A Wee1 checkpoint inhibits anaphase onset (original) (raw)
The budding yeast Wee1 kinase Swe1 restrains the metaphase-to-anaphase transition by preventing the Cdk1-dependent phosphorylation and activation of APCCdc20.
Abstract
Cdk1 drives both mitotic entry and the metaphase-to-anaphase transition. Past work has shown that Wee1 inhibition of Cdk1 blocks mitotic entry. Here we show that the budding yeast Wee1 kinase, Swe1, also restrains the metaphase-to-anaphase transition by preventing Cdk1 phosphorylation and activation of the mitotic form of the anaphase-promoting complex/cyclosome (APCCdc20). Deletion of SWE1 or its opposing phosphatase MIH1 (the budding yeast cdc25+) altered the timing of anaphase onset, and activation of the Swe1-dependent morphogenesis checkpoint or overexpression of Swe1 blocked cells in metaphase with reduced APC activity in vivo and in vitro. The morphogenesis checkpoint also depended on Cdc55, a regulatory subunit of protein phosphatase 2A (PP2A). cdc55Δ checkpoint defects were rescued by mutating 12 Cdk1 phosphorylation sites on the APC, demonstrating that the APC is a target of this checkpoint. These data suggest a model in which stepwise activation of Cdk1 and inhibition of PP2ACdc55 triggers anaphase onset.
Introduction
Cyclin-dependent kinase 1 (Cdk1) activity drives many mitotic events, including spindle assembly during mitotic entry and chromosome segregation at anaphase onset (Morgan, 2007). The ordering of mitotic events therefore requires the ordered and coordinated phosphorylation of many Cdk1 substrates. This ordering is achieved by two mechanisms: substrate specificity conferred by different cyclin subunits whose levels rise and fall in waves during the cell cycle, and a stepwise increase in total Cdk1 activity that reaches a maximum at anaphase onset (Stern and Nurse, 1996; Uhlmann et al., 2011). Although the importance of Cdk1 activity in mitosis has long been recognized, the critical Cdk1 substrates that drive mitotic transitions are poorly defined.
Mitotic onset is regulated in all eukaryotes by an increase in Cdk1 activity caused by the dephosphorylation of Cdk1 on a conserved inhibitory tyrosine (tyrosine 19 in budding yeast; Russell and Nurse, 1986; Nurse, 1990; Dunphy and Kumagai, 1991; Gautier et al., 1991; Harvey et al., 2005). The kinase and phosphatase responsible for the modification of this residue are Wee1 and Cdc25, respectively (Gould and Nurse, 1989; Gould et al., 1990). wee1 mutants in fission yeast shorten G2 by causing premature activation of Cdk1, whereas cdc25 mutants never accumulate sufficient Cdk1 activity to enter mitosis (Nurse, 1975; Russell and Nurse, 1986, 1987). Wee1 and Cdc25 are the targets of numerous cell cycle checkpoints, all of which delay mitotic entry by activating Wee1 or inhibiting Cdc25 (Kellogg, 2003).
Budding yeast Wee1 (Swe1) and Cdc25 (Mih1) are targets of a morphogenesis checkpoint that has been shown to delay mitotic onset in response to either defects in the actin cytoskeleton, small cell size, or the extent of membrane growth (Lew and Reed, 1995; McMillan et al., 1998; Harvey and Kellogg, 2003; McNulty and Lew, 2005; Anastasia et al., 2012). Although multiple signals may activate this checkpoint, all of its effects depend on phosphorylation and inhibition of Cdk1 (encoded by the yeast CDC28 gene; Lew and Reed, 1995). Most work has focused on a role for this checkpoint in blocking mitotic entry; however, there is evidence that the checkpoint also causes delays during mitosis (Carroll et al., 1998; Barral et al., 1999; Sreenivasan and Kellogg, 1999; Theesfeld et al., 1999; Chiroli et al., 2007). The downstream targets of Cdk1 whose reduced phosphorylation is responsible for checkpoint arrest have not been identified.
Cdk1 activity is required for anaphase onset. Mutants in Cdk1 delay in metaphase, and mutation of two mitotic cyclins, CLB1 and CLB2, causes a permanent arrest before anaphase initiation (Rudner et al., 2000; Rahal and Amon, 2008), suggesting that only a low level of Cdk1 activity is needed for cells to reach metaphase. Cdk1 regulation of anaphase onset is thought to occur by the phosphorylation and activation of the anaphase-promoting complex or cyclosome (APC; Félix et al., 1990; King et al., 1995; Lahav-Baratz et al., 1995; Patra and Dunphy, 1998; Shteinberg et al., 1999; Rudner and Murray, 2000; Kraft et al., 2003).
The APC is a multi-subunit E3 ubiquitin ligase (King et al., 1995; Sudakin et al., 1995; Peters, 2006) that targets Securin (Pds1 in budding yeast) for degradation, relieving its repression of Separase (Esp1), the protease that triggers sister chromatid separation (Cohen-Fix et al., 1996; Funabiki et al., 1996; Ciosk et al., 1998). In mitotic cell cycles two sub-stoichiometric activators, Cdc20 and Cdh1, activate the APC. Cdc20 activates the APC in mitosis and catalyzes Pds1 and mitotic cyclin destruction (Visintin et al., 1997; Yeong et al., 2000). Cdh1 is activated late in mitosis, completes the destruction of APC substrates, and remains active through G1 (Schwab et al., 1997; Visintin et al., 1997; Zachariae et al., 1998). Cdk1 activates APCCdc20 and inhibits APCCdh1 (Zachariae et al., 1998; Jaspersen et al., 1999; Kramer et al., 2000; Rudner and Murray, 2000). This opposing regulation by Cdk1 allows the transfer of ubiquitination activity from Cdc20 to Cdh1 as cells transition from mitosis to G1 (Cross, 2003).
Cdk1 activates vertebrate APC in vitro, and mutation of 12 Cdk1 sites on the TPR subunits Cdc16, Cdc23, and Cdc27 in budding yeast (hereafter called apc-12A) reduces APC phosphorylation in vivo and in vitro, and APC activity in vivo (Lahav-Baratz et al., 1995; Shteinberg et al., 1999; Kramer et al., 2000; Rudner and Murray, 2000; Golan et al., 2002; Kraft et al., 2003). The apc-12A mutations cause a delay in mitosis and weaken the APC–Cdc20 interaction, but have no effect on APCCdh1 activity (Rudner and Murray, 2000).
Recent work has begun to characterize the phosphatases that dephosphorylate Cdk1 substrates, and how their inactivation promotes mitosis and their activation assists entry into G1 (Stegmeier and Amon, 2004; Uhlmann et al., 2011; Mochida and Hunt, 2012). In budding yeast the phosphatase Cdc14 is released from the nucleolus in early anaphase and dephosphorylates many Cdk1 substrates (Visintin et al., 1998; Jaspersen et al., 1999; Bouchoux and Uhlmann, 2011). Although the CDC14 gene is highly conserved, the reversal of mitotic phosphorylation in other eukaryotes depends primarily on other phosphatases such as protein phosphatase 1 (PP1) and protein phosphatase 2A (PP2A; Wurzenberger and Gerlich, 2011). PP2A opposes Cdk1 phosphorylation of Wee1 and Cdc25, and is also believed to be the major phosphatase that antagonizes Cdk1 during mitosis (Kumagai and Dunphy, 1992; Chen et al., 2007; Castilho et al., 2009; Mochida et al., 2009; Harvey et al., 2011; Wicky et al., 2011; Labit et al., 2012).
Similar to swe1Δ, deletion of CDC55, a B-regulatory subunit of PP2A, allows bypass of the morphogenesis checkpoint (Chiroli et al., 2007). cdc55Δ cells also bypass the spindle checkpoint, which monitors bipolar attachment of chromosomes to the mitotic spindle (Minshull et al., 1996; Wang and Burke, 1997). These phenotypes suggest that in cdc55Δ cells a mitotic Cdk1 substrate may be inappropriately activated, resulting in a checkpoint defect.
Previously identified mitotic functions of PP2ACdc55 cannot explain its requirement in preventing anaphase onset during morphogenesis and spindle checkpoint activation. cdc55Δ cells exhibit elevated tyrosine 19 phosphorylation on Cdk1 due to misregulation of Swe1 and/or Mih1 (Minshull et al., 1996; Wang and Burke, 1997; Yang et al., 2000; Pal et al., 2008). Increased phosphotyrosine, however, would strengthen, not bypass, the morphogenesis checkpoint, and changes in inhibitory phosphorylation on Cdk1 do not cause spindle checkpoint defects (Rudner et al., 2000). PP2ACdc55 also inhibits Cdc14 release from the nucleolus after anaphase onset, and early release of Cdc14 has been proposed to cause the cdc55Δ spindle checkpoint defect (Wang and Ng, 2006; Yellman and Burke, 2006). This model, however, does not explain cdc55Δ bypass of the morphogenesis checkpoint, as deleting CDC55 causes checkpoint bypass even in the absence of Cdc14 (Chiroli et al., 2007).
Because Cdk1 activity is needed to initiate the metaphase-to-anaphase transition we wondered if Swe1 inhibition of Cdk1 regulates anaphase onset. Here we show that deletion of SWE1 and MIH1 alters the length of mitosis, and inducing high levels of inhibitory phosphorylation on Cdk1 causes a prolonged metaphase arrest, suggesting that although the morphogenesis checkpoint delays mitotic entry its primary arrest point is in metaphase. During this arrest the APC is dephosphorylated, and the APCCdc20 is inhibited both in vivo and in vitro. We also show that PP2ACdc55 counteracts Cdk1-dependent phosphorylation of the APC, and misregulation of APC phosphorylation in cdc55Δ cells partly explains their checkpoint defects. This data confirms that the APC is a target of a Wee1-dependent checkpoint, and suggests the model that dephosphorylation of the inhibitory tyrosine on Cdk1 during mitosis is needed to accumulate sufficient Cdk1 activity to trigger anaphase.
Results
Swe1 inhibition of Cdk1 occurs during mitosis
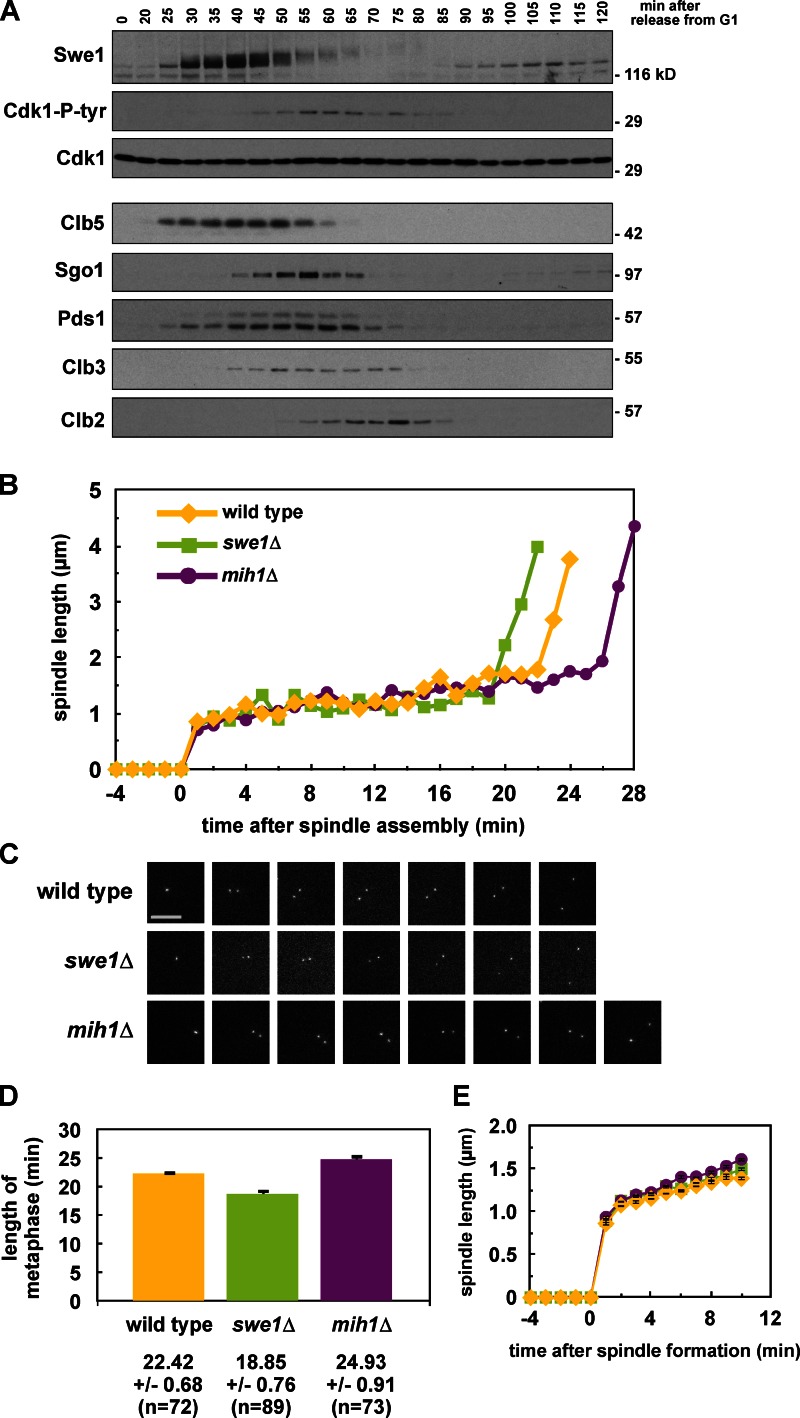
Swe1 has been shown to function in G2, before spindle assembly (Harvey and Kellogg, 2003; Crasta et al., 2006, 2008; Rahal and Amon, 2008), but we wanted to determine whether tyrosine 19 phosphorylation regulates Cdk1 during mitosis. We monitored the sum of Swe1 and Mih1 activity by blotting for tyrosine 19 phosphorylation on Cdk1 during a synchronous cell cycle. Peak Cdk1-Y19 phosphorylation correlates with peak levels of Securin (Pds1), Shugoshin (Sgo1), and the mitotic cyclin Clb3; and just before the peak of the mitotic cyclin Clb2 (Fig. 1 A; see Fig. S1 A for evidence that Sgo1 degradation depends on the APC; Karamysheva et al., 2009). This peak occurs after spindle assembly (Fig. S1, B and C) and begins to drop only as the APC becomes active, suggesting that Swe1 may inhibit Cdk1 until the metaphase-to-anaphase transition. Additionally, the peak levels of Clb2 and Pds1 occur 5 min earlier in swe1Δ relative to wild-type cells, and 5–10 min later in mih1Δ cells despite having no effect on bud emergence, suggesting that both Swe1 and Mih1 regulate anaphase onset (Fig. S1 B).
Figure 1.
Swe1 and Mih1 regulate anaphase onset. (A) Swe1 is present and active during mitosis. Wild-type (ADR4009) cells were arrested in G1 with 100 ng/ml α-factor, and released into the cell cycle (t = 0). 800 ng/ml α-factor was re-added at t = 55 min to arrest cells in the following G1. Samples for immunoblotting were taken at the indicated time points. (B–D) Swe1 inhibits and Mih1 promotes anaphase onset. Wild-type (ADR4009), _swe1_Δ (ADR4015), and _mih1_Δ (ADR4012) cells were arrested in G1 with 25 ng/ml α-factor, washed, and resuspended in synthetic media. After 25 min cells were placed on synthetic media agar pads and imaged using fluorescence microscopy. All strains were imaged on the same day and imaged on three separate occasions. Spindle length was determined by measuring the distance between spindle pole bodies tagged with Spc42-eGFP. Representative spindle measurements for cells progressing through mitosis are shown in B. Spc42-eGFP images for the spindles graphed in B are shown in C. (D) The length of metaphase (average ± SEM) was calculated for wild-type, _swe1_Δ, and _mih1_Δ cells by measuring the time spent between spindle formation and anaphase onset. Each pairwise comparison is significantly different (ANOVA, P < 0.0001, Tukey’s HSD = 2.1878 for P < 0.05). (E) Swe1 and Mih1 do not regulate spindle formation. The spindle lengths (average ± SEM) every minute after spindle formation were calculated from the wild-type, _swe1_Δ, and _mih1_Δ time courses in B.
Swe1 and Mih1 regulate anaphase onset in unperturbed mitosis
To determine if Cdk1-Y19 phosphorylation regulates anaphase onset we monitored the length of time that wild-type, swe1Δ, and mih1Δ cells spend in metaphase—defined here as the time between formation of a short metaphase spindle and rapid spindle elongation at anaphase—by imaging cells with Spc42-GFP–labeled spindle poles (Fig. 1, B and C). The average length of metaphase in wild-type (22.42 min) is significantly longer than in swe1Δ (18.85 min), and shorter than in mih1Δ (24.93 min), suggesting that tyrosine phosphorylation on Cdk1 regulates the timing of anaphase onset (Fig. 1 D).
Consistent with prior reports (Harvey and Kellogg, 2003; Rahal and Amon, 2008; Oikonomou and Cross, 2011), we observe that swe1Δ cells also form metaphase spindles significantly earlier than wild-type cells after being released from G1 (48.55 min compared with 54.79 min; Fig. S1 D), showing that Swe1 regulates both mitotic entry and anaphase onset. The length of the metaphase spindle and its rate of assembly are unchanged in swe1Δ and mih1Δ cells, suggesting that inhibitory phosphorylation on Cdk1 does not affect the mechanics or kinetics of bipolar spindle formation (Fig. 1 E and Fig. 2 C).
Figure 2.
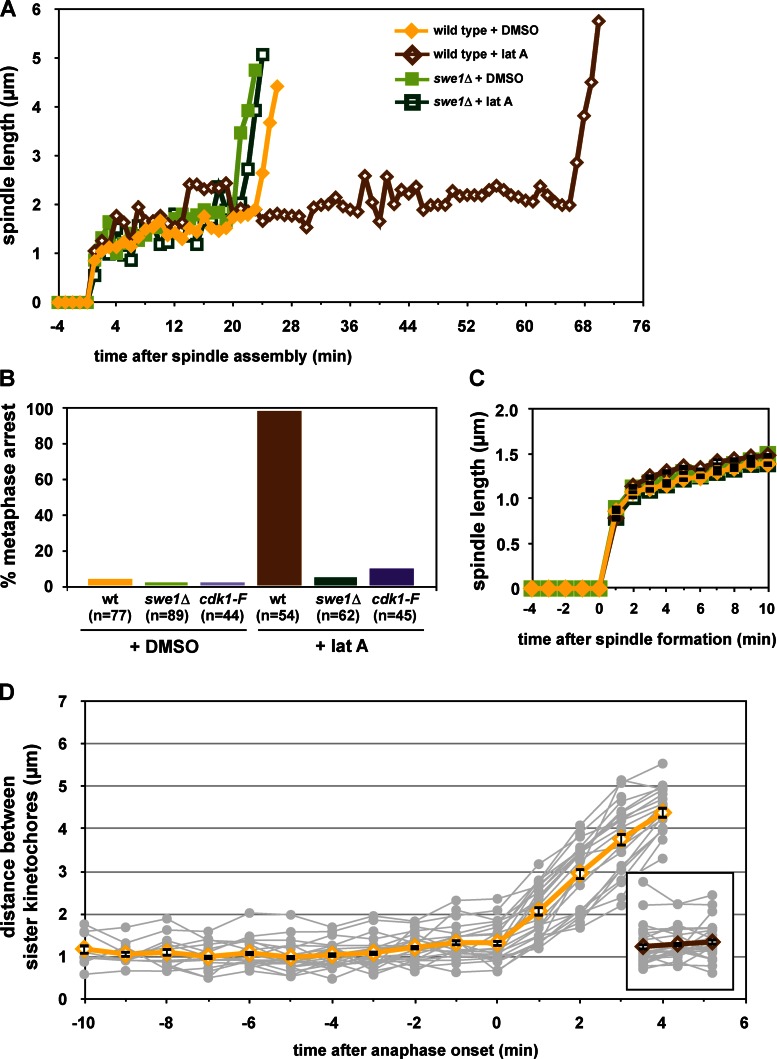
The morphogenesis checkpoint regulates anaphase onset. (A and B) Morphogenesis checkpoint activation delays anaphase onset. Wild-type (ADR4009), _swe1_Δ (ADR4015), and cdk1-Y19F (ADR4313) cells were arrested in G1 with 25 ng/ml α-factor, washed, and resuspended in synthetic media. After 25 min cells were plated on synthetic media agar pads containing 2.5 µM latA or DMSO and imaged as in Fig. 1 B. (A) Spindle measurements for single representative wild-type and _swe1_Δ cells progressing through mitosis. (B) The length of time spent from spindle formation to anaphase onset was calculated for wild-type, _swe1_Δ, and cdk1-Y19F cells. Cells that spent >60 min with short spindles are characterized as “metaphase arrested,” whereas cells that spent <60 min with short spindles are characterized as “unarrested.” Wild-type and _swe1_Δ ± latA were imaged on three separate occasions and cdk1-Y19F ± latA were imaged on two separate occasions. The graph combines all data from all experiments. (C) Morphogenesis checkpoint activation does not perturb mitotic spindle formation. The spindle length (average ± SEM) every minute after spindle formation was calculated for all wild-type, _swe1_Δ, and cdk1-Y19F cells from B. (D) Morphogenesis checkpoint activation does not prevent spindle bipolarity. NDC80-eGFP (ADR5026) cells were grown and imaged as in A. The distance between separated Ndc80-eGFP foci was measured in cells entering anaphase (DMSO) and 150 min after α-factor wash-out (latA, inset). Gray lines are individual cell measurements; yellow and brown lines are average (±SEM) measurements.
We also examined anaphase spindle elongation in swe1Δ and mih1Δ cells. Other than a small but significant increase in the rate of slow anaphase movement in mih1Δ cells, we see little difference between swe1Δ, mih1Δ, and wild-type cells (Fig. S2).
The morphogenesis checkpoint arrests cells at metaphase
Inhibitory phosphorylation on Cdk1 is the target of the morphogenesis checkpoint (Lew and Reed, 1995; Harvey and Kellogg, 2003), so we investigated if this checkpoint arrests cells in metaphase. Past work has shown that this checkpoint slows the cell cycle before nuclear division (Lew and Reed, 1995; McMillan et al., 1998), but whether the checkpoint functions before and/or after spindle assembly has been debated (Lim et al., 1996; Harvey and Kellogg, 2003; Crasta et al., 2006, 2008; Chiroli et al., 2007).
Using SPC42-eGFP we observed that checkpoint activation with the actin-depolymerizing drug latrunculin A (latA) causes more than 95% of wild-type cells to arrest for greater than 60 min with short metaphase spindles (Fig. 2, A and B). swe1Δ and cdk1-Y19F cells completely bypass this arrest (Fig. 2 B).
As in unperturbed swe1Δ and mih1Δ cells, we see no defect in the assembly of a bipolar spindle when the morphogenesis checkpoint is activated (Fig. 1 E and Fig. 2 C). To confirm that these cells are indeed arresting at metaphase we monitored kinetochore biorientation using a fluorescently tagged component of the kinetochore, NDC80-GFP. Ndc80-GFP foci are on average 1.13 ± 0.08 µm apart in control cells before anaphase onset, and rapidly separate at the metaphase-to-anaphase transition (Fig. 2 D). Ndc80-GFP foci are similarly separated in cells that are arrested by the morphogenesis checkpoint (Fig. 2 D, inset).
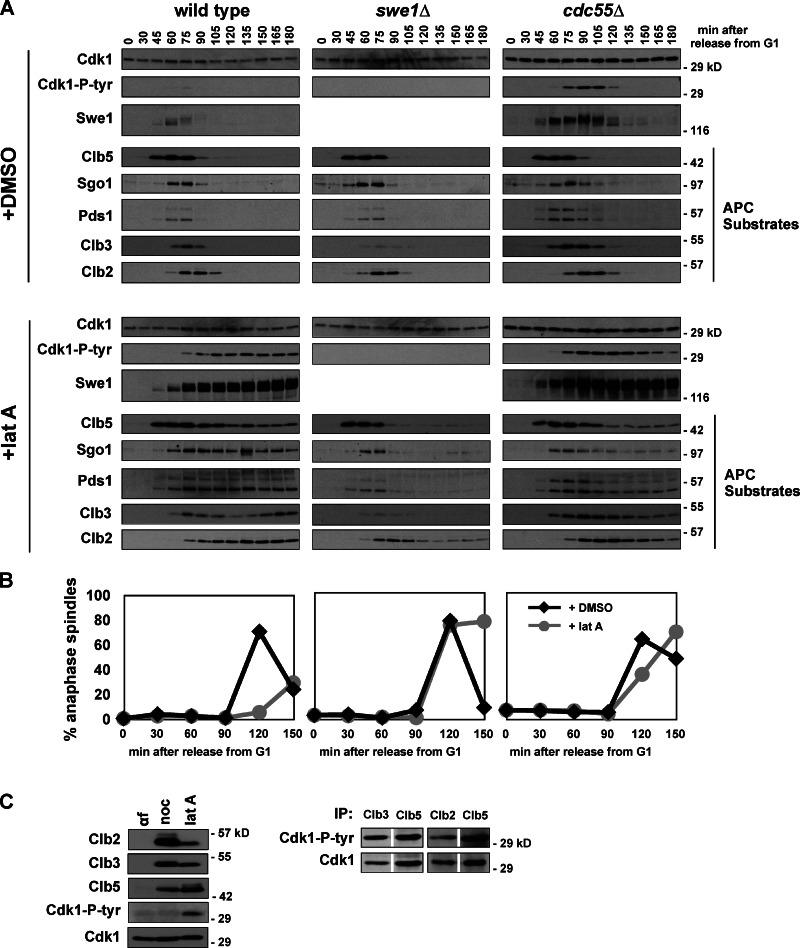
During morphogenesis checkpoint arrest Swe1 protein is stabilized and phosphorylated, tyrosine phosphorylation accumulates on Cdk1, and APC substrates are stabilized (Fig. 3 A). swe1Δ and cdk1-Y19F cells treated with latA accumulate and degrade APC substrates with similar kinetics as untreated wild-type, swe1Δ, and cdk1-Y19F cells (Fig. 3 A and Fig. S3 A). As in our measurements of live cells (Fig. 2, A and B), wild-type cells treated with latA delay anaphase by at least 60 min, while the kinetics of anaphase onset is identical in treated or untreated swe1Δ cells (Fig. 3 B). Deletion of CDC55 has also been reported to bypass the morphogenesis checkpoint (Chiroli et al., 2007). Here, cdc55Δ cells treated with latA also initiate anaphase with similar kinetics as untreated cells, but in these cells inhibitory phosphorylation on Cdk1 persists and APC substrates are degraded slowly (Fig. 3, A and B). The elevated levels of tyrosine-phosphorylated Cdk1 and Swe1 in cdc55Δ cells has been reported previously (Minshull et al., 1996; Pal et al., 2008; Harvey et al., 2011) and is due to PP2ACdc55 activation of Mih1 and inhibition of Swe1. The phenotype of cdc55Δ will be discussed in greater detail below.
Figure 3.
The morphogenesis checkpoint stabilizes APC substrates. (A and B) Activation of the morphogenesis checkpoint stabilizes APC substrates. Wild-type (ADR4009), _swe1_Δ (ADR4015), and _cdc55_Δ (ADR4738) cells were arrested in G1 with 100 ng/ml α-factor, washed, and released from the arrest (t = 0). Cells were treated with 2.5 µM latA or DMSO at t = 25 and 800 ng/ml α-factor was re-added at t = 65 to arrest cells in the following G1. Samples for immunoblotting (A) and fluorescence microscopy (B) were taken at the indicated time points. Anaphase onset was determined by scoring the distance between Spc42-eGFP–labeled SPBs in fixed cells. At least 200 cells were scored for each time point. (C) LatA-activated Swe1 does not preferentially phosphorylate a particular Cdk1–cyclin complex. Wild-type (ADR4006) cells were arrested with 100 ng/ml α-factor, 10 µg/ml nocodazole, or 2.5 µM latA. Cells were harvested for immunoblotting (left), or, for the latA-arrested cells, immunoprecipitation. Cdk1–Clb2, –Clb3 or –Clb5 complexes were immunoprecipitated, normalized for Cdk1 levels, and blotted for Cdk1 and Cdk1–P-tyr to compare the relative stoichiometry of tyrosine-phosphorylated Cdk1. Two independent experiments are shown, one that compares Clb3 and Clb5 immunoprecipitates (left) and one that compares Clb2 and Clb5 immunoprecipitates.
We wondered whether morphogenesis checkpoint activation causes preferential phosphorylation of specific Cdk1–Clb complexes. We therefore immunoprecipitated different Clb–Cdk1 complexes during a morphogenesis checkpoint arrest and found that the ratio of tyrosine-phosphorylated Cdk1 to total Cdk1 is nearly identical for each Clb complex. Consistent with a previous report, this shows that in vivo Swe1 has no obvious preference for a specific Cdk1–Clb complex during checkpoint arrest (Fig. 3 C; Keaton et al., 2007).
Swe1 phosphorylation of Cdk1 inhibits the APC in vivo
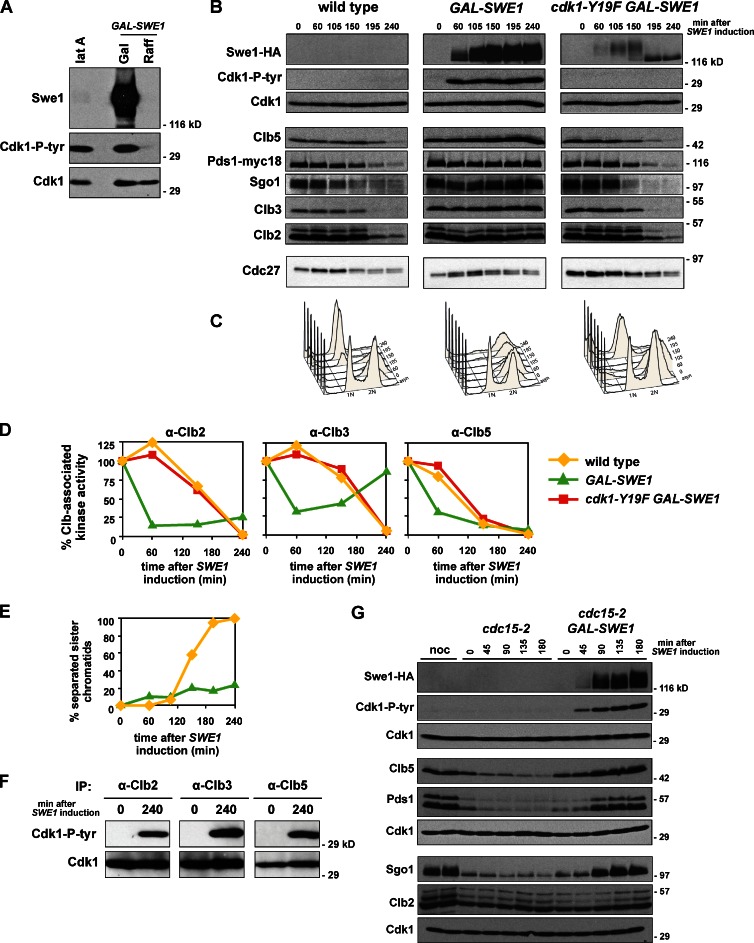
The stabilization of APC substrates during morphogenesis checkpoint arrest suggests that the APC may be a target of this checkpoint. To avoid the expense and variability of latA, and evaluate the effects of inhibitory phosphorylation on Cdk1 in additional conditions, we tested if overexpression of Swe1 might serve as a valid model for checkpoint arrest (Booher et al., 1993; Crasta et al., 2006). After overexpression of Swe1 (GAL-SWE1), inhibitory phosphorylation on Cdk1 accumulates to levels nearly identical to that seen during a latA arrest despite an enormous difference in the levels of Swe1 protein (Fig. 4 A). Because the effects of latA depend fully on tyrosine 19 on Cdk1 (Fig. S3 A), we further characterized the effects of Swe1 overexpression.
Figure 4.
Overexpression of Swe1 blocks cells in metaphase. (A) Swe1 overexpression and latA induce similar levels of tyrosine phosphorylation on Cdk1. Wild-type (ADR4009) cells were treated with latA and GAL-SWE1 (ADR4289) cells were grown in YEP + 2% raffinose (Raff) and induced by addition of 2% galactose (Gal). Cells were harvested and immunoblotted for Swe1, Cdk1, and Cdk1-P-tyr. (B and C) Overexpression of Swe1 in mitosis stabilizes APC substrates and arrests cells with 2N DNA content. Wild-type (ADR2617), GAL-SWE1 (ADR3871), and GAL-SWE1 cdk1-Y19F (ADR4228) cells were grown in YEP + 2% raffinose, arrested with 10 µg/ml nocodazole, and induced with 2% galactose (t = 0) for 1 h. Nocodazole was washed out, and cells released into YEP + 2% galactose. 25 ng/ml α-factor was added (t = 150) to arrest cells in the following G1. Samples were taken for immunoblotting (A) and flow cytometry (B) at the indicated time points. (D) Swe1 inhibits Clb-associated kinase activity. Cells were grown as in A, and Cdk1–Clb2, –Clb3 or –Clb5 complexes were immunoprecipitated with anti-Clb antibodies and their histone H1 kinase activity was measured to assess Cdk1 inhibition. (E) Overexpression of Swe1 impairs sister chromatid separation. Wild-type (ADR1393) and GAL-SWE1 (ADR1395) strains were grown as in A, except 1 mM CuSO4 was added at t = −30 to induce expression of GFP-lacI, and the separation of sister lacO arrays at TRP1 (∼12 kb from CENIV) was visualized by fluorescence microscopy. The graphs in D and E are representative of one of three repeats. (F) Overexpressed Swe1 does not preferentially phosphorylate a particular Cdk1–cyclin complex. GAL-SWE1 cells (ADR3871) were grown as in A, and Cdk1–Clb2, –Clb3, or –Clb5 complexes were immunoprecipitated at the indicated time points and analyzed by immunoblotting with anti-Cdk1 and anti–Cdk1-P-tyr antibodies. (G) Overexpression of Swe1 in anaphase restabilizes APCCdc20 substrates. cdc15-2 (ADR4252) and cdc15-2 GAL-SWE1 (ADR4245) cells were grown in YEP + 2% raffinose and arrested in anaphase by temperature shift to 35°C, followed by induction with 2% galactose (t = 0). Parallel cultures were arrested in 10 µg/ml nocodazole (noc) at 25°C to illustrate peak levels of APC substrates. Cdk1 was used as a loading control for three independent membranes loaded with identical samples.
To examine the effect of Swe1 overexpression during mitosis we induced GAL-SWE1 during a nocodazole arrest, washed away the nocodazole allowing cell cycle progression, and monitored cell cycle progression by immunoblot. Control cells release from the nocodazole arrest and reenter G1 after 2 h (Fig. 4, B and C). Similar to latA treatment (Fig. 3 A), when Swe1 is induced to high levels, inhibitory phosphorylation accumulates on Cdk1, the APC substrates Clb2, Clb3, Clb5, Pds1, and Sgo1 remain stable, and DNA content remains at 2N. These cells rebuild short metaphase spindles at a slightly slower rate than wild-type cells (Fig. S4 A), but these spindles do not elongate and sister chromatid separation is blocked (Fig. 4 E), indicating that these cells remain arrested in metaphase. This arrest is not caused by activation of the spindle or DNA damage checkpoints as mad2Δ and mec1Δ cells, which are defective in these checkpoints, arrest in mitosis when Swe1 is overexpressed (Fig. S4, B and C). cdk1-Y19F cells overexpressing Swe1 release normally from the mitotic arrest and degrade APC substrates as they enter G1 (Fig. 4, B and C). Additionally, overexpression of a kinase-dead version of Swe1, swe1-N584A, also does not cause mitotic arrest, ruling out the possibility that high levels of Swe1, whose degradation depends on the APC (Thornton and Toczyski, 2003), competitively inhibits the APC (Fig. S4 D).
As expected, Cdk1 activity falls as tyrosine phosphorylation accumulates on Cdk1. After 60 min of Swe1 induction, the Clb2-, Clb3- and Clb5-associated histone H1 kinase activity falls to 60–80% of its starting activity (Fig. 4 D, green lines), compared with wild-type cells whose Clb-associated activity falls to undetectable levels as they reenter G1 (yellow lines). When these precipitates are probed for tyrosine-phosphorylated Cdk1 all three cyclins are associated with phosphorylated Cdk1, revealing that as during latA treatment (Fig. 3 C), Swe1 does not preferentially target a particular Cdk1–Clb complex (Fig. 4 F).
Expression of Swe1 during anaphase, after the APCCdc20 has been activated, is also sufficient to turn off the APCCdc20. When Swe1 is overexpressed in anaphase-arrested cdc15-2 cells, Clb5, Pds1, and Sgo1 are restabilized, accumulating to levels as high as seen during a nocodazole arrest (Fig. 4 G).
Swe1 phosphorylation of Cdk1 inhibits APC phosphorylation and activity
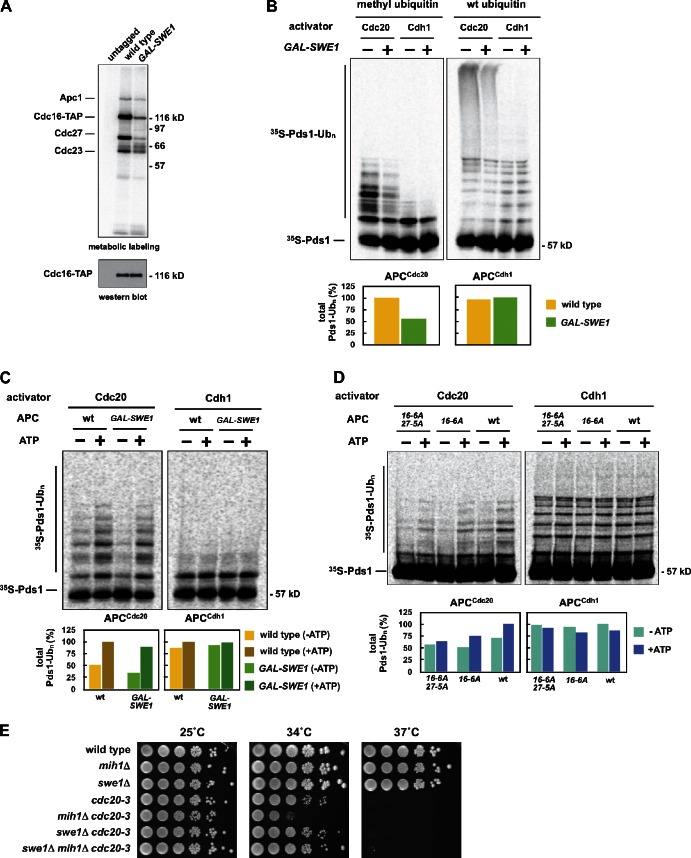
Cdk1-dependent phosphorylation of the APC activates the APCCdc20 in vivo (Rudner and Murray, 2000), so we investigated if high levels of Swe1 reduce APC phosphorylation. To observe the phosphorylation status of the entire APC we monitored phosphorylation of purified APC after metabolic labeling of cells with inorganic 32P-labeled orthophosphate. Induction of high levels of Swe1 causes a quantitative loss of phosphorylation on five APC subunits: Apc1, Cdc16, Cdc27, Cdc23, and Apc9 (Fig. 5 A; Fig. S5 A). The rapid dephosphorylation of Cdc27 is also observed by immunoblotting (Fig. 4 B).
Figure 5.
Overexpression of Swe1 in mitosis inhibits APC phosphorylation in vivo and APCCdc20 activity in vitro. (A) Wild-type (ADR22), CDC16-TAP (ADR3877), and GAL-SWE1 CDC16-TAP (ADR3858) cells were grown in YEP + 2% raffinose, arrested in mitosis with 10 µg/ml nocodazole, and induced with 2% galactose for 90 min. Cells were then washed in medium lacking phosphate, and grown for 45 min in the presence of [32P]orthophosphate. The APC was purified, run on a polyacrylamide gel, and exposed to a phosphorimager screen or immunoblotted. (B) Overexpression of Swe1 inhibits ubiquitination of Pds1 in vitro. swe1Δ CDC16-TAP (ADR3877) and GAL-SWE1 CDC16-TAP (ADR3859) cells were grown in YEP + 2% raffinose, arrested in mitosis with 30 µg/ml benomyl, and induced with 2% galactose. The APC was purified and its activity assayed in an in vitro ubiquitination assay. Reactions were run on a polyacrylamide gel and exposed to a phosphorimager screen. The average activity (±SEM) of three independent experiments is plotted below a representative assay. APCCdc20 activity was quantified from assays containing methylated ubiquitin (Ub) and APCCdh1 activity was quantified from assays containing wild-type ubiquitin. (C) Cdk1–Clb2 kinase treatment reactivates Swe1-inhibited APC. APC was purified from swe1Δ CDC16-TAP and GAL-SWE1 CDC16-TAP cells as described in B. The APC was incubated with purified Cdk1–Clb2 complexes ± ATP before cleavage from IgG-coupled magnetic beads. The activity of the purified APC was then measured as in B. Quantification of a representative assay is shown below. Parallel samples were treated with γ-[32P]ATP to confirm efficient phosphorylation (Fig. S5 H). (D) CDC16-TAP (ADR3089), cdc16-6A-TAP (ADR3822), and cdc16-6A-TAP cdc27-5A (ADR3891) cells were arrested in mitosis with 30 µg/ml benomyl. Purified APC (Fig. S5 I) was phosphorylated by purified Cdk1–Clb2 complexes as in C and its activity measured. Quantification of a representative assay is shown below. Parallel samples were treated with γ-[32P]ATP to confirm efficient phosphorylation (Fig. S5 J). The experiments shown in B, C, and D are representative of one of two repeats. (E) mih1Δ lowers the maximum permissive temperature of _cdc20_-3. Eightfold serial dilutions of wild-type (ADR22), mih1Δ (ADR3178), swe1Δ (ADR3170), cdc20-3 (ADR3161), mih1Δ cdc20-3 (ADR3149), swe1Δ cdc20-3 (ADR3155), and swe1Δ mih1Δ cdc20-3 (ADR3141) cells were spotted onto YEPD plates and grown at the indicated temperatures.
We directly assayed APC activity from cells overexpressing Swe1 to test if Swe1-dependent inhibition of APC phosphorylation lowers APCCdc20 activity in vitro. APC purified from cells overexpressing Swe1 has reduced APCCdc20 activity against Pds1 (Fig. 5 B; Fig. S5, B and C) and Clb5 (Fig. S5 D). Each ubiquitinated species is reduced approximately twofold (Fig. 5 B), with little difference between individual ubiquitinated species (Fig. S5 E).
When the Swe1-inhibited APC is assayed using the activator Cdh1, there is no change in APCCdh1 activity relative to control APC (Fig. 5 B; Fig. S5 C). This is consistent with past data that suggest APCCdh1 activity is unaffected by the phosphorylation of core APC subunits (Zachariae et al., 1998; Rudner and Murray, 2000).
If the loss of APCCdc20 activity is due solely to a reduction in Cdk1-dependent phosphorylation of the APC, we reasoned we could reactivate the APC by phosphorylating it in vitro with purified Cdk1–Clb2 complexes (Fig. S5 F). After phosphorylation by Cdk1–Clb2, the Swe1-inhibited APC is phosphorylated primarily on Cdc16 and Cdc27 (Fig. S5, G and H), and this phosphorylation greatly increases APCCdc20 activity (Fig. 5 C, +ATP). Mutating Cdk1 sites on Cdc16 (cdc16-6A) partially blocks reactivation, whereas mutating sites on both Cdc16 and Cdc27 (cdc16-6A cdc27-5A) further prevents this reactivation (Fig. 5 D).
Reinforcing this in vitro data we see a strong genetic interaction between mih1Δ and mutants in the APC activators, cdh1Δ and cdc20-3. mih1Δ and cdh1Δ are synthetically lethal (Fig. S5 K), and mih1Δ and cdc20-3 are synthetically sick, with a lower maximum permissive temperature (Fig. 5 E). Both of these interactions are fully rescued by swe1Δ.
PP2ACdc55 regulates APC phosphorylation and activity
PP2A is a heterotrimeric complex composed of a catalytic subunit, an A-regulatory or scaffolding subunit, and one of two B-regulatory subunits (CDC55 or RTS1; Healy et al., 1991; Lin and Arndt, 1995; Stark, 1996; Shu et al., 1997; Wurzenberger and Gerlich, 2011). The B-regulatory subunits provide specificity to the two forms of the phosphatase, PP2ACdc55 and PP2ARts1. Cells deleted for CDC55 also bypass the morphogenesis checkpoint (Chiroli et al., 2007). As mentioned above, cdc55Δ and swe1Δ cells initiate anaphase and reenter S phase with similar kinetics after treatment with latA (Fig. 3 B; Fig. S3 B). However, in cdc55Δ cells Swe1 is stabilized, tyrosine phosphorylation on Cdk1 remains high, and APC substrates, including Pds1, are degraded very slowly (Fig. 3 A). The marked difference between swe1Δ and cdc55Δ suggests that cdc55Δ may bypass the checkpoint through a different mechanism than swe1Δ, and we hypothesized its checkpoint defect may be caused by counteracting Cdk1 activation of the APC.
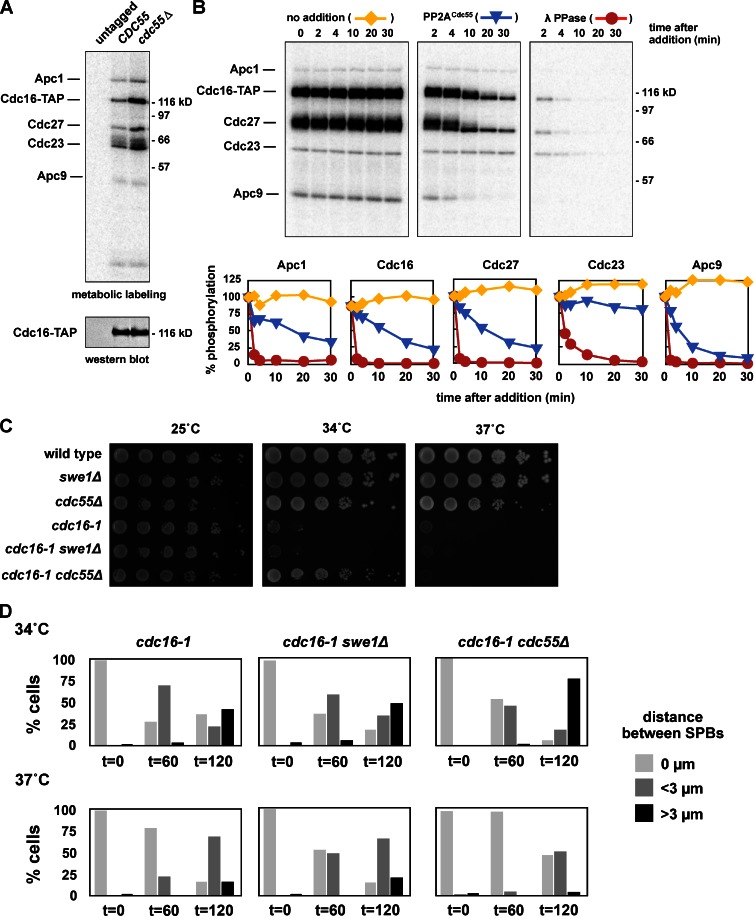
We therefore examined if APC phosphorylation is altered in cdc55Δ cells using metabolic labeling. Deletion of CDC55 causes a dramatic increase in the phosphorylation of Cdc16, Cdc27, and Cdc23, as well as reproducible changes in the phosphorylation of Apc1, Apc4, and Apc5 (Fig. 6 A). Purified PP2ACdc55 can also dephosphorylate purified APC that has been phosphorylated in vitro by Cdk1–Clb2 (Fig. 6 B; Fig. S5 L). The dephosphorylation of Apc1, Cdc16, Cdc27, and Apc9 occur at similar rates (blue lines), while Cdc23 is not dephosphorylated. Cdc23 is also poorly phosphorylated by Cdk1–Clb2, suggesting that it may be inaccessible in these in vitro reactions. The dephosphorylation of the APC in vitro by PP2ACdc55 is blocked by the addition of 2 µM okadaic acid, confirming that it is performed by a PP2A complex and not a contaminating phosphatase (Fig. S5 M).
Figure 6.
PP2ACdc55 regulates APC phosphorylation and activity in vivo. (A) The APC is hyper-phosphorylated in vivo in cdc55Δ cells. cdc15-2 (ADR1168), cdc15-2 CDC16-TAP (ADR5137), and cdc15-2 cdc55Δ CDC16-TAP (ADR4060) cells were arrested in G1 with 100 ng/ml α-factor, washed, and released at 37°C. After 2.5 h cells were washed in medium lacking phosphate, and grown for 45 min in the presence of [32P]orthophosphate at 37°C. The APC was purified, run on a polyacrylamide gel, and exposed to a phosphorimager screen or immunoblotted. (B) PP2ACdc55 dephosphorylates the APC in vitro. The APC was purified from CDC16-TAP (ADR3089) cells and phosphorylated with purified Cdk1/Clb2 and γ-[32P]ATP while immobilized on IgG-coupled magnetic beads. The beads were washed and then incubated for the indicated times at room temperature with no addition (yellow lines), TAP-purified PP2ACdc55 (blue lines), or lambda phosphatase (red lines). The three reactions share a t = 0 sample that was taken before additions. The dephosphorylation of individual APC subunits was quantified and graphed relative to t = 0. The experiment shown is representative of one of three repeats. (C) cdc55Δ, but not swe1Δ, suppresses the temperature sensitivity of an APC mutant. Eightfold serial dilutions of wild-type (ADR4009), swe1Δ (ADR4015), cdc55Δ (ADR4738), cdc16-1 (ADR4979), cdc16-1 swe1Δ (ADR4984), and cdc16-1 cdc55Δ (ADR5020) cells were spotted onto plates and grown at the indicated temperatures. (D) cdc55Δ, but not swe1Δ, allows premature anaphase spindle elongation in cells with an impaired APC, but does not bypass APC function. cdc16-1 (ADR4979), _cdc16-1 swe1_Δ (ADR4984), and _cdc16-1 cdc55_Δ (ADR5020) cells were arrested in G1 with 100 ng/ml α-factor. 1 h before α-factor wash-out, cells were shifted to 34 or 37°C to inactivate Cdc16. Cells were then washed and released from G1 arrest (t = 0) at either 34 or 37°C. Samples for fluorescence microscopy were taken at the indicated time points and the distance between Spc42-eGFP–labeled SPBs was measured in at least 100 cells. Anaphase spindles were defined as spindles >3 µm in length.
If PP2ACdc55 inhibits the APC by dephosphorylation, we wondered if cdc55Δ cells may be able to suppress mutations in APC subunits. The temperature-sensitive mutants cdc16-1 and cdc23-1 are both partially suppressed by cdc55Δ but not by swe1Δ (Fig. 6 C; Fig. S3 D). When cdc55Δ cdc16-1 cells are grown at a semi-restrictive temperature of 34°C a large fraction of cells proceed into anaphase (Fig. 6 D), while cdc16-1 and swe1Δ cdc16-1 cells remain arrested with short metaphase spindles. However, cdc55Δ does not bypass APC function, as cdc55Δ cdc16-1 cells grown at the fully restrictive temperature of 37°C behave indistinguishably from swe1Δ cdc16-1 and cdc16-1 cells. A similar result has been reported in Drosophila, where a mutation in Cdc27 (mks) is suppressed by a mutation of a PP2A B-subunit (twins/aar; Deak et al., 2003).
Blocking APC phosphorylation suppresses checkpoint defects
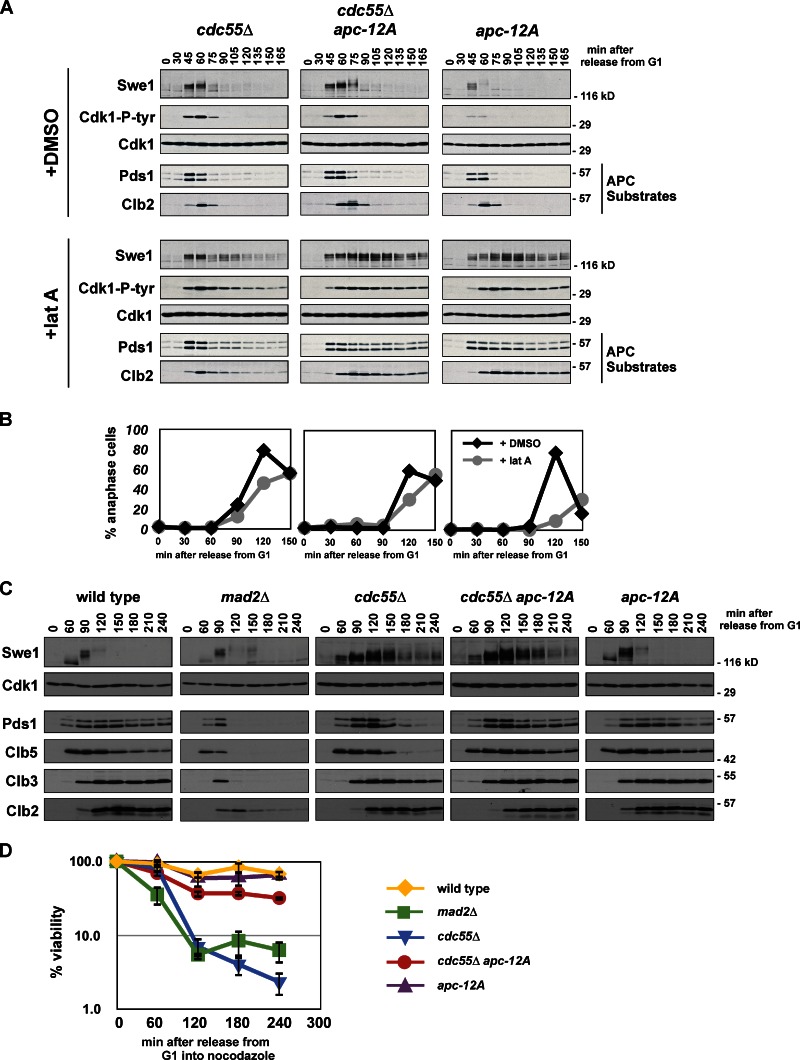
Because cdc55Δ cells increase APC phosphorylation, we tested if the morphogenesis checkpoint defect of cdc55Δ is suppressed by mutating the known Cdk1 phosphorylation sites on the APC. Combining apc-12A with cdc55Δ completely rescues the slow degradation of Pds1 and Clb2 seen in cdc55Δ cells (Fig. 7 A). Swe1 and tyrosine phosphorylation on Cdk1 are also stabilized in cdc55Δ apc-12A. Consistent with APC substrate stabilization, latA-treated cdc55Δ apc-12A cells only slowly reinitiate DNA replication (Fig. S3 E).
Figure 7.
PP2ACdc55-dependent dephosphorylation of Cdk1 sites on the APC inhibits anaphase onset. (A and B) Mutation of 12 Cdk1 phosphorylation sites on the APC in morphogenesis checkpoint-activated cells blocks _cdc55_Δ-dependent activation of the APC. _cdc55_Δ (ADR4738), _cdc55_Δ apc-12A (ADR4902), and apc-12A (ADR4973) cells were grown and treated with latA as in Fig. 3 A and samples for immunoblotting (A) and fluorescence microscopy (B) were taken at the indicated time points. More than 200 cells were scored for each data point in B. The data shown are from one representative experiment out of two repeats. (C and D) Mutation of 12 Cdk1 phosphorylation sites on the APC in spindle checkpoint-activated cells blocks _cdc55Δ_-dependent activation of the APC and rapid loss of viability. Wild-type (ADR4009), _mad2_Δ (ADR4099), _cdc55_Δ (ADR4738), _cdc55_Δ apc-12A (ADR4902), and apc-12A (ADR4973) cells were arrested in G1 with 100 ng/ml α-factor, released from G1, 10 µg/ml nocodazole was added at t = 35, and 800 ng/ml α-factor was re-added at t = 65 to arrest cells in the following G1. Samples for immunoblotting (C) and viability assays (D) were taken at the indicated time points. Viability (average ± SEM) was calculated from three independent experiments relative to viability at t = 0.
The spindle checkpoint also acts through APC inhibition (Hwang et al., 1998; Kim et al., 1998), so we wondered if cdc55Δ bypasses both the morphogenesis and spindle checkpoints through increased APC phosphorylation. Similar to the effect of swe1Δ on the morphogenesis checkpoint (Fig. 3 A), mad2Δ bypasses the spindle checkpoint and activates APC-dependent proteolysis with normal timing (Fig. 7 C; Minshull et al., 1996). Like the morphogenesis checkpoint and in contrast to mad2Δ cells, cdc55Δ cells bypass the spindle checkpoint despite slow activation of the APC, and this slow activation is completely blocked in cdc55Δ apc-12A cells (Fig. 7 C). The rapid loss of viability that accompanies cdc55Δ checkpoint bypass (Fig. 7 D, blue line) is also significantly suppressed in cdc55Δ apc-12A cells (red line).
Discussion
The morphogenesis checkpoint arrests cells at metaphase
We have shown that chronic tyrosine phosphorylation of Cdk1 triggered by the morphogenesis checkpoint arrests cells at metaphase with short metaphase spindles that have made bipolar attachments to sister chromatids (Fig. 2 and Fig. 8). During this arrest Cdk1–cyclin complexes are inhibited, showing that metaphase can occur during periods of relatively low Cdk1 activity. Additionally, in unperturbed cell cycles swe1Δ mutants shorten and mih1Δ mutants lengthen metaphase, suggesting that the removal of inhibitory phosphorylation on Cdk1 is an important trigger of anaphase onset in every cell cycle.
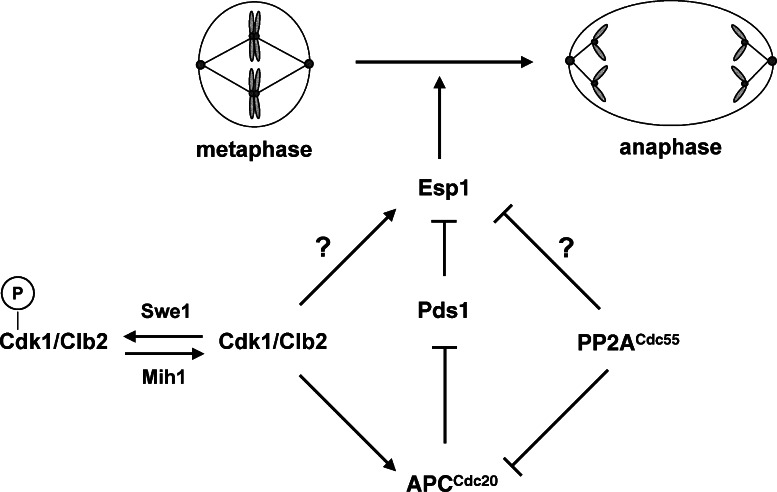
Figure 8.
Cdk1 activates and PP2ACdc55 inhibits anaphase onset. Our data support a model in which Cdk1 and PP2ACdc55 regulate phosphorylation of the APC and Esp1. In this model, Cdk1 regulates Esp1 directly by phosphorylation, and indirectly through modulation of APCCdc20 activity and Pds1 degradation. Dephosphorylation of tyrosine 19 on Cdk1 by the Mih1 (Cdc25) phosphatase allows normal anaphase onset by activating Cdk1, whereas Swe1 (Wee1) phosphorylation slows anaphase onset by phosphorylating and inhibiting Cdk1. During a morphogenesis checkpoint arrest, Swe1 is inhibited and Mih1 is activated, thereby blocking anaphase. Deletion of CDC55 allows premature activation of the APC and Esp1, which bypasses the morphogenesis checkpoint arrest despite continued signaling through inhibitory phosphorylation of Cdk1. Premature activation of the APC and Esp1 in cdc55Δ mutants could also explain cdc55Δ bypass of the spindle and DNA damage checkpoints, which block anaphase by inhibiting Cdc20 activity and Pds1 destruction, respectively (Minshull et al., 1996; Wang and Burke, 1997; Hwang et al., 1998; Kim et al., 1998; Sanchez et al., 1999; Tang and Wang, 2006).
The literature on the morphogenesis checkpoint has been equivocal on where the checkpoint arrests the cell cycle. Although some work has pointed to a function in mitosis (Carroll et al., 1998; Barral et al., 1999; Sreenivasan and Kellogg, 1999; Theesfeld et al., 1999; Chiroli et al., 2007), most have focused on the G2/M transition, presumably because of past work on fission yeast and vertebrate Wee1 and Cdc25 (Lew and Reed, 1995; McMillan et al., 1998; Kellogg, 2003). An additional difficulty has been historical and semantic: the description of many budding yeast mutants that arrest with short mitotic spindles and bipolar attachments, such as APC mutants, have been described as having a G2/M arrest despite the evidence that these cells arrest in metaphase (Hartwell et al., 1973). Here we define G2 and mitosis in universal terms: G2 is the time before spindle assembly and kinetochore capture by the mitotic spindle, and mitosis begins when spindle poles separate and the spindle assembles.
swe1Δ cells initiate mitotic entry 5–10 min earlier than wild-type cells after release from a G1 arrest (Fig. S1 D; Harvey and Kellogg, 2003; Rahal and Amon, 2008; Oikonomou and Cross, 2011), suggesting that Swe1 also functions in G2. One report has suggested that the delay in spindle assembly is due to the destabilization of the microtubule motors Cin8 and Kip1 (Crasta et al., 2006). Our data and others (Chee and Haase, 2010) do not agree with this interpretation, as Cin8 and Kip1 levels peak before Cdk1 is dephosphorylated, showing that Cin8 and Kip1 abundance is not regulated by Cdk1 activity (Fig. S1 E). Additionally, our analysis of Cdk1 phosphotyrosine levels through the cell cycle differs dramatically from that published in the above report (Fig. 1 A). These authors based many of their conclusions about the role of inhibited Cdk1 on the phenotype of a cdk1-Y19E mutant. Although the Y19E substitution may mimic tyrosine phosphorylation and decrease Cdk1 activity (Lim et al., 1996), it is likely that this mutation has additional defects as has been shown in the cdk1-T18V, Y19F and cdk1-Y19F mutants (Rudner et al., 2000).
Stepwise activation of Cdk1 triggers anaphase
Numerous studies have suggested that inhibitory phosphorylation regulates Cdk1 activation in a stepwise manner in both vertebrates and yeast (Stern and Nurse, 1996; Pomerening et al., 2003, 2005; Deibler and Kirschner, 2010; Harvey et al., 2011). These studies have primarily focused on how Cdk1 dephosphorylation by Cdc25 allows cells to transition from G2 to mitosis. Other work has suggested a second step in Cdk1 activation during mitosis that regulates anaphase onset (Rudner et al., 2000; Lindqvist et al., 2007; Rahal and Amon, 2008). Mutation of two yeast mitotic cyclins, CLB1 and CLB2, arrests cells in metaphase, showing that a low level of Cdk1 activity can drive spindle assembly, but additional activity is needed to trigger anaphase (Rahal and Amon, 2008). Our work suggests that Cdk1 dephosphorylation may cause the increase in Cdk1 activity that triggers anaphase onset, in part by activation of the APC (Fig. 8). In fission yeast and vertebrates high levels of tyrosine-phosphorylated Cdk1 arrests cells in G2, and mitotic entry requires a large change in Cdk1 activity. Budding yeast may use different Cdk1 activity thresholds, rendering mitotic onset relatively impervious to Swe1-dependent Cdk1 inhibition, and instead use this inhibition to control anaphase onset.
After overexpression of Swe1, residual Cdk1–cyclin activity remains in cells (Fig. 4 D). Because tyrosine phosphorylation of Cdk1 has been shown to completely inhibit Cdk1 activity (Parker et al., 1992), we believe only a fraction of Cdk1 is targeted by Swe1, and this pool of Cdk1 is equally phosphorylated when the morphogenesis checkpoint is activated, despite far lower levels of Swe1 (Fig. 4 A). How cells restrict Swe1 activity even when it is overexpressed is unknown, but may reflect limited import of Swe1 into the nucleus where its function is needed to activate the morphogenesis checkpoint (Keaton et al., 2008).
If dephosphorylation of Cdk1 is a primary trigger for anaphase onset, why does deletion of MIH1, unlike mutants of cdc25+ in fission yeast, have only a mild phenotype (Nurse et al., 1976; Russell et al., 1989)? Although Mih1 is the only Cdc25 homologue in budding yeast at least one other phosphatase dephosphorylates Cdk1, as Cdk1 is dephosphorylated in mih1Δ cells as Clb5 and Pds1 destruction initiates (Fig. S1 B; Pal et al., 2008). PP2ACdc55 has been proposed to be a second Cdk1 phosphatase because mih1Δ cdc55Δ cells are inviable (Pal et al., 2008), but this synthetic interaction could be explained instead by PP2ACdc55 regulation of Swe1 (Harvey et al., 2011).
Do Wee1 and Cdc25 in other organisms also influence anaphase onset? One study in unperturbed human cells correlated mitotic progression with Cdk1 tyrosine phosphorylation and showed that spindle assembly begins before any appreciable change in Cdk1 tyrosine phosphorylation (Lindqvist et al., 2007). In addition, Cdk1 dephosphorylation is completed just before maximal APC3 (the human Cdc27 homologue) phosphorylation and cyclin B proteolysis. This work suggests that as in yeast (Rudner et al., 2000; Rahal and Amon, 2008), vertebrates have a Cdk1 activity threshold that is required to initiate anaphase, and the complete dephosphorylation of Cdk1 during mitosis may assist in achieving this threshold.
The APC is a target of the morphogenesis checkpoint
Swe1-dependent inhibition of Cdk1 reduces APC activity in vivo and in vitro and this inhibition is accompanied by dephosphorylation of the APC, suggesting the APC is a target of the morphogenesis checkpoint (Fig. 5 and Fig. 8). To prove that APC phosphorylation is a target of the checkpoint we examined the unusual checkpoint defect of cdc55Δ cells, which is characterized by checkpoint bypass despite relatively slow degradation of APC substrates (Fig. 3 A; Minshull et al., 1996; Wang and Burke, 1997; Chiroli et al., 2007). Deletion of CDC55 increases APC phosphorylation in vivo (Fig. 6 A and Fig. 8) and purified PP2ACdc55 dephosphorylates the APC in vitro (Fig. 6 B) suggesting a model that in cdc55Δ cells checkpoint inhibition of the APC is countered by increased phosphorylation and activation of the APC. Consistent with this model, mutation of 12 APC phosphorylation sites in cdc55Δ cells suppresses premature APC activation during both morphogenesis and spindle checkpoint activation, and prevents rapid lethality after exposure to nocodazole (Fig. 7). These results are consistent with prior studies that have suggested that PP2A may dephosphorylate and inhibit the APCCdc20 in yeast, insect, and human cells (Deak et al., 2003; Burgess et al., 2010; Mui et al., 2010; Voets and Wolthuis, 2010).
APC inhibition is sufficient but may not be necessary to block anaphase
We have also demonstrated that in budding yeast, as in other eukaryotes (Lahav-Baratz et al., 1995; Shteinberg et al., 1999; Kramer et al., 2000; Kraft et al., 2003), Cdk1–Clb2 can activate the APCCdc20 in vitro, and this activation requires the phosphorylation sites in Cdc16 and Cdc27. This is the first demonstration that defects caused by APC phosphorylation site mutants in vivo correlate with changes in APC activity in vitro, and confirms that these sites contribute to APC–Cdc20 binding (Rudner and Murray, 2000). Similar to mutations in Cdc23 and Cdc27 that weaken activator binding (Matyskiela and Morgan, 2009), decreased phosphorylation of the APC reduces all ubiquitinated species equally (Fig. S4 E), arguing that phosphorylation does not affect APC processivity.
It remains unclear if APC phosphorylation is an essential function of Cdk1, as it has not been shown that mutating APC subunits to prevent APC phosphorylation blocks anaphase. Although past work has suggested that the TPR subunits are the primary phosphorylated subunits of the APC (Rudner and Murray, 2000; Yoon et al., 2006; Steen et al., 2008), we have shown that Apc1 also contains Cdk1-dependent phosphorylation (Fig. 4 A) and there may also exist additional unidentified Cdk1 sites on the APC. Phosphorylation of the APC may be essential in multicellular organisms—a single study has shown that mutating two sites in the Drosophila cdc27 cannot rescue a lethal P-element insertion into the endogenous cdc27 (Huang et al., 2007).
One study has shown that rapid and complete inhibition of the analogue-sensitive Cdk1-as1 causes a dramatic inhibition of CDC20 transcription and drop in protein levels (Liang et al., 2011). Though this is an attractive model for how morphogenesis checkpoint activation delays anaphase onset, it appears that regulation of CDC20 transcription is not the key target of the morphogenesis checkpoint for two reasons: (1) In contrast to Cdk1-as1 inhibition, Cdc20 is stable during morphogenesis checkpoint activation (Fig. S5 C), and (2) restoring Cdc20 levels after inhibiting Cdk1-as1 does not reverse a block to anaphase onset (Liang et al., 2011). Cdc20 regulation appears to be more complicated in Xenopus egg extracts where Cdk1-dependent phosphorylation of Cdc20 inhibits its binding to the APC, and regulated dephosphorylation by PP2A promotes anaphase onset (Kramer et al., 2000; Labit et al., 2012).
If Cdk1 regulated anaphase onset solely through the APCCdc20 then deletion of APC substrates should be sufficient to bypass a morphogenesis checkpoint arrest. Cells lacking PDS1 and CLB5 (as well as SGO1), the two essential APCCdc20 substrates (Shirayama et al., 1999), retain the ability to block anaphase in response to latA treatment, suggesting that APC inhibition may not be necessary for cell cycle arrest during checkpoint activation (Fig. S3, G and H; Chiroli et al., 2007). Inhibition of the APC does, however, prevent checkpoint bypass of swe1Δ cells (Fig. S3 I), showing APC inhibition is sufficient to block anaphase. These results suggest that inhibition of Cdk1 phosphorylation of a second target acts redundantly to maintain sister chromatid cohesion during morphogenesis checkpoint arrest (Fig. 8).
Cdk1 and PP2ACdc55 regulate anaphase onset by two mechanisms
Although the apc-12A mutations suppress some of the cdc55Δ defects, the timing of anaphase onset is similar in cdc55Δ and cdc55Δ apc-12A cells after latA treatment (Fig. 7 B). In addition, despite premature anaphase onset in cdc55Δ cdc16-1 cells grown at 34°C (Fig. 6 D), there is little difference in the kinetics of APC substrate degradation compared with swe1Δ cdc16-1 and cdc16-1 cells (Fig. S3 F). These data argue strongly that anaphase onset can be regulated independently of APC activation, and that PP2ACdc55 regulates a second target downstream of the APC. This conclusion is supported by past work showing that cdc55Δ mutants initiate anaphase in cells expressing a stabilized form of Pds1, which normally blocks sister chromatid separation (Tang and Wang, 2006).
Other reports have shown that PP2ACdc55 inhibits activation of the FEAR pathway and release of Cdc14 from the nucleolus (Wang and Ng, 2006; Yellman and Burke, 2006). Although premature Cdc14 release might allow derepression of the APCCdh1 and some APC proteolysis, this model doesn’t satisfactorily explain how cdc55Δ cells prematurely initiate sister chromatid separation in the absence of Pds1 degradation (Fig. 5, A and B; Fig. 7 B; Tang and Wang, 2006; Chiroli et al., 2007). PP2ACdc55 could function downstream of Pds1 destruction by regulating Separase activity directly, or regulate Cohesin cleavage, and prior data supports both models (Queralt et al., 2006; Yaakov et al., 2012). We favor a model whereby Cdk1 activates and PP2ACdc55 inhibits both the APC and Esp1 (Fig. 8). This redundancy would reinforce the switch-like onset of anaphase and could assist cells during recovery from checkpoint arrests when the APC may become active more slowly than in an unperturbed mitosis.
Materials and methods
Strain and plasmid construction
Table I lists the strains used in this work. All strains are derivatives of the W303 strain background (W303-1a; see Table 1 for complete genotype). All deletions and replacements were confirmed by immunoblotting, phenotype, or PCR. The sequences of all primers used in this study are available upon request. The bacterial strains TG1 and DH5α were used for amplification of DNA, and Rosetta (EMD Millipore) was used for protein purification.
Table 1.
Strain list
| Strain | Mating type | Genotype |
|---|---|---|
| ADR22 | _MAT_α | wild type W303-1aa |
| ADR477 | MATa | CDC28-HA-URA3 |
| ADR797 | MATa | leu2::pGAL-SWE1-HA-LEU2 bar1Δ::LEU2 pep4Δ::TRP1 |
| ADR806 | MATa | ura3::pGAL-SIC1-Δ3P-HA-His6 bar1Δ::LEU2 pep4Δ::TRP1 |
| ADR809 | MATa | bar1Δ::LEU2 pep4Δ::TRP1 |
| ADR877 | MATa | mih1Δ::LEU2 |
| ADR1168 | MATa | cdc15-2 |
| ADR1373 | MATa | leu2::pGAL-SWE1-HA-LEU2 mih1Δ::LEU2 CDC28-HA-URA3 |
| ADR1393 | MATa | trp1::lacO-256X::TRP1 his3::pCup1-GFP12-lacI12::HIS3 bar1Δ |
| ADR1395 | MATa | trp1::lacO-256X::TRP1 his3::pCup1-GFP12-lacI12::HIS3 mih1Δ::LEU2 leu2::pGAL-SWE1-HA-LEU2 CDC28-HA-URA3 bar1Δ |
| ADR1435 | MATa | cdh1Δ::HIS3 |
| ADR2260 | _MAT_α | mih1Δ::LEU2 |
| ADR2617 | MATa | PDS1-myc18X-leu2::HIS3 bar1Δ |
| ADR3089 | MATa | CDC16-TAP-HIS3 bar1Δ |
| ADR3141 | MATa | swe1Δ::TRP1 mih1Δ::LEU2 cdc20-3 |
| ADR3149 | MATa | mih1Δ::LEU2 cdc20-3 |
| ADR3155 | MATa | swe1Δ::TRP1 cdc20-3 |
| ADR3168 | _MAT_α | mih1Δ::LEU2 swe1Δ::TRP1 |
| ADR3161 | MATa | cdc20-3 |
| ADR3170 | MATa | swe1Δ::TRP1 |
| ADR3178 | MATa | mih1Δ::LEU2 |
| ADR3738 | MATa | PDS1-myc18X-leu2::HIS3 mih1Δ::KANR leu2::pGAL-swe1-N584A-HA-LEU2 bar1Δ |
| ADR3740 | MATa | PDS1-myc18X-leu2::HIS3 mih1Δ::KANR leu2::pGAL-SWE1-HA-LEU2 bar1Δ |
| ADR3822 | _MAT_α | cdc16-6A-TAP-KlURA3 |
| ADR3858 | MATa | CDC16-TAP-HIS3 mih1Δ::KANR leu2::pGAL-SWE1-HA-LEU2 bar1Δ |
| ADR3859 | MATa | CDC16-TAP-HIS3 mih1Δ::KANR leu2::pGAL-SWE1-HA-LEU2 bar1Δ |
| ADR3871 | MATa | PDS1-myc18X-leu2::HIS3 mih1Δ::KANR leu2::pGAL-SWE1-HA-LEU2 bar1Δ |
| ADR3877 | _MAT_α | CDC16-TAP-HIS3 swe1Δ::TRP1 mih1Δ::LEU2 |
| ADR3891 | MATa | cdc16-6A-TAP-KlURA3 cdc27-5A-KANR |
| ADR3919 | MATa | PDS1-myc18X-leu2::HIS3 mih1Δ::KANR mec1Δ::TRP1 sml1Δ::NATR leu2::pGAL-SWE1-HA-LEU2 bar1Δ |
| ADR3921 | MATa | PDS1-myc18X-leu2::HIS3 mec1Δ::TRP1 sml1Δ::NATR bar1Δ |
| ADR3938 | MATa | PDS1-myc18X-leu2::HIS3 mad2Δ::URA3 |
| ADR3940 | MATa | PDS1-myc18X-leu2::HIS3 mad2Δ::URA3 mih1Δ:KANR leu2::pGAL-SWE1-HA-LEU2 |
| ADR4006 | MATa | bar1Δ |
| ADR4009 | MATa | SPC42-eGFP-KANR bar1Δ |
| ADR4012 | MATa | mih1Δ::LEU2 SPC42-eGFP-KANR bar1Δ |
| ADR4015 | MATa | swe1Δ::TRP1 SPC42-eGFP-KANR bar1Δ |
| ADR4060 | MATa | cdc15-2 cdc55Δ::LEU2 CDC16-TAP-HIS3 |
| ADR4099 | MATa | mad2Δ::URA3 SPC42-eGFP-KANR bar1Δ |
| ADR4169 | MATa | PDS1-myc18X-leu2::HIS3 SPC42-GFP-HYGR bar1Δ |
| ADR4171 | MATa | PDS1-myc18X-leu2::HIS3 SPC42-GFP-HYGR mih1Δ::KANR leu2::pGAL-SWE1-HA-LEU2 bar1Δ |
| ADR4191 | MATa | _KIP1-myc13X-NATR CIN8-GFP-HYGR bar1_Δ |
| ADR4193 | MATa | _PDS1-myc18X-leu2::HIS3 CIN8-GFP-HYGR bar1_Δ |
| ADR4197 | MATa | mih1Δ::LEU2 swe1Δ::TRP1 APC1-TAP-URA3 |
| ADR4198 | MATa | mih1Δ::LEU2 leu2::pGAL-SWE1-HA-LEU2 APC1-TAP-URA3 |
| ADR4228 | MATa | PDS1-myc18X-leu2::HIS3 mih1Δ::KANR HYGR -cdc28-Y19F leu2::pGAL-SWE1-HA-LEU2 bar1Δ |
| ADR4245 | MATa | PDS1-myc18X-leu2::HIS3 mih1Δ::KANR cdc15-2 leu2::pGAL-SWE1-HA-LEU2 |
| ADR4252 | MATa | PDS1-myc18X-leu2::HIS3 cdc15-2 |
| ADR4289 | MATa | _mih1Δ::KANR leu2::pGAL-SWE1-HA-LEU2 CIN8-eGFP-HYGR bar1_Δ |
| ADR4313 | MATa | KANR-cdc28-Y19F SPC42-eGFP-HYGR bar1Δ |
| ADR4647 | MATa | sgo1Δ::HYGR pds1_Δ::NATR pMET-CLB5-KANR SPC42-eGFP-Sphis5+ bar1_Δ |
| ADR4738 | MATa | cdc55_Δ::HIS3 SPC42-eGFP-KANR bar1_Δ |
| ADR4902 | MATa | cdc16-6A-TRP1 cdc27-5A-KANR cdc23-A-HA cdc55_Δ::his3::LEU2 SPC42-eGFP-HYGR bar1_Δ |
| ADR4909 | MATa | mih1Δ::KANR swe1Δ::TRP1 HA3X-Cdh1 bar1Δ |
| ADR4910 | MATa | mih1Δ::KANR swe1Δ::TRP1 his3::pGAL-SWE1-HIS3 HA3X-CDH1 bar1Δ |
| ADR4973 | MATa | _cdc16-6A-TRP1 cdc27-5A-KANR cdc23-A-HA SPC42-eGFP-HYGR bar1_Δ |
| ADR4979 | MATa | _cdc16-1 SPC42-eGFP-KANR bar1_Δ |
| ADR4984 | MATa | _swe1Δ::TRP1 cdc16-1 SPC42-eGFP-KANR bar1_Δ |
| ADR5020 | MATa | cdc16-1 cdc55Δ::HIS3 SPC42-eGFP-KANR bar1Δ |
| ADR5026 | MATa | _NDC80-eGFP-Sphis5+ bar1_Δ |
| ADR5137 | MATa | cdc15-2 cdc55Δ::LEU2 swe1Δ::TRP1 CDC16-TAP-HIS3 bar1Δ |
| ADR5297 | MATa | RTS1-TAP-HIS3 bar1Δ |
| ADR5465 | MATa | CDC55-TAP-klTRP1 bar1Δ |
| ADR5599 | MATa | cdc23-1 SPC42-eGFP-KANR bar1Δ |
| ADR5600 | MATa | _swe1Δ::TRP1 cdc23-1 SPC42-eGFP-KANR bar1_Δ |
| ADR5623 | MATa | _cdc55Δ::HIS3 cdc23-1 SPC42-eGFP-KANR bar1_Δ |
CDC16-TAP-HIS3 strains were made by crossing TC80 (a gift of Christopher Carroll and David Morgan, UCSF, San Francisco, CA; Carroll et al., 2005) to the appropriate strains. The CDC55-TAP-klTRP1 strain was made using pBS1479 (Rigaut et al., 1999) and the appropriate oligonucleotides. cdc15-2, cdc20-3, and PDS1-myc18X-leu2::HIS3 strains were made by crossing K1993, K8029, and K6445 (gifts of Kim Nasmyth, University of Oxford, Oxford, UK) to the appropriate strains and the LEU2 converted to HIS3 with pLH7 (Cross, 1997). The plasmids used to make leu2::pGAL-SWE1-HA-LEU2 and leu2::pGAL-swe1-N584A-HA-LEU2 are pSwe1-41 (a gift of Bob Booher, Onyx Pharmaceuticals, Richmond, CA; Booher et al., 1993) and pSH14 (a gift of Stacey Harvey and Douglas Kellogg, UCSC, Santa Cruz, CA; Harvey et al., 2005), respectively. HA3X-CDH1 strains were made by crossing A1576 (a gift of Angelika Amon, MIT, Cambridge, MA) to the appropriate strains. ura3::pGAL-SIC1-Δ3P-HA-HIS6 was derived from RJD961 (a gift of Raymond Deshaies and Rati Verma, California Institute of Technology, Pasadena, CA; Verma et al., 1997).
BAR1 was deleted using pJGsst1 (a gift of Jeremy Thorner, University of California, Berkeley, CA). his3::pCup1-GFP12-lacI12::HIS3 and trp1::lacO-256X::TRP1 were made by integrating pSB116 (Biggins et al., 1999) and pAFS59 (Straight et al., 1996), respectively. MIH1 was deleted using pIP33 (a gift of Peter Sorger, Massachusetts Institute of Technology, Cambridge, MA). swe1Δ::TRP1 strains were made by crossing JM449 (a gift of Jeremy Minshull, DNA2.0, Menlo Park, CA) to the appropriate strains. cdc55Δ::HIS3 was created using pJM6 (Minshull et al., 1996) and cdc55Δ::his3::LEU2 converted to LEU2 using pHL3 (Cross, 1997). CDC28-HA-URA3 was made as described previously (Booher et al., 1993). cdc16-1 and cdc23-1 are derived from the A364A strains H16C1B1 and H23C1A1 (Hartwell et al., 1973) and have been backcrossed at least five times to W303-1a.
CDH1 was deleted using pAR127. An EcoRI–HindIII fragment of the CDH1 locus was amplified by PCR and cloned into pSK(−) (Agilent Technologies) to create pAR125. An XbaI–SmaI fragment of the HIS3 gene was then cloned into pAR125 cut with SpeI–XmnI, to create pAR127, which replaces the entire CDH1 open reading frame with HIS3. The EcoRI–NotI fragment of pAR127 was used to transform yeast.
cdc16-6A-TRP1, cdc23-A-HA, and cdc27-5A-KANR alleles were made as described previously (Rudner and Murray, 2000). Alanine-substituted mutants in CDC16, CDC23, and CDC27 were made using site-directed mutagenesis (Kunkel, 1985). Mutations were confirmed by the introduction of new restriction enzyme sites and by sequencing (ABI). For CDC16, the EcoR1–Xho1 fragment of pWAM10 (Lamb et al., 1994) was cloned into KS(−) (Agilent Technologies) to create pAR290. pAR290 was mutagenized to create pAR293, which contains all six serine/threonine-to-alanine substitutions. pAR294 was cut with EcoRI and NotI, and ligated to a EcoRI–PstI PCR fragment that contains the 3′ end of CDC16, a PstI–SpeI PCR fragment that contains the TRP1 gene, and a SpeI–NotI PCR fragment that contains the 3′ untranslated region of the CDC16 gene. The resultant plasmid, pAR303, was cut with XhoI and NotI, and integrated at the CDC16 locus. The TRP1 transformants were screened by PCR for the presence of all mutations. For CDC23, the BamHI–NotI fragment of pRS239 (Lamb et al., 1994) was cloned into KS(−) to create pAR228. pAR228 was mutagenized to create pAR240, which contains the single serine-to-alanine substitution in CDC23. pAR228 was cut with BamHI and NotI, transformed into cdc23-1 cells, and selected for growth at 37°C. Transformants were screened by Western blot for the HA tag present at the 3′ end of the gene, and by PCR for the presence of the alanine substitution. For CDC27, the PstI–NotI fragment of pJL25 (Lamb et al., 1994) was cloned into KS(−) to create pAR201. pAR201 was mutagenized to create pAR203, which contains all five serine/threonine-to-alanine substitutions in CDC27. pAR203 was cut with NdeI and NotI, and ligated to a NdeI–XbaI PCR fragment that contains the KANR gene and a XbaI–NotI PCR fragment containing the 3′ untranslated region of CDC27. The resultant plasmid, pAR271, was cut with KpnI and NotI, and integrated at the CDC27 locus. Transformants were screened by PCR for the presence of all mutants. The cdc16-6A-TAP-klURA3 allele was made by amplifying a TAP-klURA3 cassette from pBS1539 (Rigaut et al., 1999), integrating it into the _cdc16-6A_-TRP1 strain, and selecting for presence of the TAP tag and URA3 and loss of TRP1.
CIN8-GFP-HYGR, KIP1-myc13X-NATR, and SGO1-myc13X-HIS3 were created using PCR-targeted recombination using pFA6a-GFP(S65T)-kanMX6 and pFA6a-13Myc-kanMX6 (Longtine et al., 1998) and gene-specific primers and the marker was switched by homologous recombination using a HYGR or NATR cassette amplified off pAG32 or pAG25 (Goldstein and McCusker, 1999). SPC42-eGFP-KANR, SPC42-eGFP-Sphis5+, and NDC80-eGFP-Sphis5+ were created by amplifying eGFP and a marker off pKT127 and pKT128 (Sheff and Thorn, 2004), and SPC42-eGFP-HYGR was made by switching KANR to HYGR using pAG32. PDS1, SGO1, MEC1, SML1, and MIH1 were deleted using cassettes amplified from pAG32, pAG25, pRS404, and directly from the MATa yeast deletion array (Thermo Fisher Scientific).
KANR- pMET3-CLB5 was created by amplifying KANR- pMET3 cassette from pAR704 with CLB5 specific primers. pAR704 was made by inserting a PCR-amplified pMET3 (from −626 to −1) into the BglII and SalI sites of pFA6a-kanMX (Longtine et al., 1998). his3::pGAL-SWE1-HIS3 was created from pAR707. The full-length SWE1 gene was amplified from genomic DNA and cloned into pAR121 as a Xho1–BamH1 fragment. pAR121 is pRS303 with the GAL1-10 promoter cloned between Kpn1–Xho1. pAR707 was integrated at the his3-11 locus after restriction digestion with Nhe1.
KANR (or HYGR)-CDC28-Y19F was created from two overlapping PCR fragments: KANR or HYGR (amplified from pFA6a-kanMX6 and pAG32) and the promoter (from −334 to −1) and 5′ region of the CDC28-Y19F allele, including the mutation (amplified from pSF38; a gift of Peter Sorger, Harvard Medical School, Boston, MA). These two fragments were used to create a single long cassette that was integrated by homologous recombination. Positive transformations were determined by amplifying the 5′ region of CDC28 and digesting this fragment with NdeI, which was inserted near the Y19F substitution in pSF38. Additionally, positive _KANR_-CDC28-Y19F strains have no detectable Cdk1-Y19 phosphorylation as determined by immunoblotting using an α-Cdk1-P-Tyr antibody.
Physiology
Unless noted in the figure legend, cells were grown in yeast extract peptone media + 2% dextrose (YEPD) at 25 or 30°C. Cell cycle arrests were performed with 10 µg/ml nocodazole (Sigma-Aldrich), 25–100 ng/ml α-factor (Biosynthesis), and 2.5–5 µM latA (Sigma-Aldrich or Tocris Bioscience), or at 35°C for 2.5–3 h. LatA efficacy varied between batches and companies so the amount necessary to induce a fully Swe1-dependent arrest was determined empirically.
To fix cells for microscopy, ∼2.0 × 106 cells were harvested and fixed with 4% paraformaldehyde in PBS, pH 7.5, for 15 min, washed in 100 mM KPO4/1.2 M sorbitol, pH 7.5, sonicated, and resuspended in KPO4/sorbitol. Samples were imaged directly using a microscope (Ti; Nikon) with a Plan Apo 60× 1.4 NA objective (Nikon) and FITC filter set (Chroma Technology Corp.) at room temperature with a camera (CoolSnap HQ2; Photometrics). A minimum of 200 cells was visually scored per time point.
Live microscopy
Live imaging pads were made by adding 25% gelatin (wt/vol) to SC media at 56°C, pipetting 50 µl between two microscope slides, and allowing it to cool. 1–2 µl of cultures were pipetted onto live imaging pads, covered by a coverslip and sealed with VALAP (1:1:1 Vaseline/lanolin/petroleum jelly). Strains were imaged at 25°C for 2 h using bright-field and a FITC filter set (Chroma Technology Corp.) on either a fluorescence microscope (model DMI6000B; Leica) with a Plan Apo 63× 1.40 NA objective (Leica) and a camera (Orca AG; Hamamatsu Photonics) or on a microscope (Ti; Nikon) with a Plan Apo 60× 1.40 NA objective (Nikon) and a camera (CoolSnap HQ2; Photometrics). 17 Z-slices, spaced every 0.5 µm were imaged at each time point. Fluorescence excitation was attenuated using neutral density filters and 100–200-ms exposure times were used for both GFP and bright-field acquisition. Measurements were made using Volocity (PerkinElmer) or NIS-Elements software (Nikon). Look-up tables were manually adjusted linearly. Example images were prepared using ImageJ software (National Institutes of Health).
FACS analysis
For each time point 1.0 × 107 cells were harvested and resuspended in 70% ethanol. Samples were rotated overnight at 4°C, pelleted, and treated with 0.25 mg/ml RNase (Thermo Fisher Scientific) for 1 h at 50°C. After RNase treatment, cells were treated with 0.25 mg/ml proteinase K (Thermo Fisher Scientific) for 1 h at 60°C, stained with Sytox Green (Molecular Probes), and incubated for 30 min at room temperature. Samples were sonicated before analysis and analyzed using either an FC500 or CyAn ADP (Beckman Coulter) flow cytometer using a 488-nm laser.
Western blots and immunoprecipitation
These methods have been described previously (Rudner and Murray, 2000; Rudner et al., 2000). Immunoprecipitations were performed in APC lysis buffer (50 mM Hepes-KOH, pH 7.8, 700 mM NaCl, 150 mM NaF, 150 mM Na-β-glycerophosphate pH 8.3, 1 mM EDTA, 1 mM EGTA, 5% glycerol, 0.25% NP-40, 1 mM DTT, 1 mM PMSF, 1 mM Na3VO4, 1 mM benzamidine, and leupeptin, bestatin, pepstatin A, and chymostatin all at 1 mM).
The following antibodies were used for Western blots and immunoprecipitations: The use of 9E10 ascites (Covance), α-Clb2, α-Clb3, 12CA5 ascites (Covance), and α-Cdc27 have been described previously (Rudner and Murray, 2000; Rudner et al., 2000). Rabbit polyclonal α-Sgo1, α-Pds1, α-Clb5, α-Cdk1, α-Swe1, and α-Cdc20 were used at 1:1,000; α-Orc6 at 1:2,000; and α-GFP at 1:4,000 in TBS-T with 4% fat-free milk powder, 5% glycerol, and 0.02% NaN3. α-P-cdc2 (Tyr15-4539; Cell Signaling Technology) was used at a dilution of 1:1,000 in TBS-T, 4% BSA, and 0.02% NaN3. An autoclaved solution of 5% milk was used to make the 4% milk dilution buffer to increase the longevity of the antibody solution. Membranes were preblocked with TBS-T with 4% fat-free milk powder before incubation with all primary antibodies.
α-Clb5, α-Pds1, and α-Sgo1 antibodies were generated as follows. Coding sequences for the truncated proteins Clb52-137, Pds1178-373, and Sgo1129-326 were amplified using PCR and cloned into pGEX6P-1 (Promega) as BamH1–EcoR1 fragments to create pAR627, pAR624, and pAR717, respectively. 1 mg of each GST fusion protein was injected into rabbits every 4 wk for 8–16 wk (uOttawa Animal Facility, Ottawa, Ontario, Canada). Rabbit serum was harvested, clarified by centrifugation, and loaded on Affigel-10 (Bio-Rad Laboratories) columns coupled to purified malE-Clb5, malE-Pds1, or malE-Sgo1, respectively. malE-fusion proteins were expressed from the plasmids pAR651, pAR652, and pAR724, which contain the same fragments listed above cloned as BamH1–Sal1 fragments into pMAL-c2 (New England Biolabs, Inc.). Antibody was eluted from Affigel columns with either 100 mM triethylamine, pH 11.5, or 100 mM glycine, pH 2.3. The triethylamine and glycine elutions were neutralized, dialyzed in PBS + 50% glycerol, and stored at −80°C.
Plasmids GST-Swe12-312 (in pGEX-4T3, pAR622), malE-Swe12-819 (in pMAL-c2, pAR623), and His6-Cdk12-298 (pAR727; gifts of Vu Thai and Doug Kellogg, UCSC, Santa Cruz, Santa Cruz, CA; Sreenivasan and Kellogg, 1999) were used to produce GST-Swe1 and His6-Cdk1 protein for injection into rabbits as described above. α-Swe1 antibodies were purified on Affigel-10 (Bio-Rad Laboratories) columns coupled to purified malE-Swe1. α-Cdk1 antibodies were purified using an Affigel-15 (Bio-Rad Laboratories) column coupled to purified His6-Cdk1, which is insoluble, so the coupling was done in the presence of 0.3% SDS.
α-GFP serum (a gift of Aaron Straight [Stanford University, Stanford, CA] and Andrew Murray [Harvard University, Cambridge, MA]) was generated by immunizing rabbits with bacterially expressed His6-GFP (pAFS97), and purified as above using an Affigel-10 (Bio-Rad Laboratories) column coupled to purified His6-GFP.
α-Cdc20 antibody was generated as described previously (Camasses et al., 2003). Rabbit sera was purified as above using an Affigel-15 (Bio-Rad Laboratories) column coupled to purified His6-Cdc20470-610, which was expressed from a BamH1–Not1 fragment cloned into pHIS-parallel2 to create pAR784. His6-Cdc20470-610 is insoluble, so the coupling was done in the presence of 0.3% SDS.
α-Orc6 antibody was a kind gift of Joachim Li (UCSF, San Francisco, CA).
Ubiquitination assay
Purification of the APC using a C-terminal TAP tag on Cdc16 and ubiquitination assays were performed as described previously (Carroll and Morgan, 2002; Matyskiela and Morgan, 2009). Harvested cells were lysed in APC lysis buffer (see above). The clarified lysate was incubated with IgG-coupled magnetic beads (Invitrogen) for 1 h at 4°C. 35S-methionine labeled substrates Pds1-ZZ, Pds1-dbkb-ZZ and Clb5-HA-ZZ, Clb5-dbΔ-HA-ZZ, and unlabeled activators ZZ-Cdc20 and ZZ-Cdh1 (gifts of Maria Enquist, Scott Foster, and David Morgan, UCSF, San Francisco, CA) were made in vitro (TnT T7 Quick Coupled Transcription/Translation System; Promega), and purified with IgG-coupled magnetic beads. Proteins were cleaved from beads for 30 min at 25°C with TEV protease. His6-Uba1 was purified from yeast, and His6-Ubc1 and His6-Ubc4 were purified from bacteria as described previously (Carroll and Morgan, 2005; Rodrigo-Brenni and Morgan, 2007). Ubiquitination (and phosphorylation, below) was quantified using a phosphorimager (Molecular Dynamics) or a Typhoon Trio phosphorimager and ImageQuant software (GE Healthcare).
In vivo labeling of the APC
CDC16-TAP cells were arrested in mitosis by spindle checkpoint activation with nocodazole or in anaphase by temperature shift. Once the cells were arrested at the indicated stage of the cell cycle, 10 × 107 cells were harvested by centrifugation and labeled in 2 ml phosphate-free synthetic medium containing 0.5–1 mCi 32PO4 (GE Healthcare or PerkinElmer) as described previously (Rudner and Murray, 2000). Uptake of label was monitored by scintillation counting (TriCarb 2910TR; PerkinElmer) of the cells and media, and exceeds 98%.
Purification of Cdk1–Clb2 and PP2ACdc55 complexes
pRS326 containing pGAL-CLB-TAP, pAR546, was created by in vivo recombination in yeast by cotransforming the CLB2 gene with overlapping homology to a BamH1–HindIII-digested pRS-AB1234 (pGAL-TAP) plasmid (a gift of Christopher Carroll and David O. Morgan, UCSF, San Francisco, CA) and Clb2-TAP was overexpressed by growth in galactose. Cdc55–CBP complexes were purified from asynchronously growing CDC55-TAP (ADR 5465) cells.
Harvested cells were lysed in APC lysis buffer and purified as described previously (Rudner et al., 2005). The activity of purified Cdk1–Clb2-CBP complexes was assayed with histone H1 kinase assays (see below).
Kinase and phosphatase assays
Phosphorylation of histone H1 and the APC has been described previously (Rudner and Murray, 2000). For histone H1 kinase assays, Clb-associated complexes were purified with 1–2 µl of α-Clb2, α-Clb3, or α-Clb5 antibodies and protein A magnetic beads (Invitrogen), and incubated with 2.5 µg histone H1 (EMD Millipore). To phosphorylate the APC, the APC was purified as described, bound to IgG-coupled beads, and treated with purified Cdk1–Clb2 complexes. Kinase reactions for both were performed with cold ATP (1 mM for cold reactions and 0.01 mM for hot reactions), and 1 µCi γ-[32P]ATP. APC phosphorylated with unlabeled ATP was washed, cleaved, and tested in the ubiquitination assay. Dephosphorylation of the APC was measured by incubating in vitro–phosphorylated APC still bound to beads with TAP-purified PP2ACdc55 complexes or lambda phosphatase (New England Biolabs, Inc.). Okadaic acid (LC Laboratories) was used at 2 µM.
Online supplemental material
Fig. S1 shows alterations in mitotic progression in swe1Δ and mih1Δ cells. Fig. S2 shows analysis of anaphase kinetics in wild-type, swe1Δ, and mih1Δ cells. Fig. S3 provides additional controls that the APC is a target of the morphogenesis checkpoint. Fig. S4 provides additional data that overexpression of Swe1 arrests cells in mitosis and does not depend on the spindle or DNA damage checkpoints. Fig. S5 provides additional controls that Swe1 regulates APCCdc20, as well as silver-stained gels of the reagents used in Figs. 4 and 6, and additional quantification. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201212038/DC1.
Supplementary Material
Supplemental Material
Acknowledgments
We thank Christopher Carroll, Maria Enquist, Mary Matyskiela, Scott Foster, David Morgan, Stacey Harvey, Vu Thai, Doug Kellogg, Bob Booher, and Joachim Li for strains, plasmids, and antibodies; and Paul Maddox, Mads Kaern, Daniel Jedrysiak, Doug Kellogg, Bodo Stern, Jeremy Minshull, Andrew Murray, Christopher Carroll, David Morgan, Mary Matyskiela, Dara Spatz Friedman, and the Rudner laboratory for invaluable discussions, technical advice, and unwavering support. FACS analysis was performed at the uOttawa Flow Cytometry Resource Center, and immunization of rabbits for antibody production was performed at the uOttawa Animal Care Facility.
This work was supported by grants from the CIHR (177774) and CFI (13119); N. Lianga and E.C. Williams are supported by CIHR Canada Graduate Fellowships and were supported by an Ontario Graduate Scholarship in Science and Technology (N. Lianga) and an Ontario Graduate Scholarship and CIHR Master’s Award (E.C. Williams); and A.D. Rudner is a CIHR New Investigator and Ontario Early Researcher.
Footnotes
Abbreviations used in this paper:
APC
anaphase-promoting complex or cyclosome
latA
latrunculin A
PP2A
protein phosphatase 2A
Ub
ubiquitin
YEPD
yeast extract peptone media + 2% dextrose
References
- Amon A. 1997. Regulation of B-type cyclin proteolysis by Cdc28-associated kinases in budding yeast. EMBO J. 16:2693–2702 10.1093/emboj/16.10.2693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastasia S.D., Nguyen D.L., Thai V., Meloy M., MacDonough T., Kellogg D.R. 2012. A link between mitotic entry and membrane growth suggests a novel model for cell size control. J. Cell Biol. 197:89–104 10.1083/jcb.201108108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barral Y., Parra M., Bidlingmaier S., Snyder M. 1999. Nim1-related kinases coordinate cell cycle progression with the organization of the peripheral cytoskeleton in yeast. Genes Dev. 13:176–187 10.1101/gad.13.2.176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins S., Severin F.F., Bhalla N., Sassoon I., Hyman A.A., Murray A.W. 1999. The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes Dev. 13:532–544 10.1101/gad.13.5.532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booher R.N., Deshaies R.J., Kirschner M.W. 1993. Properties of Saccharomyces cerevisiae wee1 and its differential regulation of p34_CDC28_ in response to G1 and G2 cyclins. EMBO J. 12:3417–3426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchoux C., Uhlmann F. 2011. A quantitative model for ordered Cdk substrate dephosphorylation during mitotic exit. Cell. 147:803–814 10.1016/j.cell.2011.09.047 [DOI] [PubMed] [Google Scholar]
- Burgess A., Vigneron S., Brioudes E., Labbé J.-C., Lorca T., Castro A. 2010. Loss of human Greatwall results in G2 arrest and multiple mitotic defects due to deregulation of the cyclin B-Cdc2/PP2A balance. Proc. Natl. Acad. Sci. USA. 107:12564–12569 10.1073/pnas.0914191107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camasses A., Bogdanova A., Shevchenko A., Zachariae W. 2003. The CCT chaperonin promotes activation of the anaphase-promoting complex through the generation of functional Cdc20. Mol. Cell. 12:87–100 10.1016/S1097-2765(03)00244-2 [DOI] [PubMed] [Google Scholar]
- Carroll C.W., Morgan D.O. 2002. The Doc1 subunit is a processivity factor for the anaphase-promoting complex. Nat. Cell Biol. 4:880–887 10.1038/ncb871 [DOI] [PubMed] [Google Scholar]
- Carroll C.W., Morgan D.O. 2005. Enzymology of the anaphase-promoting complex. Methods Enzymol. 398:219–230 10.1016/S0076-6879(05)98018-X [DOI] [PubMed] [Google Scholar]
- Carroll C.W., Altman R., Schieltz D., Yates J.R., Kellogg D. 1998. The septins are required for the mitosis-specific activation of the Gin4 kinase. J. Cell Biol. 143:709–717 10.1083/jcb.143.3.709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll C.W., Enquist-Newman M., Morgan D.O. 2005. The APC subunit Doc1 promotes recognition of the substrate destruction box. Curr. Biol. 15:11–18 10.1016/j.cub.2004.12.066 [DOI] [PubMed] [Google Scholar]
- Castilho P.V., Williams B.C., Mochida S., Zhao Y., Goldberg M.L. 2009. The M phase kinase Greatwall (Gwl) promotes inactivation of PP2A/B55delta, a phosphatase directed against CDK phosphosites. Mol. Biol. Cell. 20:4777–4789 10.1091/mbc.E09-07-0643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chee M.K., Haase S.B. 2010. B-cyclin/CDKs regulate mitotic spindle assembly by phosphorylating kinesins-5 in budding yeast. PLoS Genet. 6:e1000935 10.1371/journal.pgen.1000935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Archambault V., Kar A., Lio’ P., D’Avino P.P., Sinka R., Lilley K., Laue E.D., Deak P., Capalbo L., Glover D.M. 2007. Multiple protein phosphatases are required for mitosis in Drosophila. Curr. Biol. 17:293–303 10.1016/j.cub.2007.01.068 [DOI] [PubMed] [Google Scholar]
- Chiroli E., Rossio V., Lucchini G., Piatti S. 2007. The budding yeast PP2ACdc55 protein phosphatase prevents the onset of anaphase in response to morphogenetic defects. J. Cell Biol. 177:599–611 10.1083/jcb.200609088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciosk R., Zachariae W., Michaelis C., Shevchenko A., Mann M., Nasmyth K. 1998. An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell. 93:1067–1076 10.1016/S0092-8674(00)81211-8 [DOI] [PubMed] [Google Scholar]
- Cohen-Fix O., Peters J.M., Kirschner M.W., Koshland D. 1996. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev. 10:3081–3093 10.1101/gad.10.24.3081 [DOI] [PubMed] [Google Scholar]
- Crasta K., Huang P., Morgan G., Winey M., Surana U. 2006. Cdk1 regulates centrosome separation by restraining proteolysis of microtubule-associated proteins. EMBO J. 25:2551–2563 10.1038/sj.emboj.7601136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crasta K., Lim H.H., Giddings T.H., Jr, Winey M., Surana U. 2008. Inactivation of Cdh1 by synergistic action of Cdk1 and polo kinase is necessary for proper assembly of the mitotic spindle. Nat. Cell Biol. 10:665–675 10.1038/ncb1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross F.R. 1997. ‘Marker swap’ plasmids: convenient tools for budding yeast molecular genetics. Yeast. 13:647–653 [DOI] [PubMed] [Google Scholar]
- Cross F.R. 2003. Two redundant oscillatory mechanisms in the yeast cell cycle. Dev. Cell. 4:741–752 10.1016/S1534-5807(03)00119-9 [DOI] [PubMed] [Google Scholar]
- Deak P., Donaldson M., Glover D.M. 2003. Mutations in mákos, a Drosophila gene encoding the Cdc27 subunit of the anaphase promoting complex, enhance centrosomal defects in polo and are suppressed by mutations in twins/aar, which encodes a regulatory subunit of PP2A. J. Cell Sci. 116:4147–4158 10.1242/jcs.00722 [DOI] [PubMed] [Google Scholar]
- Deibler R.W., Kirschner M.W. 2010. Quantitative reconstitution of mitotic CDK1 activation in somatic cell extracts. Mol. Cell. 37:753–767 10.1016/j.molcel.2010.02.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunphy W.G., Kumagai A. 1991. The cdc25 protein contains an intrinsic phosphatase activity. Cell. 67:189–196 10.1016/0092-8674(91)90582-J [DOI] [PubMed] [Google Scholar]
- Félix M.A., Labbé J.C., Dorée M., Hunt T., Karsenti E. 1990. Triggering of cyclin degradation in interphase extracts of amphibian eggs by cdc2 kinase. Nature. 346:379–382 10.1038/346379a0 [DOI] [PubMed] [Google Scholar]
- Funabiki H., Yamano H., Kumada K., Nagao K., Hunt T., Yanagida M. 1996. Cut2 proteolysis required for sister-chromatid seperation in fission yeast. Nature. 381:438–441 10.1038/381438a0 [DOI] [PubMed] [Google Scholar]
- Gautier J., Solomon M.J., Booher R.N., Bazan J.F., Kirschner M.W. 1991. cdc25 is a specific tyrosine phosphatase that directly activates p34cdc2. Cell. 67:197–211 10.1016/0092-8674(91)90583-K [DOI] [PubMed] [Google Scholar]
- Golan A., Yudkovsky Y., Hershko A. 2002. The cyclin-ubiquitin ligase activity of cyclosome/APC is jointly activated by protein kinases Cdk1-cyclin B and Plk. J. Biol. Chem. 277:15552–15557 10.1074/jbc.M111476200 [DOI] [PubMed] [Google Scholar]
- Goldstein A.L., McCusker J.H. 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 15:1541–1553 [DOI] [PubMed] [Google Scholar]
- Gould K.L., Nurse P. 1989. Tyrosine phosphorylation of the fission yeast cdc2+ protein kinase regulates entry into mitosis. Nature. 342:39–45 10.1038/342039a0 [DOI] [PubMed] [Google Scholar]
- Gould K.L., Moreno S., Tonks N.K., Nurse P. 1990. Complementation of the mitotic activator, p80cdc25, by a human protein-tyrosine phosphatase. Science. 250:1573–1576 10.1126/science.1703321 [DOI] [PubMed] [Google Scholar]
- Hartwell L.H., Mortimer R.K., Culotti J., Culotti M. 1973. Genetic control of the cell division cycle in yeast: V. genetic analysis of cdc mutants. Genetics. 74:267–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey S.L., Kellogg D.R. 2003. Conservation of mechanisms controlling entry into mitosis: budding yeast wee1 delays entry into mitosis and is required for cell size control. Curr. Biol. 13:264–275 10.1016/S0960-9822(03)00049-6 [DOI] [PubMed] [Google Scholar]
- Harvey S.L., Charlet A., Haas W., Gygi S.P., Kellogg D.R. 2005. Cdk1-dependent regulation of the mitotic inhibitor Wee1. Cell. 122:407–420 10.1016/j.cell.2005.05.029 [DOI] [PubMed] [Google Scholar]
- Harvey S.L., Enciso G., Dephoure N., Gygi S.P., Gunawardena J., Kellogg D.R. 2011. A phosphatase threshold sets the level of Cdk1 activity in early mitosis in budding yeast. Mol. Biol. Cell. 22:3595–3608 10.1091/mbc.E11-04-0340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Healy A.M., Zolnierowicz S., Stapleton A.E., Goebl M., DePaoli-Roach A.A., Pringle J.R. 1991. CDC55, a Saccharomyces cerevisiae gene involved in cellular morphogenesis: identification, characterization, and homology to the B subunit of mammalian type 2A protein phosphatase. Mol. Cell. Biol. 11:5767–5780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J.-Y., Morley G., Li D., Whitaker M. 2007. Cdk1 phosphorylation sites on Cdc27 are required for correct chromosomal localisation and APC/C function in syncytial Drosophila embryos. J. Cell Sci. 120:1990–1997 10.1242/jcs.006833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang L.H., Lau L.F., Smith D.L., Mistrot C.A., Hardwick K.G., Hwang E.S., Amon A., Murray A.W. 1998. Budding yeast Cdc20: a target of the spindle checkpoint. Science. 279:1041–1044 10.1126/science.279.5353.1041 [DOI] [PubMed] [Google Scholar]
- Jaspersen S.L., Charles J.F., Morgan D.O. 1999. Inhibitory phosphorylation of the APC regulator Hct1 is controlled by the kinase Cdc28 and the phosphatase Cdc14. Curr. Biol. 9:227–236 10.1016/S0960-9822(99)80111-0 [DOI] [PubMed] [Google Scholar]
- Karamysheva Z., Diaz-Martinez L.A., Crow S.E., Li B., Yu H. 2009. Multiple anaphase-promoting complex/cyclosome degrons mediate the degradation of human Sgo1. J. Biol. Chem. 284:1772–1780 10.1074/jbc.M807083200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keaton M.A., Bardes E.S.G., Marquitz A.R., Freel C.D., Zyla T.R., Rudolph J., Lew D.J. 2007. Differential susceptibility of yeast S and M phase CDK complexes to inhibitory tyrosine phosphorylation. Curr. Biol. 17:1181–1189 10.1016/j.cub.2007.05.075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keaton M.A., Szkotnicki L., Marquitz A.R., Harrison J., Zyla T.R., Lew D.J. 2008. Nucleocytoplasmic trafficking of G2/M regulators in yeast. Mol. Biol. Cell. 19:4006–4018 10.1091/mbc.E08-03-0286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg D.R. 2003. Wee1-dependent mechanisms required for coordination of cell growth and cell division. J. Cell Sci. 116:4883–4890 10.1242/jcs.00908 [DOI] [PubMed] [Google Scholar]
- Kim S.H., Lin D.P., Matsumoto S., Kitazono A., Matsumoto T. 1998. Fission yeast Slp1: an effector of the Mad2-dependent spindle checkpoint. Science. 279:1045–1047 10.1126/science.279.5353.1045 [DOI] [PubMed] [Google Scholar]
- King R.W., Peters J.M., Tugendreich S., Rolfe M., Hieter P., Kirschner M.W. 1995. A 20S complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell. 81:279–288 10.1016/0092-8674(95)90338-0 [DOI] [PubMed] [Google Scholar]
- Kraft C., Herzog F., Gieffers C., Mechtler K., Hagting A., Pines J., Peters J.-M. 2003. Mitotic regulation of the human anaphase-promoting complex by phosphorylation. EMBO J. 22:6598–6609 10.1093/emboj/cdg627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer E.R., Scheuringer N., Podtelejnikov A.V., Mann M., Peters J.M. 2000. Mitotic regulation of the APC activator proteins CDC20 and CDH1. Mol. Biol. Cell. 11:1555–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A., Dunphy W.G. 1992. Regulation of the cdc25 protein during the cell cycle in Xenopus extracts. Cell. 70:139–151 10.1016/0092-8674(92)90540-S [DOI] [PubMed] [Google Scholar]
- Kunkel T.A. 1985. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc. Natl. Acad. Sci. USA. 82:488–492 10.1073/pnas.82.2.488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labit H., Fujimitsu K., Bayin N.S., Takaki T., Gannon J., Yamano H. 2012. Dephosphorylation of Cdc20 is required for its C-box-dependent activation of the APC/C. EMBO J. 31:3351–3362 10.1038/emboj.2012.168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahav-Baratz S., Sudakin V., Ruderman J.V., Hershko A. 1995. Reversible phosphorylation controls the activity of cyclosome-associated cyclin-ubiquitin ligase. Proc. Natl. Acad. Sci. USA. 92:9303–9307 10.1073/pnas.92.20.9303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb J.R., Michaud W.A., Sikorski R.S., Hieter P.A. 1994. Cdc16p, Cdc23p and Cdc27p form a complex essential for mitosis. EMBO J. 13:4321–4328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew D.J., Reed S.I. 1995. A cell cycle checkpoint monitors cell morphogenesis in budding yeast. J. Cell Biol. 129:739–749 10.1083/jcb.129.3.739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H., Lim H.H., Venkitaraman A., Surana U. 2011. Cdk1 promotes kinetochore bi-orientation and regulates Cdc20 expression during recovery from spindle checkpoint arrest. EMBO J. 31:403–416 10.1038/emboj.2011.385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim H.H., Goh P.Y., Surana U. 1996. Spindle pole body separation in Saccharomyces cerevisiae requires dephosphorylation of the tyrosine 19 residue of Cdc28. Mol. Cell. Biol. 16:6385–6397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F.C., Arndt K.T. 1995. The role of Saccharomyces cerevisiae type 2A phosphatase in the actin cytoskeleton and in entry into mitosis. EMBO J. 14:2745–2759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindqvist A., van Zon W., Karlsson Rosenthal C., Wolthuis R.M.F. 2007. Cyclin B1-Cdk1 activation continues after centrosome separation to control mitotic progression. PLoS Biol. 5:e123 10.1371/journal.pbio.0050123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A., III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961 [DOI] [PubMed] [Google Scholar]
- Matyskiela M.E., Morgan D.O. 2009. Analysis of activator-binding sites on the APC/C supports a cooperative substrate-binding mechanism. Mol. Cell. 34:68–80 10.1016/j.molcel.2009.02.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan J.N., Sia R.A., Lew D.J. 1998. A morphogenesis checkpoint monitors the actin cytoskeleton in yeast. J. Cell Biol. 142:1487–1499 10.1083/jcb.142.6.1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNulty J.J., Lew D.J. 2005. Swe1p responds to cytoskeletal perturbation, not bud size, in S. cerevisiae. Curr. Biol. 15:2190–2198 10.1016/j.cub.2005.11.039 [DOI] [PubMed] [Google Scholar]
- Minshull J., Straight A., Rudner A.D., Dernburg A.F., Belmont A., Murray A.W. 1996. Protein phosphatase 2A regulates MPF activity and sister chromatid cohesion in budding yeast. Curr. Biol. 6:1609–1620 10.1016/S0960-9822(02)70784-7 [DOI] [PubMed] [Google Scholar]
- Mochida S., Hunt T. 2012. Protein phosphatases and their regulation in the control of mitosis. EMBO Rep. 13:197–203 10.1038/embor.2011.263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida S., Ikeo S., Gannon J., Hunt T. 2009. Regulated activity of PP2A-B55 delta is crucial for controlling entry into and exit from mitosis in Xenopus egg extracts. EMBO J. 28:2777–2785 10.1038/emboj.2009.238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan D.O. 2007. The Cell Cycle: Principles of Control. New Science Press; Sunderland, MA: 297 pp [Google Scholar]
- Mui M.Z., Roopchand D.E., Gentry M.S., Hallberg R.L., Vogel J., Branton P.E. 2010. Adenovirus protein E4orf4 induces premature APCCdc20 activation in Saccharomyces cerevisiae by a protein phosphatase 2A-dependent mechanism. J. Virol. 84:4798–4809 10.1128/JVI.02434-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurse P. 1975. Genetic control of cell size at cell division in yeast. Nature. 256:547–551 10.1038/256547a0 [DOI] [PubMed] [Google Scholar]
- Nurse P. 1990. Universal control mechanism regulating onset of M-phase. Nature. 344:503–508 10.1038/344503a0 [DOI] [PubMed] [Google Scholar]
- Nurse P., Thuriaux P., Nasmyth K. 1976. Genetic control of the cell division cycle in the fission yeast Schizosaccharomyces pombe. Mol. Gen. Genet. 146:167–178 10.1007/BF00268085 [DOI] [PubMed] [Google Scholar]
- Oikonomou C., Cross F.R. 2011. Rising cyclin-CDK levels order cell cycle events. PLoS ONE. 6:e20788 10.1371/journal.pone.0020788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal G., Paraz M.T.Z., Kellogg D.R. 2008. Regulation of Mih1/Cdc25 by protein phosphatase 2A and casein kinase 1. J. Cell Biol. 180:931–945 10.1083/jcb.200711014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker L.L., Atherton-Fessler S., Piwnica-Worms H. 1992. p107wee1 is a dual-specificity kinase that phosphorylates p34cdc2 on tyrosine 15. Proc. Natl. Acad. Sci. USA. 89:2917–2921 10.1073/pnas.89.7.2917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patra D., Dunphy W.G. 1998. Xe-p9, a Xenopus Suc1/Cks protein, is essential for the Cdc2-dependent phosphorylation of the anaphase- promoting complex at mitosis. Genes Dev. 12:2549–2559 10.1101/gad.12.16.2549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J.-M. 2006. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat. Rev. Mol. Cell Biol. 7:644–656 10.1038/nrm1988 [DOI] [PubMed] [Google Scholar]
- Pomerening J.R., Sontag E.D., Ferrell J.E., Jr 2003. Building a cell cycle oscillator: hysteresis and bistability in the activation of Cdc2. Nat. Cell Biol. 5:346–351 10.1038/ncb954 [DOI] [PubMed] [Google Scholar]
- Pomerening J.R., Kim S.Y., Ferrell J.E., Jr 2005. Systems-level dissection of the cell-cycle oscillator: bypassing positive feedback produces damped oscillations. Cell. 122:565–578 10.1016/j.cell.2005.06.016 [DOI] [PubMed] [Google Scholar]
- Queralt E., Lehane C., Novak B., Uhlmann F. 2006. Downregulation of PP2A(Cdc55) phosphatase by separase initiates mitotic exit in budding yeast. Cell. 125:719–732 10.1016/j.cell.2006.03.038 [DOI] [PubMed] [Google Scholar]
- Rahal R., Amon A. 2008. Mitotic CDKs control the metaphase-anaphase transition and trigger spindle elongation. Genes Dev. 22:1534–1548 10.1101/gad.1638308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaut G., Shevchenko A., Rutz B., Wilm M., Mann M., Séraphin B. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030–1032 10.1038/13732 [DOI] [PubMed] [Google Scholar]
- Rodrigo-Brenni M.C., Morgan D.O. 2007. Sequential E2s drive polyubiquitin chain assembly on APC targets. Cell. 130:127–139 10.1016/j.cell.2007.05.027 [DOI] [PubMed] [Google Scholar]
- Rudner A.D., Murray A.W. 2000. Phosphorylation by Cdc28 activates the Cdc20-dependent activity of the anaphase-promoting complex. J. Cell Biol. 149:1377–1390 10.1083/jcb.149.7.1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner A.D., Hardwick K.G., Murray A.W. 2000. Cdc28 activates exit from mitosis in budding yeast. J. Cell Biol. 149:1361–1376 10.1083/jcb.149.7.1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner A.D., Hall B.E., Ellenberger T., Moazed D. 2005. A nonhistone protein-protein interaction required for assembly of the SIR complex and silent chromatin. Mol. Cell. Biol. 25:4514–4528 10.1128/MCB.25.11.4514-4528.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell P., Nurse P. 1986. cdc25+ functions as an inducer in the mitotic control of fission yeast. Cell. 45:145–153 10.1016/0092-8674(86)90546-5 [DOI] [PubMed] [Google Scholar]
- Russell P., Nurse P. 1987. Negative regulation of mitosis by wee1+, a gene encoding a protein kinase homolog. Cell. 49:559–567 10.1016/0092-8674(87)90458-2 [DOI] [PubMed] [Google Scholar]
- Russell P., Moreno S., Reed S.I. 1989. Conservation of mitotic controls in fission and budding yeasts. Cell. 57:295–303 10.1016/0092-8674(89)90967-7 [DOI] [PubMed] [Google Scholar]
- Sanchez Y., Bachant J., Wang H., Hu F., Liu D., Tetzlaff M., Elledge S.J. 1999. Control of the DNA damage checkpoint by chk1 and rad53 protein kinases through distinct mechanisms. Science. 286:1166–1171 10.1126/science.286.5442.1166 [DOI] [PubMed] [Google Scholar]
- Schwab M., Lutum A.S., Seufert W. 1997. Yeast Hct1 is a regulator of Clb2 cyclin proteolysis. Cell. 90:683–693 10.1016/S0092-8674(00)80529-2 [DOI] [PubMed] [Google Scholar]
- Sheff M.A., Thorn K.S. 2004. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast. 21:661–670 10.1002/yea.1130 [DOI] [PubMed] [Google Scholar]
- Shirayama M., Tóth A., Gálová M., Nasmyth K. 1999. APC(Cdc20) promotes exit from mitosis by destroying the anaphase inhibitor Pds1 and cyclin Clb5. Nature. 402:203–207 10.1038/46080 [DOI] [PubMed] [Google Scholar]
- Shteinberg M., Protopopov Y., Listovsky T., Brandeis M., Hershko A. 1999. Phosphorylation of the cyclosome is required for its stimulation by Fizzy/cdc20. Biochem. Biophys. Res. Commun. 260:193–198 10.1006/bbrc.1999.0884 [DOI] [PubMed] [Google Scholar]
- Shu Y., Yang H., Hallberg E., Hallberg R. 1997. Molecular genetic analysis of Rts1p, a B’ regulatory subunit of Saccharomyces cerevisiae protein phosphatase 2A. Mol. Cell. Biol. 17:3242–3253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreenivasan A., Kellogg D. 1999. The elm1 kinase functions in a mitotic signaling network in budding yeast. Mol. Cell. Biol. 19:7983–7994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark M.J. 1996. Yeast protein serine/threonine phosphatases: multiple roles and diverse regulation. Yeast. 12:1647–1675 [DOI] [PubMed] [Google Scholar]
- Steen J.A.J., Steen H., Georgi A., Parker K., Springer M., Kirchner M., Hamprecht F., Kirschner M.W. 2008. Different phosphorylation states of the anaphase promoting complex in response to antimitotic drugs: a quantitative proteomic analysis. Proc. Natl. Acad. Sci. USA. 105:6069–6074 10.1073/pnas.0709807104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmeier F., Amon A. 2004. Closing mitosis: the functions of the Cdc14 phosphatase and its regulation. Annu. Rev. Genet. 38:203–232 10.1146/annurev.genet.38.072902.093051 [DOI] [PubMed] [Google Scholar]
- Stern B., Nurse P. 1996. A quantitative model for the cdc2 control of S phase and mitosis in fission yeast. Trends Genet. 12:345–350 [PubMed] [Google Scholar]
- Straight A.F., Belmont A.S., Robinett C.C., Murray A.W. 1996. GFP tagging of budding yeast chromosomes reveals that protein-protein interactions can mediate sister chromatid cohesion. Curr. Biol. 6:1599–1608 10.1016/S0960-9822(02)70783-5 [DOI] [PubMed] [Google Scholar]
- Sudakin V., Ganoth D., Dahan A., Heller H., Hershko J., Luca F.C., Ruderman J.V., Hershko A. 1995. The cyclosome, a large complex containing cyclin-selective ubiquitin ligase activity, targets cyclins for destruction at the end of mitosis. Mol. Biol. Cell. 6:185–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X., Wang Y. 2006. Pds1/Esp1-dependent and -independent sister chromatid separation in mutants defective for protein phosphatase 2A. Proc. Natl. Acad. Sci. USA. 103:16290–16295 10.1073/pnas.0607856103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theesfeld C.L., Irazoqui J.E., Bloom K., Lew D.J. 1999. The role of actin in spindle orientation changes during the Saccharomyces cerevisiae cell cycle. J. Cell Biol. 146:1019–1032 10.1083/jcb.146.5.1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton B.R., Toczyski D.P. 2003. Securin and B-cyclin/CDK are the only essential targets of the APC. Nat. Cell Biol. 5:1090–1094 10.1038/ncb1066 [DOI] [PubMed] [Google Scholar]
- Uhlmann F., Bouchoux C., López-Avilés S. 2011. A quantitative model for cyclin-dependent kinase control of the cell cycle: revisited. Philos. Trans. R. Soc. Lond. B Biol. Sci. 366:3572–3583 10.1098/rstb.2011.0082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R., Annan R.S., Huddleston M.J., Carr S.A., Reynard G., Deshaies R.J. 1997. Phosphorylation of Sic1p by G1 Cdk required for its degradation and entry into S phase. Science. 278:455–460 10.1126/science.278.5337.455 [DOI] [PubMed] [Google Scholar]
- Visintin R., Prinz S., Amon A. 1997. CDC20 and CDH1: a family of substrate-specific activators of APC-dependent proteolysis. Science. 278:460–463 10.1126/science.278.5337.460 [DOI] [PubMed] [Google Scholar]
- Visintin R., Craig K., Hwang E.S., Prinz S., Tyers M., Amon A. 1998. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol. Cell. 2:709–718 10.1016/S1097-2765(00)80286-5 [DOI] [PubMed] [Google Scholar]
- Voets E., Wolthuis R.M.F. 2010. MASTL is the human orthologue of Greatwall kinase that facilitates mitotic entry, anaphase and cytokinesis. Cell Cycle. 9:3591–3601 10.4161/cc.9.17.12832 [DOI] [PubMed] [Google Scholar]
- Wang Y., Burke D.J. 1997. Cdc55p, the B-type regulatory subunit of protein phosphatase 2A, has multiple functions in mitosis and is required for the kinetochore/spindle checkpoint in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:620–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Ng T.-Y. 2006. Phosphatase 2A negatively regulates mitotic exit in Saccharomyces cerevisiae. Mol. Biol. Cell. 17:80–89 10.1091/mbc.E04-12-1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicky S., Tjandra H., Schieltz D., Yates J., III, Kellogg D.R. 2011. The Zds proteins control entry into mitosis and target protein phosphatase 2A to the Cdc25 phosphatase. Mol. Biol. Cell. 22:20–32 10.1091/mbc.E10-06-0487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurzenberger C., Gerlich D.W. 2011. Phosphatases: providing safe passage through mitotic exit. Nat. Rev. Mol. Cell Biol. 12:469–482 10.1038/nrm3149 [DOI] [PubMed] [Google Scholar]
- Yaakov G., Thorn K.S., Morgan D.O. 2012. Separase biosensor reveals that cohesin cleavage timing depends on phosphatase PP2A(Cdc55) regulation. Dev. Cell. 23:124–136 10.1016/j.devcel.2012.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Jiang W., Gentry M., Hallberg R.L. 2000. Loss of a protein phosphatase 2A regulatory subunit (Cdc55p) elicits improper regulation of Swe1p degradation. Mol. Cell. Biol. 20:8143–8156 10.1128/MCB.20.21.8143-8156.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yellman C.M., Burke D.J. 2006. The role of Cdc55 in the spindle checkpoint is through regulation of mitotic exit in Saccharomyces cerevisiae. Mol. Biol. Cell. 17:658–666 10.1091/mbc.E05-04-0336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeong F.M., Lim H.H., Padmashree C.G., Surana U. 2000. Exit from mitosis in budding yeast: biphasic inactivation of the Cdc28-Clb2 mitotic kinase and the role of Cdc20. Mol. Cell. 5:501–511 10.1016/S1097-2765(00)80444-X [DOI] [PubMed] [Google Scholar]
- Yoon H.-J., Feoktistova A., Chen J.-S., Jennings J.L., Link A.J., Gould K.L. 2006. Role of Hcn1 and its phosphorylation in fission yeast anaphase-promoting complex/cyclosome function. J. Biol. Chem. 281:32284–32293 10.1074/jbc.M603867200 [DOI] [PubMed] [Google Scholar]
- Zachariae W., Schwab M., Nasmyth K., Seufert W. 1998. Control of cyclin ubiquitination by CDK-regulated binding of Hct1 to the anaphase promoting complex. Science. 282:1721–1724 10.1126/science.282.5394.1721 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material