The CP110-interacting proteins Talpid3 and Cep290 play overlapping and distinct roles in cilia assembly (original) (raw)
Talpid3 and Cep290 promote proper ciliary vesicle formation by regulating centriolar satellite accretion and Rab8a localization.
Abstract
We have identified Talpid3/KIAA0586 as a component of a CP110-containing protein complex important for centrosome and cilia function. Talpid3 assembles a ring-like structure at the extreme distal end of centrioles. Ablation of Talpid3 resulted in an aberrant distribution of centriolar satellites involved in protein trafficking to centrosomes as well as cilia assembly defects, reminiscent of loss of Cep290, another CP110-associated protein. Talpid3 depletion also led to mislocalization of Rab8a, a small GTPase thought to be essential for ciliary vesicle formation. Expression of activated Rab8a suppressed cilia assembly defects provoked by Talpid3 depletion, suggesting that Talpid3 affects cilia formation through Rab8a recruitment and/or activation. Remarkably, ultrastructural analyses showed that Talpid3 is required for centriolar satellite dispersal, which precedes the formation of mature ciliary vesicles, a process requiring Cep290. These studies suggest that Talpid3 and Cep290 play overlapping and distinct roles in ciliary vesicle formation through regulation of centriolar satellite accretion and Rab8a.
Introduction
Centrosomes, composed of two cylindrical centrioles, termed the mother and daughter centrioles, and pericentriolar material (PCM), function as microtubule-organizing centers in animal cells (Bettencourt-Dias and Glover, 2007). In cycling cells, the centrosome nucleates the assembly of spindle poles in mitosis, but once cells exit from the cell cycle and enter quiescence, the mother centriole differentiates into a basal body to assemble a primary cilium (Kobayashi and Dynlacht, 2011). Primary cilia function as cellular antennae, mediating several signaling pathways essential for growth and differentiation (Ishikawa and Marshall, 2011).
Although the mechanisms underlying the transition from mother centrioles to the basal bodies of primary cilia remain largely enigmatic, it has been shown that formation of a ciliary vesicle that caps the distal end of mother centrioles occurs at a very early stage in ciliogenesis (Sorokin, 1962). It is thought that the ciliary vesicle initially appears at the distal end of mother centrioles and subsequently fuses with secondary vesicles, leading to the formation of a ciliary sheath around the assembling axoneme and eventually forming a ciliary membrane (Ghossoub et al., 2011). A small GTPase, Rab8a, has been shown to localize to centrosomes and the ciliary sheath, and this protein is required for ciliary membrane formation (Nachury et al., 2007; Kim et al., 2008; Tsang et al., 2008; Hsiao et al., 2009; Murga-Zamalloa et al., 2010). Rab8a is required for ciliogenesis, and Rabin8 is a known guanine nucleotide exchange factor (GEF) in this pathway (Hattula et al., 2002; Nachury et al., 2007; Yoshimura et al., 2007; Knödler et al., 2010; Chiba et al., 2013). Recently, it has been reported that Rab8a localizes near centrosomes during the early phase of ciliogenesis, and these Rab8a-containing structures resemble nascent ciliary sheaths, suggesting that Rab8a plays important roles in ciliary vesicle formation and elongation (Westlake et al., 2011).
Centriolar satellites are nonmembranous, electron-dense particles surrounding centrosomes in many animal cells, and they have been implicated in dynein-dependent protein trafficking (Kubo et al., 1999). PCM1, first identified as a component of centriolar satellites, interacts with many proteins, and probably functions as a scaffold protein (Dammermann and Merdes, 2002; Kubo and Tsukita, 2003). Interestingly, several proteins that localize to centriolar satellites, including PCM1, Cep290, BBS4, Ofd1, and Cep72, are required for cilia formation (Ferrante et al., 2006; Nachury et al., 2007; Kim et al., 2008; Tsang et al., 2008; Singla et al., 2010; Stowe et al., 2012), suggesting that centriolar satellites play key roles in cilia formation. Given that PCM1 and Cep290 associate with Rab8a, and loss of these proteins results in mislocalization of Rab8a (Kim et al., 2008; Tsang et al., 2008), it is possible that centriolar satellites may be necessary for Rab8a to promote cilia assembly.
We have previously identified a centrosomal protein, CP110, as a suppressor of centriole reduplication induced by prolonged S phase (Chen et al., 2002). CP110 localizes to the distal end of centrioles and disappears from basal bodies before cilia assembly (Kleylein-Sohn et al., 2007; Spektor et al., 2007). CP110 and two associated proteins, Cep97 and Kif24, suppress primary cilia formation, most likely by capping the distal end of the mother centriole and remodeling centriolar microtubules in growing cells able to form cilia (Spektor et al., 2007; Kobayashi et al., 2011). In addition, CP110 suppresses ciliogenesis through interactions with Cep290 and Rab8a (Tsang et al., 2008). These results suggest a functional network of interactions that connect CP110, Cep290, and Rab8a. However, the precise roles of Cep290 and Rab8a in ciliogenesis in mammalian cells have not yet been elucidated.
In this study, we identified KIAA0586/Talpid3 as a CP110-interacting protein. The talpid3 mutation in chickens was originally identified on the basis of its limb defects (Ede and Kelly, 1964), and later reports showed that this mutant exhibits aberrant Sonic hedgehog activity (Lewis et al., 1999; Davey et al., 2006). Chicken Talpid3 encodes a coiled-coil protein that localizes to centrosomes (Yin et al., 2009). Recently, developmental studies have shown that loss of Talpid3 in chicken, mouse, and zebrafish abolishes primary cilia formation, resulting in polydactyly, as well as facial, neural, and left/right symmetry defects (Yin et al., 2009; Bangs et al., 2011; Ben et al., 2011). However, it remained unclear how Talpid3 functions during cilia assembly. Here, we show that Talpid3 localizes to a ring around the distal end of centrioles, assembling near the distal appendages of mother centrioles. Depletion of Talpid3 results in defects in cilia formation and mislocalization of centriolar satellites and Rab8a, phenocopying the loss of Cep290 and suggesting that Talpid3 and Cep290 share overlapping roles in cilia formation. However, we found that initial ciliary vesicle formation is impaired by depletion of Talpid3, whereas subsequent enlargement of early ciliary vesicles is arrested in Cep290-ablated cells. These data suggest a model in which Talpid3 functions at an initial step of ciliary vesicle formation and Cep290 acts in the maturation of these vesicles, thereby promoting cilia assembly.
Results
Talpid3 interacts with a CP110-containing protein complex
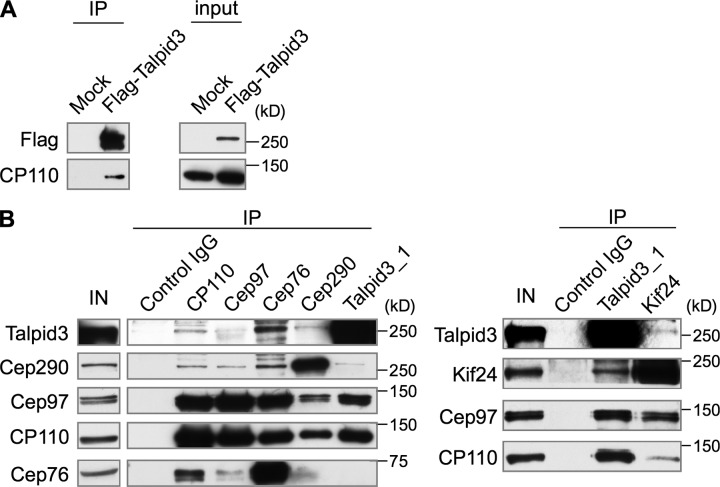
In an effort to identify distal centriolar proteins that play a role in ciliogenesis, we performed a proteomic screen for components of CP110-containing complexes (see Materials and methods). We identified a candidate interacting protein, KIAA0586, which is an orthologue of chicken Talpid3. Talpid3/KIAA0586 is conserved in vertebrates, from humans to zebrafish, but it is absent from insects and worms. Human Talpid3 contains one coiled-coil domain in the central region (Fig. S1 A). To test whether Talpid3 and CP110 interact in cells, we transfected a plasmid expressing Flag-Talpid3 in HEK293 cells, performed anti-Flag immunoprecipitations, and demonstrated that both Flag-Talpid3 and CP110 were coprecipitated (Fig. 1 A). We raised antibodies against human Talpid3 that detect the endogenous protein (Fig. S1 A) and showed that both antibodies were specific for Talpid3 by transfecting siRNAs that ablated the full-length protein from crude lysates (Fig. S1 B; and unpublished data). Next, we performed immunoprecipitations and found that endogenous Talpid3 was coimmunoprecipitated with CP110 and Kif24 antibodies, whereas considerably less Talpid3 was precipitated with antibodies against Cep97 and Cep290 (Fig. 1 B). Reciprocal immunoprecipitations with a Talpid3 antibody confirmed its interaction with CP110, Cep97, and Kif24, and, to a lesser extent, with Cep290 (Fig. 1 B). We also detected Talpid3 in Cep76 immunoprecipitates, although the association was not detected in the reverse direction (Fig. 1 B). Moreover, we observed Talpid3 binding to CP110 and Cep97 and weaker interactions with Cep290 and Kif24 in a human retinal pigment epithelial cell line immortalized with telomerase (hTERT-RPE1 or RPE1 hereafter; Fig. S1 C). Collectively, these data indicate that Talpid3 robustly binds to CP110 and more weakly associates with Cep97, Cep290, and Kif24.
Figure 1.
Talpid3 interacts with CP110, Cep97, Kif24, and Cep290. (A) Flag-tagged Talpid3 or Flag alone was expressed in HEK293 cells, and lysates were immunoprecipitated with an anti-Flag antibody. The resulting immunoprecipitates were Western blotted with anti-Flag, or anti-CP110 antibodies. (B) Western blotting of endogenous proteins after immunoprecipitation of HEK293 cell extracts with antibodies indicated at the top of panel. IN, input.
Subcellular localization of Talpid3
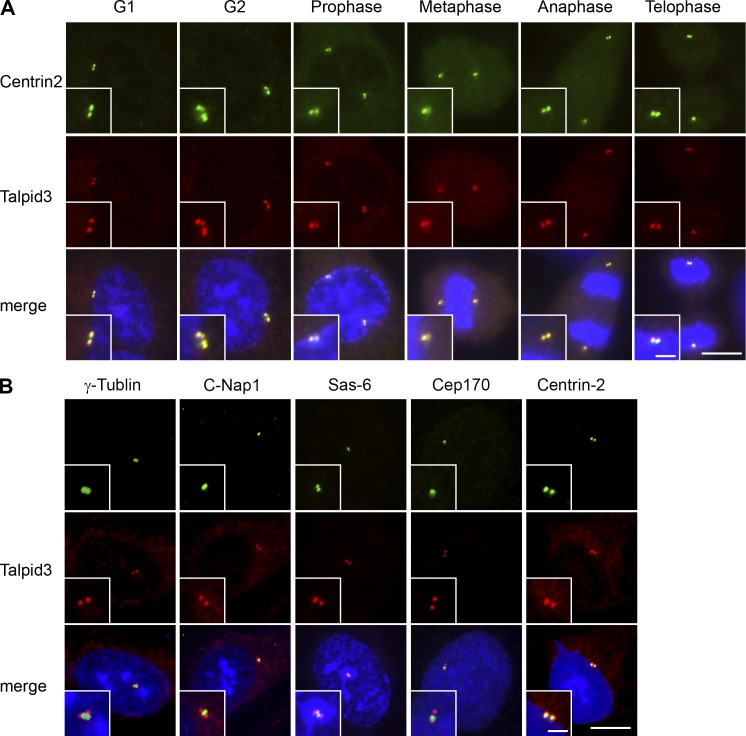
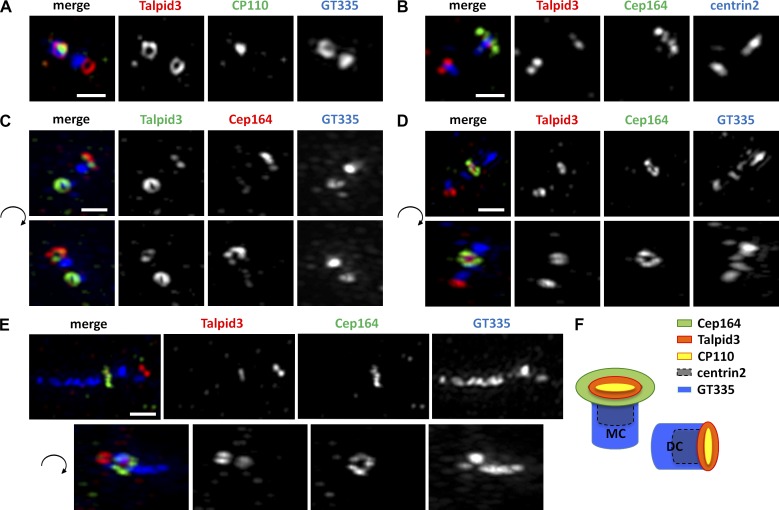
We examined the subcellular localization of Talpid3 by transfecting a plasmid expressing Flag-Talpid3 into RPE1 cells and performing immunofluorescence experiments. As shown in Fig. S1 D, Flag-Talpid3 exclusively colocalized with glutamylated tubulin at centrosomes. We then performed immunofluorescence experiments using anti-Talpid3 antibodies to assess endogenous Talpid3 localization at different stages of the cell cycle in RPE1 cells. Talpid3 antibodies stained two dots in G1 cells and four dots in G2 and mitotic cells, and in each case, staining overlapped with centrin2, a centriole marker (Fig. 2 A). We also detected Talpid3 localization to basal bodies of primary cilia and daughter centrioles in serum-starved RPE1 cells (Fig. S1 E). Talpid3 fluorescence was abolished in Talpid3-depleted cells, indicating that our antibody specifically recognized endogenous Talpid3 in cultured cells (Fig. S1 E). A second Talpid3 antibody also showed centrosomal staining in RPE1 cells (Fig. S1 F). Together, these data suggest that Talpid3 localizes to centrosomes throughout the cell cycle and during ciliogenesis. We co-stained Talpid3 with a subset of centrosome markers and found that Talpid3 staining overlapped with centrin2, a distal-end centriolar marker (Paoletti et al., 1996), but not with γ-tubulin (pericentriolar matrix), C-Nap1 (proximal region), Sas6 (pro-centrioles), or Cep170 (subdistal region; Fig. 2 B). Next, we obtained super-resolution immunofluorescence images using structured illumination microscopy. Remarkably, these experiments indicated that Talpid3 forms a ring-like structure enveloping CP110 at the extreme distal ends of centrioles (Fig. 3 A). Colocalization studies further showed that Talpid3 formed a “rim” around the centriolar barrel (visualized with anti-glutamylated tubulin [GT335] and centrin2 antibodies), and nested within, and very close to, the ring formed by Cep164 at the distal appendages in the presence or absence of an axoneme (Fig. 3, B–E). Collectively, these data confirm that Talpid3 localizes to the distal ends of centrioles, close to other components of CP110-containing protein complexes and near the distal appendages (Fig. 3 F; Kleylein-Sohn et al., 2007; Spektor et al., 2007; Tsang et al., 2008).
Figure 2.
Talpid3 localizes to the distal ends of centrioles. (A) RPE1 cells in diverse stages of the cell cycle were processed for immunofluorescence with anti-Talpid3_2 (red) and anti-centrin (green) antibodies. (B) RPE1 cells were visualized with anti-Talpid3_2 (red) and anti–γ-tubulin, C-Nap1, Sas-6, Cep170, or Centrin2 (green) antibodies. Bars: (A and B)10 µm; (insets) 2.5 µm. DNA was stained with DAPI (blue).
Figure 3.
Talpid3 forms a ring-like structure at the distal rim of both mother and daughter centrioles. (A and B) RPE1 cells were visualized using an OMX super-resolution microscope in asynchronously growing conditions stained with the indicated combinations of antibodies. (C–E) Antibodies against glutamylated tubulin (GT335), Cep164, and Talpid3_2 were used to stain RPE1 cells during growing (C), 6 h serum starvation (D), and 24 h serum starvation (E) conditions. The bottom panels of C–E show y-axis rotations of the top panels by 165° (C), 48° (D), and 150° (E) in a clockwise manner to best represent the sub-centriolar localization pattern of Talpid3. (F) Schematic representation of the centrosome illustrates the localization of Talpid3 at the distal rim of both the mother centrioles (MC) and daughter centrioles (DC). Bars, 500 nm.
To determine the centrosomal targeting domain of Talpid3, we generated a series of Flag-tagged Talpid3 deletion mutants. We expressed these mutants in RPE1 cells and performed immunofluorescence experiments. Several truncations spanning residues 466–602, which contain a coiled-coil domain (residues 466–500), localized to centrosomes (Figs. S1 G). These data suggest that a central region, including a coiled-coil domain, is important for Talpid3 targeting to centrosomes, in agreement with previous work (Yin et al., 2009). Surprisingly, we also found that an N-terminal region (residues 1–465) weakly localized to centrosomes (Fig. S1 G), raising the possibility that human Talpid3 localization requires an association with centrosome proteins via two distinct regions within Talpid3.
Depletion of Talpid3 prevents primary cilia formation
We next investigated the consequences of Talpid3 depletion. Developmental studies of Talpid3 have shown that this protein is required for cilia formation in several vertebrates, including mice, zebrafish, and chickens (Yin et al., 2009; Bangs et al., 2011; Ben et al., 2011), and therefore, we examined cilia formation in Talpid3-depleted human RPE1 cells. We transfected siRNAs and verified that the levels of Talpid3 protein were reduced by 70–90% in RPE1 cells (Fig. S2 A). When we acutely ablated Talpid3 using several distinct siRNAs in RPE1 cells and induced cilia formation by serum starvation, we found a significant reduction in primary cilia assembly using four canonical markers, glutamylated tubulin (GT335), detyrosinated tubulin, IFT88, and Arl13b (Fig. S2, B and C; and unpublished data). We also ablated Talpid3 in 3T3 cells using a pool of siRNAs or three distinct siRNAs, and observed modest but significant loss of cilia (Fig. S2, D and E). These data suggest that Talpid3 is required for cilia formation in both human and mouse cells. Because it is known that CP110 prevents unrestrained centriolar elongation (Spektor et al., 2007; Kohlmaier et al., 2009; Schmidt et al., 2009) and depletion of CP110-associated proteins, Cep76 or Neurl4, induces aberrant centriole-like structures in U2OS cells (Tsang et al., 2009; Li et al., 2012), we next tested whether loss of Talpid3 provokes these phenotypes. As shown in Fig. S2 (F and G), ablation of Talpid3 did not result in observable centriolar or centrosomal defects, suggesting that Talpid3 may be specifically required for cilia formation.
Talpid3 or Cep290 depletion affects localization of centriolar satellite proteins
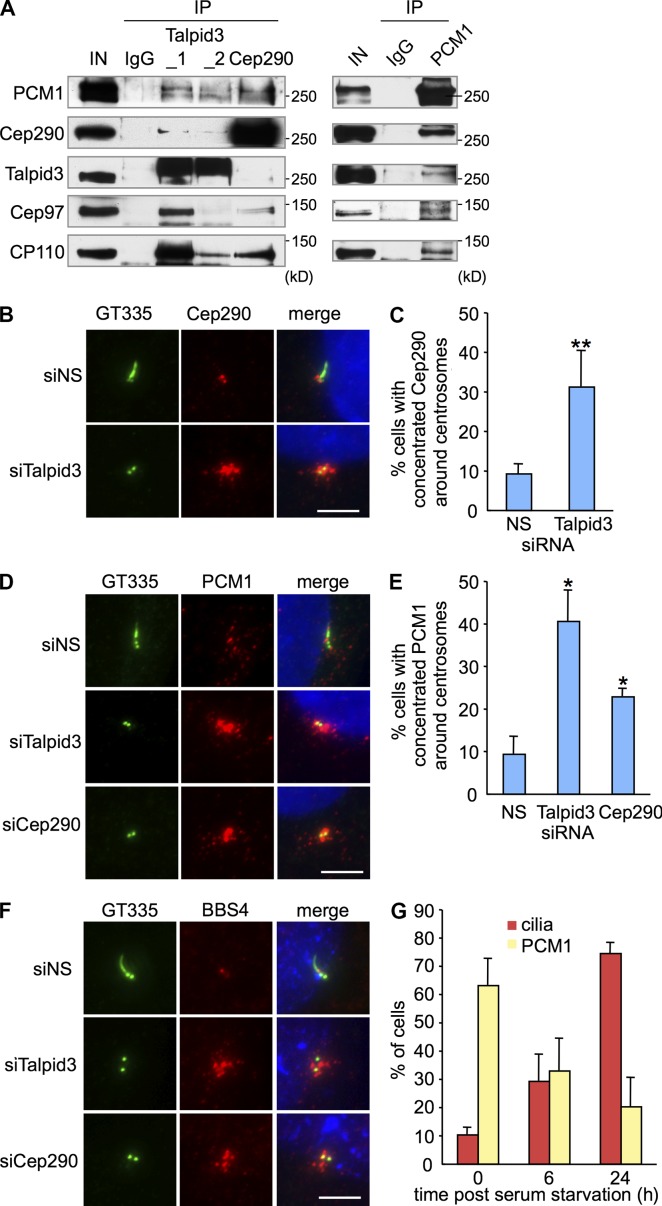
In an effort to uncover the mechanistic role of Talpid3 in cilia assembly, we performed a second proteomic screen to identify Talpid3-binding partners. Tandem mass spectrometry after immunopurification of Flag-tagged Talpid3 confirmed interactions with CP110 and Cep97 (Table S1). Interestingly, we also identified PCM1 as an interacting protein, suggesting that Talpid3, like Cep290, associates with this component of centriolar satellites (Kim et al., 2008). This result was specific because PCM1 peptides were not recovered subsequent to immunopurification of Flag-Kif24. We then determined whether endogenous Talpid3 binds to PCM1. We performed immunoprecipitations using RPE1 cell lysates and observed that PCM1 interacts with Talpid3, CP110, and Cep97 as well as Cep290 (Fig. 4 A).
Figure 4.
Talpid3 interacts with PCM1 and regulates localization of centriolar satellites markers. (A) Western blotting of endogenous PCM1, Cep290, Talpid3, Cep97, and CP110 after immunoprecipitation of RPE1 cell extracts with antibodies indicated at the top of panel. IN, input. (B, D, and F) RPE1 cells transiently transfected with control (siNS), Talpid3, or Cep290 siRNAs were induced to quiescence for 48 h and processed for immunofluorescence with anti-glutamylated tubulin (GT335, green) and (B) anti-Cep290, (D) anti-PCM1, or (F) anti-BBS4 (red) antibodies. Bars, 5 µm. DNA was stained with DAPI (blue). (C and E) The percentage of quiescent RPE1 cells with accumulations of Cep290 (C) or PCM1 (E) granules around the centrosomes are shown. Average of three independent experiments are shown. *, P < 0.01; **, P < 0.05 compared with NS. (G) RPE1 cells were induced to quiescence and processed for immunofluorescence with anti-glutamylated tubulin and anti-PCM1 antibodies at indicated times after serum starvation. The percentages of RPE1 cells with primary cilia or accumulation of PCM1 granules around centrosomes were determined. Average of two independent experiments is shown.
Next, we asked whether these Talpid3-interacting proteins are affected by the loss of Talpid3 in quiescent RPE1 cells. Although the number of CP110 foci was not significantly affected by Talpid3 depletion (Fig. S3, A and B), we found that Cep290 fluorescence became dramatically concentrated around centrosomes in Talpid3-depleted cells (Fig. 4, B and C; and Fig. S3 C). In light of our proteomic data and studies showing that Cep290 localizes to centriolar satellites as well as centrosomes (Andersen et al., 2003; Chang et al., 2006; Sayer et al., 2006; Valente et al., 2006; Kim et al., 2008; Tsang et al., 2008), we also examined the distribution of PCM1, a canonical centriolar satellite marker essential for cilia formation (Nachury et al., 2007; Kim et al., 2008). We observed that PCM1 granules accumulated around centrosomes when Talpid3 was depleted in RPE1 cells (Fig. 4, D and E; and Fig. S3 D). This phenotype was confirmed by depletion of Talpid3 using additional siRNAs (Fig. S3 E). We also observed this phenotype in Cep290-ablated cells, consistent with previous reports (Kim et al., 2008; Stowe et al., 2012). PCM1 protein levels in lysates were not affected by loss of Talpid3 or Cep290 in serum-starved RPE1 cells, nor did ablation of Talpid3 affect Cep290 abundance (Fig. S3 F). These results suggest that ablation of Talpid3 or Cep290 impacts the localization, but not the abundance, of centriolar satellite proteins. Furthermore, we confirmed this phenotype with another established centriolar satellite marker, the Bardet-Biedl Syndrome (BBS) protein, BBS4 (Fig. 4 F; Nachury et al., 2007; Lopes et al., 2011). Collectively, these data suggest that loss of Talpid3 or Cep290 in quiescent RPE1 cells causes accumulation of centriolar satellites. We next asked how this concentration of centriolar satellites around centrosomes relates to cilia formation in RPE1 cells. We found that accumulation of centriolar satellites negatively correlated with cilia assembly (Fig. 4 G). In contrast, silencing of Talpid3 or Cep290 in HeLa or U2OS cells, which do not form primary cilia, had no effect on the distribution of centriolar satellites around centrosomes (Fig. S3 G). Taken together, these data suggest that (1) defects in cilia formation in Talpid3- or Cep290-depleted RPE1 cells are associated with the inappropriate accumulation of centriolar satellites, and (2) depletion of either protein provokes the accumulation of satellites in cells uniquely able to assemble cilia.
We performed rescue experiments by expressing full-length mouse Talpid3, or several truncations thereof, in RPE1 cells depleted of endogenous Talpid3. We observed that both phenotypes provoked by Talpid3 silencing, namely defective cilia assembly and aberrant concentration of centriolar satellites around centrosomes, were largely rescued by expression of full-length Talpid3 (Fig. S3, H and I). However, no Talpid3 truncation mutants significantly rescued the phenotypes associated with Talpid3 deletion (Fig. S3, H and I). These data suggest that multiple domains of Talpid3 contribute to its role in regulating a normal distribution of centriolar satellites and cilia formation.
Our data further suggested that PCM1 associates with a CP110–Cep97–Talpid3 complex and that Talpid3 modulates the localization of centriolar satellites through this interaction. However, these findings did not rule out the possibility that Talpid3 depletion resulted in a general disruption of centrosome integrity. We therefore tested whether a subset of centrosome proteins were affected by loss of Talpid3 in quiescent RPE1 cells. We observed no differences in the staining patterns of pericentrin (pericentriolar matrix), rootletin (proximal region of centrioles), centrin (distal region of centrioles), Cep164 (distal appendages of mother centrioles), cenexin (distal and subdistal appendages of mother centrioles), or Cep170 (subdistal appendages of mother centrioles; Fig. S4 A), suggesting that targeting of these proteins to centrosomes was not impacted by the absence of Talpid3.
Talpid3 and Cep290 are required for proper localization of Rab8a
Loss of Cep290 or PCM1 has been reported to result in exclusion of Rab8a from the ciliary membrane (Kim et al., 2008; Tsang et al., 2008). In addition, another centriolar satellite protein, BBS4, is a component of the BBSome, a core complex of seven BBS proteins that was shown to associate with Rabin8, a GEF for Rab8a (Hattula et al., 2002; Nachury et al., 2007). These observations led us to hypothesize that Talpid3, like Cep290, could control localization of Rab8a during cilia formation. We therefore examined localization of Rab8a to cilia in Talpid3-ablated cells. Interestingly, Rab8a staining of cilia was dramatically reduced upon suppression of Talpid3, similar to Cep290 silencing (Fig. S4, B and C). Exclusion of Rab8a from the ciliary membrane was also observed when Talpid3 or Cep290 was depleted from RPE1 cells stably expressing GFP-Rab8a (GFP-Rab8a-RPE1; unpublished data). These data suggest that Talpid3 affects the localization of Rab8a to the ciliary membrane.
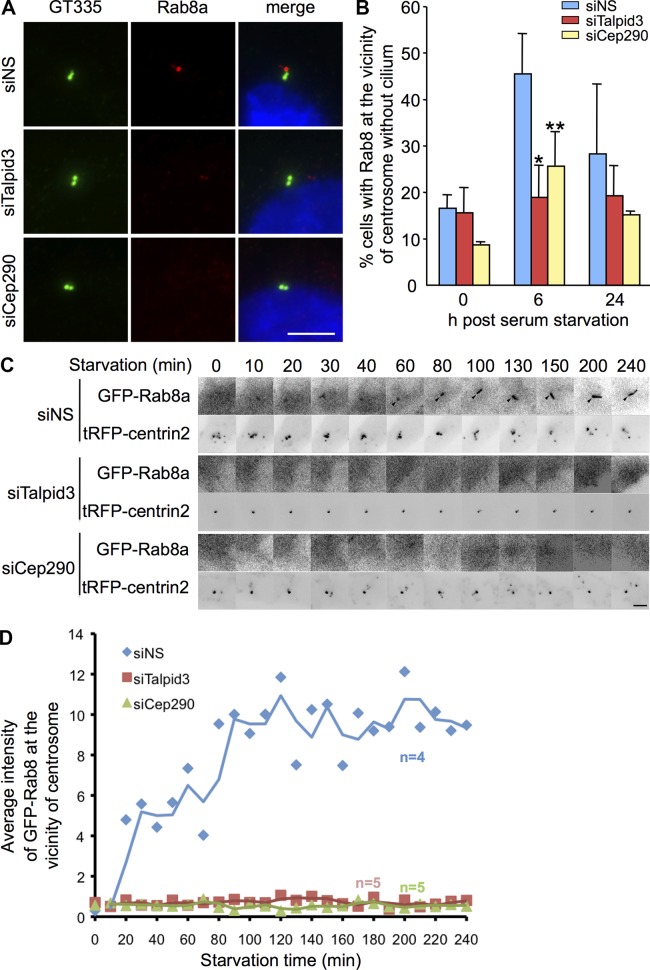
Recent studies showed that ectopically expressed Rab8a accumulates in the vicinity of centrosomes, shortly after cilia are induced to assemble and before elongation of axonemes (Westlake et al., 2011). Moreover, ultrastructural studies have shown that ciliary vesicles are formed near mother centrioles at an early stage of ciliogenesis (Sorokin, 1962). Taken together, these data suggest that Rab8a may be required for vesicle docking and ciliary vesicle formation during the early stages of ciliogenesis. We therefore examined targeting of Rab8a near centrosomes at different time points after induction of ciliogenesis by serum starvation. Although the appearance of cilia increased linearly after serum starvation (unpublished data), we found that the number of cells with Rab8a-positive foci adjacent to nonciliated centrosomes peaked at 6 h and diminished by 24 h after serum starvation in control RPE1 cells (Fig. 5, A and B). These data confirm that endogenous Rab8a localizes near centrosomes at an early stage of ciliogenesis before assembly of the ciliary axoneme. In striking contrast, Talpid3-depleted cells exhibited a significant diminution in the number of cells with Rab8a-positive foci in the vicinity of centrosomes, 6 h after serum starvation, and we observed a similar effect in Cep290-ablated cells (Fig. 5, A and B). To further confirm this phenotype, we performed live-cell imaging using GFP-Rab8a-RPE1 cells. Fluorescent signals of GFP-Rab8a near centrosomes marked by tgRFP-Centrin2 were monitored after serum starvation. We observed that GFP-Rab8a targets to the centrosome within 20 min after serum starvation in control RPE1 cells, and the GFP-Rab8a signal continues to elongate over time (Fig. 5, C and D; and Video 1), consistent with an earlier study (Westlake et al., 2011). On the other hand, GFP-Rab8a signal was not enriched near centrosomes in Talpid3- or Cep290-depleted cells within 240 min after serum starvation (Fig. 5, C and D). Our data suggest that both proteins are required to recruit GFP-Rab8a to centrosomes during the early stages of ciliogenesis before axoneme elongation in RPE1 cells, revealing a potentially critical role in ciliary vesicle formation.
Figure 5.
Talpid3 and Cep290 control Rab8 localization at early phase of ciliogenesis. (A) RPE1 cells transiently transfected with control (siNS), Talpid3, or Cep290 siRNA were induced to quiescence for 6 h and processed for immunofluorescence with anti-glutamylated tubulin (GT335, green) and anti-Rab8a (red) antibodies. DNA was stained with DAPI (blue). Bar, 5 µm. (B) RPE1 cells transiently transfected with control, Talpid3, or Cep290 siRNAs were induced to quiescence and processed for immunofluorescence with anti-glutamylated tubulin and anti-Rab8a antibodies at indicated times after serum starvation. The percentages of RPE1 cells that stain for Rab8a in the vicinity of centrosomes were determined. Average of two to four independent experiments is shown. *, P < 0.01; **, P < 0.05 compared with NS at 6 h after serum starvation. (C and D) GFP-Rab8a RPE1 cells transiently transfected with a plasmid expressing tagRFP-Centrin2 and control, Talpid3, or Cep290 siRNAs were induced to quiescence and processed for live-cell imaging. (C) Images were taken at indicated time points. Bar, 2.5 µm. (D) The average intensity of GFP-Rab8a signals at the vicinity of centrosomes was quantified. Dots and lines show average values and trend lines, respectively.
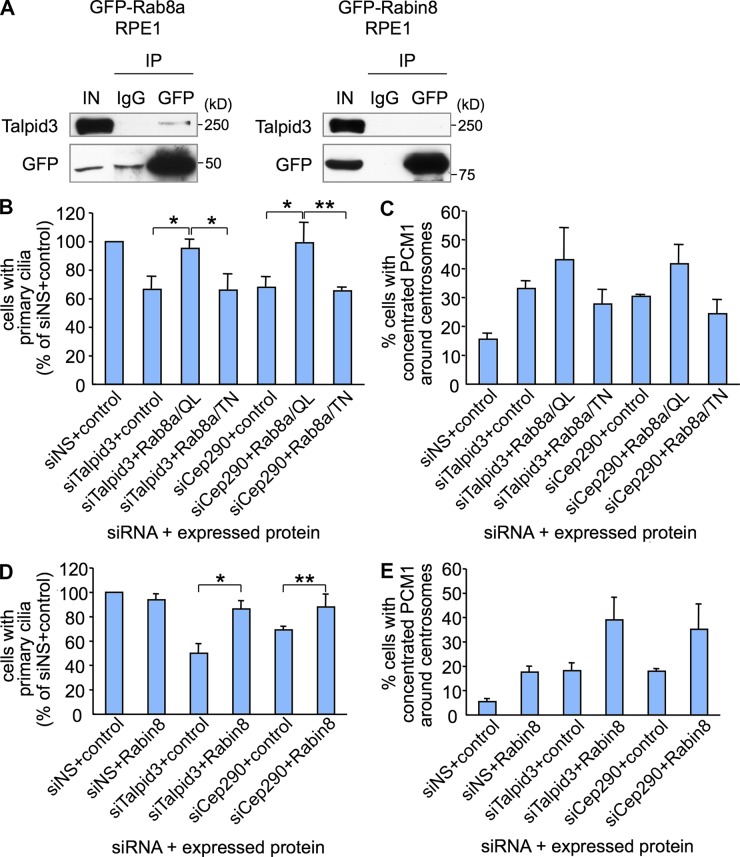
To examine how Talpid3 affects Rab8a localization in RPE1 cells, we performed immunoprecipitation assays using stable RPE1 cells expressing GFP-Rab8a or GFP-Rabin8. Talpid3 was coprecipitated with GFP-Rab8a but not with GFP-Rabin8 (Fig. 6 A). These data suggest that Rab8a localization may be regulated through an association with Talpid3.
Figure 6.
Rab8a functions downstream of Talpid3 and Cep290 in ciliogenesis. (A) Western blotting of Talpid3 and GFP after immunoprecipitation of RPE1 cell extracts stably expressing GFP-Rab8a or GFP-Rabin8 with antibodies indicated at the top of panel. IN, input. (B and D) RPE1 cells treated with control (siNS), Talpid3, or Cep290 siRNA were transiently transfected with plasmids expressing GFP and (B) Myc, Myc-Rab8a/QL, or Myc-Rab8a/TN, or (D) Flag, or Flag-Rabin8, and induced to quiescence for 48 h. Cells were processed for immunofluorescence with an anti-glutamylated tubulin (GT335) antibody. The percentages of GFP-positive RPE1 cells with primary cilia were determined. (C and E) RPE1 cells treated with control, Talpid3, or Cep290 siRNA were transiently transfected with plasmids expressing GFP-Centrin2 and (C) Myc, Myc-Rab8a/QL, or Myc-Rab8a/TN, or (E) Flag, or Flag-Rabin8, and induced to quiescence for 48 h. Cells were processed for immunofluorescence with an anti-PCM1 antibody. The percentages of GFP-Centrin2–positive RPE1 cells with concentrated PCM1 granules around centrosomes were determined. (B–E) Average of three independent experiments is shown. *, P < 0.01; **, P < 0.05.
Activation of Rab8a suppresses defects in cilia formation after Talpid3 depletion
Based on our findings, we hypothesized that Talpid3 or Cep290 depletion promotes an abnormal accumulation of centriolar satellites at centrosomes, impeding proper localization of Rab8a, which is thought to be essential for ciliary vesicle formation at the early stages of cilia formation. To test the epistatic nature of these events, we asked whether the phenotypes induced by loss of Talpid3 or Cep290 could be suppressed through activation of Rab8a. We ectopically expressed constitutively active (QL) or negative (TN) Rab8a in Talpid3- or Cep290-depleted RPE1 cells and examined cilia assembly and concentration of PCM1-positive granules around centrosomes. Significantly, we found that the constitutively active form of Rab8a rescued defects in cilia formation provoked by Talpid3 or Cep290 loss, but the inactive form of Rab8a did not (Fig. 6 B), suggesting that GTP-bound, activated Rab8a functions downstream of Talpid3 or Cep290 during cilia assembly. In striking contrast, constitutive activation of Rab8a could not suppress the abnormal accumulation of PCM1 dots at centrosomes caused by ablation of Talpid3 or Cep290 (Fig. 6 C). Furthermore, we found that expression of Rabin8, a GEF for Rab8, also significantly rescued the cilia assembly defect caused by depletion of Talpid3 or Cep290, but it did not reverse the accumulation of PCM1 dots around centrosomes (Fig. 6, D and E). These data indicate that accumulation of centriolar satellites caused by depletion of Talpid3 or Cep290 may impair downstream events associated with Rab8a function. Collectively, these data also support the idea that Talpid3 and Cep290 regulate the timely dispersal of centriolar satellites, which in turn affects Rab8a localization and/or activation during cilia formation.
Ablation of Talpid3 and/or Cep290 suppresses cilia formation through overlapping and distinct processes
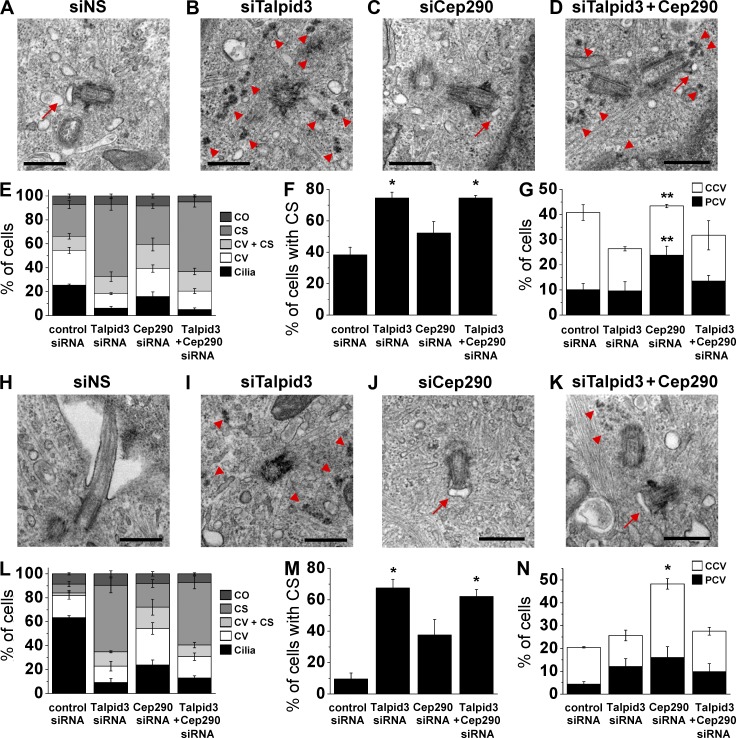
Rab8a serves as an early indicator for ciliary vesicle formation because it is strongly enriched at centrosomes within 6 h of serum starvation (Fig. 5 B). Therefore, to determine whether ciliary vesicle formation is impaired in Talpid3- or Cep290-depleted cells and to examine the relationship between vesicle assembly and centriolar satellite accumulation, we analyzed RPE1 cells at the ultrastructural level using transmission electron microscopy (TEM) at 6 or 24 h of serum starvation after treatment with siRNAs targeting each gene.
In cells depleted of Talpid3 we observed few cilia, as expected, and a reduced number of ciliary vesicles near mother centrioles, as compared with controls (Fig. 7, B and E). Remarkably, we observed an approximately twofold and sevenfold increase in electron-dense granules surrounding the centrosomes in Talpid3-ablated cells after 6 or 24 h of serum starvation, respectively (Fig. 7, B, E, F, I, L, and M). The physical features of these granules were reminiscent of centriolar satellites (Kubo et al., 1999), and the results are consistent with our immunofluorescence studies (Fig. 4, B–F). Conversely, we observed a smaller increase in the percentage of cells with centriolar satellites after Cep290 ablation (Fig. 7, E and F). The phenotype resulting from combined depletion of Talpid3 and Cep290 more closely resembled the one produced by Talpid3 ablation (Fig. 7, D–F and K–M), suggesting that Talpid3 may function upstream of Cep290.
Figure 7.
Ablation of Talpid3 or Cep290 suppresses cilia formation via distinct processes. (A–D, H–K) RPE1 cells transiently transfected with siRNA against control (siNS; A and H), Talpid3 (B and I), Cep290 (C and J), or Talpid3 and Cep290 (D and K) were serum starved for 6 h (A–D) or 24 h (H–K) and processed for TEM. Red arrows indicate ciliary vesicles and red arrowheads indicate centriolar satellites. Bars, 500 nm. (E and L) Each siRNA treated population was quantified at 6 h (E) or 24 h (L) of serum starvation with the percentages of cells that were ciliated (Cilia), contained ciliary vesicles (CV), contained ciliary vesicles and centriolar satellites (CV + CS), or centriolar satellites (CS). Centrosomes without CV and/or CS were labeled as centrosomes only (CO). (F and M) Percentages of cells with CS and CV + CS are quantified for each siRNA-treated sample at 6 h (F) or 24 h (M) of serum starvation. (G and N) Percentages of cells with CV and CV + CS are quantified at 6 h (G) or 24 h (N) of serum starvation according to the frequency of populations containing primary ciliary vesicles (PCV) or capped ciliary vesicles (CCV). (E–G and L–N) Average of three independent experiments is shown. Error bars indicate SEM. *, P < 0.01; **, P < 0.05 compared with control siRNA.
In addition to the aberrant centriolar satellite accumulation, Talpid3 and Cep290 depletion provoked defects in the assembly of ciliary vesicles. Ciliary vesicle formation is a dynamic process, which precedes the growth of the ciliary shaft (Sorokin, 1962, 1968; Schmidt et al., 2012; Kuhns et al., 2013). After 6 h of serum deprivation, we observed ciliary vesicles at the distal tip of mother centrioles in the majority of control cells, suggesting that ciliary vesicle formation is temporally associated with targeting of Rab8a to centrosomes (Fig. 5 B; and Fig. 7, A, E, and G). We observed two classes of vesicles: larger, capped ciliary vesicles (CCVs) and smaller, primary ciliary vesicles (PCVs) associated with distal appendages (Fig. S5, A and B). CCVs sub-tend the entire distal end of mother centrioles, whereas PCVs are considerably smaller and appear to associate primarily with individual (or a subset of) distal appendages. Cells containing CCVs or PCVs were thus scored based on the shape and size of these vesicles, which are sharply circumscribed by lipid bilayers (Fig. 7, A, C, D, J, and K; and Fig. S5, A and B). PCVs are likely to represent the earliest form of ciliary vesicles. Interestingly, in Cep290-depleted cells, vesicles were associated with the distal tip of mother centrioles, but their size was considerably smaller than the CCVs found in control cells (Fig. 7, compare A with C). Indeed, these PCVs were significantly enriched in Cep290-depleted cells by greater than twofold as compared with controls (Fig. 7 G).
Our previous experiments demonstrated that loss of Talpid3 and/or Cep290 significantly impaired cilia formation after 24 h of serum deprivation, a point at which >70% of wild-type RPE1 cells are ciliated (Fig. 4 G). Therefore, to determine whether the loss of Talpid3 and/or Cep290 leads to a block or delay in vesicle assembly and maturation, we analyzed this population of siRNA-transfected cells by TEM. We found that >60% of control cells were ciliated, and the number of cells with centriolar satellites or ciliary vesicles decreased concomitantly relative to the earlier time point (Fig. 7, E–G, and L–N). However, in Talpid3-ablated cells we found that the overall distribution of cells containing ciliary vesicles and primary cilia, as well as centriolar satellites, was similar to the 6-h time point, suggesting that Talpid3 regulates the dispersal of centriolar satellites, which precedes ciliary vesicle formation (Fig. 7, E–G, and L–N). In contrast, after 24 h of serum starvation there was a striking ∼2.5-fold increase in the number of Cep290-depleted cells with ciliary vesicles as compared with the control, accounting for a commensurate lack of cilia in the Cep290-depleted population (Fig. 7, L and N). Of note, in the majority of Cep290-ablated cells, ciliary vesicles persisted as CCVs (Fig. 7 N). These observations suggest that Cep290 is not rate-limiting for the early events in PCV assembly. Rather, Cep290 might participate in a series of steps associated with the conversion of PCVs to CCVs, as well as further maturation of CCVs, which would eventually encapsulate the growing axoneme by a series of vesicular fusion events (Sorokin, 1962, 1968). Co-silencing of Talpid3 and Cep290 again provoked vesicular phenotypes similar to Talpid3 knockdown, further supporting an epistatic relationship wherein Talpid3 functions before Cep290 during cilia formation (Fig. 7, E, G, L, and N). In contrast, depletion of IFT88, a protein acting at a later stage in the cilia assembly pathway, did not lead to defects in centriolar satellite dispersal or PCV-to-CCV maturation at 6 or 24 h of serum deprivation, but instead, depleted cells showed impaired axonemal elongation (unpublished data), consistent with prior studies (Joo et al., 2013). This suggests that the process of ciliogenesis is regulated by a cohort of proteins, specifically involved in distinct phases of cilia formation, and that Talpid3 and Cep290 are involved in the earliest stages.
Interestingly, in addition to cells with PCVs (Fig. 7, C and D; and Fig. S5 B) and CCVs (Fig. 7, A and J; and Fig. S5 A), a third population was associated with both ciliary vesicles and centriolar satellites (CV + CS; Fig. 7, D and K). Talpid3 depletion led to an increase in this latter population in both early (6 h) and late (24 h) stages of cilia formation, as compared with the control, which essentially lacked this population at both stages (Fig. 7, E and L). This result suggests the involvement of Talpid3 in the transition from centriolar satellite accumulation to ciliary vesicle formation, and ablation of this protein essentially inhibits this transition.
Taken together, these data suggest that Talpid3 and Cep290 have overlapping yet distinct roles in cilia assembly. Talpid3, and to a lesser degree, Cep290, are critical for the loss of centriolar satellites, which precedes the formation and/or capture of PCVs. Cep290 also acts subsequently, participating in the transition from PCV to CCV, downstream of centriolar satellite dispersal.
Discussion
A model describing the role of Talpid3 and Cep290 in early events of ciliogenesis
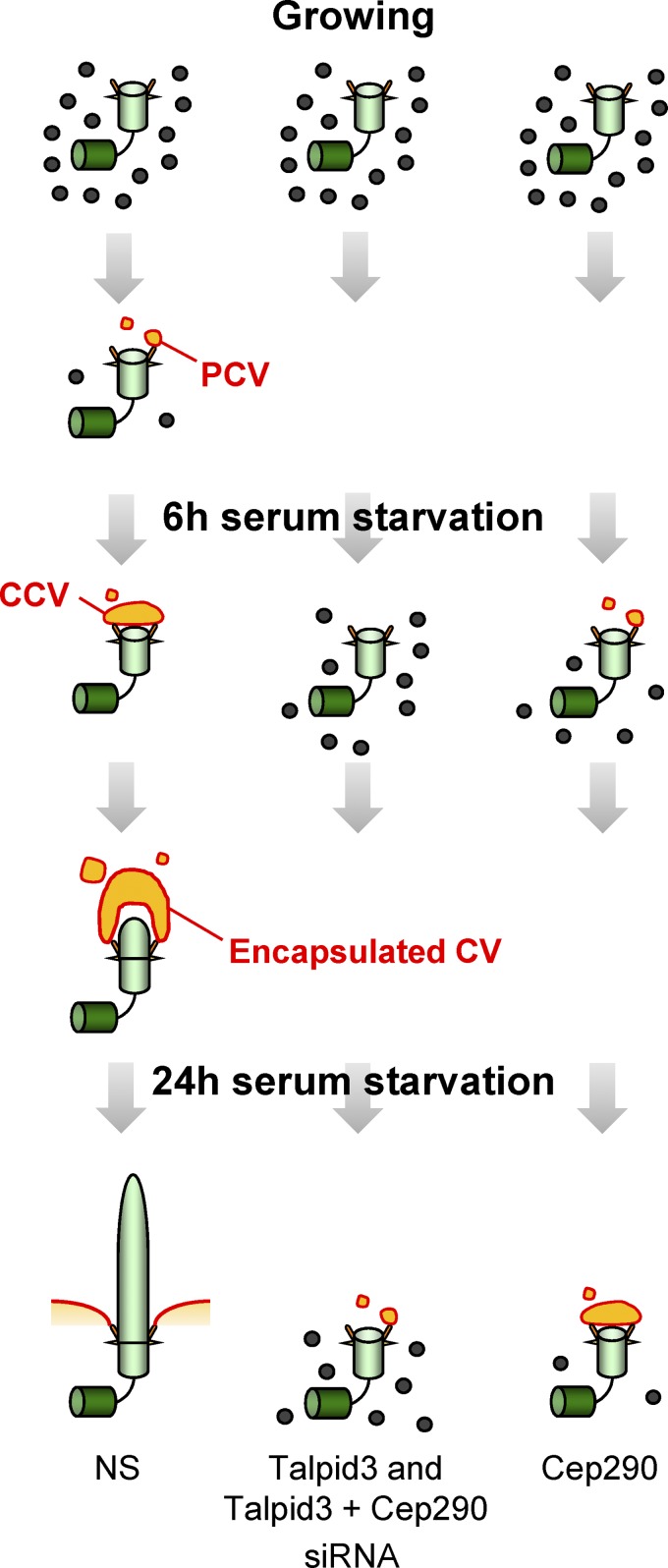
We present a model in which the accumulation of centriolar satellites around the centrosomes in growing cells decreases as cells receive the signal to ciliate (Fig. 8). The loss of satellites is associated with the formation and/or the docking of ciliary vesicles at the distal appendages of the mother centriole, and the subsequent fusion of secondary vesicles leads to the emergence of a capped basal body and a further encapsulated ciliary axoneme. In this setting, distal appendages could serve as a platform onto which vesicles first dock and then fuse. Loss of Talpid3 impairs this process, as evidenced by the accumulation of centriolar satellites and a decrease in the number of cells with ciliary vesicles. It is likely that proteins residing at the distal surface of the centriole/basal body also participate in attachment of the ciliary vesicle, and Talpid3 could play a role here. In this context, it is noteworthy that Talpid3 resides along the “rim” of the distal end of the centriolar barrel, in close proximity to the distal appendage protein, Cep164 (Fig. 3; Graser et al., 2007; Schmidt et al., 2012). Additional proteomic studies to identify Talpid3-interacting proteins could provide clues regarding the attachment of ciliary vesicles to basal bodies.
Figure 8.
Model summarizing the roles of Talpid3 and/or Cep290 during cilium assembly. Centrioles associated with ciliary vesicles are marked as PCV, CCV, and Encapsulated CV (primary, capped, and encapsulated ciliary vesicle, respectively). See text for details.
Our results also suggest the involvement of Talpid3 in the disappearance of centriolar satellites that precedes ciliary vesicle formation. Cep290 also participates in this process because assembly of CCVs and encapsulated ciliary vesicles is delayed, although “capture” of a primary ciliary vesicle is unaffected at the early stages of cilia formation (6 h after induction of quiescence). Even at later stages of cilium induction, Talpid3-ablated cells remain largely arrested in the process of transitioning from a state marked by centriolar satellite accumulation to the next phase of ciliary vesicle formation. In contrast, “capped” vesicular structures accumulated in Cep290-depleted cells during these later stages, suggesting a delay at a point subsequent to the dispersal of centriolar satellites. Thus, the early stages of ciliogenesis can be separated into distinct phases involving the loss of centriolar satellites, followed by the formation and docking of a primary ciliary vesicle at the distal appendage, and subsequent fusion with secondary vesicles to enlarge into a continuous vesicle that encapsulates the nascent ciliary axoneme. Our depletion studies thus strongly suggest that Talpid3 and Cep290 participate in overlapping but also distinct stages of cilia formation.
We note that cells from Talpid3 mutant chicken embryos did not exhibit an overt accumulation of centriolar satellites, although both mutant embryos and Talpid3-depleted cells exhibited near-complete loss of primary cilia and abnormal microtubular extensions in the absence of vesicle formation (Fig. S5, C–E; Yin et al., 2009). It is likely that divergent findings in the two studies stem from either species-specific or technical differences, such as the use of a null mutant as compared with RNAi-mediated silencing, which was robust but not absolute.
The intersecting roles of Rab8a, Talpid3, and Cep290
Our studies also suggest that the regulated targeting and activation of Rab8a to early vesicles could be a pivotal focal point for the control of onset of ciliogenesis. Given that (1) Rab8a accumulates at centrosomes shortly after serum is removed (Westlake et al., 2011); (2) Rab8a localization at the centrosome is abrogated by loss of Talpid3 or Cep290 (Fig. 5 and Video 1); (3) expression of a constitutively activated form of Rab8a or Rabin8, a GEF for Rab8, in Talpid3- or Cep290-depleted cells rescues ciliogenesis without reversing the accumulation of centriolar satellites (Fig. 6); and (4) our observation that primary ciliary vesicle docking readily occurs after Cep290 knockdown, but conversion of the ciliary vesicle to fully formed primary cilia is impaired or delayed (Fig. 7), we suggest that Rab8a acts at a stage of ciliary vesicle formation subsequent to the dispersal of centriolar satellites that could coincide with the enlargement of the ciliary vesicle at the distal tip of the mother centriole. We propose, further, that Talpid3 and Cep290 could act upstream of Rab8a localization and/or activation.
Because centriolar satellites are known to be involved in trafficking of centrosome proteins, Talpid3 and/or Cep290 might control trafficking of Rab8a to the distal tip of the mother centriole, where ciliary vesicles are formed, through interactions with Rab8a and PCM1. Although we did not detect interactions between Talpid3 or Cep290 and Rabin8 (Fig. 6 A; and unpublished data), we cannot exclude the possibility that Talpid3 and/or Cep290 regulate other upstream and/or downstream factors of Rab8a. Possible examples include the Cep290-interacting protein Retinitis pigmentosa GTPase regulator (RPGR), which is known to function as a GEF for Rab8a in vitro (Murga-Zamalloa et al., 2010), and CC2D2A, a Joubert Syndrome–associated protein that is required to localize Rab8a to the outer segment of photoreceptors (Bachmann-Gagescu et al., 2011). Additional examples could include Rkip (Raf-1 kinase inhibitory protein), Hook2, involved in controlling Rab8a localization during ciliogenesis (Baron Gaillard et al., 2011; Murga-Zamalloa et al., 2011), and the BBSome, comprised of proteins defective in Bardet-Biedl syndrome, which interact with PCM1 and Rabin8 and localize to centriolar satellites (Nachury et al., 2007). Another small GTPase, Rab11, activates Rabin8 during cilia formation (Knödler et al., 2010), and further, Sec15, which is a downstream effector of Rab8 and a component of the exocyst, an evolutionarily conserved complex implicated in intracellular vesicle transport, has been shown to be involved in ciliogenesis (Feng et al., 2012). Talpid3 and/or Cep290 might target one or more of these proteins to regulate ciliary vesicle formation. Future studies will be needed to determine how Talpid3 and Cep290 regulate Rab8a-dependent ciliary vesicle formation.
Cep290 has also been shown to localize to the transition zone of flagella in Chlamydomonas and the primary cilium in mammalian cells using immunoelectron microscopy (Craige et al., 2010; Kee et al., 2012). In this setting, it has been postulated that Cep290 tethers the ciliary membrane to transition zone microtubules in mature cilia/flagella. Thus, in addition to the roles we have described here, Cep290 plays an important role much later, after cilia have assembled. However, given that Cep290 also localizes to centriolar satellites, which are primarily observed before the formation of cilia (Fig. 4 G), this population of Cep290 is likely to participate during a step distinct from its role in tethering at the transition zone. Our findings indicate that Cep290 participates in the overall process of early ciliation, including centriolar satellite dispersal and PCV-to-CCV transition, which precedes the formation of the transition zone and subsequent elongation of the axoneme. Given that Cep290 is known to associate with a large spectrum of genetic diseases or ciliopathies, including Nephronophthisis, Joubert syndrome, Bardet-Biedl syndrome, and Meckel-Gruber syndrome, and more than 100 mutations in the Cep290 gene have been identified thus far (Coppieters et al., 2010), it is reasonable to postulate that Cep290 functions in multiple ways during cilia formation. It will be interesting to clarify whether each mutation differentially affects the early and late functions of Cep290 during cilia assembly and to determine whether the corresponding ciliopathies result from specific defects in early events in cilium assembly.
Materials and methods
Cell culture and plasmids
Human RPE1-hTERT, 3T3, 293, U2OS, HeLa, GFP-Rab8a RPE1, and GFP-Rabin8 RPE1 cells (gifts from C. Westlake and P.K. Jackson [Genentech, South San Francisco, CA]) were grown in DMEM supplemented with 10% FBS. A human Talpid3/KIAA0586 cDNA (KIAA0586) was obtained from Kazusa DNA Research Institute (Kazusa-kamatari, Chiba, Japan). To generate Flag-tagged Talpid3 proteins, human Talpid3 fragments encoding residues 1–1533, 1–465, 466–1000, 1001–1533, 1–1000, 466–1533, 162–602, and 603–1233 were amplified by PCR and subcloned into pCMV5-Flag (Kobayashi et al., 2009). To generate Flag-tagged mouse Talpid3 proteins, mouse Talpid3 fragments encoding residues 1–1520, 1–483, 484–1018, 1019–1520, 1–1018, and 484–1520 were amplified by PCR and subcloned into pCMV5-Flag. Plasmids expressing Myc-tagged Rab8a/WT, Q67L, and T22N were obtained from T. Katada (University of Colorado, Boulder, CO) and K. Kontani (University of Tokyo, Tokyo, Japan). GFP-centrin2 was obtained from M. Winey and H.A. Fisk, and Flag-tagged Rabin8 was obtained from K. Mizuno and S. Chiba (Tohoku University, Sendai, Japan). pEGFP-N1 (Invitrogen) was transfected to express GFP in cells.
Antibodies
To generate rabbit anti-Talpid3 antibodies, a GST fusion protein containing residues 1–180 (anti-Talpid3_1) or 847–1026 (anti-Talpid3_2) of human Talpid3 was expressed in Escherichia coli and purified to homogeneity. Antibodies against Talpid3 were purified by affinity chromatography. Antibodies used in this study included anti-Cep290 (Bethyl Laboratories, Inc.); rabbit anti-CP110 (antigen: 1–149 aa of human CP110; Chen et al., 2002); rabbit anti-Cep97 (antigen: 539–689 aa of human Cep97; Spektor et al., 2007); rabbit anti-Kif24 (antigen: 1208–1368 aa of human Kif24; Kobayashi et al., 2011); rabbit anti-Cep76 (antigen: 1–143 aa of human Cep76; Tsang et al., 2009); anti-glutamylated tubulin (GT335; Adipogen); anti-centrin2 (20H5; EMD Millipore); anti–α-tubulin, anti–β-actin, anti-Flag, anti-acetylated tubulin, and anti–γ-tubulin (all from Sigma-Aldrich); anti-GFP (Sigma-Aldrich and Roche); anti-detyrosinated tubulin (EMD Millipore); anti–C-Nap1 (BD); anti–Sas-6 and anti-PCM1 (Santa Cruz Biotechnology, Inc.); anti-Cep170 (Invitrogen); anti-Rab8 (a gift from J. Peranen [University of Helsinki, Helsinki, Finland] and BD); anti-pericentrin (Covance); anti-rootletin and anti-Cep164 (gifts from E. Nigg and E. Lee [University of Basel, Basel, Switzerland] and Novus Biologicals); anti-cenexin (a gift from K. Lee, National Institutes of Health, Bethesda, MD); and anti-BBS4 (a gift from A.M. Fry, University of Leicester, Leicester, England, UK).
RNAi
Synthetic siRNA oligonucleotides were obtained from Thermo Fisher Scientific. Transfection of siRNAs using Lipofectamine 2000 or Lipofectamine RNAiMAX (Invitrogen) was performed according to the manufacturer’s instructions. The 21-nucleotide siRNA sequence for the nonspecific control was 5′-AATTCTCCGAACGTGTCACGT-3′. The 21-nucleotide siRNA sequences for human Talpid3 was 5′-CAAAGTTACCTACGTGTTATT-3′. Non-overlapping siRNA oligos targeting human Talpid3 were: #1, 5′-GTTAAAGGCACTAAGGTAA-3′; #2, 5′-GGGACTAGTTTGAATGGAA-3′; #3, 5′-GATATGGATCGGACACAAA-3′; and #4, 5′-CCGCAGCTCCGTTGATAAA-3′. The 21-nucleotide siRNA sequences for mouse Talpid3 (SmartPool and individual oligos) were: #1, 5′-GGATGGAGCTGCTATGTATTT-3′; #2, 5′-GCAGTTACCTTTGAATAAATT-3′; #3, 5′-GATATTACCTAGTGTGGATTT-3′; and #4, 5′-CCACAACGTTGCAAGACGATT-3′. The siRNA for Cep290 was 5′-AAATTAAGATGCTCACCGATT-3′ (Tsang et al., 2008).
Immunoprecipitation
Cells were lysed with ELB buffer (50 mM Hepes, pH 7, 250 mM NaCl, 5 mM EDTA, pH 8, 0.1% NP-40, 1 mM DTT, 0.5 mM AEBSF, 2 µg/ml leupeptin, 2 µg/ml aprotinin, 10 mM NaF, 50 mM β-glycerophosphate, and 10% glycerol) at 4°C for 30 min. For immunoprecipitation, 2 mg of the resulting supernatant after centrifugation was incubated with an appropriate antibody at 4°C for 1 h and collected using protein A– or G–Sepharose. The resin was washed with ELB buffer, and the bound polypeptides were analyzed by SDS-PAGE and immunoblotting. 50 µg of lysate was typically loaded in the input (IN) lane. For Flag-Talpid3 immunoprecipitation, pCMV5-Flag-Talpid3 or pCMV5-Flag was transfected into HEK293 cells using calcium phosphate. Cells were harvested 48–72 h after transfection. Flag beads (Sigma-Aldrich) were incubated with cell extract at 4°C for 2 h and subsequently washed and processed as described above.
Purification and identification of Talpid3 and Talpid3-binding proteins
In effort to purify the compendium of proteins associated with CP110 and its interactors, we identified proteins associated with Cep76 (Tsang et al., 2009). Flag (vector only control) or Flag-Cep76 was transfected into HEK293 cells using calcium phosphate, and lysates were immuno-affinity purified with anti-Flag agarose beads as described previously (Spektor et al., 2007). Cells were lysed in buffer containing 50 mM Hepes, pH 7, 250 mM NaCl, 5 mM EDTA, pH 8, 0.1% NP-40, 1 mM DTT, 0.2 mM AEBSF, 2 µg/ml leupeptin, 2 µg/ml aprotinin, 10 mM NaF, 50 mM glycerophosphate, and 10% glycerol at 4°C for 30 min. 100 mg of each lysate were incubated with anti-Flag (M2) agarose beads (Sigma-Aldrich) for 3 h at 4°C. The resin was washed with lysis buffer, and the bound polypeptides were eluted with Flag peptide for 30 min at 4°C. The resultant eluates were subjected to mass spectrometric analysis. 42 peptides corresponding to Talpid3/KIAA05686 were identified in the mass spectrometric analysis of Flag-Cep76–associated polypeptides, whereas no Talpid3 peptides were identified in the control. Mass spectrometric identification of Talpid3-associated proteins was performed in a similar manner, with slight modifications (Kobayashi et al., 2011). Flag-Talpid3 was expressed in HEK293 cells and immunoprecipitated with anti-Flag beads for 2 h at 4°C. Bound proteins were eluted with Flag peptide for 30 min, and the resulting eluates were TCA precipitated and fractionated by SDS-PAGE until the dye front reached 1 cm into the resolving gel. A gel slice containing polypeptides was excised after Coomassie staining and subjected to mass spectrometric analysis.
Immunofluorescence microscopy
Cells were fixed with cold methanol for 5 min or with 10% formalin solution (Sigma-Aldrich) for 8 min, and permeabilized with 0.2% Triton X-100/PBS for 10 min. Slides were blocked with 5% BSA in PBS before incubation with primary antibodies. Secondary antibodies used were Cy3-conjugated (Jackson ImmunoResearch Laboratories, Inc.) or Alexa Fluor 488–conjugated (Invitrogen) donkey anti–mouse, anti–rabbit, or anti–rat IgG. Cells were stained with DAPI, and slides were mounted, observed, and photographed using a microscope (63× or 100×, NA 1.4; Axiovert 200M, Carl Zeiss) equipped with a cooled CCD (Retiga 2000R; QImaging) and MetaMorph Software (Molecular Devices). Super-resolution microscopy was performed using a structured-illumination microscopy system (DeltaVision OMX 3D; Applied Precision). A 100×, 1.4 NA oil objective (Olympus) was used with 405 nm, 488 nm, and 593 nm laser illumination and standard excitation and emission filter sets. 125-nm z-steps were applied to acquire raw images, which were reconstructed in 3D using SoftWoRx software (Applied Precision). Image analysis was performed using Photoshop (Adobe).
Live-cell imaging
GFP-Rab8a RPE1 cells were cotransfected with a plasmid expressing tagRFP-Centrin2 and indicated siRNAs for 48 h. The culture media was changed to serum-free DMEM right before live-cell imaging. The fluorescence images were taken with a 40× oil objective (Olympus) mounted on an inverted microscope (model IX71; Olympus) equipped with an on-stage environmental chamber kept at 37°C and 5% CO2 (Tokai Hit). Image analysis was performed using MetaMorph Software.
Transmission electron microscopy (TEM)
EM was performed as before with slight modification (Li et al., 2012). RPE1 cells were washed with PBS followed by fixation with 0.1 M sodium cacodylate buffer (pH 7.4) supplemented with 2% paraformaldehyde, 2.5% glutaraldehyde, and 0.1% Ruthenium red. The cells were post-fixed with 1% osmium tetroxide for 1.5 h at room temperature, and stained with 1% uranyl acetate, then processed in a standard manner and embedded in EMbed 812 (Electron Microscopy Sciences) for TEM. Serial thin (60 nm) sections were cut, mounted on 200 mesh or slotted copper grids, and stained with uranyl acetate and lead citrate. Stained grids were examined using an electron microscope (model CM-12; Philips/FEI) and photographed with a 4-k × 2.7-k digital camera (Gatan, Inc.). TEM experiments were independently repeated three times. Cells with centrioles or cilia were observed in adjacent sections to score for centriolar satellites and/or ciliary vesicle phenotypes. Representative images were processed using Adobe Photoshop. In Fig. 7, the total numbers of cells for each indicated knockdown are: A (n = 91); B (n = 84); C (n = 118), D (n = 79); F (n = 93); G (n = 75); H (n = 102); and I (n = 69).
Online supplemental material
Fig. S1 shows that Talpid3 interacts with CP110-containing protein complex and localizes to the centrosome (related to Figs. 1–3). Fig. S2 shows that ablation of Talpid3 results in cilia assembly defects. Fig. S3 shows that depletion of Talpid3 causes accumulation of centriolar satellite markers without affecting protein levels (related to Fig. 4). Fig. S4 shows that Talpid3 and Cep290 regulate targeting of Rab8a to the ciliary axoneme (related to Fig. 5). Fig. S5 shows that ablation of Talpid3 leads to elongated centrioles without ciliary vesicles (related to Fig. 7). Table S1 shows data of tandem mass spectrometry after immuno-affinity chromatography (related to Fig. 4). Video 1 shows the recruitment and elongation of GFP-Rab8a in control (NS), Talpid3-, or Cep290 siRNA–treated RPE1 cells (related to Fig. 5). Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201304153/DC1.
Supplementary Material
Supplemental Material
Acknowledgments
We thank all members of the Dynlacht laboratory for constructive advice and encouragement. We are grateful to W. Lane and H. Zheng for assistance with mass spectrometric identification of Talpid3- and CP110-associated proteins; K. Dancel, C. Petzold, and F. Liang at the NYU Imaging Center for help with electron microscopy; and the Rockefeller Bio-Imaging Resource Center for help with the OMX microscope. We thank C. Westlake, P.K. Jackson, J. Peranen, E. Nigg, E. Lee, K. Lee, A.M. Fry, M. Winey, H.A. Fisk, T. Katada, K. Kontani, K. Mizuno, and S. Chiba for providing cell lines, antibodies, and plasmids.
T. Kobayashi was supported by grants from JSPS (25860048) and The Kurata Memorial Hitachi Science and Technology Foundation. T. Inoue was supported by a grant from the National Institutes of Health (DK090868). B.D. Dynlacht was supported by grants from the March of Dimes and the National Institutes of Health (1R01HD069647).
Footnotes
Abbreviations used in this paper:
CCV
capped ciliary vesicle
GEF
guanine nucleotide exchange factor
PCM
pericentriolar material
PCV
primary ciliary vesicle
TEM
transmission electron microscopy
References
- Andersen J.S., Wilkinson C.J., Mayor T., Mortensen P., Nigg E.A., Mann M. 2003. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 426:570–574 10.1038/nature02166 [DOI] [PubMed] [Google Scholar]
- Bachmann-Gagescu R., Phelps I.G., Stearns G., Link B.A., Brockerhoff S.E., Moens C.B., Doherty D. 2011. The ciliopathy gene cc2d2a controls zebrafish photoreceptor outer segment development through a role in Rab8-dependent vesicle trafficking. Hum. Mol. Genet. 20:4041–4055 10.1093/hmg/ddr332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bangs F., Antonio N., Thongnuek P., Welten M., Davey M.G., Briscoe J., Tickle C. 2011. Generation of mice with functional inactivation of talpid3, a gene first identified in chicken. Development. 138:3261–3272 10.1242/dev.063602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron Gaillard C.L., Pallesi-Pocachard E., Massey-Harroche D., Richard F., Arsanto J.P., Chauvin J.P., Lecine P., Krämer H., Borg J.P., Le Bivic A. 2011. Hook2 is involved in the morphogenesis of the primary cilium. Mol. Biol. Cell. 22:4549–4562 10.1091/mbc.E11-05-0405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben J., Elworthy S., Ng A.S., van Eeden F., Ingham P.W. 2011. Targeted mutation of the talpid3 gene in zebrafish reveals its conserved requirement for ciliogenesis and Hedgehog signalling across the vertebrates. Development. 138:4969–4978 10.1242/dev.070862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt-Dias M., Glover D.M. 2007. Centrosome biogenesis and function: centrosomics brings new understanding. Nat. Rev. Mol. Cell Biol. 8:451–463 10.1038/nrm2180 [DOI] [PubMed] [Google Scholar]
- Chang B., Khanna H., Hawes N., Jimeno D., He S., Lillo C., Parapuram S.K., Cheng H., Scott A., Hurd R.E., et al. 2006. In-frame deletion in a novel centrosomal/ciliary protein CEP290/NPHP6 perturbs its interaction with RPGR and results in early-onset retinal degeneration in the rd16 mouse. Hum. Mol. Genet. 15:1847–1857 10.1093/hmg/ddl107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Indjeian V.B., McManus M., Wang L., Dynlacht B.D. 2002. CP110, a cell cycle-dependent CDK substrate, regulates centrosome duplication in human cells. Dev. Cell. 3:339–350 10.1016/S1534-5807(02)00258-7 [DOI] [PubMed] [Google Scholar]
- Chiba S., Amagai Y., Homma Y., Fukuda M., Mizuno K. 2013. NDR2-mediated Rabin8 phosphorylation is crucial for ciliogenesis by switching binding specificity from phosphatidylserine to Sec15. EMBO J. 32:874–885 10.1038/emboj.2013.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppieters F., Lefever S., Leroy B.P., De Baere E. 2010. CEP290, a gene with many faces: mutation overview and presentation of CEP290base. Hum. Mutat. 31:1097–1108 10.1002/humu.21337 [DOI] [PubMed] [Google Scholar]
- Craige B., Tsao C.C., Diener D.R., Hou Y., Lechtreck K.F., Rosenbaum J.L., Witman G.B. 2010. CEP290 tethers flagellar transition zone microtubules to the membrane and regulates flagellar protein content. J. Cell Biol. 190:927–940 10.1083/jcb.201006105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammermann A., Merdes A. 2002. Assembly of centrosomal proteins and microtubule organization depends on PCM-1. J. Cell Biol. 159:255–266 10.1083/jcb.200204023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey M.G., Paton I.R., Yin Y., Schmidt M., Bangs F.K., Morrice D.R., Smith T.G., Buxton P., Stamataki D., Tanaka M., et al. 2006. The chicken talpid3 gene encodes a novel protein essential for Hedgehog signaling. Genes Dev. 20:1365–1377 10.1101/gad.369106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ede D.A., Kelly W.A. 1964. Developmental abnormalities in the trunk and limbs of the Talpid3 mutant of the fowl. J. Embryol. Exp. Morphol. 12:339–356 [PubMed] [Google Scholar]
- Feng S., Knödler A., Ren J., Zhang J., Zhang X., Hong Y., Huang S., Peränen J., Guo W. 2012. A Rab8 guanine nucleotide exchange factor-effector interaction network regulates primary ciliogenesis. J. Biol. Chem. 287:15602–15609 10.1074/jbc.M111.333245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrante M.I., Zullo A., Barra A., Bimonte S., Messaddeq N., Studer M., Dollé P., Franco B. 2006. Oral-facial-digital type I protein is required for primary cilia formation and left-right axis specification. Nat. Genet. 38:112–117 10.1038/ng1684 [DOI] [PubMed] [Google Scholar]
- Ghossoub R., Molla-Herman A., Bastin P., Benmerah A. 2011. The ciliary pocket: a once-forgotten membrane domain at the base of cilia. Biol. Cell. 103:131–144 10.1042/BC20100128 [DOI] [PubMed] [Google Scholar]
- Graser S., Stierhof Y.D., Lavoie S.B., Gassner O.S., Lamla S., Le Clech M., Nigg E.A. 2007. Cep164, a novel centriole appendage protein required for primary cilium formation. J. Cell Biol. 179:321–330 10.1083/jcb.200707181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattula K., Furuhjelm J., Arffman A., Peränen J. 2002. A Rab8-specific GDP/GTP exchange factor is involved in actin remodeling and polarized membrane transport. Mol. Biol. Cell. 13:3268–3280 10.109h1/mbc.E02-03-0143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao Y.C., Tong Z.J., Westfall J.E., Ault J.G., Page-McCaw P.S., Ferland R.J. 2009. Ahi1, whose human ortholog is mutated in Joubert syndrome, is required for Rab8a localization, ciliogenesis and vesicle trafficking. Hum. Mol. Genet. 18:3926–3941 10.1093/hmg/ddp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H., Marshall W.F. 2011. Ciliogenesis: building the cell’s antenna. Nat. Rev. Mol. Cell Biol. 12:222–234 10.1038/nrm3085 [DOI] [PubMed] [Google Scholar]
- Joo K., Kim C.G., Lee M.S., Moon H.Y., Lee S.H., Kim M.J., Kweon H.S., Park W.Y., Kim C.H., Gleeson J.G., Kim J. 2013. CCDC41 is required for ciliary vesicle docking to the mother centriole. Proc. Natl. Acad. Sci. USA. 110:5987–5992 10.1073/pnas.1220927110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee H.L., Dishinger J.F., Blasius T.L., Liu C.J., Margolis B., Verhey K.J. 2012. A size-exclusion permeability barrier and nucleoporins characterize a ciliary pore complex that regulates transport into cilia. Nat. Cell Biol. 14:431–437 10.1038/ncb2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Krishnaswami S.R., Gleeson J.G. 2008. CEP290 interacts with the centriolar satellite component PCM-1 and is required for Rab8 localization to the primary cilium. Hum. Mol. Genet. 17:3796–3805 10.1093/hmg/ddn277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleylein-Sohn J., Westendorf J., Le Clech M., Habedanck R., Stierhof Y.D., Nigg E.A. 2007. Plk4-induced centriole biogenesis in human cells. Dev. Cell. 13:190–202 10.1016/j.devcel.2007.07.002 [DOI] [PubMed] [Google Scholar]
- Knödler A., Feng S., Zhang J., Zhang X., Das A., Peränen J., Guo W. 2010. Coordination of Rab8 and Rab11 in primary ciliogenesis. Proc. Natl. Acad. Sci. USA. 107:6346–6351 10.1073/pnas.1002401107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Dynlacht B.D. 2011. Regulating the transition from centriole to basal body. J. Cell Biol. 193:435–444 10.1083/jcb.201101005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Hori Y., Ueda N., Kajiho H., Muraoka S., Shima F., Kataoka T., Kontani K., Katada T. 2009. Biochemical characterization of missense mutations in the Arf/Arl-family small GTPase Arl6 causing Bardet-Biedl syndrome. Biochem. Biophys. Res. Commun. 381:439–442 10.1016/j.bbrc.2009.02.087 [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Tsang W.Y., Li J., Lane W., Dynlacht B.D. 2011. Centriolar kinesin Kif24 interacts with CP110 to remodel microtubules and regulate ciliogenesis. Cell. 145:914–925 10.1016/j.cell.2011.04.028 [DOI] [PubMed] [Google Scholar]
- Kohlmaier G., Loncarek J., Meng X., McEwen B.F., Mogensen M.M., Spektor A., Dynlacht B.D., Khodjakov A., Gönczy P. 2009. Overly long centrioles and defective cell division upon excess of the SAS-4-related protein CPAP. Curr. Biol. 19:1012–1018 10.1016/j.cub.2009.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo A., Tsukita S. 2003. Non-membranous granular organelle consisting of PCM-1: subcellular distribution and cell-cycle-dependent assembly/disassembly. J. Cell Sci. 116:919–928 10.1242/jcs.00282 [DOI] [PubMed] [Google Scholar]
- Kubo A., Sasaki H., Yuba-Kubo A., Tsukita S., Shiina N. 1999. Centriolar satellites: molecular characterization, ATP-dependent movement toward centrioles and possible involvement in ciliogenesis. J. Cell Biol. 147:969–980 10.1083/jcb.147.5.969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhns S., Schmidt K.N., Reymann J., Gilbert D.F., Neuner A., Hub B., Carvalho R., Wiedemann P., Zentgraf H., Erfle H., et al. 2013. The microtubule affinity regulating kinase MARK4 promotes axoneme extension during early ciliogenesis. J. Cell Biol. 200:505–522 10.1083/jcb.201206013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis K.E., Drossopoulou G., Paton I.R., Morrice D.R., Robertson K.E., Burt D.W., Ingham P.W., Tickle C. 1999. Expression of ptc and gli genes in talpid3 suggests bifurcation in Shh pathway. Development. 126:2397–2407 [DOI] [PubMed] [Google Scholar]
- Li J., Kim S., Kobayashi T., Liang F.X., Korzeniewski N., Duensing S., Dynlacht B.D. 2012. Neurl4, a novel daughter centriole protein, prevents formation of ectopic microtubule organizing centres. EMBO Rep. 13:547–553 10.1038/embor.2012.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes C.A., Prosser S.L., Romio L., Hirst R.A., O’Callaghan C., Woolf A.S., Fry A.M. 2011. Centriolar satellites are assembly points for proteins implicated in human ciliopathies, including oral-facial-digital syndrome 1. J. Cell Sci. 124:600–612 10.1242/jcs.077156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murga-Zamalloa C.A., Atkins S.J., Peranen J., Swaroop A., Khanna H. 2010. Interaction of retinitis pigmentosa GTPase regulator (RPGR) with RAB8A GTPase: implications for cilia dysfunction and photoreceptor degeneration. Hum. Mol. Genet. 19:3591–3598 10.1093/hmg/ddq275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murga-Zamalloa C.A., Ghosh A.K., Patil S.B., Reed N.A., Chan L.S., Davuluri S., Peränen J., Hurd T.W., Rachel R.A., Khanna H. 2011. Accumulation of the Raf-1 kinase inhibitory protein (Rkip) is associated with Cep290-mediated photoreceptor degeneration in ciliopathies. J. Biol. Chem. 286:28276–28286 10.1074/jbc.M111.237560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachury M.V., Loktev A.V., Zhang Q., Westlake C.J., Peränen J., Merdes A., Slusarski D.C., Scheller R.H., Bazan J.F., Sheffield V.C., Jackson P.K. 2007. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 129:1201–1213 10.1016/j.cell.2007.03.053 [DOI] [PubMed] [Google Scholar]
- Paoletti A., Moudjou M., Paintrand M., Salisbury J.L., Bornens M. 1996. Most of centrin in animal cells is not centrosome-associated and centrosomal centrin is confined to the distal lumen of centrioles. J. Cell Sci. 109:3089–3102 [DOI] [PubMed] [Google Scholar]
- Sayer J.A., Otto E.A., O’Toole J.F., Nurnberg G., Kennedy M.A., Becker C., Hennies H.C., Helou J., Attanasio M., Fausett B.V., et al. 2006. The centrosomal protein nephrocystin-6 is mutated in Joubert syndrome and activates transcription factor ATF4. Nat. Genet. 38:674–681 10.1038/ng1786 [DOI] [PubMed] [Google Scholar]
- Schmidt K.N., Kuhns S., Neuner A., Hub B., Zentgraf H., Pereira G. 2012. Cep164 mediates vesicular docking to the mother centriole during early steps of ciliogenesis. J. Cell Biol. 199:1083–1101 10.1083/jcb.201202126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt T.I., Kleylein-Sohn J., Westendorf J., Le Clech M., Lavoie S.B., Stierhof Y.D., Nigg E.A. 2009. Control of centriole length by CPAP and CP110. Curr. Biol. 19:1005–1011 10.1016/j.cub.2009.05.016 [DOI] [PubMed] [Google Scholar]
- Singla V., Romaguera-Ros M., Garcia-Verdugo J.M., Reiter J.F. 2010. Ofd1, a human disease gene, regulates the length and distal structure of centrioles. Dev. Cell. 18:410–424 10.1016/j.devcel.2009.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin S. 1962. Centrioles and the formation of rudimentary cilia by fibroblasts and smooth muscle cells. J. Cell Biol. 15:363–377 10.1083/jcb.15.2.363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin S.P. 1968. Reconstructions of centriole formation and ciliogenesis in mammalian lungs. J. Cell Sci. 3:207–230 [DOI] [PubMed] [Google Scholar]
- Spektor A., Tsang W.Y., Khoo D., Dynlacht B.D. 2007. Cep97 and CP110 suppress a cilia assembly program. Cell. 130:678–690 10.1016/j.cell.2007.06.027 [DOI] [PubMed] [Google Scholar]
- Stowe T.R., Wilkinson C.J., Iqbal A., Stearns T. 2012. The centriolar satellite proteins Cep72 and Cep290 interact and are required for recruitment of BBS proteins to the cilium. Mol. Biol. Cell. 23:3322–3335 10.1091/mbc.E12-02-0134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang W.Y., Bossard C., Khanna H., Peränen J., Swaroop A., Malhotra V., Dynlacht B.D. 2008. CP110 suppresses primary cilia formation through its interaction with CEP290, a protein deficient in human ciliary disease. Dev. Cell. 15:187–197 10.1016/j.devcel.2008.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang W.Y., Spektor A., Vijayakumar S., Bista B.R., Li J., Sanchez I., Duensing S., Dynlacht B.D. 2009. Cep76, a centrosomal protein that specifically restrains centriole reduplication. Dev. Cell. 16:649–660 10.1016/j.devcel.2009.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente E.M., Silhavy J.L., Brancati F., Barrano G., Krishnaswami S.R., Castori M., Lancaster M.A., Boltshauser E., Boccone L., Al-Gazali L., et al. International Joubert Syndrome Related Disorders Study Group 2006. Mutations in CEP290, which encodes a centrosomal protein, cause pleiotropic forms of Joubert syndrome. Nat. Genet. 38:623–625 10.1038/ng1805 [DOI] [PubMed] [Google Scholar]
- Westlake C.J., Baye L.M., Nachury M.V., Wright K.J., Ervin K.E., Phu L., Chalouni C., Beck J.S., Kirkpatrick D.S., Slusarski D.C., et al. 2011. Primary cilia membrane assembly is initiated by Rab11 and transport protein particle II (TRAPPII) complex-dependent trafficking of Rabin8 to the centrosome. Proc. Natl. Acad. Sci. USA. 108:2759–2764 10.1073/pnas.1018823108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y., Bangs F., Paton I.R., Prescott A., James J., Davey M.G., Whitley P., Genikhovich G., Technau U., Burt D.W., Tickle C. 2009. The Talpid3 gene (KIAA0586) encodes a centrosomal protein that is essential for primary cilia formation. Development. 136:655–664 10.1242/dev.028464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura S., Egerer J., Fuchs E., Haas A.K., Barr F.A. 2007. Functional dissection of Rab GTPases involved in primary cilium formation. J. Cell Biol. 178:363–369 10.1083/jcb.200703047 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material