma exerts neuroprotective and antiapoptotic effect via 14-3-3 epsilon upregulation |
| [26]/2009 | Xiong et al. | Pituitary, adrenal gland, and splenic lymphocytes of rats with MCAO | 2DE-MS | 13 proteins, including STAR, closely related to the immune and/or the neuroendorine system changed |
| [27]/2009 | Mizutani et al. | Cerebellum of rats with and without treadmill training (2.5 h post-MCAO) | 2DE-MS | 25-kDa SAP, GFAP, enhanced in the cerebellum with treadmill training |
| [28]/2009 | Yao et al. | Brain of rats with/without reperfusion in core versus penumbra at 6, 12, 24 h postphotothrombotic MCAO) | SELDI-TOF-MS | 36 peaks downregulated, seven upregulated, dynamic changes of ischemic core and penumbra |
| [29]/2010 | Sung et al. | Cerebral cortex adult female rats treated with oil or estradiol (24 h post-MCAO) | 2DE-MS | PP2A, astrocytic phosphoprotein PEA-15 decreased, estradiol prevented injury-induced increase of HSP60 |
| [30]/2011 | Koh et al. | Cerebral cortex of EGb761 or vehicle treated rats (24 h post-MCAO) | 2DE-MS | EGb 761 protect neuronal cells through up- or downregulation of proteins such as Peroxiredoxin-2 and protein phosphatase 2A subunit B, etc. |
| [31]/2011 | Sung et al. | Cerebral cortex of rats with nicotinamide treatment (24 h post-MCAO) | 2DE-MS | Enolase, PP2A subunit B, Prx-2 decreased in control, nicotinamide prevented injury-induced HSP60 inc |
| [32]/2012 | Domenico et al. | Brain tissue of MCAO inWT and STAT3 KO mice | 2DE-MS | STAT3 as a mediator of neuronal function by controlling metabolic, synaptic, structural and transcriptional pathways |
| Animal cells or cell lines (Section 2.2) | ||||
| [40]/2009 | Dowell et al. | Secretome of primary murine astrocyte cultures | 1D/2D-LC-MS | 420 proteins including 187 secreted proteins identified in normal |
| [41]/2010 | Greco et al. | SILAC, multi-LC-MS | 516 proteins including 92 differential expressed in normal secretome | |
| [47]/2008 | Lu et al. | Microvascular endothelium cells of mouse brain | SDS-PAGE-LC-MS | 881 proteins found in laser capture microdissection (LCM) identified endothelial cells |
| [49]/2009 | Datta et al. | Rat B104 neuroblastoma cell line | LC-MS (iTRAQ) | Proteins involved in the chronic neurological disorders also round in acute neuronal injury |
| [51]/2011 | Zhou et al. | Murine neuronal culture | LC-MS “click- chemistry” | Nascent proteome characterized using “click-chemistry” in ischemic (OGD) preconditioned, ischemic-injured and ischemic-tolerant conditions |
Table 2.
Proteomics studies of neurovascular injury using human samples
| Ref./year | Authors | Samples | Methods | Results |
|---|---|---|---|---|
| Human cell/cell line (Section 2.3.1) | ||||
| [53]/2009 | Minagar et al. | Cerebral endothelial cells stimulated by glutamate, with and without NMDA receptor antagonist MK-801 | 2DE-MS and LC-MS | Cerebral endothelial cells respond to glutamate (e.g., glutamate dehydrogenase, protein 14-3-3 epsilon, etc.) and response blocked by glutamate antagonist |
| [54] | Ning et al. | Conditioned media from human brain endothelial treated with sodium nitroprusside for oxidative stress and plasma from human ischemic stroke | SDS-PAGE and LC-MS | 277 proteins secreted by brain endothelial cells in response to oxidative stress show coordinated temporal profile – also serve as screening tool for biomarkers discovery. Candidate such as TSP-1 elevated in stroke patients (<8hr). |
| Human brain tissue (Section 2.3.2) | ||||
| [68]/2009 | Cuadrado et al. | Postmortem brain tissue/cell from infarct core and the contralateral hemisphere | Protein array/laser capture microdissection | MMP-1, MMP-2, MMP-3, MMP-8, MMP-9, MMP-10, MMP-13, and TIMP-1 upregulated in the infracted tissue compared to healthy control areas and it is cell specific |
| [69] | Cuadrado et al. | Postmortem human brain tissue after ischemic stroke | DIGE-MS | Identified 39 differential expressed proteins related to cerebral ischemia |
| Body fluids (Section 2.3.3) | ||||
| Urine | ||||
| [77]/2012 | Dawson et al. | Urine of patients with stoke and controls | CE-TOF-MS | Build a 35 biomarkers-classifier model for diagnosis of stroke |
| CSF | ||||
| [83]/2003 | Zimmermann-Ivol et al. | Postmortem CSF (as model of “massive brain injury” versus healthy control CSF | 2DE-MS | H-FABP as serum biomarker of stroke |
| [84]/2004 | Lescuyer et al. | Postmortem CSF (as model of “massive brain injury”) versus healthy control CSF | 2DE-MS | 13 differentially expressed proteins as potential candidate for stroke |
| [85]/2008 | Dayon et al. | Postmortern versus antemortem CSF | TMT, LC-MS | 78 proteins increased in postmortem CSF samples versus control |
| Microdialysate | ||||
| [86]/2007 | Maurer et al. | Microdialysates of fronto- temporal brain tissue in vasospastic/nonvasospastic aneurysmal subarachnoid hemorrhage (sSAH) patients | 2DE-MS | GAPDH and HSP7C as early markers of vasospasm after aneurismal subarachnoid hemorrhage |
| [87]/2011 | Dayon et al. | Microdialysates from the infarct core (IC), penumbra (P), unaffected contralateral (CT) brain of ischemic stroke patients | iTRAQ, LC-MS | 53 proteins increased in microdialysate of IC or P with respect to the CT. Candidate GSTP1, PRDX1, S100-B were significantly increased in blood of stroke patients |
| Plasma/serum | ||||
| [90]/2004 | Allard et al. | Plasma of patients with ischemic or hemorrhagic stoke | SELDI and SDS- PAGE-LC-MS | ApoC-I and ApoC-III as plasma markers To distinguish ischemic and hemorrhagic stroke |
| [91]/2008 | Zhang et al. | Plasma of patients with acute cerebral infarction and control subjects | SELDI-TOF-MS | Panel of 13 markers for diagnosis of stroke |
| [92]/2009 | Huang et al. | Serum of ischemic stroke patients and nonstroke patients | MALDI-TOF | Hemoglobin as a potential biomarker for the diagnosis of ischemic stroke |
| [123]2010 | Ning et al. | Plasma of ischemic stroke patients, control, and post-tPA treatment (tx) | LC-MS (degradomics) | Degradomics and targeted MMP array increase in multiple MMPs and distinct degradeomic pattern emerge post tPA tx, lasting 3–5 days posttreatment |
| [93]/2011 | Pan et al. | Serum of stroke patients treated with and without electroacupuncture (EA) | 2DE-MS | SerpinG1 downregulated and gelsolin, complement component I, C3, C4B, β2-glycoprotein I proteins upregulated post EA |
| [94]/2012 | Lopez et al. | Plasma of patients with acute ischemic and hemorrhagic stokes | SRM | Ischemic versus hemorrhagic stroke versus normal differentiated by a panel combination of apolipoproteins - ApoA-1, A-II, B, C-1, C-II, C-III, D, E, H |
| [127]2012 | Lopez et al. | Plasma of patients with and without ischemic PFO related stroke; pre- and post-PFO endovascular repair | LC-MS (two pass) | Post-PFO repair acute inflammatory response and coagulation signaling are decreased |
2.1 Animal models
Animal models including those from cerebral tissue, cell lines, cerebrospinal fluid (CSF), and plasma will be discussed in Section 2.1.1. Various animal models of stroke have been studied using proteomic technology. Since early proteomic instrumentation required large amounts of samples with high protein concentration, in vivo brain tissue of several animal models have been studied extensively to explore mechanisms of cerebrovascular injury and repair.
2.1.1 Hypertensive rat strains
Hypertensive rat strains, models for small and large vessel ischemic stroke, have been used to investigate acute phase protein before imaging evident cerebral ischemia [19–21]. Sironi et al. used classical proteomic methods (2DE-MS) to study the effect of salt loading on the detailed protein pattern of serum and urine in three different hypertensive rat strains: Wistar-Kyoto, spontaneously hypertensive rats, and spontaneously hypertensive stroke-prone rats [19]. In their study, protein patterns in serum and urine were assessed over time by 2DE-MS analysis with magnetic resonance imaging and histology as outcome. They showed that prior to clinically apparent stroke, several proteins were excreted in urine after several weeks of salt loading, including transferrin, hemopexin, albumin, a2-HS-glycoprotein, kallikrein-binding protein, a1-antitrypsin, Gc-globulin, and transthyretin. Markers of an inflammatory response, including very high levels of thiostatin, were detected in the serum of spontaneously hypertensive stroke-prone rats at least 4 wk before a stroke occurred. In a follow-up study 3 years later, they used 2DE-MS to analyze protein content in CSF as imaging evidence of BBB impairment progressed (gadolinium diffusion magnetic resonance imaging) after 42 ± 3 days from the start of salt loading. Interestingly, high levels of relatively large (>130 kDa) plasma-derived proteins were found in the CSF, pointing toward early effects of systemic inflammation on the brain and BBB function. Together with previous data from serum and urine, these proteomic studies not only provide data for early changes in biological fluids of cerebrovascular injury, but also demonstrate the importance of central-peripheral interaction as the BBB becomes more permeable during early stages of injury [20]. Understanding the sequence of inflammatory events prior to clinically apparent stroke is important in finding new targets for stroke prevention.
2.1.2 Middle cerebral artery occlusion models
Middle cerebral artery occlusion (MCAO) in rat or mouse is a well-characterized model of focal cerebral ischemia and have been the most extensively studied using proteomic techniques [22–32]. The MCAO model has been used to study the effects of ischemia itself and the effects of various neuroprotective processes such as preconditioning in focal ischemia. While earlier investigations used surface-enhanced laser desorption/ionization (SELDI) and antibody-based Western methodology, most studies employ traditional gel-based 2DE-MS as discussed below (Table 1).
2.1.2.1 Global analysis of ischemic injury
Global analyses of the effects of focal ischemia were conducted using a targeted antibody approach, SELDI, and 2DE-MS for discovery of unknown proteins at various ischemic time windows. Focking et al. performed multi-Western blots on more than 400 proteins in a transient focal ischemia model 1 h post-MCAO, and found that hypoxia-inducible factor-1, phosphotyrosine, cyclin, and STAT3 were detected to be upregulated during early ischemic brain damage [24]. They showed that compared to previous screenings of gene expression, a proteomic approach can help to identify subtle changes in acute transient ischemic injury. Utilizing 2DE-MS, Chen et al. studied a permanent rat MCAO model (post-24 h) and found more than 1500 protein spots with 12 proteins significantly upregulated 3- to 46-fold compared to sham. In addition to finding various phosphorylated states in these proteins, they found robust increases in several proteins, including dihydropyrimidinase-related protein 2, spectrin α II chain, heat-shock cognate protein 70 pseudogene 1, and tropomodulin 2, some of which are related to acute axonal and neuronal injury and repair [25]. Besides changes in the ischemic core studied above, more subtle changes in the penumbra can be detected by proteomic methodology as well. For example, Yao et al. studied reperfusion injury in ischemic core versus penumbra using SELDI-TOF-MS in a photothrombotic MCAO model. At 6, 12, and 24 h after photothrombotic MCAO with or without reperfusion, dynamic changes of protein profiles were detected in both ischemic core and penumbra, although changes postreperfusion were observed mostly in the penumbra [28]. These studies probe the complex pathophysiology of cerebrovascular disease with respect to the mechanism, severity, and location of injury. For comprehensive proteomic studies, animal models can provide standardized platforms for developing methods and testing novel neuroprotective strategies such as those discussed in Section 2.1.2.2.
2.1.2.2 Evaluation of neuroprotective strategies
The effects of various neuroprotective strategies, such as preconditioning and the use of potential neuroprotective compounds (i.e., estradiol, ginko, etc.), have been investigated by 2DE-MS in the MCAO model. Dhodda et al. combined both genomic (GeneChip) and proteomic 2DE-MS methods to study preconditioning (10-min transient MCAO) at 3–72 h after treatment. In addition to finding an early increase in HSP70 at 24 h post-preconditioning, they found a temporal relationship between altered gene expression and brain damage after focal ischemia, suggesting that preconditioning-induced neuroprotection against focal ischemia may act at the level of gene expression [22].
An innovative study evaluated the neuroprotective effect of rosiglitazone by combining various molecular biology techniques (siRNA and transgenic knockin PPAR-gamma dominant-negative mutant) and proteomic technology to probe underlying mechanisms [33]. Through proteomic screening, they found that 14-3-3 epsilon was highly up-regulated in rats treated with rosiglitazone, and identified the antiapoptotic and neuroprotective effect of the PPAR-gamma pathway to be related to 14-3-3 epsilon transcription. This type of hypothesis-driven approach, complemented by discovery proteomic screening and rigorous molecular biological validation, is a promising model for future studies.
To study the effect of estradiol, Sung et al. performed protein analysis on the cerebral cortex of female rats 24 h post-MCAOusing 2DE-MS, and found changes in levels of various proteins such as phosphatase 2A and astrocytic phosphoprotein PEA-15, and that estradiol prevented the injury-induced increase of HSP60 [29]. Using a similar approach a year later, they also tested the neuroprotective effect of nicotinamide treatment and found it to also ameliorate ischemic-induced increase of HSP60 [31]. Coordinated changes in proteomic profiles were reported in testing other potential neuroprotectants such as ginkgo biloba extract (EGb761) on a similar MCAO model post-24 h [30]. Other animal models, such as neuron-specific STAT3 knockout transgenic mice, were used to investigate the role of various pathways in gender-specific ischemia, and downstream mediators of neuronal function were found in metabolic, synaptic, structural, and transcriptional pathways [32]. In addition to cortical brain tissue, ischemia has been examined in other brain locations. Mizutani et al. compared protein expression of cerebellum tissue after 2.5 hrs of transient MCAO between rats with and without treadmill training, and found that 25-kDa synaptosomal-associated protein and glial fibrillary acidic protein were significantly enhanced in the cerebellum of rats with treadmill training [27]. Ultimately, proteomic analyses of neuroprotection may reveal common mechanisms of brain tissue salvage regardless of the specific pathways being targeted.
2.1.2.3 CSF and other tissue
CSF has also been studied after ischemic insult in MCAO models. Suzuyama et al. studied the temporal changes in the CSF proteome of a rat transient MCAO model using SELDI-TOF MS and 2DE-MALDI-MS [23]. Their results showed that monomeric transthyretinmay represent an ischemia-specific CSF marker in the acute stage of ischemia.
Protein profiles of other organ tissue, such as rat pituitary, adrenal gland, and splenic lymphocyte, have been investigated using proteomic techniques, to elucidate potential changes in the immune neuroendocrine system following cerebral ischemia injury based on MCAO rat model. Xiong et al. identified 13 proteins that were closely related to the immune and/or the neuroendocrine system using 2DE-MS proteomic technique, demonstrating immune neuroendocrine system changes postcerebral ischemia [26].
For animal studies, due to the amount of injured tissue required, most early studies have used permanent MCAO rat models. The MCAO model has been very useful in testing the effects of various neuroprotective strategies. Several studies discussed above detected more subtle changes at the protein expression level using transient ischemic models in various locations (core versus penumbra) and tissue types (cortex, cerebellum, CSF). The candidates found in predisease and injury states have clinical application for stroke prevention and treatment. While the majority of studies have been done in ischemic animal models, hemorrhagic models have also been studied recently. For example, Chiu et al. used an ICHmodel of stereotaxic infusion of collagenase/heparin into the right striatum and studied the effect of hyperglycemia in the perihematomal region—the most important area for hematoma expansion and clinical deterioration. In addition to identifying eight differentially expressed proteins and more significant apoptosis in the perihematomal region of hyperglycemic rats, they found a significant increase in albumin from nor-moglycemic rats compared to hyperglycemic rats—probing into the neuroprotective role of albumin in acute ICH and various glycemic states [34].
In summary, in both ischemic and hemorrhagic animal models, proteomic profiling has been shown to be a promising tool to be used in conjunction with other molecular biochemistry techniques to explore mechanistic pathways and discover targets of injury and repair. However, the complexity of cortical tissue has also encouraged studies in cell culture models, a simpler and more focused system for proteomic analysis, which will be discussed in Section 2.2.
2.2 Animal cells or cell lines
Primary central nervous system (CNS) cells or cell lines are valuable in vitro models to study stroke in the context of the neurovascular unit (NVU)—a concept that emphasizes the importance of dynamic interactions between endothelium, astrocytes, neurons, and the extracellular matrix [9, 35, 36]. For proteomic studies, cell cultures are simpler and more homogenous than in vivo tissue, and have been very useful for the study of protein–protein interaction [36]. Both cell and conditioned media can be studied in stimulus-response experiments to catalog proteins of interest and probe biologically relevant pathways. Section 2.2.1 will review studies done on various cell types with particular attention to components of the NVU.
2.2.1 Astrocytes
Astrocytes are important active participants in nervous system development, neurovascular metabolic coupling, and neurological disease progression [38, 39]. Astrocyte-secreted proteins in normal condition have been studied by Dowell et al. using shotgun proteomic approaches of 1D and 2D LC-MS/MS [40]. A total of 420 proteins from conditioned media of primary murine astrocyte cultures were identified and 187 were thought to be secreted proteins based on bioinformatic analysis by SignalP, SecretomeP, and gene ontology.To refine the secretome, Greco et al. utilized the stable isotope labeling with amino acids in cell culture quantitative method to find relative protein abundances between the primary murine astrocyte proteome and its secretome [41]. In their works, multi-LC-MS/MS analysis of astrocyte conditioned media and cell lysates resulted in the relative quantification of 516 proteins, 92 of which were greater than 1.5-fold enriched in astrocyte-conditioned media. These studies have contributed to the understanding of normal astrocytic secretory pathways and extracellular signals. Ultimately, combination and correlation with genomic databases of astrocyte gene expression may help us more fully understand how glial responses influence the pathophysiology of cerebral ischemia and injury [42].
2.2.2 Brain endothelium
Human brain endothelial cells, a major component of the BBB, serve as critical regulators of neuronal integrity with important trophic and signaling roles [43–46]. To catalog normal mouse brain microvascular endothelial cells (BMEC) in situ, Lu et al. used a laser capture microdissection method to isolate endothelial cells from frozen mouse brain tissue and SDS-PAGE-LC-MS to separate and identify specific proteins [47]. Using linear IT coupled with a Fourier transform MS, they were able to identify 881 BMEC proteins captured by immunoguided laser capture microdissection, laying the foundation for BMEC-specific proteome in normal condition and establishing a database for normal microvascular endothelial proteins. Eventually, many of these studies can potentially be connected with multiple transcriptome analyses of the BBB to help us understand how neurovascular homeostasis is altered after stroke [48].
2.2.3 Neuroblastoma cells and neurons
Neurons are the pivotal and most vulnerable component of the NVU in the CNS during ischemia. To characterize the molecular events of the dynamic penumbra, Datta et al. applied quantitative isobaric tags for relative and absolute quantification based shotgun proteomics in an in vitro neuronal model using B104 neuroblastoma cell line, to compare protein expressions in four in vitro hypoxia–ischemia models— cells subjected to normal growth media; low serum control; hypoxia; and oxygen-glucose deprivation [49]. In this model of acute neuronal injury, they found increased expression of proteins involved in ammoniagenesis, antiapoptotic, anti-inflammatory, and mitochondrial heat-shock response; and downregulation of proteins related to antioxidative defense and protein metabolism. They also identified differential expression of proteins involved in chronic neurological disorders such as Alzheimer’s disease, Parkinson’s disease, or bipolar disorder. In another study, Zhou et al. utilized innovative “click chemistry”—where an azide-containing amino acid is incorporated into newly synthesized proteins, enabling it to be purified by alkyne-conjugated probes—to quantitatively characterize newly synthesized proteins [50, 51]. They characterized a murine neuronal “nascent proteome”— newly synthesized proteins at the onset of ischemic insult in ischemic (oxygen–glucose deprivation treated) preconditioned, ischemic-injured, and ischemic-tolerant conditions [51]. Combining various functional/cytometric in vitro assays and quantitative proteomic screening, they advanced the search for novel targets of neuronal injury in hyperacute ischemia.
2.3 Human samples
Clinical proteomic study is very promising in providing important disease phenotypic snapshot in real time, but the challenges need to be met with innovative methodology developed for specific clinical questions [52]. While earlier stroke clinical studies have focused on biomarker discovery for diagnosis and prognosis, emerging research using innovative neuroproteomics methodology has the potential to directly probe physiology in real time at the bedside to help gauge therapeutic response [10]. Clinical proteomic studies utilizing cell, tissue, and body fluids (urine, CSF/microdialysate, plasma/serum) are listed in Table 2. In Section 2.3.1, we outline and summarize specific studies according to the sample—from cell to tissue, to bodily fluids.
2.3.1 Human cells/cell line
As a model for studying human diseases, cell culture is easily used to screen for effects on disease development utilizing a stimulus–response model. For example, to better understand neurotransmitter glutamate mediated early excitotoxic injury, combining both 2DE-MALDI-MS and LC-ESI-MS methods, Minagar et al. examined exposure of human cerebral endothelial cells to glutamate and found an important endothelial response blocked by _N_-methyl-d-aspartate receptor antagonist MK-801 [53].
Moreover, cell cultures also help to simplify and screen for better clinical targets and overcome challenges due to the complexity and heterogeneity of clinical samples [52, 54]. In screening conditioned media in an oxidative stress model of neurovascular injury in human brain endothelial cells, we found that while healthy endothelial cells displayed interaction and crosstalk involving secreted factors, membrane receptors and matrix components over time, oxidatively challenged endothelial cells not only have different protein expression, but lacked this important networking. After proteinase degradomic profiling and analysis of these secreted factors with respect to time, the high-ranking candidate thrombospondin-1, found in the endothelia in vitro screening, was tested directly in stroke patients and found to be elevated in plasma of acute ischemic stroke patients within 8 h of symptom onset compared to controls with similar clinical risk factors. This type of initial in vitro screening of cell culture systems may help to discover cell-specific novel biomarkers in clinical stroke [54].
2.3.2 Human sample—Tissue
Human stroke brain tissue is difficult to obtain, so studies have been done in postmortem samples. Targeted protein arrays have been utilized to study well-characterized proteins such as matrix metalloproeinases (MMPs), which are upregulated after cerebral ischemia and reperfusion, and are associated with brain tissue damage and hemorrhagic transformation. In animal models of cerebral ischemia, expression of several MMPs is significantly increased in both injury and repair phases of ischemia [55–67]. To assess the global expression of MMP family proteins directly in the human brain after stroke, Cuadrado et al. combined laser microdissection and targeted protein array methods to analyze six postmortem brains (3 men, 3 women), within four days of stroke (range 10–87 h) [68]. In comparison to control patients who died of noninflammatory and nonneurological disease, they found that MMP-1, MMP-2, MMP-3, MMP-8, MMP-9, MMP-10, MMP-13, and TIMP-1 to be upregulated in the infarcted tissue, and the distribution of specific MMPs to be cell-type (microvascular versus neuronal) and injury dependent. In an important follow-up study, the Montaner group also used discovery methods to analyze ischemic brain in six deceased stroke patients and three control subjects by 2D DIGE. They identified 39 proteins, validated 10 proteins by Western blot, and found the cellular location of 3 of these proteins (dihydropyrimidinase-related protein 2, vesicle-fusing ATPase, and Rho dissociation inhibitor 1) [69]. These studies advanced the understanding of human ischemic injury and validated candidate markers of cortical injury from animal models of ischemia.
2.3.3 Body fluids—Urine, CSF/microdialysate, and serum/plasma
To be useful for bedside diagnosis, prognosis, and monitoring of cerebrovascular disease, clinically accessible body fluids such as blood and urine are needed rather than brain tissue. Analysis of human plasma and body fluid proteomes has become a promising approach to discovery of biomarkers for human diseases [70–74]. Section 2.3.3.1 will focus on stroke-related studies in urine, CSF/microdialystes and serum/plasma—arranged in order of the protein content and complexity of fluids.
2.3.3.1 Urine
Compared to CSF and blood, urine has the advantage of being the most noninvasively available, abundant bodily fluid; and as a result of being a filtrate of serum, it is relatively simple in its composition [75,76]. However, urine is also most easily influenced and confounded by transient food, medication, and fluid intake. Dawson et al. explored the urinary proteome in 69 patient cases with acute stroke or transient ischemic attack without clear time of onset, against 33 controls using a CE-TOF-MS method. They built a 35-biomarker classifier model for the diagnosis of transient ischemic attack/stroke, with sensitivity of 56%, specificity 93%, and the AUC (area under the curve) on receiver operating characteristic of 0.86 [77]. This study maybe confounded by an imbalance in stroke risk factors that affect renal function and renally secreted medications in cases and controls (hypertension 53% versus 85%, p < 0.001; diabetes 6% versus 19%, p = 0.058; calcium channel blocker therapy 33.8% versus 54.8%, p = 0.045; alpha-blocker therapy 3.1% versus 8%; p = 0.013). Although the biomarker differences maybe attributed to difference in the cohort itself rather than stroke, this is an important first step in investigating the potential of urinary proteomic biomarkers for cerebrovasclar disease.
2.3.3.2 CSF and microdialysate
CSF, the fluid surrounding the brain, is commonly recognized as the sample of choice for biomarker discovery in neurodegenerative diseases, and it was one of the first CNS samples to be studied [78]. While less complex than cortex tissue or plasma, CSF remains a rich source of protein, with peptides representative of both sides of the BBB—this is especially true in the context of BBB damage during stroke. However, CSF sampling requires relatively invasive procedures, such as lumbar puncture, which are not part of routine clinical care for most strokes. Thus, only neurovascular disease subtypes (e.g., subarachnoid hemorrhage (SAH), traumatic brain injury) for which CSF can be clinically available have been more extensively studied. Recent proteomic advances in CSF for traumatic brain injury and SAH have been reviewed in detail by Wang et al., Lad et al., and Kobeissy et al. [79–82]. Since CSF is not part of routine clinical sampling for the majority of cerebrovascular disease subtypes such as ischemic stroke, initial investigation used a target approach in postmortem CSF—in particular using postmortem status as a model “massive brain insult.” Zimmermann-Ivol et al. studied heart fatty acid binding protein (H-FABP) as a diagnostic biomarker for stroke in comparison to neuron-specific enolase and S100B proteins using 2DE separation of CSF proteins and found that FABP was elevated in deceased patients [83]. Lescuyer et al. compared protein expression between postmortem CSF samples and healthy subjects by 2DE-MS and identified 13 differentially expressed proteins previously reported to be associated with brain destruction or neurodegenerative conditions—demonstrating that CSF is a rich reservoir for injured brain proteins [84]. Dayon et al. applied the six-plex isobaric tandem mass tagging quantitative proteomics approach to investigate human CSF samples and found 78 identified proteins increased in postmortem CSF samples compared to antemortem [85]. Some of these proteins, such as GFAP, protein S100B, and PARK7, have been previously described as brain damage biomarkers, supporting postmortem CSF as a model of brain insult utilizing quantitative MS-based methodology.
In contrast to postmortem CSF from the studies above, cerebral microdialysate has been studied during active disease states—in particular in patients with hemorrhage, since CSF sampling is sometimes part of clinical care [86, 87]. The study of cerebral microdialysate in acute brain injury has been reviewed in detail by Hillered et al. [88]. Maurer et al. conducted a proteome-wide screening using a 2DE-MS method in cerebral microdialysate post-SAH, and found that GAPDH and heat-shock cognate 71-kDa protein are two early markers predicting SAH-related symptomatic vasospasm to help stratify therapeutic intervention in these high-risk patients [86]. Dayon et al. investigated microdialysates from various infarct locations in ischemic stroke patients (n = 6) using a shotgun proteomic approach with quantitative isobaric tagging, and found 53 proteins increased in the ischemic core or penumbra in comparison to the contralateral unaffected side [87]. They tested the most significantly changed proteins from microdialysates in stroke patient blood, and found several proteins to be increased significantly after ischemic stroke (glutathione _S_-transferase P, 8×, p = 0.0002; peroxiredoxin-1, 20×, p = 0.0001; S100-B, 11×, p = 0.0093; n = 14). This study highlighted the value of cerebral microdialysate in screening for blood stroke markers.
2.3.3.3 Plasma/serum
Blood is the most clinically useful diagnostic fluid and therefore major effort has been devoted to the plasma proteome in search of biomarkers [70–74]. Due to the complexity and dynamic range of blood, it is also one of the most challenging [89]. Various technologies (Fig. 1) have been used to identify both low- and high-abundance proteins [90–94]. Sections 2.3.3.3.1–2.3.3.3.4 discuss various methods of plasma analysis to address important clinical questions by (i) validating stroke diagnostic biomarkers using candidate-based antibody methods; (ii) utilizing discovery proteomics to find new candidate markers in both disease and predisease states; (iii) implementing novel pharmacoproteomic approaches to monitor therapeutic efficacy at the bedside; (iv) distinguishing stroke subtypes such as ischemia versus hemorrhage. These studies address clinical needs in the diagnosis and management of cerebrovascular disease with high potential for translation to clinical use.
2.3.3.3.1 Antibody-based studies for diagnosis of stroke
Detailed specific protein and peptide biomarkers for cerebrovascular disease, including ischemic and hemorrhagic strokes, are beyond the scope of this publication and have been reviewed elsewhere [10, 95]. Here, we will briefly review biomarkers that are related to recent proteomic efforts. Initial interest in blood biomarkers of stroke has been based on well-characterized protease candidates from preclinical studies—for example, MMPs that degrade the BBB during ischemia, playing important roles between the vascular and CNS compartment [66, 96–100, 100]. Traditional antibody-based ELISA and zymography demonstrated that not only are plasmaMMP-9 levels elevated in patients with acute ischemic and hemorrhagic stroke, elevation in total plasma MMP-9 level also predicts cerebral ischemic hemorrhagic transformation and tPA-related hemorrhage [66, 96–100]. While these early clinical studies support the use of MMP-9 as a promising marker of stroke outcome, individual variability and protein–protein interaction in a noisy clinical system make a single marker difficult to translate to the bedside [67, 101]. To overcome these difficulties, several studies have utilized multimarker panels with protein array or ELISAs [10]. For example, Montaner et al. have examined a panel of markers using ELISA—including C-reactive protein, D-dimer, sRAGE, MMP-9, S100B, BNP, NT-3, caspase-3, chimerin-II, secretagogin, cerebellin, and NPY—to distinguish ischemic versus hemorrhagic stroke within 6 h of symptom onset (AUC = 0.762) [10]. Turck et al. explored the blood level of 29 proteins with immunoassays. They found that glutathione _S_-transferase-π can help to predict early stroke onset time (<3 h) in over 50% of ischemic strokes to help triage thrombolysis, and they went on to test this marker in a separate patient cohort [103].
Similar strategies of looking at selected proteins have also been used to study the evolution of hemorrhagic strokes [10]. For example, Foerch et al. measured promising single biomarkers such as GFAP and S100b to monitor ischemic hemorrhagic transformation or diagnose primary ICH [104–107]. They found GFAP, a glial protein, to be a promising candidate in the early phase of ICH compared to other markers – with the potential to triage decision for prompt treatment – and validated their finding in several studies [104–107]. Turck et al. used a combination of clinical scores and brain injury-related biomarkers (H-FABP, NDKA, UFD1, S100b, troponin I) on admission to predict poor clinical outcome in aneurysmal SAH [108]. Numerous other candidate peptide and protein biomarkers such as plasma BNP, VEGF, sphingolipids, MMPs, gelsolin, anti-TNF, neutrophils and many others have been studied to help to predict clinical outcome and other co-morbidities associated with aneurysmal SAH–a subject beyond the scope of this paper [109–117].
These types of studies demonstrate that even in this era of high-throughput proteomics, traditional antibody based protein quantification methodologies remain great complimentary resources—especially when focused on biologically relevant pathways and coupled with clinical data. These panel approaches also highlight the synergy of monitoring multiple pathways and their interactions. The success of a single biomarker such as troponin for cardiac injury is difficult to match in cerebrovascular disease, due to a wider range of pathology, participating cell types, and stroke subtypes in comparison to other vascular injuries. Real-time clinical assessment and imaging can add to the predict value of individual biomarkers in focused clinical questions. As more innovative blood proteomic technology becomes available, it can help to advance the field not only in discovering novel targets, but also in individualizing clinical care. Some of these findings are discussed in the following sections.
2.3.3.3.2 Proteomic studies for diagnosis of cerebrovascular disease
Initial plasma proteomic studies focused on the diagnosis of acute stroke. For example, using SELDI-TOF-MS, Zhang et al. screened for potential biomarkers to diagnose acute stroke and found 13 biomarkers with sensitivity of 84.4% and specificity of 95.0%, respectively (n = 32 acute stroke; n = 92 control) [91]. While this type of study can obtain rapid results and have promising clinical applicability, no specific proteins or biological pathways can be named. Using MALDI-TOF MS, Huang et al. identified serum protein biomarkers for ischemic stroke and found hemoglobin (Hb) α-chain and β-chain to be differentially expressed between stroke patients and controls (p <0.0001) with sensitivity of 70.2% and the specificity is 85.3% (n = 47 within 3 days of stroke; n = 34 control) [91]. Large-scale collaborative studies utilizing samples from major epidemiologic studies such as Women’s Health Initiative yielded biomarkers for stroke and coronary artery disease risk factors. In this study, Prentice et al. pooled samples from 800 women who developed heart disease, 800 women who developed stroke, and 1600 controls. After immunodepletion, the samples underwent quantitative analysis using “light” C12 or the “heavy” C13 acrylamide isotope labeling. Of the 37 proteins for CHD and 47 proteins for stroke, several proteins including insulin-like growth factor binding protein 4 were noted to be statistically significant risk markers for stroke. This study had one of the largest pooled sample sets investigating plasma stroke and heart disease risk factors in patients who later developed the disease— laying the foundation in studying predisease state in human plasma [118].
2.3.3.3.3 Pharmacoproteomic approaches to monitor therapeutic efficacy
Unlike the in vitro and in vivo models discussed in previous sections, clinical samples, especially blood, are more heterogeneous since no two patients are quite alike—especially in risk factors, medication intake, or even food intake. To reduce confounders in an inherently complex system, the most robust clinical proteomic comparisons are those of profiles taken over time from the same individual. Pragmatically and methodologically, since proteomics technology can afford the sensitivity of studying a multitude of markers at the same time, can one potentially leverage this power to study a smaller number of individuals over time and disease states, utilizing each individual as their own control? Accordingly, applying novel proteomic techniques in specific bedside models, where measurements can be made before and after a specific intervention that triggers systemic changes in plasma signaling, helps to minimize confounders and monitor therapeutic efficacy. This new field of “pharmacoproteomics,” matching the right technology to monitor critical clinical interventions or drug development, has the potential to help to “individualize” treatment by maximizing therapeutic efficacy and minimizing the risk of side effects in real time [52, 119].
One method in particular, developed by the Overall group and others [120, 120], is coined “degradomics”—the investigation of, and techniques for characterizing, the “substrate repertoire” of proteases of interest. In a way, this method allows one to look through the “trash” or “chewed-up bits” of proteinase degradation products as a measure of composite protease activity—offering more biologically relevant information on the synergistic and functional redundancy of different proteases with respect to a particular physiologic challenge [120, 122]. This methodology is especially attractive in studying protein therapeutics (such as tPA—a serine protease), and other proteases important in neurovascular injury such as MMPs. Indeed, we found that in the plasma of patients given tPA for ischemic stroke, there are robust differential protease substrate patterns over time, some of which are brain-specific, giving a dynamic view into the therapeutic efficacy of tPA [123]. Combining degradomics and a targeted protein array approach (MMP-1, MMP-2, MMP-3, MMP-7, MMP-8, MMP-9, MMP-10, MMP-13, and their inhibitors – TIMP-1, TIMP-2), tPA-treated patients not only had increases in multiple MMPs and corresponding inhibitors, but they also had a distinct degradomic pattern that persisted even up to 3–5 days after stroke onset, whereas control patients had little change over time [123]. These findings suggest that beyond a single or even a panel of proteins (such as MMP-2 and MMP-9), systemic therapy such as tPA triggers complex and dynamic responses that can be detected at the bedside. These coordinated signaling responses after stroke may eventually impact the balance between treatment efficacy and associated risks of thrombolysis-related complications such as hemorrhage and edema.
Another understudied area of clinical proteomics has been the characterization of PTM in clinical fluids. For example, one of the most successful clinical biomarkers in medicine is Hb A1c, the nonspecific glycation product of Hb. This marker has revolutionized the care of diabetes by reliably monitoring cumulative disease burden. In particular, PTMs detected in plasma may carry important organ–organ signaling in various disease states and may serve as “tags” for various enrichment and separation techniques. For example, utilizing lectin array and lectin blotting for glycan profiling to evaluate plasma glycosylation patterns, glycoproteins of interest can be enriched and profiled using various lectin affinity chromatography and LC-MS/MS platforms [124]. To this end, we studied the utility of glycosylation profiling in global hypoxic ischemic brain injury triggered by sudden cardiac arrest—a highly prevalent (>300 000/year) and devastating condition with only 3–7% of survivors return to their previous level of functioning [125, 126]. Therapeutic hypothermia— keeping a postcardiac arrest patient in hypothermic condition for 24 h—is an efficacious neuroprotective treatment that is grossly underutilized due to strict exclusion criteria for potential side effects. Analyzing glycosylation patterns is an ideal place to start since, in addition to being one of the most important extracellular PTMs, it is relevant in immunity and coagulation—paramount in hypothermia-related side effects such as sepsis and bleeding—and it occurs rapidly over the treatment time-window. We found significantly different glycosylation patterns between control and cooled postcardiac arrest patients with respect to clinical outcomes in ConA and MAL arrays and blotting; postlectin enrichment, more than a thousand proteins were identified [125,126]. Thus,in addition to MS advances, existing proteomic separation, enrichment, and other front end preparation techniques are crucial to help to identify plasma factors with potential clinical utility to triage clinical treatment and prognosticate outcome.
This type of pharmacoproteomic approach can also be used to study other stroke subtype intervention, such as endovascular closure of patent foramen ovale (PFO)—an invasive procedure that requires better risk stratification and monitoring of therapeutic efficacy to individualize treatment. PFO, a common congenital cardiac abnormality in adults, is an opening between the right and left atria in the heart. As a “back door to the brain,” PFO can serve as a conduit for paradoxical embolism in adults, allowing venous clots to enter the arterial circulation, avoiding filtration by the lungs, and causing ischemic stroke. The anatomy of PFO suggests that, in addition to clots, it can also allow other harmful circulatory factors to travel directly from the venous to the arterial circulation, a concept ideal for discovery proteomic exploration. Lopez et al. applied a novel quantitative two-pass discovery workflow using high-resolution LC-MS/MS coupled with label-free analysis to follow protein expression in PFO patients before and after PFO closure [127]. In this study, we were able to identify quantitative differences in protein expression not only before and after PFO closure, but also in long-term follow-up to evaluate the change in plasma phenotype and help monitor therapeutic efficacy of various treatments [127]. The studies above are only a few examples, which demonstrate the rewards of coupling bedside intervention models with innovative proteomic technology to help to select and monitor patients for the appropriate clinical therapy in real time—a major need for new and ongoing cerebrovascular disease treatments.
2.3.3.3.4 Diagnosis of ischemia versus hemorrhage
Another major focus in neurovascular disease has been the search for biomarkers to distinguish various stroke subtypes. To date, many advances have been made in terms of using differential gene expression patterns of white blood cells [15, 17, 18]. However, since gene expression and protein levels do not always match in stroke, recent works utiltizing protein biomarkers are of special relevance [14]. For example, major effort has been utilized in distinguishing between hemorrhagic versus ischemic stroke subtypes. This is a serious clinical challenge, since ischemic and hemorrhagic strokes require completely divergent treatment— blood-thinning agents, the proper treatment for ischemic stroke, can be lethal if given to a hemorrhagic stroke patient. Current clinical protocols to distinguish between the two stroke subtypes rely on imaging, such as computed tomography (CT) of the brain, which is costly and exposes patients to radiation. Over the last few years, the use of CT has been under much scrutiny in the medical community as it has been reported that approximately 2% of all cancer diagnoses in the United States are attributable to CT use [128, 129]. Thus, the search for blood biomarkers and other clinical measures to diagnosis and follow patients with cerebrovascular disease has gained much attention [10]. Early advances utilizing strong anionic exchange SELDI profiles of plasma samples from 21 stroke patients compared to equal number of controls found seven differentially and statistically significantly expressed peaks (p <0.05). From these peaks, four candidate proteins were identified: apolipoprotein CI (ApoC-I), apolipoprotein CIII (ApoC-III), serum amyloid A, and antithrombin-III fragment. This study found ELISA levels of ApoC-I and ApoC-III to be capable of discriminating between hemorrhagic (n = 15) and ischemic (n = 16) strokes (p <0.001) [90]. Recently, using SRM, Lopez et al. were able to apply a novel multimarker receiver operating characteristic algorithm to select from a panel of apolipoproteins (including ApoA-I, A-II, B, C-I, C-II, C-III, D, E, H)—those markers that not only distinguish between ischemic and hemorrhagic stroke, but also among ischemic and hemorrahagic stoke patients and normal controls (n = 111) [94]. In this study, we found that the ischemic versus hemorrhagic groups were differentiated best by ApoC-III and ApoA-I with an AUC value of 0.92, ischemic versus normal groups by ApoC-III and ApoC-I with AUC 0.93, and hemorrhagic versus normal by ApoC-I and ApoA-II with a resulting AUC value of 0.98. These results highlight the versatility and high-throughput nature of a selective panel of highly abundant apolipoproteins not only in distinguishing various stroke subtypes, but also potentially diagnosing stroke compared to controls, bringing blood biomarkers a step closer to stroke clinical application.
3 Rewards, challenges, and future directions
With the increasing application of proteomics to stroke, many important proteins and pathways have been identified both from the preclinical and clinical sides as summarized above. Advances have been made in both methodology and the fine tuning of clinical questions, as the demand for proteomics technology in stroke research and clinical application continues to grow [8, 52, 130].
It is important to note that while we made every attempt to include all literature available on Medline since the introduction of the word “proteome,” the study of multiple proteins and their interactions even predates the discovery of DNA. Larger scale protein studies have been ongoing even before the word “proteome” existed in published literature [131–134]. This is therefore still a very limited overview of relatively recent advances to apply newer proteomics technology to cerebrovascular disease. Each day exciting new studies are published as the field rapidly advances. However, it is interesting to note that proteomic investigations of cerebrovascular disease have been far fewer than of other vascular or CNS diseases reviewed elsewhere [74, 80, 135–143]. Several challenges exist to the study of neurovascular disease, some related to the heterogeneity of disease itself and some to the availability of high-end instrumentation, and others relate to sampling as discussed previously [52, 144, 145]. For example, human stroke antimortem brain tissue and even CSF are not readily available for research. Blood has been the most readily available clinical sample, and traditionally it has been thought that proteomic technologies have had limitations for research on human body fluids such as plasma, due to high-abundance proteins—plasma contains about ten proteins that together represent about 98% of the total protein content—masking the more clinically relevant lower abundance proteins [146] [89]. While this has been an important limitation, emerging data show that, not just the low-abundance, but high-abundance proteins too have critical roles in human disease pathophysiology, and can be good biomarkers. In fact, currently FDA-approved clinical markers such as C-reactive protein for the prediction of cardiac risk factors, prostate-specific antigen for prostate cancer, and various immunoglobulins for collagen vascular disease, are all higher-abundance markers. So stroke has a lot to learn from these other fields. For example, the posttranslational modified Hb A1c has revolutionized the care of diabetes as it can reliably monitor cumulative disease burden and effectively follow the treatment efficacy of insulin and other medications. Perhaps not just cataloguing markers, but the PTMs of various important high-abundance proteins, focused sub-proteomic analysis, or protein–small molecule interactions would be of interest.
Novel technology with SRM, in vitro cell culture screening, quantitative labeling, and functional protease analysis demonstrate that not only are studies in more complex media such as blood feasible, but both low- and high-abundance proteins can be of immense importance in clinical application. A pharmacoproteomic approach to obtain a “composite” glimpse of the important signaling cascade directly at the bedside in real time may help to monitor therapeutic efficacy, improve patient selection, and ensure more precise clinical phenotyping for clinical trials. A bedside “stimulus-response model” of monitoring therapeutic efficacy pre- and posttreatment may help to minimize confounders by using patient’s pretherapy state as a baseline. For example, currently we carry our own blood type to ensure proper match in emergencies—in the future can we also carry our proteomic baseline, such that, our response to therapeutic intervention can be measured against this known background to monitor therapeutic efficacy in disease states? While larger clinical trials are warranted to establish the sensitivity and specificity of biomarkers for routine use from the current vast candidate pool, smaller well-designed and well-controlled preclinical, translational, and clinical bedside models are direly needed to investigate the underlying mechanisms and expand the field to understand cerebrovascular disease in a dynamic state.
4 Conclusion
The application of proteomics to the investigation of cerebrovascular disease is just beginning to reap rewards. Many advances described above have been made in a short time utilizing proteomics methodology still in development. While larger collaborative clinical trials are needed to refine and test these promising methodologies, small focused translational studies both at the bench and directly at the bedside are also crucial to identifying physiologically relevant candidates for clinical validation and gauge therapeutic response. Future integration of proteomics with genomic, transcriptomic, and metabolomic studies may help to better understand the complexities of neurovascular injury and gain new perspectives. A translational targeted and discovery approach complementing innovative proteomic technology with existing molecular biology models in preclinical studies, and a pharmacoproteomic approach to directly probe clinical physiology and gauge therapeutic efficacy at the bedside, are key to capture the dynamic range of cerebrovascular injury and repair.
Acknowledgments
This work was supported by the NIH/NINDS: R01 NS067139 (Ning) and P01-NS55104 (Lo).
Abbreviations
BBB
blood brain barrier
BMEC
brain microvascular endothaila cells
CNS
central nervous system
CSF
cerebrospinal fluid
CT
computed tomography
Hb
hemoglobin
ICH
intracranial hemorrhage
MCAO
middle cerebral artery occlusion
MMP
matrix metalloproteinases
NVU
neurovascular unit
PFO
patent foramen ovale
SAH
subarachnoid hemorrhage
SELDI
surface-enhanced laser desorption/ionization
SRM
selected reaction monitoring
tPA
tissue plasminogen activator
Footnotes
The authors have declared no conflict of interest.
5 References
- 1.Group NINDS. N. Engl. J. Med. 1995;333:1581–1587. doi: 10.1056/NEJM199512143332401. [DOI] [PubMed] [Google Scholar]
- 2.Reed SD, Cramer SC, Blough DK, Meyer K, Jarvik JG. Stroke. 2001;32:1832–1840. doi: 10.1161/01.str.32.8.1832. [DOI] [PubMed] [Google Scholar]
- 3.Morgenstern LB, Bartholomew LK, Grotta JC, Staub L, King M, Chan W. Arch. Intern. Med. 2003;163:2198–2202. doi: 10.1001/archinte.163.18.2198. [DOI] [PubMed] [Google Scholar]
- 4.Hacke W, Donnan G, Fieschi C, Kaste M, von Kummer R, Broderick JP, Brott T, Frankel M, Grotta JC, Haley EC, Jr, Kwiatkowski T, Levine SR, Lewandowski C, Lu M, Lyden P, Marler JR, Patel S, Tilley BC, Albers G, Bluhmki E, Wilhelm M, Hamilton S. Lancet. 2004;363:768–774. doi: 10.1016/S0140-6736(04)15692-4. [DOI] [PubMed] [Google Scholar]
- 5.Hacke W, Kaste M, Bluhmki E, Brozman M, Davalos A, Guidetti D, Larrue V, Lees KR, Medeghri Z, Machnig T, Schneider D, von Kummer R, Wahlgren N, Toni D. N. Engl. J. Med. 2008;359:1317–1329. doi: 10.1056/NEJMoa0804656. [DOI] [PubMed] [Google Scholar]
- 6.Lees KR, Bluhmki E, von Kummer R, Brott TG, Toni D, Grotta JC, Albers GW, Kaste M, Marler JR, Hamilton SA, Tilley BC, Davis SM, Donnan GA, Hacke W, Allen K, Mau J, Meier D, del Zoppo G, De Silva DA, Butcher KS, Parsons MW, Barber PA, Levi C, Bladin C, Byrnes G. Lancet. 2010;375:1695–1703. [Google Scholar]
- 7.Bluhmki E, Chamorro A, Davalos A, Machnig T, Sauce C, Wahlgren N, Wardlaw J, Hacke W. Lancet Neurol. 2009;8:1095–1102. doi: 10.1016/S1474-4422(09)70264-9. [DOI] [PubMed] [Google Scholar]
- 8.Iadecola C, Goldman SS, Harder DR, Heistad DD, Katusic ZS, Moskowitz MA, Simard JM, Sloan MA, Traystman RJ, Velletri PA. Stroke. 2006;37:1578–1581. doi: 10.1161/01.STR.0000221297.57305.8a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lo EH, Dalkara T, Moskowitz MA. Nat. Rev. Neurosci. 2003;4:399–415. doi: 10.1038/nrn1106. [DOI] [PubMed] [Google Scholar]
- 10.Foerch C, Montaner J, Furie KL, Ning MM, Lo EH. Neurology. 2009;73:393–399. doi: 10.1212/WNL.0b013e3181b05ef9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Z, Chan DW. Cancer Epidemiol. Biomarkers Prev. 2005;14:2283–2286. doi: 10.1158/1055-9965.EPI-05-0774. [DOI] [PubMed] [Google Scholar]
- 12.Weston AD, Hood L. J. Proteome Res. 2004;3:179–196. doi: 10.1021/pr0499693. [DOI] [PubMed] [Google Scholar]
- 13.Arrell DK, Neverova I, Van Eyk JE. Circ. Res. 2001;88:763–773. doi: 10.1161/hh0801.090193. [DOI] [PubMed] [Google Scholar]
- 14.Heiss WD. Stroke. 1992;23:1668–1672. doi: 10.1161/01.str.23.11.1668. [DOI] [PubMed] [Google Scholar]
- 15.Sharp FR, Xu H, Lit L, Walker W, Apperson M, Gilbert DL, Glauser TA, Wong B, Hershey A, Liu DZ, Pinter J, Zhan X, Liu X, Ran R. Arch. Neurol. 2006;63:1529–1536. doi: 10.1001/archneur.63.11.1529. [DOI] [PubMed] [Google Scholar]
- 16.Liu DZ, Tian Y, Ander BP, Xu H, Stamova BS, Zhan X, Turner RJ, Jickling G, Sharp FR. J. Cereb. Blood Flow Metab. 2010;30:92–101. doi: 10.1038/jcbfm.2009.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jickling GC, Stamova B, Ander BP, Zhan X, Liu D, Sison SM, Verro P, Sharp FR. Stroke. 2012;43:2036–2041. doi: 10.1161/STROKEAHA.111.648725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jickling GC, Stamova B, Ander BP, Zhan X, Tian Y, Liu D, Xu H, Johnston SC, Verro P, Sharp FR. Ann. Neurol. 2011;70:477–485. doi: 10.1002/ana.22497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sironi L, Tremoli E, Miller I, Guerrini U, Calvio AM, Eberini I, Gemeiner M, Asdente M, Paoletti R, Gianazza E. Stroke. 2001;32:753–760. doi: 10.1161/01.str.32.3.753. [DOI] [PubMed] [Google Scholar]
- 20.Sironi L, Guerrini U, Tremoli E, Miller I, Gelosa P, Lascialfari A, Zucca I, Eberini I, Gemeiner M, Paoletti R, Gianazza E. J. Neurosci. Res. 2004;78:115–122. doi: 10.1002/jnr.20219. [DOI] [PubMed] [Google Scholar]
- 21.Bergerat A, Decano J, Wu CJ, Choi H, Nesvizhskii AI, Moran AM, Ruiz-Opazo N, Steffen M, Herrera VL. Mol. Med. 2011;17:588–598. doi: 10.2119/molmed.2010.00228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dhodda VK, Sailor KA, Bowen KK, Vemuganti R. J. Neurochem. 2004;89:73–89. doi: 10.1111/j.1471-4159.2004.02316.x. [DOI] [PubMed] [Google Scholar]
- 23.Suzuyama K, Shiraishi T, Oishi T, Ueda S, Okamoto H, Furuta M, Mineta T, Tabuchi K. Brain Res. Mol. Brain Res. 2004;129:44–53. doi: 10.1016/j.molbrainres.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 24.Focking M, Besselmann M, Trapp T. Acta Neurobiol. Exp. (Wars) 2006;66:273–278. doi: 10.55782/ane-2006-1616. [DOI] [PubMed] [Google Scholar]
- 25.Chen A, Liao WP, Lu Q, Wong WS, Wong PT. Neurochem. Int. 2007;50:1078–1086. doi: 10.1016/j.neuint.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 26.Xiong X, Liang Q, Chen J, Fan R, Cheng T. Biosci. Biotechnol. Biochem. 2009;73:657–664. doi: 10.1271/bbb.80717. [DOI] [PubMed] [Google Scholar]
- 27.Mizutani K, Sonoda S, Hayashi N, Takasaki A, Beppu H, Saitoh E, Shimpo K. Am. J. Phys. Med. Rehabil. 2010;89:107–114. doi: 10.1097/PHM.0b013e3181b3323b. [DOI] [PubMed] [Google Scholar]
- 28.Yao H, Nakahara T, Nakagawa N, Hashimoto K, Kuroki T. Neurochem. Res. 2009;34:1999–2007. doi: 10.1007/s11064-009-9988-6. [DOI] [PubMed] [Google Scholar]
- 29.Sung JH, Cho EH, Min W, Kim MJ, Kim MO, Jung EJ, Koh PO. Neurosci. Lett. 2010;477:66–71. doi: 10.1016/j.neulet.2010.04.028. [DOI] [PubMed] [Google Scholar]
- 30.Koh PO. Am. J. Chin. Med. 2011;39:315–324. doi: 10.1142/S0192415X11008841. [DOI] [PubMed] [Google Scholar]
- 31.Sung JH, Kim MO, Koh PO. Neuroscience. 2011;174:171–177. doi: 10.1016/j.neuroscience.2010.11.021. [DOI] [PubMed] [Google Scholar]
- 32.Di Domenico F, Casalena G, Jia J, Sultana R, Barone E, Cai J, Pierce WM, Cini C, Mancuso C, Perluigi M, Davis CM, Alkayed NJ, Butterfield AD. J. Neurochem. 2012;121:680–692. doi: 10.1111/j.1471-4159.2012.07721.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu JS, Cheung WM, Tsai YS, Chen YT, Fong WH, Tsai HD, Chen YC, Liou JY, Shyue SK, Chen JJ, Chen YE, Maeda N, Wu KK, Lin TN. Circulation. 2009;119:1124–1134. doi: 10.1161/CIRCULATIONAHA.108.812537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chiu CD, Chen TY, Chin LT, Shen CC, Huo J, Ma SY, Chen HM, Chu CH. Proteomics. 2012;12:113–123. doi: 10.1002/pmic.201100256. [DOI] [PubMed] [Google Scholar]
- 35.Lo EH, Rosenberg GA. Stroke. 2009;40:S2–S3. doi: 10.1161/STROKEAHA.108.534404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lo EH, Wang X, Cuzner ML. J. Neurosci. Res. 2002;69:1–9. doi: 10.1002/jnr.10270. [DOI] [PubMed] [Google Scholar]
- 37.Gagne JP, Isabelle M, Lo KS, Bourassa S, Hendzel MJ, Dawson VL, Dawson TM, Poirier GG. Nucleic Acids Res. 2008;36:6959–6976. doi: 10.1093/nar/gkn771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arai K, Lo EH. J. Neurosci. Res. 2010;88:758–763. doi: 10.1002/jnr.22256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo S, Arai K, Stins MF, Chuang DM, Lo EH. Stroke. 2009;40:652–655. doi: 10.1161/STROKEAHA.108.524504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dowell JA, Johnson JA, Li L. J. Proteome Res. 2009;8:4135–4143. doi: 10.1021/pr900248y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Greco TM, Seeholzer SH, Mak A, Spruce L, Ischiropoulos H. J. Proteome Res. 2010;9:2764–2774. doi: 10.1021/pr100134n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zamanian JL, Xu L, Foo LC, Nouri N, Zhou L, Giffard RG, Barres BA. J. Neurosci. 2012;32:6391–6410. doi: 10.1523/JNEUROSCI.6221-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hawkins BT, Davis TP. Pharmacol. Rev. 2005;57:173–185. doi: 10.1124/pr.57.2.4. [DOI] [PubMed] [Google Scholar]
- 44.Zlokovic BV. Neuron. 2008;57:178–201. doi: 10.1016/j.neuron.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 45.Guo S, Kim WJ, Lok J, Lee SR, Besancon E, Luo BH, Stins MF, Wang X, Dedhar S, Lo EH. Proc. Natl. Acad. Sci. U.S.A. 2008;105:7582–7587. doi: 10.1073/pnas.0801105105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lok J, Sardi SP, Guo S, Besancon E, Ha DM, Rosell A, Kim WJ, Corfas G, Lo EH. J. Cereb. Blood Flow Metab. 2009;29:39–43. doi: 10.1038/jcbfm.2008.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu Q, Murugesan N, Macdonald JA, Wu SL, Pachter JS, Hancock WS. Electrophoresis. 2008;29:2689–2695. doi: 10.1002/elps.200700936. [DOI] [PubMed] [Google Scholar]
- 48.Enerson BE, Drewes LR. J. Cereb. Blood Flow Metab. 2006;26:959–973. doi: 10.1038/sj.jcbfm.9600249. [DOI] [PubMed] [Google Scholar]
- 49.Datta A, Park JE, Li X, Zhang H, Ho ZS, Heese K, Lim SK, Tam JP, Sze SK. J. Proteome Res. 2010;9:472–484. doi: 10.1021/pr900829h. [DOI] [PubMed] [Google Scholar]
- 50.Dieterich DC, Hodas JJ, Gouzer G, Shadrin IY, Ngo JT, Triller A, Tirrell DA, Schuman EM. Nat. Neurosci. 2010;13:897–905. doi: 10.1038/nn.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou A, Simon RP, David L. Int. J. Comput. Biol. Drug. Des. 2011;4:40–55. doi: 10.1504/IJCBDD.2011.038656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ning M, Lo EH. Transl. Stroke Res. 2010;1:233–237. doi: 10.1007/s12975-010-0048-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Minagar A, Alexander JS, Kelley RE, Harper M, Jennings MH. J. Mol. Neurosci. 2009;38:182–192. doi: 10.1007/s12031-008-9149-4. [DOI] [PubMed] [Google Scholar]
- 54.Ning M, Sarracino DA, Kho AT, Guo S, Lee SR, Krastins B, Buonanno FS, Vizcaino JA, Orchard S, McMullin D, Wang X, Lo EH. Stroke. 2010;42:37–43. doi: 10.1161/STROKEAHA.110.585703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fujimura M, Gasche Y, Morita-Fujimura Y, Massengale J, Kawase M, Chan PH. Brain Res. 1999;842:92–100. doi: 10.1016/s0006-8993(99)01843-0. [DOI] [PubMed] [Google Scholar]
- 56.Asahi M, Wang X, Mori T, Sumii T, Jung JC, Moskowitz MA, Fini ME, Lo EH. J. Neurosci. 2001;21:7724–7732. doi: 10.1523/JNEUROSCI.21-19-07724.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Asahi M, Asahi K, Jung JC, del Zoppo GJ, Fini ME, Lo EH. J. Cereb. Blood Flow Metab. 2000;20:1681–1689. doi: 10.1097/00004647-200012000-00007. [DOI] [PubMed] [Google Scholar]
- 58.Heo JH, Lucero J, Abumiya T, Koziol JA, Copeland BR, del Zoppo GJ. J. Cereb. Blood Flow Metab. 1999;19:624–633. doi: 10.1097/00004647-199906000-00005. [DOI] [PubMed] [Google Scholar]
- 59.Mun-Bryce S, Rosenberg GA. J. Cereb. Blood Flow Metab. 1998;18:1163–1172. doi: 10.1097/00004647-199811000-00001. [DOI] [PubMed] [Google Scholar]
- 60.Rosenberg GA, Navratil M, Barone F, Feuerstein G. J. Cereb. Blood Flow Metab. 1996;16:360–366. doi: 10.1097/00004647-199605000-00002. [DOI] [PubMed] [Google Scholar]
- 61.Anthony DC, Miller KM, Fearn S, Townsend MJ, Opdenakker G, Wells GM, Clements JM, Chandler S, Gearing AJ, Perry VH. J. Neuroimmunol. 1998;87:62–72. doi: 10.1016/s0165-5728(98)00046-0. [DOI] [PubMed] [Google Scholar]
- 62.Rosenberg GA, Estrada EY, Dencoff JE. Stroke. 1998;29:2189–2195. doi: 10.1161/01.str.29.10.2189. [DOI] [PubMed] [Google Scholar]
- 63.Romanic AM, White RF, Arleth AJ, Ohlstein EH, Barone FC. Stroke. 1998;29:1020–1030. doi: 10.1161/01.str.29.5.1020. [DOI] [PubMed] [Google Scholar]
- 64.Jiang X, Namura S, Nagata I. Neurosci. Lett. 2001;305:41–44. doi: 10.1016/s0304-3940(01)01800-6. [DOI] [PubMed] [Google Scholar]
- 65.Zhao BQ, Wang S, Kim HY, Storrie H, Rosen BR, Mooney DJ, Wang X, Lo EH. Nat. Med. 2006;12:441–445. doi: 10.1038/nm1387. [DOI] [PubMed] [Google Scholar]
- 66.Ning M, Furie KL, Koroshetz WJ, Lee H, Barron M, Lederer M, Wang X, Zhu M, Sorensen AG, Lo EH, Kelly PJ. Neurology. 2006;66:1550–1555. doi: 10.1212/01.wnl.0000216133.98416.b4. [DOI] [PubMed] [Google Scholar]
- 67.Ning M, Wang X, Lo EH. Handb. Clin. Neurol. 2008;92:117–136. doi: 10.1016/S0072-9752(08)01906-4. [DOI] [PubMed] [Google Scholar]
- 68.Cuadrado E, Rosell A, Penalba A, Slevin M, Alvarez-Sabin J, Ortega-Aznar A, Montaner J. J. Proteome Res. 2009;8:3191–3197. doi: 10.1021/pr801012x. [DOI] [PubMed] [Google Scholar]
- 69.Cuadrado E, Rosell A, Colome N, Hernandez-Guillamon M, Garcia-Berrocoso T, Ribo M, Alcazar A, Ortega-Aznar A, Salinas M, Canals F, Montaner J. J. Neuropathol. Exp. Neurol. 2010;69:1105–1115. doi: 10.1097/NEN.0b013e3181f8c539. [DOI] [PubMed] [Google Scholar]
- 70.Hu S, Loo JA, Wong DT. Proteomics. 2006;6:6326–6353. doi: 10.1002/pmic.200600284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Veenstra TD, Conrads TP, Hood BL, Avellino AM, Ellenbogen RG, Morrison RS. Mol. Cell Proteomics. 2005;4:409–418. doi: 10.1074/mcp.M500006-MCP200. [DOI] [PubMed] [Google Scholar]
- 72.Ping P, Vondriska TM, Creighton CJ, Gandhi TK, Yang Z, Menon R, Kwon MS, Cho SY, Drwal G, Kellmann M, Peri S, Suresh S, Gronborg M, Molina H, Chaerkady R, Rekha B, Shet AS, Gerszten RE, Wu H, Raftery M, Wasinger V, Schulz-Knappe P, Hanash SM, Paik YK, Hancock WS, States DJ, Omenn GS, Pandey A. Proteomics. 2005;5:3506–3519. doi: 10.1002/pmic.200500140. [DOI] [PubMed] [Google Scholar]
- 73.Omenn GS, States DJ, Adamski M, Blackwell TW, Menon R, Hermjakob H, Apweiler R, Haab BB, Simpson RJ, Eddes JS, Kapp EA, Moritz RL, Chan DW, Rai AJ, Admon A, Aebersold R, Eng J, Hancock WS, Hefta SA, Meyer H, Paik YK, Yoo JS, Ping P, Pounds J, Adkins J, Qian X, Wang R, Wasinger V, Wu CY, Zhao X, Zeng R, Archakov A, Tsugita A, Beer I, Pandey A, Pisano M, Andrews P, Tammen H, Speicher DW, Hanash SM. Proteomics. 2005;5:3226–3245. doi: 10.1002/pmic.200500358. [DOI] [PubMed] [Google Scholar]
- 74.Berhane BT, Zong C, Liem DA, Huang A, Le S, Edmondson RD, Jones RC, Qiao X, Whitelegge JP, Ping P, Vondriska TM. Proteomics. 2005;5:3520–3530. doi: 10.1002/pmic.200401308. [DOI] [PubMed] [Google Scholar]
- 75.Kentsis A, Monigatti F, Dorff K, Campagne F, Bachur R, Steen H. Proteomics Clin. Appl. 2009;3:1052–1061. doi: 10.1002/prca.200900008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kentsis A, Lin YY, Kurek K, Calicchio M, Wang YY, Monigatti F, Campagne F, Lee R, Horwitz B, Steen H, Bachur R. Ann. Emerg. Med. 2010;55:62–70. e4. doi: 10.1016/j.annemergmed.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dawson J, Walters M, Delles C, Mischak H, Mullen W. PLoS One. 2012;7:e35879. doi: 10.1371/journal.pone.0035879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Romeo MJ, Espina V, Lowenthal M, Espina BH, Petricoin EF, 3rd, Liotta LA. Expert Rev. Proteomics. 2005;2:57–70. doi: 10.1586/14789450.2.1.57. [DOI] [PubMed] [Google Scholar]
- 79.Lad SP, Hegen H, Gupta G, Deisenhammer F, Steinberg GK. J. Stroke Cerebrovasc. Dis. 2012;21:30–41. doi: 10.1016/j.jstrokecerebrovasdis.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 80.Wang KK, Ottens AK, Liu MC, Lewis SB, Meegan C, Oli MW, Tortella FC, Hayes RL. Expert Rev. Proteomics. 2005;2:603–614. doi: 10.1586/14789450.2.4.603. [DOI] [PubMed] [Google Scholar]
- 81.Kobeissy FH, Guingab-Cagmat JD, Razafsha M, O’Steen L, Zhang Z, Hayes RL, Chiu WT, Wang KK. Pm R. 2011;3:S139–S147. doi: 10.1016/j.pmrj.2011.02.012. [DOI] [PubMed] [Google Scholar]
- 82.Kobeissy FH, Sadasivan S, Oli MW, Robinson G, Larner SF, Zhang Z, Hayes RL, Wang KK. Proteomics Clin. Appl. 2008;2:1467–1483. doi: 10.1002/prca.200800011. [DOI] [PubMed] [Google Scholar]
- 83.Zimmermann-Ivol CG, Burkhard PR, Le Floch-Rohr J, Allard L, Hochstrasser DF, Sanchez JC. Mol. Cell Proteomics. 2004;3:66–72. doi: 10.1074/mcp.M300066-MCP200. [DOI] [PubMed] [Google Scholar]
- 84.Lescuyer P, Allard L, Zimmermann-Ivol CG, Burgess JA, Hughes-Frutiger S, Burkhard PR, Sanchez JC, Hochstrasser DF. Proteomics. 2004;4:2234–2241. doi: 10.1002/pmic.200300822. [DOI] [PubMed] [Google Scholar]
- 85.Dayon L, Hainard A, Licker V, Turck N, Kuhn K, Hochstrasser DF, Burkhard PR, Sanchez JC. Anal. Chem. 2008;80:2921–2931. doi: 10.1021/ac702422x. [DOI] [PubMed] [Google Scholar]
- 86.Maurer MH, Haux D, Sakowitz OW, Unterberg AW, Kuschinsky W. J. Cereb. Blood Flow Metab. 2007;27:1675–1683. doi: 10.1038/sj.jcbfm.9600466. [DOI] [PubMed] [Google Scholar]
- 87.Dayon L, Turck N, Garci-Berrocoso T, Walter N, Burkhard PR, Vilalta A, Sahuquillo J, Montaner J, Sanchez JC. J. Proteome Res. 2011;10:1043–1051. doi: 10.1021/pr101123t. [DOI] [PubMed] [Google Scholar]
- 88.Hillered L, Vespa PM, Hovda DA. J. Neurotrauma. 2005;22:3–41. doi: 10.1089/neu.2005.22.3. [DOI] [PubMed] [Google Scholar]
- 89.Anderson NL, Anderson NG. Mol. Cell Proteomics. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 90.Allard L, Lescuyer P, Burgess J, Leung KY, Ward M, Walter N, Burkhard PR, Corthals G, Hochstrasser DF, Sanchez JC. Proteomics. 2004;4:2242–2251. doi: 10.1002/pmic.200300809. [DOI] [PubMed] [Google Scholar]
- 91.Zhang X, Guo T, Wang H, He W, Mei H, Hong M, Yu J, Hu Y, Song S. Am. J. Clin. Pathol. 2008;130:299–304. doi: 10.1309/CA242R5VY14HUGE8. [DOI] [PubMed] [Google Scholar]
- 92.Huang P, Lo LH, Chen YC, Lin RT, Shiea J, Liu CK. J. Neurol. 2009;256:625–631. doi: 10.1007/s00415-009-0133-x. [DOI] [PubMed] [Google Scholar]
- 93.Pan S, Zhan X, Su X, Guo L, Lv L, Su B. Exp. Biol. Med. (Maywood) 2011;236:325–333. doi: 10.1258/ebm.2011.010041. [DOI] [PubMed] [Google Scholar]
- 94.Lopez MF, Sarracino DA, Prakash A, Athanas M, Krastins B, Rezai T, Sutton JN, Peterman S, Gvozdyak O, Chou S, Lo E, Buonanno F, Ning M. Proteomics Clin. Appl. 2012;6:190–200. doi: 10.1002/prca.201100041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kernagis DN, Laskowitz DT. Ann. Neurol. 2012;71:289–303. doi: 10.1002/ana.22553. [DOI] [PubMed] [Google Scholar]
- 96.Montaner J, Alvarez-Sabin J, Molina C, Angles A, Abilleira S, Arenillas J, Gonzalez MA, Monasterio J. Stroke. 2001;32:1759–1766. doi: 10.1161/01.str.32.8.1759. [DOI] [PubMed] [Google Scholar]
- 97.Abilleira S, Montaner J, Molina CA, Monasterio J, Castillo J, Alvarez-Sabin J. J. Neurosurg. 2003;99:65–70. doi: 10.3171/jns.2003.99.1.0065. [DOI] [PubMed] [Google Scholar]
- 98.Castellanos M, Leira R, Serena J, Pumar JM, Lizasoain I, Castillo J, Davalos A. Stroke. 2003;34:40–46. [PubMed] [Google Scholar]
- 99.Montaner J, Molina CA, Monasterio J, Abilleira S, Arenillas JF, Ribo M, Quintana M, Alvarez-Sabin J. Circulation. 2003;107:598–603. doi: 10.1161/01.cir.0000046451.38849.90. [DOI] [PubMed] [Google Scholar]
- 100.Montaner J, Molina CA, Arenillas JF, Ribó M, Huertas R, Fernández-Cadenas I, Rosell A, Penalba A, Monasterio J, Alvarez-Sabín J. Stroke. 2004;35:339. [Google Scholar]
- 101.Wijman CA, Smirnakis SM, Vespa P, Szigeti K, Ziai WC, Ning MM, Rosand J, Hanley DF, Geocadin R, Hall C, Le Roux PD, Suarez JI, Zaidat OO. Neurocrit. Care. 2012;16:42–54. doi: 10.1007/s12028-011-9609-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Montaner J, Mendioroz M, Delgado P, Garcia-Berrocoso T, Giralt D, Merino C, Ribo M, Rosell A, Penalba A, Fernandez-Cadenas I, Romero F, Molina C, Alvarez-Sabin J, Hernandez-Guillamon M. J. Proteomics. 2012;75:4758–4765. doi: 10.1016/j.jprot.2012.01.033. [DOI] [PubMed] [Google Scholar]
- 103.Turck N, Robin X, Walter N, Fouda C, Hainard A, Sztajzel R, Wagner G, Hochstrasser DF, Montaner J, Burkhard PR, Sanchez JC. PLoS One. 2012;7:e43830. doi: 10.1371/journal.pone.0043830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Foerch C, Wunderlich MT, Dvorak F, Humpich M, Kahles T, Goertler M, Alvarez-Sabin J, Wallesch CW, Molina CA, Steinmetz H, Sitzer M, Montaner J. Stroke. 2007;38:2491–2495. doi: 10.1161/STROKEAHA.106.480111. [DOI] [PubMed] [Google Scholar]
- 105.Foerch C, Curdt I, Yan B, Dvorak F, Hermans M, Berkefeld J, Raabe A, Neumann-Haefelin T, Steinmetz H, Sitzer M. J. Neurol. Neurosurg. Psychiatry. 2006;77:181–184. doi: 10.1136/jnnp.2005.074823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Foerch C, Otto B, Singer OC, Neumann-Haefelin T, Yan B, Berkefeld J, Steinmetz H, Sitzer M. Stroke. 2004;35:2160–2164. doi: 10.1161/01.STR.0000138730.03264.ac. [DOI] [PubMed] [Google Scholar]
- 107.Foerch C, Singer OC, Neumann-Haefelin T, du Mesnil de Rochemont R, Steinmetz H, Sitzer M. Arch. Neurol. 2005;62:1130–1134. doi: 10.1001/archneur.62.7.1130. [DOI] [PubMed] [Google Scholar]
- 108.Turck N, Vutskits L, Sanchez-Pena P, Robin X, Hainard A, Gex-Fabry M, Fouda C, Bassem H, Mueller M, Lisacek F, Puybasset L, Sanchez JC. Intensive Care Med. 2010;36:107–115. doi: 10.1007/s00134-009-1641-y. [DOI] [PubMed] [Google Scholar]
- 109.Chou SH, Feske SK, Atherton J, Konigsberg RG, De Jager PL, Du R, Ogilvy CS, Lo EH, Ning M. J. Investig. Med. 2012;60:1054–1058. doi: 10.231/JIM.0b013e3182686932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chou SH, Feske SK, Simmons SL, Konigsberg RG, Orzell SC, Marckmann A, Bourget G, Bauer DJ, De Jager PL, Du R, Arai K, Lo EH, Ning MM. Transl. Stroke Res. 2011;2:600–607. doi: 10.1007/s12975-011-0117-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chou SH, Lee PS, Konigsberg RG, Gallacci D, Chiou T, Arai K, Simmons S, Bauer D, Feske SK, Lo EH, Ning M. Stroke. 2011;42:3624–3627. doi: 10.1161/STROKEAHA.111.631135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Johnston SC, Selvin S, Gress DR. Neurology. 1998;50:1413–1418. doi: 10.1212/wnl.50.5.1413. [DOI] [PubMed] [Google Scholar]
- 113.Provencio JJ, Fu X, Siu A, Rasmussen PA, Hazen SL, Ransohoff RM. Neurocrit. Care. 2010;12:244–251. doi: 10.1007/s12028-009-9308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Macdonald RL, Pluta RM, Zhang JH. Nat. Clin. Pract. Neurol. 2007;3:256–263. doi: 10.1038/ncpneuro0490. [DOI] [PubMed] [Google Scholar]
- 115.Testai FD, Hillmann M, Amin-Hanjani S, Gorshkova I, Berdyshev E, Gorelick PB, Dawson G. Stroke. 2012;43:2066–2070. doi: 10.1161/STROKEAHA.112.650390. [DOI] [PubMed] [Google Scholar]
- 116.Testai FD, Aiyagari V, Hillmann M, Amin-Hanjani S, Dawson G, Gorelick P. Neurocrit. Care. 2011;15:416–420. doi: 10.1007/s12028-011-9559-y. [DOI] [PubMed] [Google Scholar]
- 117.Tung PP, Olmsted E, Kopelnik A, Banki NM, Drew BJ, Ko N, Lawton MT, Smith W, Foster E, Young WL, Zaroff JG. Stroke. 2005;36:1567–1569. doi: 10.1161/01.STR.0000170699.59783.d6. [DOI] [PubMed] [Google Scholar]
- 118.Prentice RL, Paczesny S, Aragaki A, Amon LM, Chen L, Pitteri SJ, McIntosh M, Wang P, Buson Busald T, Hsia J, Jackson RD, Rossouw JE, Manson JE, Johnson K, Eaton C, Hanash SM. Genome Med. 2010;2:48. doi: 10.1186/gm169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yu LR. J. Proteomics. 2011;74:2549–2553. doi: 10.1016/j.jprot.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 120.Lopez-Otin C, Overall CM. Nat. Rev. Mol. Cell Biol. 2002;3:509–519. doi: 10.1038/nrm858. [DOI] [PubMed] [Google Scholar]
- 121.Liotta LA, Ferrari M, Petricoin E. Nature. 2003;425:905. doi: 10.1038/425905a. [DOI] [PubMed] [Google Scholar]
- 122.Kobeissy FH, Sadasivan S, Liu J, Gold MS, Wang KK. Expert Rev. Proteomics. 2008;5:293–314. doi: 10.1586/14789450.5.2.293. [DOI] [PubMed] [Google Scholar]
- 123.Ning M, Sarracino DA, Buonanno FS, Krastins B, Chou S, McMullin D, Wang X, Lopez M, Lo EH. Transl. Stroke Res. 2010;1:268–275. doi: 10.1007/s12975-010-0047-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Selvaraju S, Rassi ZE. Electrophoresis. 2012;33:74–88. doi: 10.1002/elps.201100431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Cao J, Guo S, Arai K, Lo EH, Ning MM. Methods Mol. Biol. Chemokine. 2013:1–10. doi: 10.1007/978-1-62703-426-5_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Cao J, Lopez M, Sarracino D, Elia M, Feeney K, Buonanno FS, Lo EH, Ning MM. HUPO. 2012 [Google Scholar]
- 127.Lopez MFSD, Vogelsang M, Sutton JN, Athanas M, Krastins B, Garces A, Prakash A, Peterman S, Demirjian Z, Inglessis I, Feeney K, Elia M, Mc-Mullin D, Dec GW, Palacios I, Lo EH, Buonanno FS, Ning MM. J. Investig. Med. 2012;60:1122–1130. doi: 10.231/JIM.0b013e318276de0e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Brenner DJ, Hall EJ. N. Engl. J. Med. 2007;357:2277–2284. doi: 10.1056/NEJMra072149. [DOI] [PubMed] [Google Scholar]
- 129.Smith-Bindman R, Lipson J, Marcus R, Kim KP, Mahesh M, Gould R, Berrington de Gonzalez A, Miglioretti DL. Arch. Intern. Med. 2009;169:2078–2086. doi: 10.1001/archinternmed.2009.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Stroke Progress Review Omics Working Group. NINDS. 2012 http://www.ninds.nih.gov/find_people/groups/stroke_prg/2012-stroke-prg-full-report.htm#OM
- 131.Wasinger VC, Cordwell SJ, Cerpa-Poljak A, Yan JX, Gooley AA, Wilkins MR, Duncan MW, Harris R, Williams KL, Humphery-Smith I. Electrophoresis. 1995;16:1090–1094. doi: 10.1002/elps.11501601185. [DOI] [PubMed] [Google Scholar]
- 132.Pasquali C, Frutiger S, Wilkins MR, Hughes GJ, Appel RD, Bairoch A, Schaller D, Sanchez JC, Hochstrasser DF. Electrophoresis. 1996;17:547–555. doi: 10.1002/elps.1150170325. [DOI] [PubMed] [Google Scholar]
- 133.Anderson NL, Anderson NG. Electrophoresis. 1998;19:1853–1861. doi: 10.1002/elps.1150191103. [DOI] [PubMed] [Google Scholar]
- 134.Blackstock WP, Weir MP. Trends Biotechnol. 1999;17:121–127. doi: 10.1016/s0167-7799(98)01245-1. [DOI] [PubMed] [Google Scholar]
- 135.Alberio T, Fasano M. J. Biotechnol. 2010;156:325–337. doi: 10.1016/j.jbiotec.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 136.Hwang H, Zhang J, Chung KA, Leverenz JB, Zabetian CP, Peskind ER, Jankovic J, Su Z, Hancock AM, Pan C, Montine TJ, Pan S, Nutt J, Albin R, Gearing M, Beyer RP, Shi M. Mass Spectrom. Rev. 2010;29:79–125. doi: 10.1002/mas.20221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Maurer MH. Mass Spectrom. Rev. 2010;29:17–28. doi: 10.1002/mas.20213. [DOI] [PubMed] [Google Scholar]
- 138.Oli MW, Hayes RL, Robinson G, Wang KK. Methods Mol. Biol. 2009;566:293–302. doi: 10.1007/978-1-59745-562-6_19. [DOI] [PubMed] [Google Scholar]
- 139.Drake TA, Ping P. J. Lipid Res. 2007;48:1–8. doi: 10.1194/jlr.R600027-JLR200. [DOI] [PubMed] [Google Scholar]
- 140.Mayr M, Zhang J, Greene AS, Gutterman D, Perloff J, Ping P. Mol. Cell Proteomics. 2006;5:1853–1864. doi: 10.1074/mcp.R600007-MCP200. [DOI] [PubMed] [Google Scholar]
- 141.Fu Q, Schoenhoff FS, Savage WJ, Zhang P, Van Eyk JE. Proteomics Clin. Appl. 2010;4:271–284. doi: 10.1002/prca.200900217. [DOI] [PubMed] [Google Scholar]
- 142.Gerszten RE, Carr SA, Sabatine M. Clin. Chem. 2010;56:194–201. doi: 10.1373/clinchem.2009.127878. [DOI] [PubMed] [Google Scholar]
- 143.Stevens SM, Jr, Duncan RS, Koulen P, Prokai L. J. Proteome Res. 2008;7:1046–1054. doi: 10.1021/pr7006279. [DOI] [PubMed] [Google Scholar]
- 144.Leker RR, Constantini S. Acta Neurochir. Suppl. 2002;83:55–59. doi: 10.1007/978-3-7091-6743-4_10. [DOI] [PubMed] [Google Scholar]
- 145.Rynkowski MA, Kim GH, Komotar RJ, Otten ML, Ducruet AF, Zacharia BE, Kellner CP, Hahn DK, Merkow MB, Garrett MC, Starke RM, Cho BM, Sosunov SA, Connolly ES. Nat. Protoc. 2008;3:122–128. doi: 10.1038/nprot.2007.513. [DOI] [PubMed] [Google Scholar]
- 146.Garbis S, Lubec G, Fountoulakis M. J. Chromatogr. A. 2005;1077:1–18. doi: 10.1016/j.chroma.2005.04.059. [DOI] [PubMed] [Google Scholar]