Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex (original) (raw)
. Author manuscript; available in PMC: 2015 May 5.
Published in final edited form as: Nature. 2014 Dec 10;517(7536):583–588. doi: 10.1038/nature14136
Abstract
Systematic interrogation of gene function requires the ability to perturb gene expression in a robust and generalizable manner. We describe structure-guided engineering of a CRISPR-Cas9 complex to mediate efficient transcriptional activation at endogenous genomic loci. We use these engineered Cas9 activation complexes to investigate sgRNA targeting rules for effective transcriptional activation, demonstrate multiplexed activation of 10 genes simultaneously, and upregulate long intergenic non-coding RNA (lincRNA) transcripts. We also synthesize a library consisting of 70,290 guides targeting all human RefSeq coding isoforms to screen for genes which, upon activation, confer resistance to a BRAF inhibitor. Expected and potentially novel resistance genes are enriched in the top hits and are validated using individual sgRNA as well as cDNA overexpression. The signature of our top screening hits is significantly correlated with gene expression data from clinical melanoma samples. These results collectively demonstrate the potential of Cas9-based activators as a powerful genetic perturbation technology.
Achieving systematic, genome-scale perturbations within intact biological systems is important for elucidating gene function and epigenetic regulation. Genetic perturbations can be broadly classified as either loss-of-function or gain-of-function (GOF) based on their mode of action. To date, various genome-scale loss-of-function screening methods have been developed, including approaches employing RNA interference1,2 and the RNA-guided endonuclease Cas9 from the microbial CRISPR (clustered regularly interspaced short palindromic repeat) adaptive immune system3,4. Genome-scale GOF screening approaches have largely remained limited to the use of cDNA library overexpression systems. However, it is difficult to capture the complexity of transcript isoform variance using these libraries, and large cDNA sequences are often difficult to clone into size-limited viral expression vectors. The cost and complexity of synthesizing and using pooled cDNA libraries have also limited their use. Novel technologies that overcome such limitations would enable systematic, genome-scale GOF perturbations at endogenous loci.
Programmable DNA binding proteins have emerged as an exciting platform for engineering synthetic transcription factors for modulating endogenous gene expression5–11. Among the established custom DNA binding domains, Cas9 is most easily scaled to facilitate genome-scale perturbations3,4 due to its simplicity of programming relative to zinc finger proteins and transcription activator-like effectors (TALEs). Cas9 nuclease can be converted into an RNA-guided DNA binding protein (dCas9) via inactivation of its two catalytic domains12,13 and then fused to transcription activation domains. These dCas9-activator fusions targeted to the promoter region of endogenous genes can then modulate gene expression7–11. Although the current generation of dCas9-based transcription activators is able to achieve up-regulation of some endogenous loci, the magnitude of transcriptional up-regulation achieved by individual single-guide RNAs (sgRNAs)12 typically ranges from low to ineffective8–11. Tiling a given promoter region with several sgRNAs can produce more robust transcriptional activation9–11, but this requirement presents enormous challenges for scalability, and in particular for establishing pooled, genome-wide GOF screens.
In order to improve and expand applications of Cas9, we recently undertook crystallographic studies to elucidate the atomic structure of the Cas9-sgRNA-target DNA tertiary complex14, thus enabling rational engineering of Cas9 and sgRNA. Here we report a series of structure-guided engineering efforts to create a potent transcription activation complex capable of mediating robust up-regulation with a single sgRNA. Using this new activation system, we demonstrate activation of endogenous genes as well as non-coding RNAs, elucidate design rules for effective sgRNA target sites, and establish and apply genome-wide dCas9-based transcription activation screening to study drug resistance in a melanoma model. These results collectively demonstrate the broad applicability of CRISPR-based GOF screening for functional genomics research.
Structure-guided design of Cas9 complex
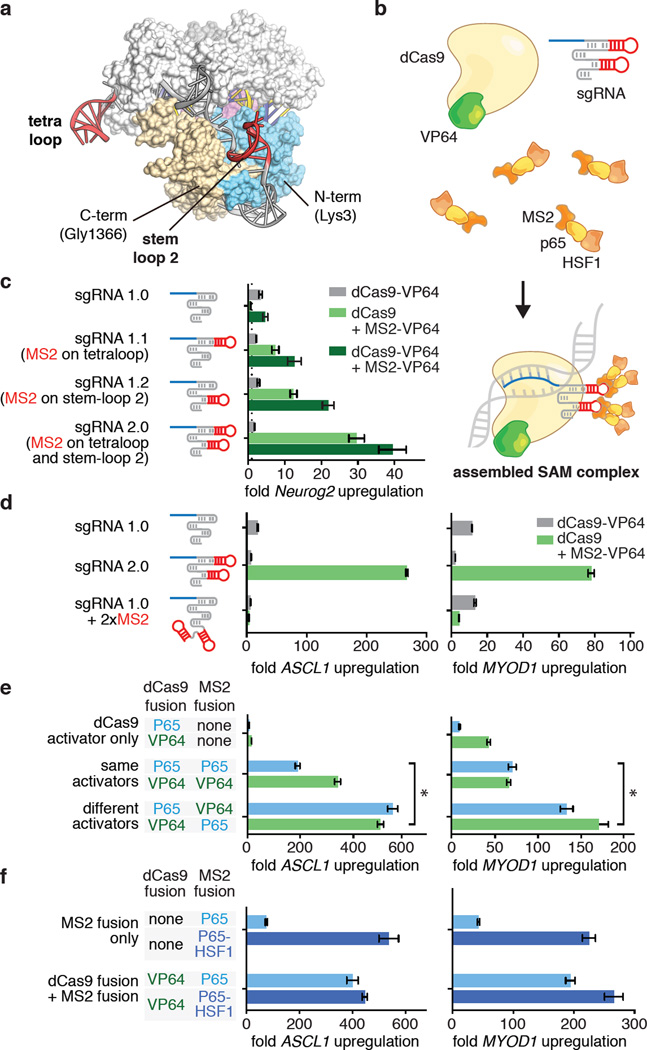
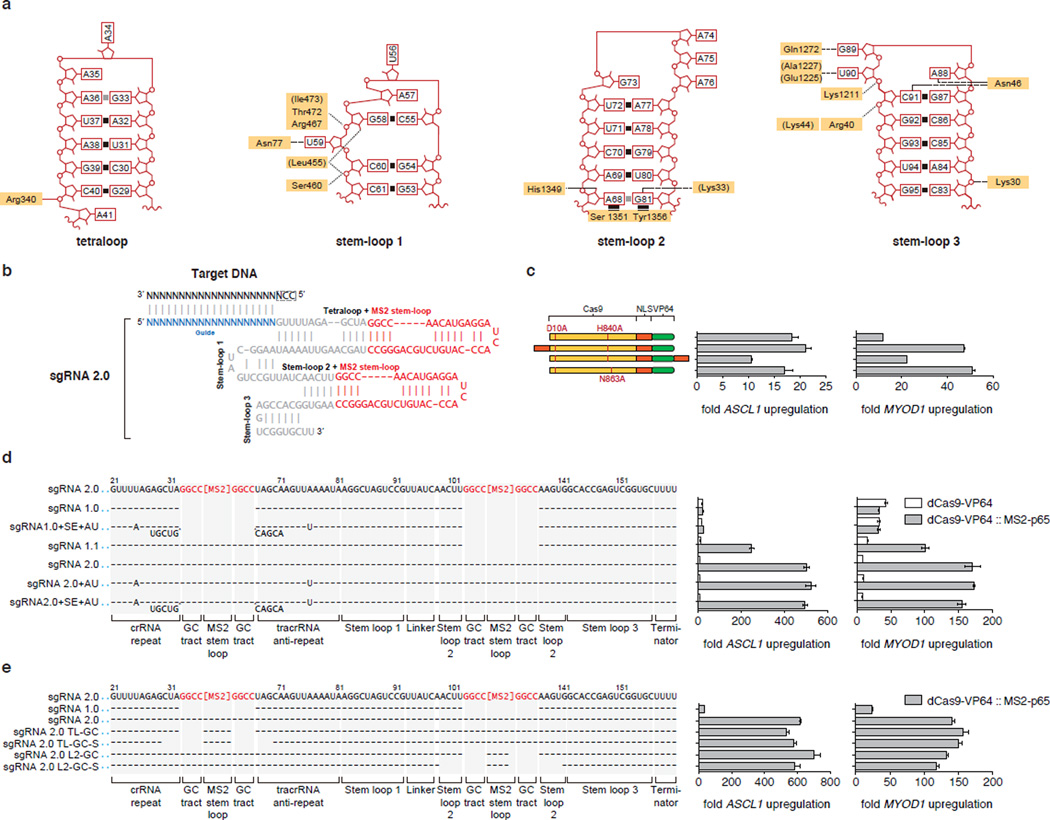
Transformation of the Cas9-sgRNA complex into an effective transcriptional activator requires finding optimal anchoring positions for the activation domains. Previous designs of dCas9-based transcription activators have relied on fusion of transactivation domains to either the N- or C-terminus of the dCas9 protein. To explore whether alternate anchoring positions would improve performance, we examined our previously determined crystal structure of the Streptococcus pyogenes dCas9 (D10A/H840A) in complex with a single guide RNA (sgRNA) and complementary target DNA14. We observed that the tetraloop and stem-loop 2 of the sgRNA protrude outside of the Cas9-sgRNA ribonucleoprotein complex, with the distal 4 bp of each stem completely free of interactions with Cas9 amino acid sidechains (Extended Data Fig. 1a). Based on these observations, along with functional data demonstrating that substitutions and deletions in the tetraloop and stem-loop 2 regions of the sgRNA sequence do not affect Cas9 catalytic function14 (Fig. 1a), we reasoned that the tetraloop and stem-loop 2 could tolerate the addition of protein-interacting RNA aptamers to facilitate the recruitment of effector domains to the Cas9 complex (Fig. 1b).
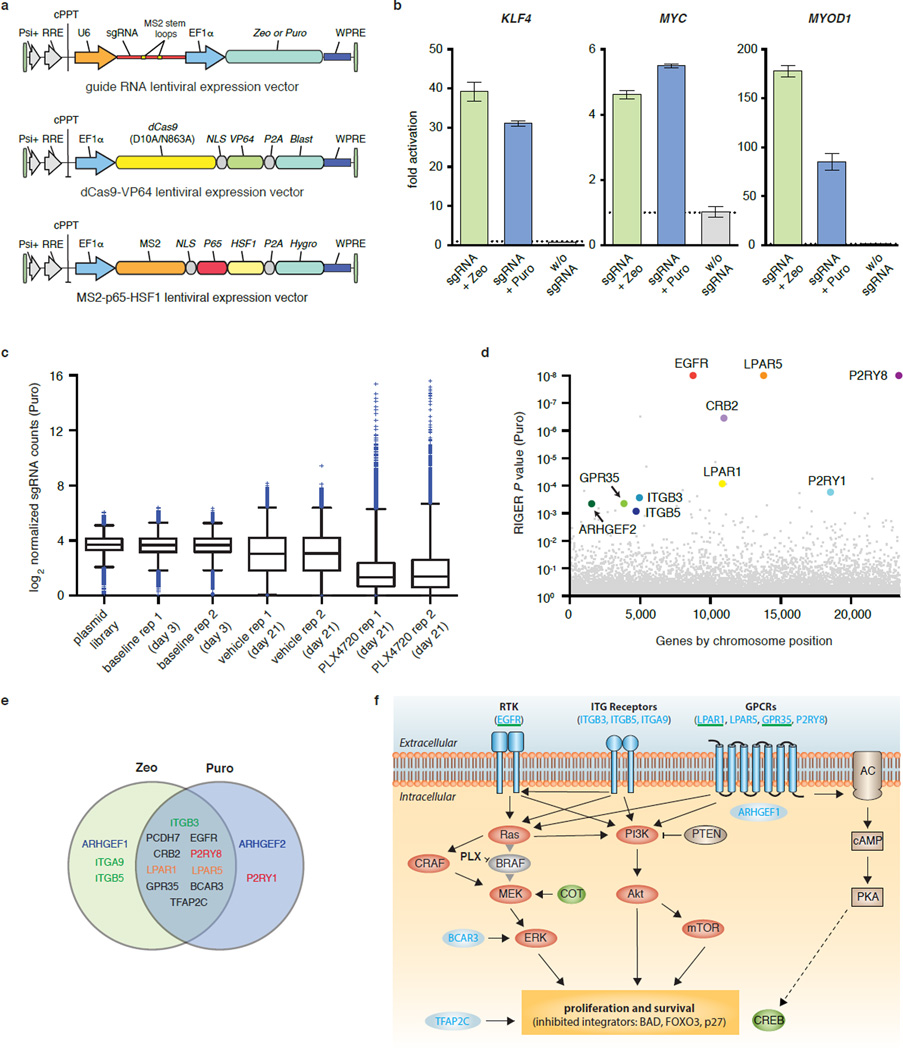
Figure 1. Structure-guided design and optimization of an RNA-guided transcription activation complex.
a, A crystal structure of the Cas9-sgRNA-target DNA tertiary complex (PDB ID: 4OO8)14 reveals that the sgRNA tetraloop and stem loop 2 are exposed. b, Schematic of the three-component SAM system. c, Design and optimization of sgRNA scaffolds for optimal recruitment of MS2-VP64 transactivators in Neuro-2a cells. d, MS2 stem-loop placement within the sgRNA significantly affects transcription activation efficiency. e, Combinations of different activation domains act in synergy to enhance the level of transcription activation. f, Addition of the HSF1 transactivation domain to MS2-p65 further increases the efficiency of transcription activation. Experiments for d-f were performed in 293FT cells. All values are mean ± SEM with n = 3. * indicates p <0.05 based on Student’s t-test.
We selected a minimal hairpin aptamer, which selectively binds dimerized MS2 bacteriophage coat proteins in mammalian cells, and appended it to the sgRNA tetraloop and stem-loop 215 (Extended Data Fig. 1b). We next tested whether MS2-mediated recruitment of VP64 to the tetraloop and stem-loop 2 could mediate transcriptional up-regulation more efficiently than a dCas9-VP64 fusion. As predicted, aptamer-mediated recruitment of MS2-VP64 to either tetraloop (sgRNA1.1) or stem-loop 2 (sgRNA1.2) mediated 3- and 5-fold higher levels of Neurog2 up-regulation than a dCas9-VP64 fusion (sgRNA 1.0), respectively. Recruitment of VP64 to both positions (sgRNA 2.0) resulted in an additive effect, leading to 12-fold increase over dCas9-VP64 (sgRNA 1.0). Combining sgRNA 2.0 with dCas9-VP64 instead of dCas9 provided an additional 1.3-fold increase in Neurog2 up-regulation (Fig. 1c). We further compared sgRNA 2.0 to a previously-described sgRNA bearing two MS2-binding stem-loops at the 3` end (sgRNA + 2×MS2)11 and found that sgRNA 2.0 drove 14- and 8.5-fold higher levels of transcription activation than sgRNA + 2×MS2 for ASCL1 and MYOD1, respectively (Fig. 1d). This difference could be due to either improved positioning of MS2 stemloops or to dCas9 protection of internal MS2 stemloops from exonuclease degradation.
To further improve the potency of Cas9-mediated gene activation, we considered how transcriptional activation is achieved in natural contexts, where endogenous transcription factors generally act in synergy with co-factors16. We thus hypothesized that combining VP64 with additional, distinct activation domains could improve activation efficiency. We chose the NF-κB trans-activating subunit p65, which, while sharing some common co-factors with VP64, recruits a distinct subset of transcription factors and chromatin remodeling complexes. For example, p65 has been shown to recruit AP-1, ATF/CREB, and SP117, whereas VP64 recruits PC418, CBP/p30019, and the SWI/SNF complex20.
We then varied the effector domain fused to dCas9 or MS2. Hetero-effector pairing of dCas9 and MS2 fusion proteins (e.g. dCas9-VP64 paired with MS2-p65 or dCas9-p65 with MS2-VP64) provided over 2.5-fold higher transcription activation for both ASCL1 and MYOD1 than homo-effector pairing (e.g. dCas9-VP64 paired with MS2-VP64 or dCas9-p65 with MS2-p65) (Fig. 1e). We further explored this concept of domain synergy by introducing the activation domain from human heat-shock factor 1 (HSF1)21 as a third activation domain, and found that an MS2-p65-HSF1 fusion protein further improved transcriptional activation of ASCL1 (12%) and MYOD1 (37%) (Fig. 1f). Additional modifications to the sgRNA as well as Cas9 protein, including varying the nuclear localization signal (NLS), provided only minor improvements (Extended Data Fig. 1c–e). Based on these collective results, we concluded that the combination of sgRNA 2.0, NLS-dCas9-VP64, and MS2-p65-HSF1 comprises the most effective transcription activation system, and designated it synergistic activation mediator (SAM). For simplicity, we will refer to sgRNA 2.0 as sgRNA in subsequent discussions, unless noted otherwise.
Design rules for efficient sgRNAs
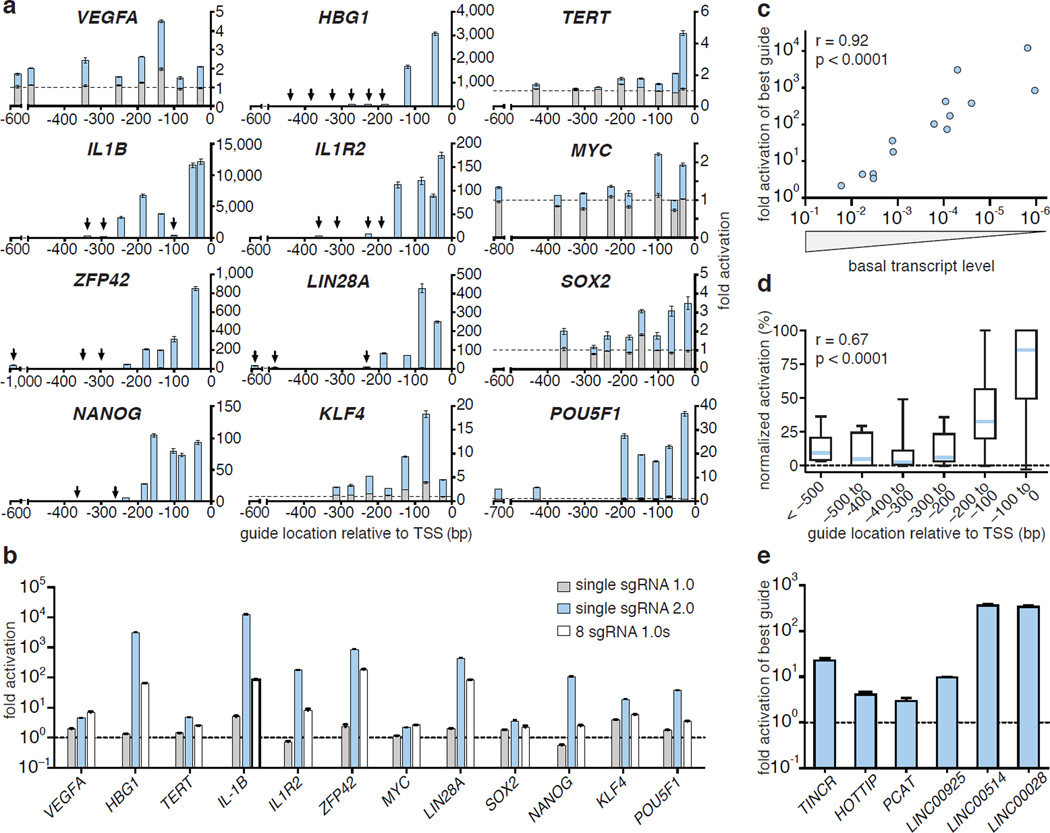
To thoroughly evaluate the effectiveness of SAM for activating endogenous gene transcription, we chose 12 genes that were previously found by several groups to be difficult to activate using dCas9-VP64 and individual sgRNA 1.0 guides8,10,11. For each gene, we selected 8 sgRNA target sites spread across the proximal promoter between −1000bp and the +1 transcription start site (TSS). For 9 out of 12 genes, the maximum level of activation achieved using dCas9-VP64 with any of the 8 sgRNA 1.0 guides was less than 2-fold, while the remaining three genes (ZFP42, KLF4 and IL1B) were maximally activated between 2- and 5-fold (Fig. 2a). In contrast, SAM stimulated transcription at least 2-fold for all genes and more than 15-fold for 8 out of 12 genes. Consistently, SAM performed better than sgRNA 1.0 + dCas9-VP64 for all 96 guides, with a median gain of 105-fold greater up-regulation across all 12 genes (activation by SAM divided by activation by sgRNA 1.0).
Figure 2. Characterization of SAM-mediated gene and lincRNA activation and derivation of selection rules for efficient sgRNAs.
a, Fold activation of 12 different genes plotted against the sgRNA location. sgRNA 1.0 with dCas9-VP64 (grey), sgRNA 2.0 with dCas9-VP64 and MS2-p65-HSF1 (blue). b, Comparison of activation efficiency of 12 target genes: dCas9-VP64 and a single sgRNA 1.0; dCas9-VP64 with a single sgRNA 2.0 and MS2-p65-HSF1, and dCas9-VP64 with a mixture of 8 sgRNA 1.0s. c, Efficiency of target gene activation as a function of baseline expression levels. d, Correlation of gene activation efficiency with sgRNA targeting position. Activation efficiency of each sgRNA for the same target gene is normalized against the highest-activating sgRNA. e, Fold activation of six lincRNA transcripts by SAM (best sgRNA out of 8 tested). All experiments were performed in 293FT cells. All values are mean ± SEM with n = 3.
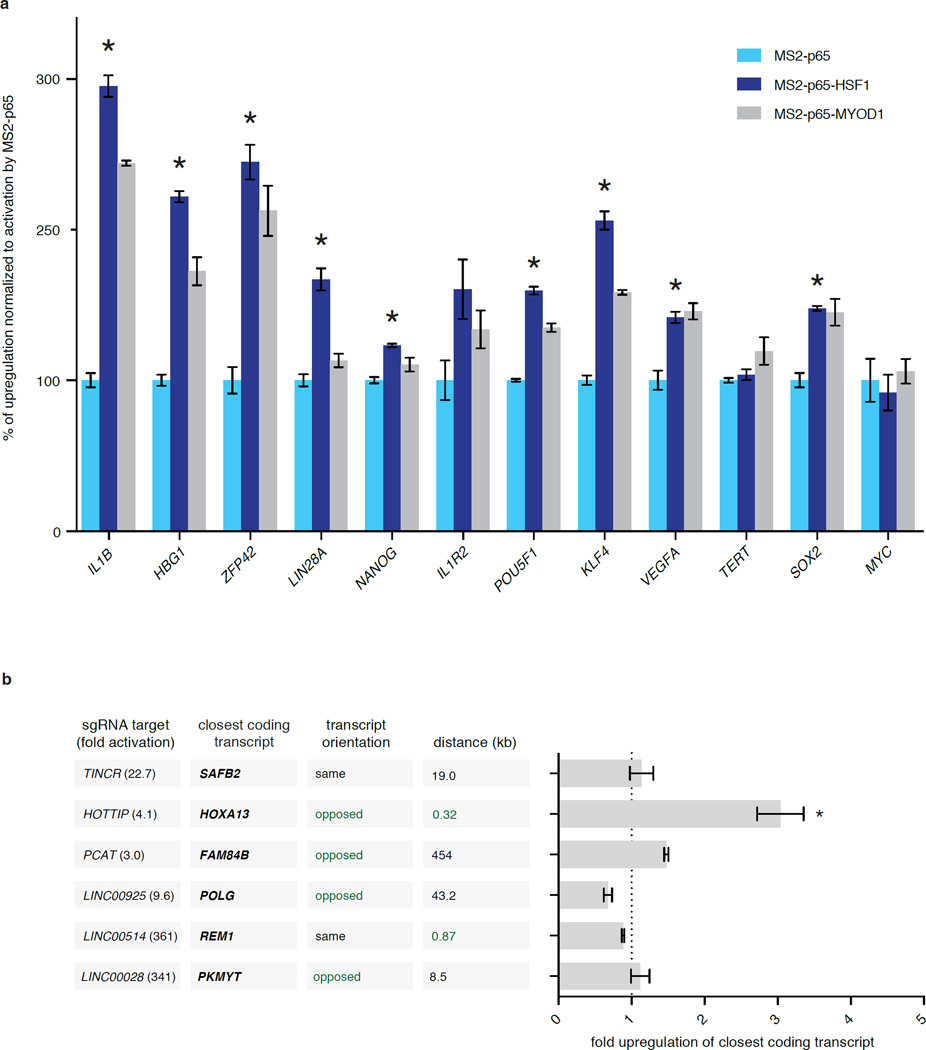
Previous studies have demonstrated that the poor activation efficiency of single sgRNAs can be overcome by combining dCas9-VP64 with a pool of sgRNAs tiling the proximal promoter region of the target gene9–11. Therefore we compared the single sgRNA activation efficiency of SAM against dCas9-VP64 combined with a pool of 8 sgRNA 1.0 guides and MS2-effector fusions, all targeting the same gene. For most genes, SAM with a single sgRNA performed more robustly than dCas9-VP64 with pools of 8 sgRNA 1.0 guides (Fig. 2b). For 9 out of 12 genes, MS2-p65-HSF1 outperformed MS2-p65 alone, and addition of another activation domain MyoD1 (MS2-p65-MyoD1) also improved performance (Extended Data Fig. 2a)
Next, we sought to determine factors that contribute to inter- and intragenic variability of activation efficiency by different sgRNAs. For intergene variability, differences in activation magnitudes could be due to epigenetic factors and/or variation in basal transcription levels. We were thus interested in correlating basal transcription with the level of transcription activation achieved using SAM. Using the relative transcriptional levels of target genes in control samples, we observed a highly significant correlation between the inverse of basal transcript level and the fold up-regulation achieved using SAM (Fig. 2c; r = 0.94, p < 0.0001). This suggests that the basal expression level of each gene largely determines the level of activation.
To study the intragenic variability of SAM activity, we aggregated the activation data for all 96 guides and found the distance between the guide RNA target site and the TSS to be the strongest predictor of activation efficiency (Fig. 2d; r = 0.67, p < 0.0001). For all genes, the highest levels of activation were consistently achieved by targeting within the −200 bp to +1 bp window. This simple design guideline can inform the selection of efficient sgRNAs for gene activation.
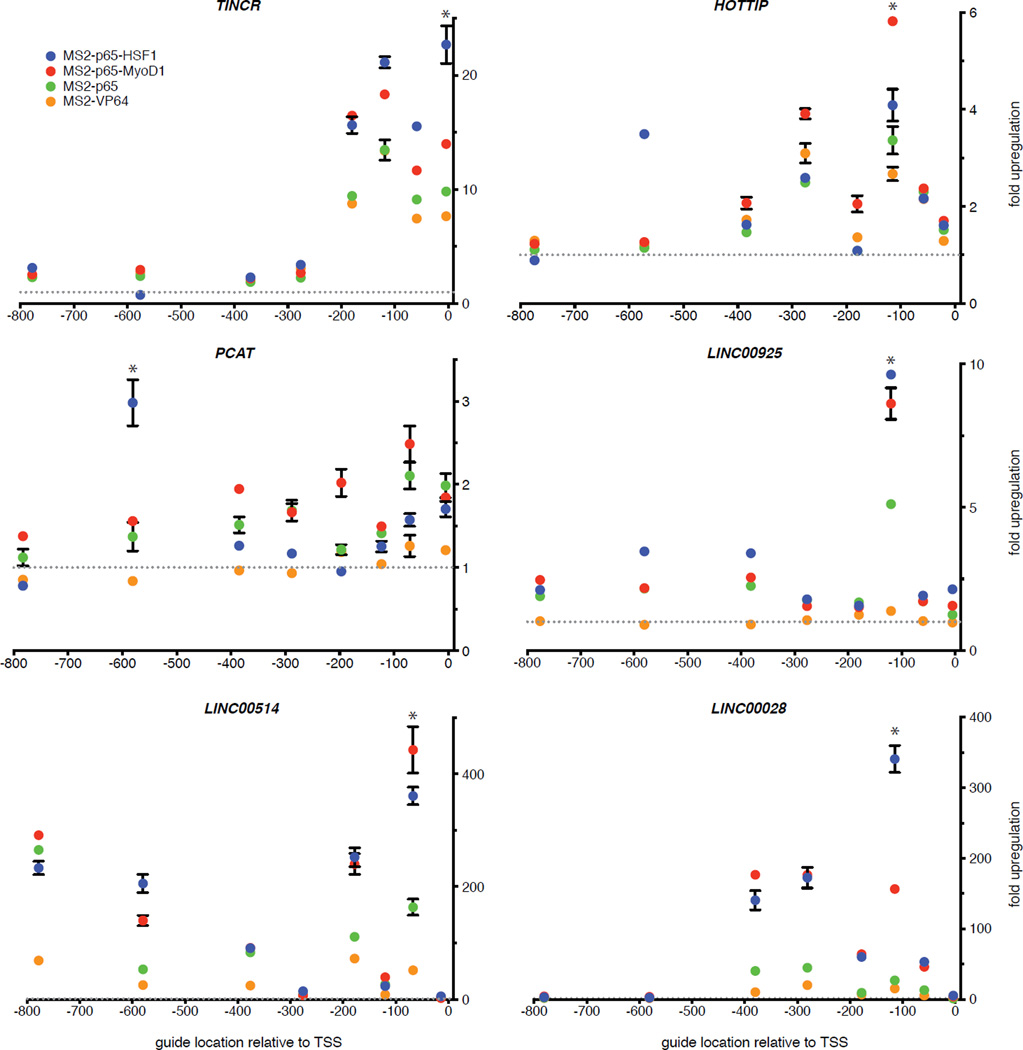
We also sought to test whether SAM is able to activate non-coding elements in addition to protein-coding genes. We chose a diverse set of 6 lincRNAs and found that SAM mediated significant up-regulation of each target (Fig. 2e), with MS2-p65-HSF1 or MS2-p65-MyoD1 leading to the highest levels of activation for each lincRNA (p < 0.01) (Extended Data Fig. 3). We also examined the effect of the most potent sgRNA for each lincRNA on the transcription of the nearest coding gene. Of all sgRNAs tested, only the sgRNA targeting HOTTIP – the only sgRNA located within 500bp of the neighboring gene’s TSS – led to significant activation of its neighbor (Extended Data Fig. 2b).
Multiplex gene activation
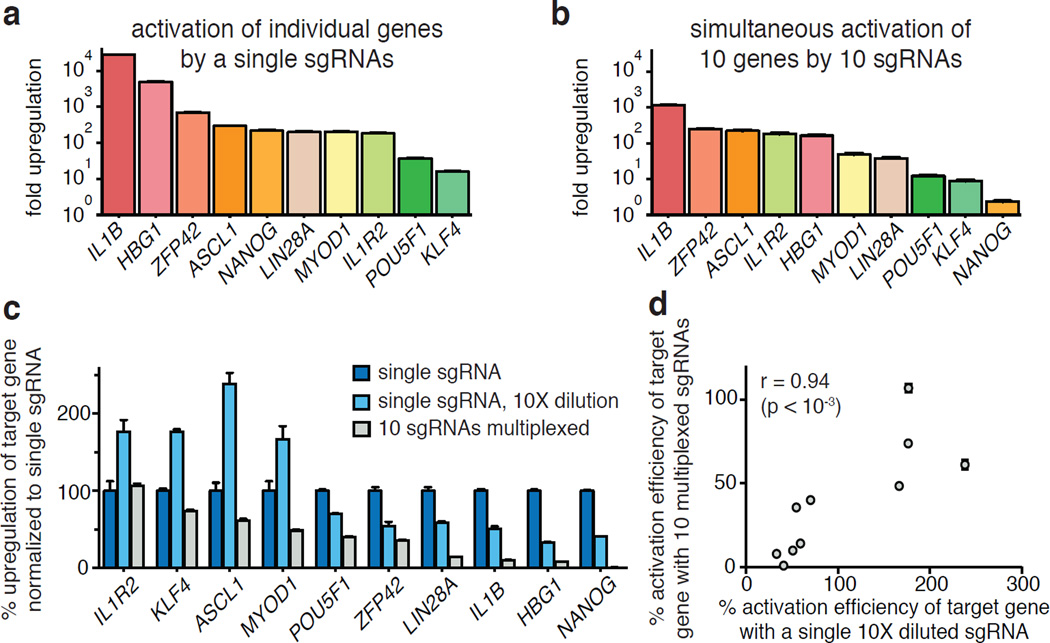
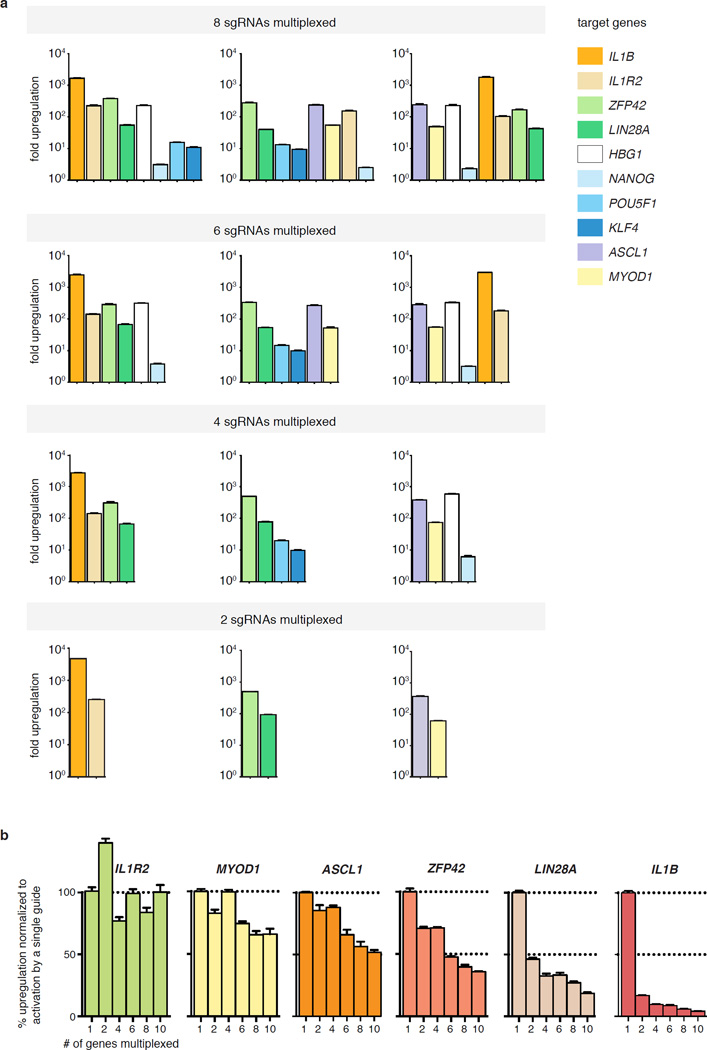
The ability to simultaneously modulate gene expression at multiple loci would allow a better understanding of complex genetic and regulatory networks. Using sets of 2 to 10 sgRNAs, we observed successful activation of all target genes (>2-fold) within all sgRNA combinations (Fig. 3a and 3b and Extended Data Fig. 4). As expected, most genes (excluding IL1R2) exhibited a decrease in the amount of up-regulation achieved when concurrently targeted with 9 other genes. Interestingly, the relative activation levels of each gene changed between multiplex activation and single-gene activation experiments (Fig. 3a and b).
Figure 3. Simultaneous activation of endogenous genes using multiplexed sgRNA expression.
a, Activation of individual genes by single sgRNAs with dCas9-VP64 and MS2-p65-HSF1. b, Simultaneous activation of the same ten genes using a mixture of ten sgRNAs each targeting a different gene. c, Effect of sgRNA dilution on gene activation efficiency. d, Correlation between the activation efficiency of a single 10-fold diluted sgRNA and that of the same sgRNA delivered within a mixture of ten different-gene targeting sgRNAs. All values are mean ± SEM with n = 3.
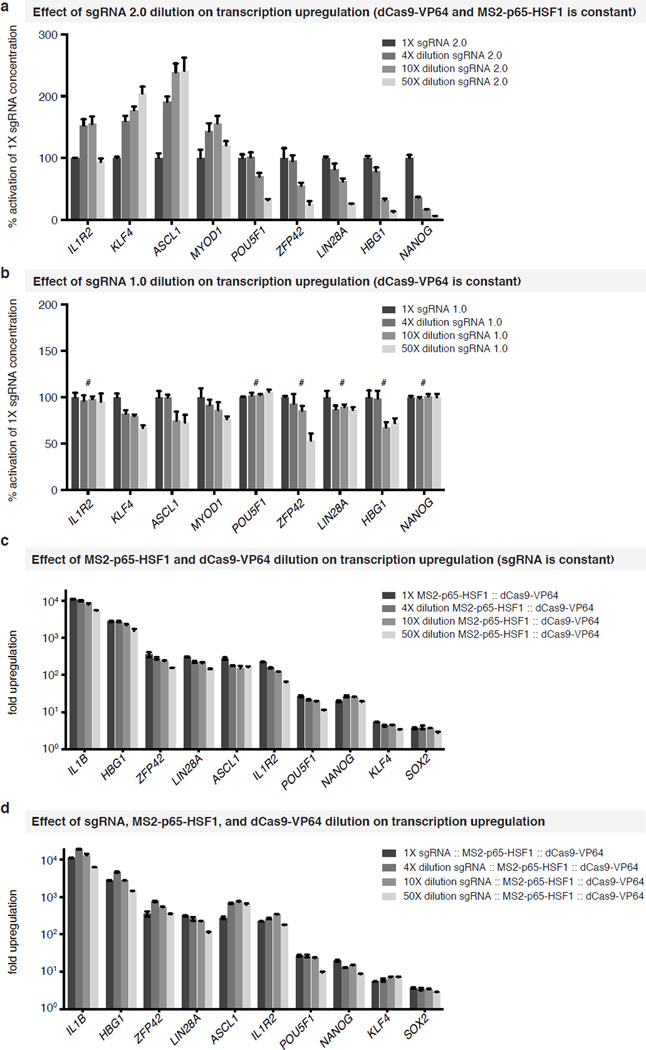
We asked if reduced activation of targets during multiplexing was due to the reduced amounts of sgRNA or SAM protein components. Surprisingly, diluting the sgRNA expression plasmid by 10-fold in single-gene activation experiments did not reduce activation for all genes (Fig. 3c). We found that genes whose levels of activation are reduced upon sgRNA dilution also exhibited dampened levels of activation when multiplexed (Fig. 3d; r = 0.94, p<0.001). In contrast, the activation efficiency of SAM was generally unperturbed by dilution of its protein components (dCas9-VP64 and MS2-p65-HSF1) (Extended Data Fig. 5). Activation efficiency remained stable particularly when all three components were diluted, retaining on average 90% activation efficiency across a 50-fold dilution range (Extended Data Fig. 5). This finding was particularly promising for genome-scale pooled screening applications, which rely on single-copy lentiviral integration.
Specificity of SAM-mediated activation
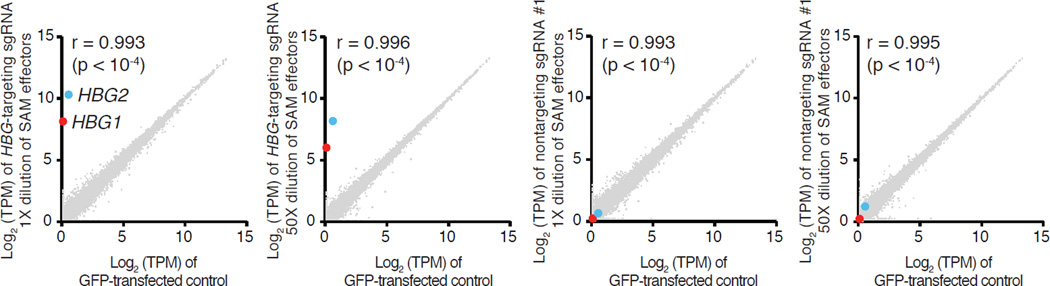
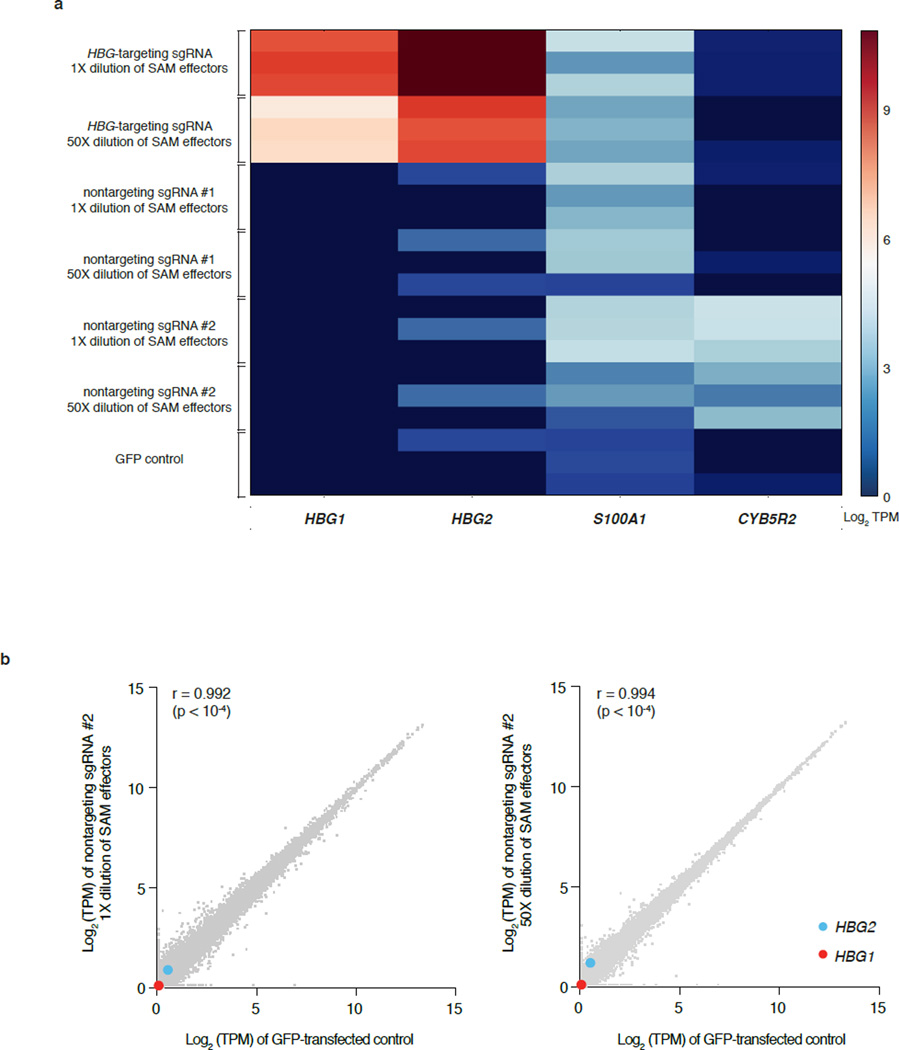
An important consideration for SAM use is its targeting specificity. Recent analysis of genome-wide dCas9-binding revealed significant concentration-dependent off-target binding22, yet its effect on the specificity of transcription modulation remains unclear. To assess SAM specificity, we chose HBG1/2 as our target gene, reasoning that globin genes would have few downstream targets that could confound our specificity analysis. We found that SAM specifically activated both HBG1 and HBG2 isoforms (p < 0.05, T-test after 0.01 FDR correction), which share the same TSS (Fig. 4 and Extended Data Fig. 6), while no other genes were found to be differentially expressed. We also tested two additional non-targeting sgRNAs with guide sequences that do not share perfect homology with the human genome. We found only two genes, S100A1 and CYB5R2, to be differentially expressed (p < 0.05, T-test after 0.01 FDR correction for multiple hypothesis testing) compared with GFP-expressing control (Extended Data Fig. 6) for both non-targeting guides. These results suggest that SAM-mediated gene activation is specific with minimal off-target activity.
Figure 4. Evaluation of SAM specificity.
Expression levels in log(TPM) values of all detected genes in RNA-seq libraries of GFP-transfected controls (x-axis of all graphs) compared to (from left to right): SAM targeting HBG1/2 genes in 1× dilution and 50× dilution, non-targeting control sgRNAs in 1× dilution and 50× dilution (y-axis). Marked are the two statistically significant differentially expressed genes (T-test q-value < 0.05 with FDR correction): HBG1 (red) and HGB2 (blue). The average from n = 3 is shown.
Genome-scale gene activation screen
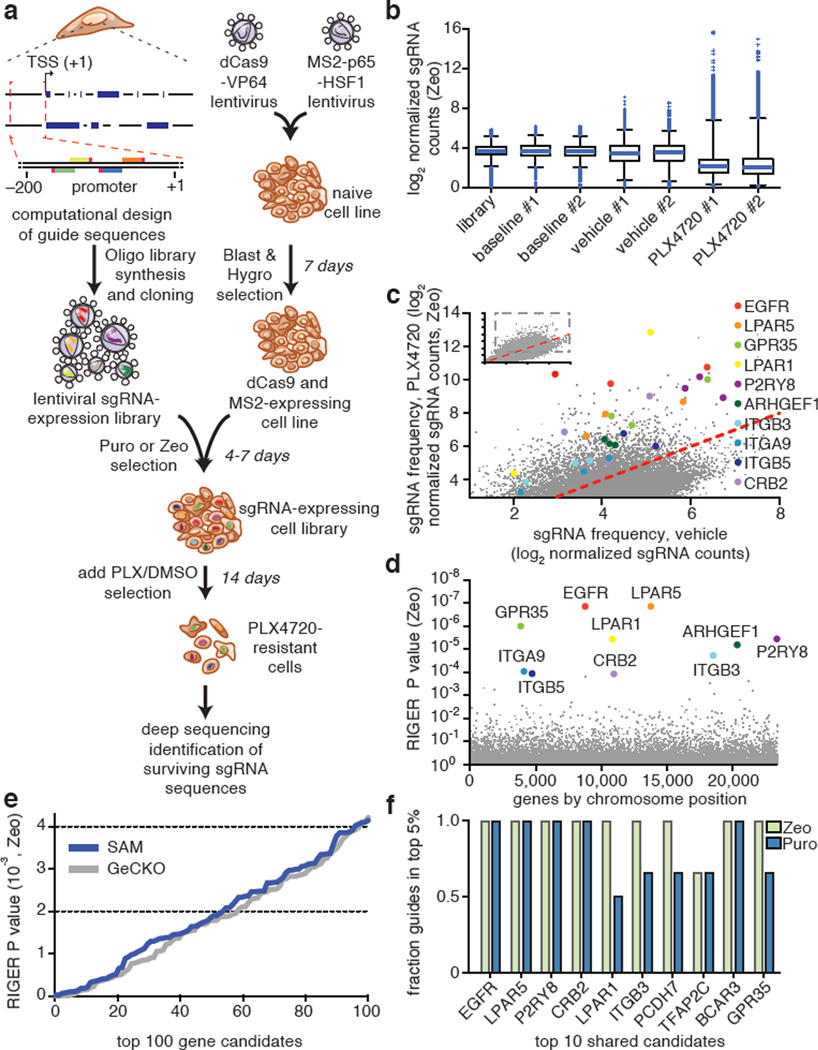
The ability to activate target genes using individual sgRNAs greatly facilitates the development of pooled, genome-scale transcriptional activation screening. To develop a SAM-based screening system, we generated lentiviral expression vectors that are able to drive robust transcription activation at low multiplicity of infection (MOI) (Extended Data Fig. 7a,b). Using this lentiviral system, we generated a genome-scale sgRNA library consisting of 70,290 guides, targeting every coding isoform from the RefSeq database (23,430 isoforms). For each gene, 3 sgRNAs were chosen to target sites within 200 bp upstream of the TSS, which was previously determined to provide more efficient activation (Fig. 2d and Fig. 5a).
Figure 5. Genome-scale gene activation screening identifies mediators of BRAF inhibitor resistance.
a, Flow chart of transcription activation screening using SAM. b, Box plot showing the distribution of sgRNA frequencies post lentiviral transduction for baseline (day 3), vehicle (day 21), and PLX-4720 (day 21) conditions. c, Scatterplot showing enrichment of specific sgRNAs after PLX-4720 treatment. d, Identification of top candidate genes using the RIGER P value analysis based on the average of both infection replicates. e, Comparison of RIGER P values for the top 100 hits from SAM and GeCKO3 PLX-4720 resistance screens. f, Consistency of sgRNAs for top screening hits. Fraction of unique sgRNAs targeting each gene that are in the top 5% of all sgRNAs is plotted.
Previously we applied genome-scale CRISPR knockout (GeCKO) library3 in A375 (BRAFV600E) melanoma cells to identify loss-of-function mutations capable of mediating resistance against the BRAF inhibitor PLX-4720. Here we sought to use the new SAM sgRNA library to identify a complementary set of gain-of-function changes that can confer BRAF inhibitor resistance (Fig. 5a). At 14-days post drug treatment, the sgRNA distribution was significantly different between cells treated with PLX-4720 and with vehicle, with the majority of sgRNAs exhibiting a reduced representation and a small set of guides showing high enrichment for PLX-4720 treated cells (Fig. 5b and Extended Data Fig. 7c). For a number of gene targets, multiple sgRNAs targeting the same gene were enriched in PLX-4720-treated cells (Fig. 5c) and the 10 most significant hits were distributed throughout the genome (Fig. 5d and Extended Data Fig. 7d). The significance of the _p_-values of our top 100 RIGER hits (Supplementary Table 1 and 2) was comparable to those observed for GeCKO screening3 (Fig. 5e). In addition, for the top 10 shared hits between two independent screens (Zeo and Puro selection for sgRNA expression), the fraction of effectively enriched guides per gene (present in the top 5% of all guides) was very high with 97% for Zeo and 81% for Puro (89% ± 10.7% overall, compared to 78% ± 27% for the top 10 GECKO hits, Fig. 5f and Extended Data Fig. 7e).
Our screen results highlight a number of gene candidates that both confirm known PLX-4720 resistance pathways and suggest new mechanisms (Extended Data Fig. 7f). First, reactivation of the ERK pathway is one of the main known resistance mechanisms23,24, and two of our screening hits, BCAR3 and EGFR, likely modulate downstream and upstream nodes of this pathway, respectively25,26. EGFR has been previously validated as a mediator of resistance to PLX-4720 through PI3K and AKT, in addition to ERK26,27. These two pathways are thought to be alternative routes of PLX-4720 resistance24,28,29. Furthermore, four out of the top 10 hits from our screen belong to the family of G protein-coupled receptors (GPCRs: GPR35, LPAR1, LPAR5, and P2RY8), which emerged as the top-ranked protein class conferring resistance to multiple MAP kinase inhibitors in melanoma cells in a recent screen using cDNA overexpression30. GPCRs signal through multiple downstream pathways including ERK, PI3K, as well as cAMP and PKA31,32. The final class of protein candidates from our screen belongs to the ITG receptor family, which is thought to interact with RTK and activate both ERK and PI3K pathways33,34.
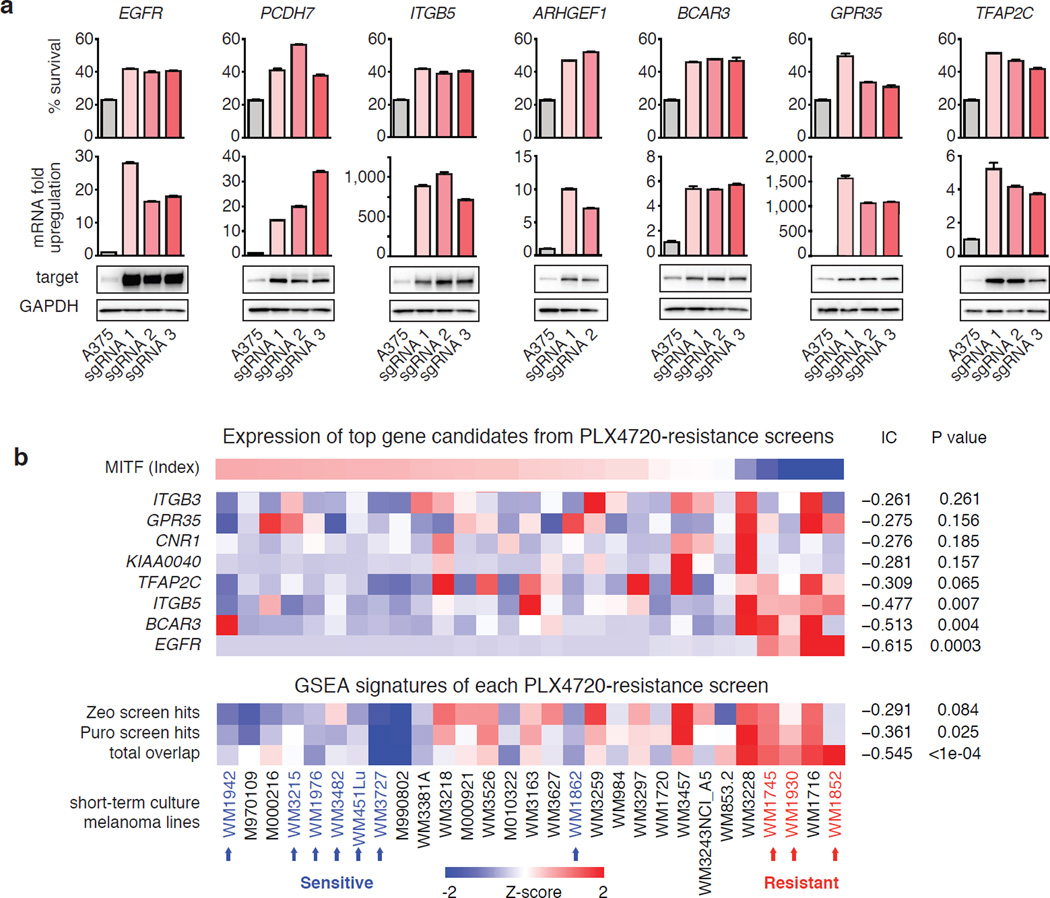
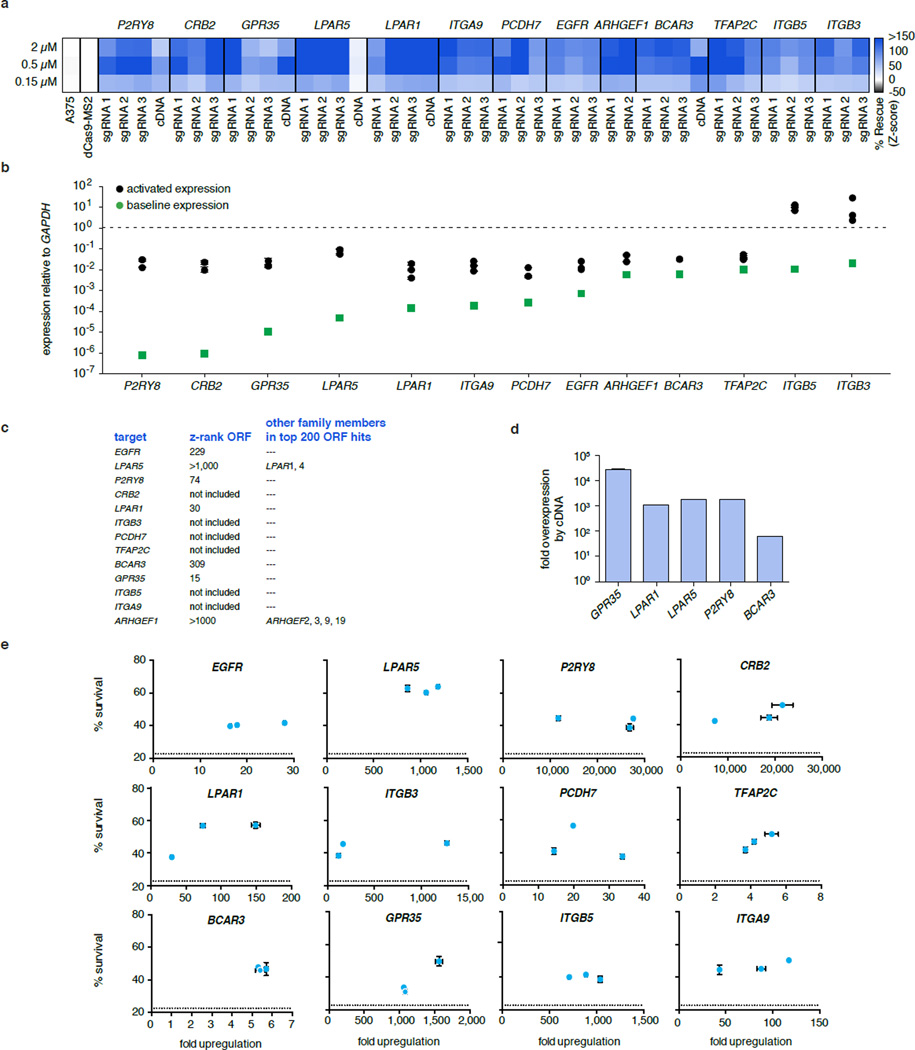
To verify the results from the PLX-4720 resistance screen, we validated each of the top 13 genes. All sgRNAs from the screen that targeted these 13 genes conferred PLX-4720 resistance when individually expressed in A375 along with SAM (Fig. 6a and Extended Data Fig. 8a). We also verified that SAM was able to facilitate robust increase in target transcript (Fig. 6a and Extended Data Fig. 8b) and protein levels (Fig. 6a). Since 5 of our top candidates from the pooled SAM screen overlapped with hits from a previously conducted arrayed cDNA screen30 (Extended Data Fig. 8c), we compared the relative efficacy of cDNA overexpression with SAM-mediated transcription activation. Interestingly, for these 5 targets, SAM led to at least similar levels of PLX-4720 resistance when compared with corresponding cDNA overexpression conditions (Extended Data Fig. 8a), despite cDNA leading to higher transcript levels (Extended Data Fig. 8d). Furthermore, we found that, for most genes, the levels of PLX-4720 resistance mediated by all three sgRNAs were comparable (Extended Data Fig. 8e).
Figure 6. Validation of top hits from genome-scale gene activation screen for PLX-4720 resistance mediators.
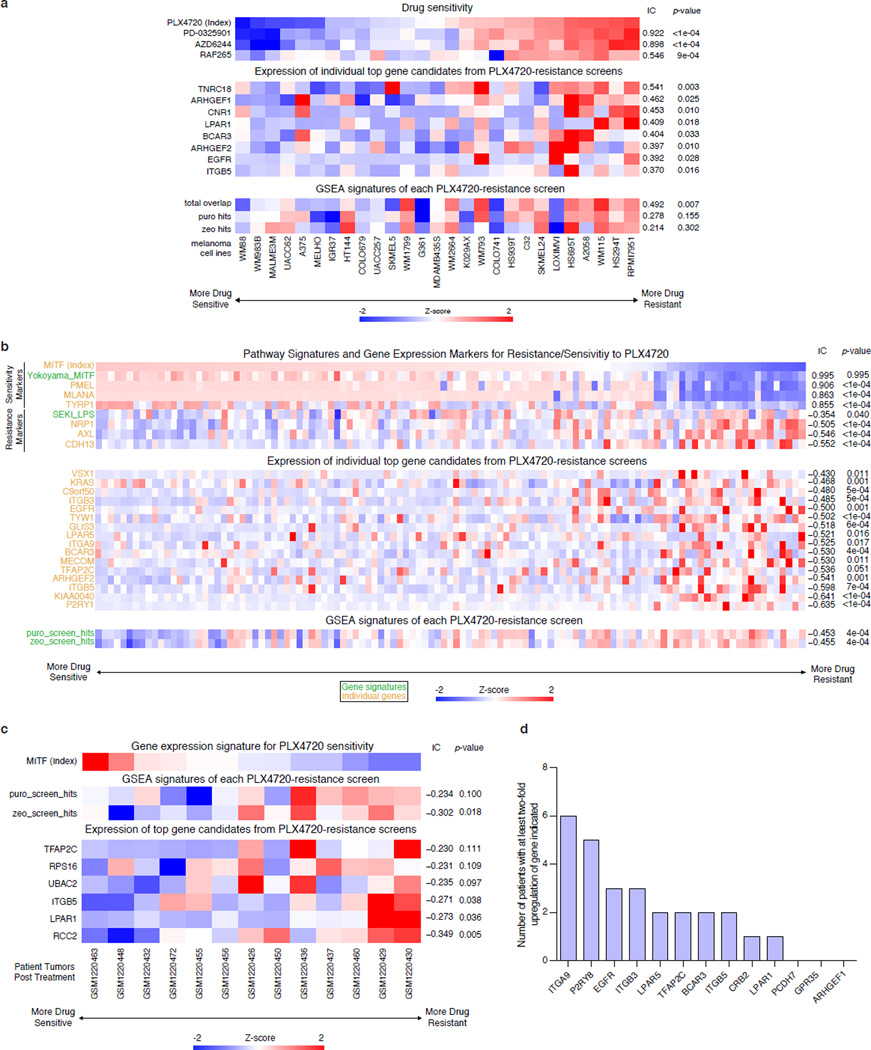
a, Comparison of PLX-4720 resistance, transcription activation and protein upregulation in A375 cells for top screening hits. b, Expression levels of top hits and screen signatures are elevated in the resistant state of short-term BRAFV600 melanoma cultures (see Methods for signature generation). The subset of samples which were previously tested for PLX-4720 sensitivity and resistance are indicated by blue and red arrows respectively39. IC: Information Coefficient. All values are mean ± SEM with n = 3.
In addition to validating our top screening hits through individual sgRNA or cDNA overexpression, we analyzed the expression profile of our screening hits using four different datasets (CCLE35, TCGA: https://tcga-data.nci.nih.gov/tcga/, short-term melanoma36,37, and pre/post treatment patient samples38). As shown previously39, a distinct transcriptional state defines BRAF-inhibition sensitive and resistant states as described by activation of endogenous MITF/associated markers (e.g. PMEL) and NF-κB-pathway activity/associated markers (e.g. AXL), respectively (Fig. 6b and Extended Data Fig. 9b). Based on short-term melanoma data fom the Cancer Genomics Hub37,39, we found that the expression of our top screening hits was significantly increased in the resistant state. Correspondingly, a gene expression signature derived from the top screening hits was correlated with BRAF-inhibitor resistance (Fig. 6b; total overlap, p < 0.0001). Further analysis performed using the CCLE, TCGA, and pre/post treatment data set also revealed similar correlations (Extended Data Fig. 9).
Discussion
In summary, we have taken a structure-guided approach to design a dCas9-based transcription activation system for achieving robust, single sgRNA-mediated gene up-regulation. By engineering the sgRNA to incorporate protein-interacting aptamers, we assembled a synthetic transcription activation complex consisting of multiple distinct effector domains modeled after natural transcription activation processes. Here we have shown that the SAM system is robust, specific, and can facilitate genome-scale gain-of-function screening when combined with a compact pooled sgRNA library. Our SAM-mediated screens exhibited a high degree of consistency and validation, with 80% effectively enriched guides per gene hit, and 100% validation of the top 10 hits.
Future engineering of the Cas9 complex based on structural information14,40 will further expand the Cas9 toolbox41. Additional developments of the SAM system may be able to take advantage of the modularity and customizability of the sgRNA scaffold to establish a series of sgRNA scaffolds bearing different aptamers for recruiting distinct types of effectors in an orthogonal manner. For instance, replacement of the MS2 stem-loops with PP7-interacting stem-loops may be used to recruit repressive elements, potentially enabling multiplexed bidirectional transcriptional control. Although we have taken initial steps toward defining selection rules for potent sgRNAs, future studies will reveal additional selection criteria that are critical for guide efficacy, such as sequence-intrinsic properties (Extended Data Fig. 10a–d). Applications of dCas9-based transcription modulators in positive and negative selection screens (Extended Data Fig. 10e–f)42 will enable the dissection of many types of genetic elements, ranging from protein-coding genes to non-coding lincRNA elements. Furthermore, combining wildtype Cas9-mediated genome modifications with SAM-mediated recruitment of epigenetic modifiers will constitute powerful approaches for studying genome organization and regulation in diverse biological processes.
Methods
Sequences
DNA sequences for SAM components sgRNA scaffolds are provided in Supplementary Sequences. sgRNA target sequences for characterization and optimization of SAM are listed in Supplementary Table 4.
Transient transfection experiments
Neuro-2a cells (Sigma-Aldrich) were grown in media containing 1:1 ratio of OptiMEM (Life Technologies) to high-glucose DMEM with GlutaMax and sodium pyruvate (Life Technologies) supplemented with 5% HyClone heat-inactivated FBS (Thermo Scientific), 1% penicillin/streptomycin (Life Technologies), and passaged at 1:5 every 2 days.
HEK293FT cells (Life Technologies) were maintained in high-glucose DMEM with GlutaMax and sodium pyruvate (Life Technologies) supplemented with 10% heat-inactivated characterized HyClone fetal bovine serum (Thermo Scientific) and 1% penicillin/streptomycin (Life Technologies). Cells were passaged daily at a ratio 1:2 or 1:2.5. For gene activation experiments, 20,000 HEK293FT cells/well were plated in 100 µL media in poly-D-lysine coated 96-well plates (BD BioSciences). 24 hours after plating, cells were transfected with a 1:1:1 mass ratio of:
- sgRNA plasmid with gene-specific targeting sequence or pUC19 control plasmid
- MS2-effector plasmid or pUC19.
- dCas9 plasmid, dCas9-effector plasmid, or pUC19.
A total plasmid mass of 0.3 µg/well was transfected using 0.6 µL/well Lipofectamine 2000 (Life Technologies) according to the manufacturer’s instructions. Culture medium was changed 5 hours after transfection. 48 hours after transfection, cell lysis and reverse transcription were performed using a Cells-to-Ct kit (Life Technologies). Relative RNA expression levels were quantified by reverse transcription and quantitative PCR (qPCR) using TaqMan qPCR probes (Life Technologies, Supplementary Table 5) and Fast Advanced Master Mix (Life Technologies). qPCR was carried out in 5 µL multiplexed reactions and 384-well format using the LightCycler 480 Instrument II. Data was analyzed by the ΔΔCt method: target Ct values (FAM dye) were normalized to GAPDH Ct values (VIC dye), and fold changes in target gene expression were determined by comparing to GFP-transfected experimental controls.
Lentivirus production
HEK293T cells (Life Technologies) were cultured as described above for HEK293FT cells. 1 day prior to transfection, cells were seeded at ~40% confluency (12×T225 flasks for library scale production, 1×T25 flask for individual guide production). Cells were transfected the next day at ~80–90% confluency. For each flask, 10 µg of plasmid containing the vector of interest, 10 µg of pMD2.G, and 15 µg of psPAX2 (Addgene) were transfected using 100 µL of Lipofectamine 2000 and 200 µL Plus Reagent (Life Technologies). 5h after transfection the media was changed. Virus supernatant was harvested 48h post-transfection, filtered with a 0.45 µm PVDF filter (Millipore), aliquoted, and stored at −80 °C.
Lentiviral transduction
A375 cells (ATCC) were cultured in RPMI 1640 (Life Technologies) supplemented with 10% FBS (Seradigm) and 1% penicillin/streptomycin (Life Technologies) and passaged every other day at a 1:4 ratio. Cells were transduced with lentivirus via spinfection in 12-well plates. 3 × 106 cells in 2 mL of media supplemented with 8 µg/mL polybrene (Sigma) were added to each well, supplemented with lentiviral supernatant and centrifuged for 2h at 1000g. 24h after spinfection, cells were detached with TrypLE (Life Technologies) and counted. Cells were replated at low density (7.5 × 106 cells per T225 Flask) and a selection agent was added either immediately (Zeocin, Blasticidin and Hygromycin, all Life Technologies) or 3h after plating (Puromycin). Concentrations for selection agents we determined using a kill curve: 0.5 µg/ml Puromycin, 200 µg/mL Zeocin, 10 µg/mL Blasticidin, and 300 µg/mL Hygromycin. Media was refreshed on day 2 and cells were passaged every other day starting on day 4 after replating. The duration of selection was 4 days for Puromycin and 7 days for Zeocin, Hygromycin and Blasticidin. Lentiviral titers were determined by spinfecting cells with 6 different volumes of lentivirus ranging from 0 to 600 µL and counting the number of surviving cells after a complete selection (3–6 days).
Design and Cloning of SAM library
RefSeq coding gene isoforms with a unique TSS (total of 23’430 isoforms) were targeted with three guides each for a total library of 70,290 guides (Supplementary Table 6). Guides were designed to target the first 200 bp upstream of each TSS and subsequently filtered for GC content >25% and minimal overlap of the target sequence. After filtering, the remaining guides were scored according to predicted off-target matches as described previously43, and three guides with the best off-target scores were selected. Cloning of the SAM sgRNA libraries was performed as previously described3 with a minimum representation of 100 transformed colonies/guide.
Depletion and PLX-4720 Screen
A375 cells stably integrated with SAM Cas9 and effector components were transduced with SAM sgRNA libraries as described above at an MOI of 0.2, with a minimal representation of 500 transduced cells/guide. Cells were maintained at >500 cells/guide during subsequent passaging. At 7 DPI (complete selection, see above), cells were split into vehicle (DMSO) and PLX-4720 conditions (2 µM PLX-4720 dissolved in DMSO, Selleckchem). Cells were passaged every 2 days for a total of 14 days of drug treatment. >500 cells/guide were harvested as a baseline at 3 DPI (4 days before treatment) and at 21 DPI (after 14 days of treatment) for gDNA extraction. Genomic DNA was extracted using the Zymo Quick-gDNA midi kit (Zymo Research). PCR of the virally integrated guides was performed on gDNA at the equivalent of >500 cells/guide in 96 parallel reactions using NEBnext High Fidelity 2× Master Mix (New England Biolabs) in a single-step reaction of 22 cycles. Primers are listed below:
PCR products from all 96 reactions were pooled, purified using Zymo-Spin™ V with Reservoir (Zymo research) and gel extracted using the Zymoclean™ Gel DNA Recovery Kit (Zymo research). Resulting libraries were deep-sequenced on Illumina MiSeq and HiSeq platforms with a total coverage of >35 million reads passing filter per library.
NGS and screen hits analysis
NGS data were de-multiplexed using unique index reads. Guide counts (Supplementary Table 7) were determined based on perfectly matched sequencing reads only. For each condition, guide counts were normalized to the total number of counts per condition, and log2 counts were calculated based on these values. Ratios of counts between conditions were calculated as log2((count 1 + 1)/(count 2 +1)) based on normalized counts.
RIGER analysis was performed using GENE-E based on the normalized day 14 log2 ratios (PLX-4720/DMSO) averaged over two independent infection replicates. All RIGER analysis used the Kolmogorov-Smirnov method as described previously44, except for Fig. 6c, where the weighed average method was used in order to enable comparison to GeCKO values determined by that method.
Gene expression and Pharmacological Validation Analysis
Gene expression data (CCLE, TCGA, short-term cultures, patient melanoma biopsies) and pharmacological data (CCLE, short-term cultures) were analyzed to better understand the biological relevance of the top gene hits from the SAM screens. In the CCLE dataset35, gene expression data (RNA-sequencing, GCHub: https://cghub.ucsc.edu/datasets/ccle.html) and pharmacological data (activity area for MAPK pathway inhibitors) from BRAFV600 mutant melanoma cell lines were used to compute the association between PLX-4720 resistance and the gene expression of each of the top hits. Additionally, gene expression signatures comprised of the top hits were generated using single-sample Gene Set Enrichment Analysis (ssGSEA)45, and the associations between PLX-4720 resistance and these signatures were computed.
Gene expression data (Affymetrix GeneChip HT-HGU133) and PLX-4720 pharmacological data (GI50; only for a subset of the samples) from short term melanoma cultures (STC)36 was also used for plotting the gene expression of top hits and their ssGSEA signature scores. Expression data for the STC samples were collapsed to maximum probe value per gene and preprocessed using robust spline normalization.
Gene expression (RNA-sequencing) and genotyping data were collected from 113 BRAFV600-mutant primary and metastatic patient tumors from The Cancer Genome Atlas (https://tcga-data.nci.nih.gov/tcga/) and this data was similarly used for determining the association between resistance and the expression of top hits/ssGSEA signature scores. Because pharmacological data was not available for the STCs (only a subset had PLX-4720 data) and the TCGA melanoma samples, a transcriptional state was plotted using marker genes and signatures39 in order to identify samples resistant to BRAF-inhibition.
Gene expression data from 13 patients with BRAFV600E melanomas38 was used for analyzing the relationship between resistance and the expression of our top hits/ssGSEA signature scores. Because all the post-treatment tumors were resistant and not every sample had a paired on-treatment biopsy, we decided to order the samples by MITF expression in the pre-treatment samples to reflect the original PLX-4720 sensitivity state of the tumors. We then used the expression data in the post-treatment resistant tumors to plot the expression of top hits/ssGSEA signature scores. We also calculated the log2-fold change between each patient’s post/pre paired samples and determined the number of patients that had at least a log2-fold change of 2 per top screen hit.
Single Sample Gene Set Enrichment Analysis
While there was a significant association between the overexpression of some of our top individual SAM screen hits and resistance in three external cancer datasets, we sought a more robust scoring system independent of any single gene. Gene expression signatures were generated based on the set of top hits from each of the two SAM screens and for the overlap between them. Using single-sample Gene Set Enrichment analysis (ssGSEA), a score was generated for each sample that represents the enrichment of the SAM screen gene expression signature in that sample and the extent to which those genes are coordinately up- or down-regulated. Additionally, signature gene sets from the Molecular Signature Database (MSigDB)46 were used in order to fully map the transcriptional BRAF-inhibitor resistant/sensitive states in the short-term culture and TCGA datasets as previously described39.
Information Coefficient for Measuring Associations in External Datasets
To measure correlations between different features (signature scores, gene expression, or drug-resistance data) in the external cancer datasets, an information-theoretic approach (Information Coefficient; IC) was used and significance was measured using a permutation test (n=10,000), as previously described39. The IC was calculated between the feature used to sort the samples (columns) in each dataset and each of the features plotted in the heatmap (pharmacological data, gene expression, and signature scores).
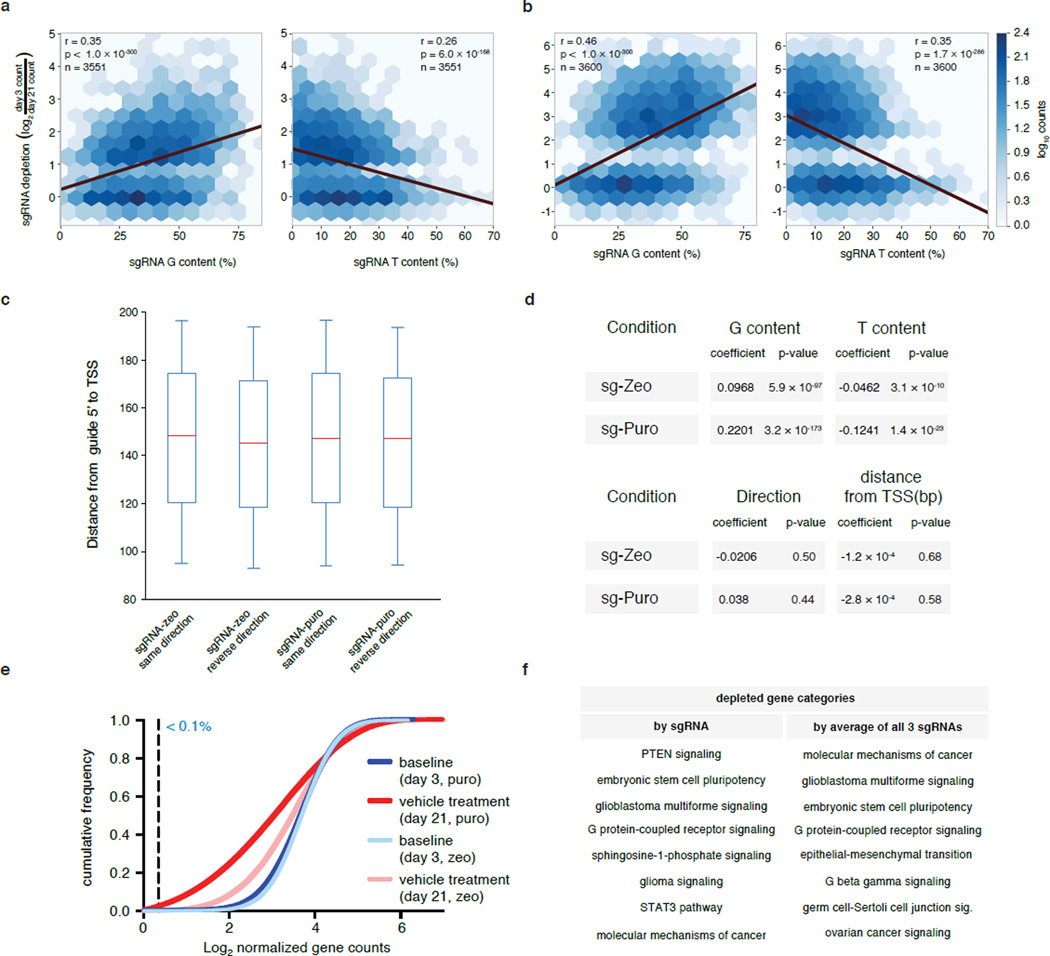
sgRNA sequence analysis
Depletion for each sgRNA was calculated as the ratio of counts (see “NGS and screen hits analysis”) between day 3 and day 21. The sgRNAs corresponding to genes with significant depletion (p < 0.05 by RIGER analysis) in sgRNA-Puro and sgRNA-Zeo libraries were selected for analyses. These sgRNAs were analyzed for nucleotide occurrence in the sgRNA sequence, distance from TSS, and guide strand relative to transcript orientation. For each variable, the correlation and significance with the sgRNA ratio was calculated by Ordinary Least Squares linear regression.
PLX-4720 Survival Assay
A375 cells stably integrated with dCas9-VP64 and MS2-p65-HSF1 were transduced with individual guides from the top screening hits of the Zeocin and Puromycin screens (13 genes total, 3 sgRNAs per gene) as well as available cDNA at an MOI of <0.2 as described above. Cells were selected for guide expression with Zeocin (Life Technologies) for 5 days and replated at low density (3 × 103 cells per well in a 96-well plate). A375 cells and A375 cells expressing dCas9-VP64 and MS2-p65-HSF1 were plated as controls. Different concentrations of PLX-4720 (2µM, 0.5µM, 0.15µM) or vehicle (DMSO) were added 3h after plating. Cells were treated with PLX-4720 for 4 days before cell viability was measured using CellTiter-Glo Luminescent Cell Viability Assay (Promega). For qPCR quantification of target gene upregulation, cells were also plated at 5 DPI (3 × 104 cells per well in a 96-well plate) and harvested for mRNA 24h after plating.
Western Blot
Protein lysates were prepared with RIPA lysis buffer (Cell Signaling Technologies) containing a protease inhibitor cocktail (Roche). Samples standardized for protein with the Pierce BCA protein assay (Thermo Scientific) were boiled at 95°C for 5 mins under reducing conditions (except for GPR35 samples, which were incubated at 37°C for 30 mins). After denaturation, samples for probing proteins with lower or higher molecular weight were separated by 10–20% or 4–15% Criterion Tris-HCl gels (Bio-Rad) and electrotransferred onto a 0.2µm or 0.45µm polyvinylidene difluoride membrane (Millipore) respectively. Blots were blocked with 5% BLOT-QuickBlocker (VWR) and probed with different primary antibodies [anti-EGFR (rabbit polyclonal, SC-03, Santa Cruz Biotechnology, 1:1000 dilution), anti-PCDH7 (rabbit polyclonal, HPA011866, Sigma-Aldrich, 1:1000 dilution), anti-ITGB5 (rabbit polyclonal, SC-14010, Santa Cruz Biotechnology, 1:500 dilution), anti-ARHGEF1 (rabbit polyclonal, 11363-1-AP, Proteintech, 1:5000 dilution), anti-BCAR3 (rabbit polyclonal, A301-671A, Bethyl Laboratories, 1:2000 dilution), anti-GPR35 (rabbit polyclonal, 10007660, Cayman Chemical, 1:1000 dilution), anti-TFAP2C (rabbit polyclonal, 2320, Cell Signaling Technology, 1:1000 dilution, 2.5% bovine serum albumin, Sigma-Aldrich)] in 2.5% BLOT-QuickBlocker (VWR) unless noted otherwise overnight at 4°C. Blots were then incubated with secondary antibody HRP-conjugated goat anti-rabbit IgG (7074, Cell Signaling Technology, 1:1000 dilution) and HRP-conjugated GAPDH (rabbit monoclonal, 3683, Cell Signaling Technology, 1:2000 dilution) in 2.5% BLOT-QuickBlocker (VWR) for 1hr at room temperature. Proteins with molecular weights similar to GAPDH (GPR35 and TFAP2C) were stripped with Restore Plus Western Blot Stripping Buffer (Thermo Scientific) before probing for GAPDH. SuperSignal West Pico and Femto Chemiluminescent Substrates (Thermo Scientific) were used for detection.
RNA Sequencing and Data Analysis
Samples harvested for RNA sequencing were prepped with TruSeq Stranded mRNA Sample Prep Kit (Illumina) and deep-sequenced on the Illumina MiSeq platform (>9 Mio reads per condition). Bowtie247 index was created based on the human hg19 UCSC genome and known gene transcriptome, and paired-end reads were aligned directly to this index using Bowtie2 with command line options “-q --phred33-quals -n 2 -e 99999999 -l 25 -I 1 -X 1000 -a -m 200 –p 4 --chunkmbs 512”. Next, RSEM v1.2748 was run with default parameters on the alignments created by Bowtie2 to estimate expression levels. RSEM’s gene level expression estimates (tau) were multiplied by 1,000,000 to obtain transcript per million (TPM) estimates for each gene, and TPM estimates were transformed to log-space by taking log2(TPM+1). The normalization between libraries was tested using an MA plot (mairplot function in Matlab V2013b). Genes were considered detected if their transformed expression level was equal to or above 1 (in log2(TPM+1) scale). All genes detected in at least one library (out of three libraries per condition) were used to construct scatter plots comparing each of the six conditions to the control GFP condition, using the average across biological replicates with >80% alignment to the hg19 UCSC known gene transcriptome (log2(mean(TPM)+1) value per gene).
To find differentially expressed genes, we performed Student’s _t_-test on each of the six conditions against the GFP condition. The _t_-test was run on all genes that had expression levels above log2(TPM+1)>2.5 in at least two libraries. This threshold was chosen as the minimal threshold for which the number of detected genes across all libraries was constant. Only genes that were significant (_p_-value pass 0.01 FDR correction) and had at least 1.5 fold change were reported and visualized using a heatmap.
Extended Data
Extended Data Figure 1. Structure-guided engineering of Cas9 sgRNA.
a, Schematic of the sgRNA stem-loops showing contacts between each stem-loop and Cas9. Contacting amino acid residues are highlighted in yellow. Tetraloop and stem-loop 2 do not make any contacts with Cas9 whereas stem-loops 1 and 3 share extensive contacts with Cas9. b, sgRNA 2.0 with MS2 stem-loops inserted into the tetraloop and stem-loop 2. c, Addition of a second NLS or an alternative HNH domain inactivating point mutation in Cas9 improve efficiency of transcription activation for MYOD1 moderately. d, dCas9-VP64 activators exhibit improved performance by recruitment of MS2-p65 to the tetraloop and stem-loop 2. Addition of an AU flip or extension in the tetraloop does not increase the effectiveness of dCas9-mediated transcription activation. e, Tetraloop and stem-loop 2 are amenable to replacement with MS2 stem-loops. Base changes from the sgRNA 2.0 scaffold are shown at the respective positions, with dashes indicating unaltered bases and bases below dashes indicating insertions. Deletions are indicated by absence of dashes at respective positions. All figures are n = 3 and mean ± SEM.
Extended Data Figure 2. SAM mediates efficient activation of a panel of 12 coding genes and 6 lincRNAs.
a, Comparison of the activation levels of 12 genes with dCas9-VP64 in combination with MS2-p65, MS2-p65-HSF1, or MS2-p65-MyoD1. MS2-p65-HSF1 mediated significantly higher levels of activation than MS2-p65 alone for 9 out of 12 genes. The best guide out of 8 tested for each gene (Fig. 2a) was used in this experiment. Activation levels for each type of MS2-fusion is presented as a percentage relative to the activation achieved using MS2-p65. b, Investigation of transcriptional changes in the closest coding transcripts for SAM-mediated activation of 6 lincRNAs. Direction of the coding transcript relative to the lincRNA and distance between transcription start sites are shown. Only targeting of HOTTIP resulted in a significant change in the levels of the closest coding transcript (HOXA13). The best guide out of 8 tested for each gene (Fig. 2e) in combination with dCas9-VP64 and MS2-p65-HSF1 was used in this experiment. All figures are n = 3 and mean ± SEM.
Extended Data Figure 3. Activation of lincRNAs by SAM.
Six lincRNAs, three characterized and three uncharacterized, were targeted using SAM. For each lincRNA, 8 sgRNAs were designed to target the proximal promoter region (+1 to −800bp from the TSS) with 4 different MS2 activators (MS2-p65-HSF1, MS2-p65-MyoD1, MS2-p65, and MS2-VP64) in combination with dCas9-VP64. MS2 activators with a combination of 2 different domains (MS2-p65-HSF1 or MS2-p65-MyoD1) consistently provided the highest activation for each lincRNA, * denotes p < 0.01 for MS2-p65-HSF1 or MS2-p65-MyoD1 vs. MS2-p65. N = 3 and mean ± SEM is shown.
Extended Data Figure 4. Multiplexed activation using SAM and activation of a panel of 10 genes as a function of SAM component dosage.
a, Activation of a panel of 10 genes by combinations of 2, 4, 6, or 8 sgRNAs simultaneously. The mean fold up-regulation is shown on a log10 scale. MS2-p65-HSF1 and dCas9-VP64 were used in this experiment. b, The relative activation efficiency of individual sgRNAs varies depending on the target gene and the degree of multiplexing. N = 3 and mean ± SEM is shown.
Extended Data Figure 5. The effect of guide and SAM-component dilution on target activation.
a, The results for dilution of sgRNA 2.0 on target activation. b, The result for dilution of sgRNA 1.0 on target activation. # denotes an activation of < 2-fold at 1× guide dilution. c, Effect of MS2-p65-HSF1 and dCas9-VP64 dilution, at 1:1, 1:4, 1:10, and 1:50 of the original dosage for each component, on the effectiveness of transcription up-regulation. The amount of sgRNA expression plasmid was kept constant. d, Effect of diluting all three SAM components (dCas9-VP64, MS2-p65-HSF1, and sgRNA) at 1:4, 1:10, and 1:50 of the original dosage for each component. Fold up-regulation is calculated using GFP-transfected cells as the baseline. Error bars indicate S.E.M. and N = 3 for all figures.
Extended Data Figure 6. RNA-seq analysis of transcriptome changes mediated by SAM.
a, A heat map of log(TPM) expression values of all statistically significant differentially expressed genes (T-test q-value < 0.05 adjusted with FDR multiple hypothesis correction) found in any of the six experimental conditions compared to the GFP-transfected control. b, Expression levels in log(TPM) values of all detected genes in RNA-seq libraries of GFP-transfected controls (x-axis of all graphs) compared to (from left to right): non-targeting control sgRNA #2 in 1× dilution and 50× dilution (y-axis). Marked are HBG1 (red) and HGB2 (blue).
Extended Data Figure 7. Genome-scale lentiviral screen using Puromycin-resistant SAM sgRNA library.
a, Design of three lentiviral vectors for expressing sgRNA, dCas9-VP64, and MS2-p65-HSF1. Each vector contains a distinct selection marker to enable co-selection of cells expressing all three vectors. b, Lentiviral delivery of SAM components was tested by first generating 293FT cell lines stably integrated with dCas9-VP64 and MS2-p65-HSF1, and subsequently transducing these cells with single-gene targeting lentiviral sgRNAs at MOI <0.2. Transcription activation efficiency is measured 4 days post sgRNA lentivirus transduction and selection with Zeocin or Puromycin. Activation is at least as effective as previously observed with transient transfection in all three cases. c, Box plot showing the distribution of sgRNA frequencies at different time points post lentiviral transduction with the Puromycin library, after treatment with DMSO vehicle or PLX-4720. Two infection replicates are shown. d, Identification of top candidate genes using the RIGER P value analysis (KS method) based on the average of both infection replicates. Genes are organized by positions within chromosomes. e, Overlap between the top 20 hits from the Zeo and Puro screens. Genes belonging to the same family are indicated by the same color. There is a 50% overlap between the top hits of each screen as shown in the intersection of the Venn diagram. f, Relevant signaling pathways in BRAF inhibitor resistance. Reactivation of the Ras-ERK pathway as well as the parallel PI3K-Akt pathway have previously been implicated as two alternative resistance mechanisms to BRAF inhibitors23,24,26–29. Both pathways have been described as stimulating proliferation and survival49. BAD, FOXO and p27 are common inhibited downstream targets49. Recently, stimulation of the cAMP - CREB pathway by GPCRs has been described as a potential additional resistance mechanism30. Top candidates from our screen are indicated in blue and putative connections to all three pathways are shown25,50,51. Candidates previously validated to mediate PLX-4720 resistance are underlined in green26,30. COT and CREB are independently validated mediators of resistance23,30.
Extended Data Figure 8. Individual validation of PLX-4720 resistance mediation by top screen hits.
a, Validation of the top 10 Zeo screen hits and the top 10 shared hits (13 genes total). Every gene was independently activated by all three guides from the screen and tested for the ability to increase survival of A375 cells treated with three different concentrations of PLX-4720 (2µM, 0.5µM and 0.15µM). The z-score based on the % increase in survival relative to control (A375 cells transduced with dCas9-VP64 and MS2-p65-HSF1 alone) is shown for each guide and PLX-4720 concentration. Five cDNAs available from a previous large-scale gain-of-function PLX-4720 resistance screen were also included30. Every guide for each top hit mediates significant PLX-4720 resistance. b, The same panel of top hits exhibits a large range of basal expression levels and is effectively activated by all guides. The expression level relative to the housekeeping gene GAPDH is shown both at baseline as well as after activation by each individual guide. c, Ranks of the validated set of genes in the previous ORF screen. Six genes were not part of the cDNA library, five hits are shared (present in the top 3%) and only LPAR5 and ARHGEF1 were present but not highly ranked. Both of these genes had highly ranked members of the same family. d, Levels of overexpression from the five tested cDNA constructs. Transcript levels were higher for these five cDNAs than those mediated by SAM for the same genes. e, Correlation of survival at 2µM PLX-4720 treatment and transcript upregulation achieved by individual guides. For most genes (9 out of 12 shown), the percent survival is very similar across transcript levels achieved by all three guides. Dotted lines indicate control survival.
Extended Data Figure 9. Expression of top hits and screen signatures are elevated in PLX-4720 resistant melanoma cell lines and patient samples.
a, Heat map showing sensitivity to different drugs (top), expression of SAM top screen hits (middle), and SAM screen signature scores (bottom; see Online Methods for signature generation) in Cancer Cell Line Encyclopedia cell lines35. Drug sensitivities are measured as Activity Areas (AA). The melanoma cell lines are sorted by PLX-4720 drug sensitivity. RAF inhibitors: PLX-4720 and RAF265; MEK inhibitors: AZD6244 and PD-0325901. b, Heat map showing expression of gene/signature markers for BRAF-inhibitor sensitivity (top), expression of SAM top screen hits (middle) and screen signature scores (bottom) in different BRAFV600 patient melanoma samples (primary or metastatic) from The Cancer Genome Atlas. c, Heat map showing MITF expression (top), screen signature scores (middle), and expression of SAM top screen hits (bottom) in different BRAFV600E patient melanoma biopsies post-treatment with BRAF inhibitors38. d, Bar chart showing the number of patients from (c) with at least a two-fold change (post/pre treatment) in gene expression of the top PLX-4720 screen hits in the post-treatment samples. All associations are measured using the information coefficient (IC) between the index and each of the features and P values are determined using a permutation test. All heat maps show z-scores.
Extended Data Figure 10. Guide depletion analysis to identify gene set enrichment and guide efficiency parameters.
a, Heat maps of sgRNA nucleotide content versus depletion after 21 days. sgRNA targeting significantly depleted genes (from RIGER analysis) in sgRNA-zeo (a) or sgRNA-puro (b) screens were analyzed for trends based on G or T content in the sgRNA sequence. sgRNA depletion is positively correlated with G content and negatively correlated with T content. Other bases analyzed (A and C) had significant (p < 0.0007) but weak (r < 0.2) negative correlation. c, 90% of guides analyzed fall within a 100bp window <200bp from the TSS. Boxplots of distance from 5’ end of the guide to the TSS for sgRNA-zeo and sgRNA-puro in same and reverse direction (relative to target transcription). Whiskers span 5th to 95th quartile. d, Coefficients and P values for ordinary least squares predicting sgRNA depletion of significantly depleted genes from G content, T content, distance from 5’ end of the guide to the TSS and direction of guide. Only nucleotide content has a significant effect on depletion in this model, consistent with a high efficiency of guides within 200bp of the TSS regardless of strand orientation (Fig. 2d). e, The cumulative frequency of sgRNAs 3 and 21 days after transduction in A375 cells is shown. Shift in the 21-day curve represents the depletion in a subset of sgRNAs. Less than 0.1% of all guides are not detected at day 3 (detected by less than 10 reads). f, Depleted guides (Supplementary Table 3) can be analyzed for significant clustering of gene categories. Gene categories exhibiting significant depletion based on Ingenuity Pathway Analysis (p<0.01 after B-H FDR correction) are shown. Categories based on the 1000 most depleted guides individually (left) and the average of all 3 guides/gene (right). These categories include either positive or negative regulators of each pathway that reduce proliferation and survival.
Supplementary Material
SuppInfo
SuppTable6
SuppTable7
Acknowledgements
We would like to thank S. Shehata, K. Zheng, C. Johannessen, L. Garraway, and O. Shalem, and members of the Zhang lab for assistance and helpful discussions. O.O.A is supported by a NSF Graduate Research Fellowship, J.S.G. is supported by a D.O.E. Computational Science Graduate Fellowship, and F.Z. is supported by the NIMH (DP1-MH100706), the NINDS (R01-NS07312401), NSF, the Keck, Searle Scholars, Klingenstein, Vallee, and Simons Foundations, and Bob Metcalfe. CRISPR reagents are available to the academic community through Addgene, and associated protocols, support forum, and computational tools are available via the Zhang lab website (http://www.genome-engineering.org).
Footnotes
Author Contributions
S.K. and F.Z. conceived the project. S.K., M.D.B., A.T., and F.Z. designed the experiments. S.K., M.D.B., A.T., C.B., P.D.H., J.J. performed experiments and analysed data. H.N. and O.N. provided structural consultation. N.H. performed the RNAseq analysis. J.S.G. performed the depletion guide efficacy analysis. O.O.A. performed the analysis of clinical datasets. S.K., A.T., P.D.H., and F.Z. wrote the paper with help from all authors.
All reagents described in this manuscript have been deposited with Addgene (Plasmid IDs: 61422-61427 for SAM component plasmid and 61597 for the human SAM guide RNA library). The authors have filed a patent application related to this work. RNA-Seq data are available at the Sequence Read Archive under accession number PRJNA269048.
References
- 1.Berns K, et al. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature. 2004;428:431–437. doi: 10.1038/nature02371. [DOI] [PubMed] [Google Scholar]
- 2.Boutros M, et al. Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science. 2004;303:832–835. doi: 10.1126/science.1091266. [DOI] [PubMed] [Google Scholar]
- 3.Shalem O, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beerli RR, Segal DJ, Dreier B, Barbas CF., 3rd Toward controlling gene expression at will: specific regulation of the erbB-2/HER-2 promoter by using polydactyl zinc finger proteins constructed from modular building blocks. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:14628–14633. doi: 10.1073/pnas.95.25.14628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang F, et al. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nature biotechnology. 2011;29:149–153. doi: 10.1038/nbt.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gilbert LA, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Konermann S, et al. Optical control of mammalian endogenous transcription and epigenetic states. Nature. 2013;500:472–476. doi: 10.1038/nature12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maeder ML, et al. CRISPR RNA-guided activation of endogenous human genes. Nature methods. 2013;10:977–979. doi: 10.1038/nmeth.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perez-Pinera P, et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nature methods. 2013;10:973–976. doi: 10.1038/nmeth.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mali P, et al. Barcoding cells using cell-surface programmable DNA-binding domains. Nature methods. 2013;10:403–406. doi: 10.1038/nmeth.2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jinek M, et al. A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nishimasu H, et al. Crystal structure of cas9 in complex with guide RNA and target DNA. Cell. 2014;156:935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peabody DS. The RNA binding site of bacteriophage MS2 coat protein. The EMBO journal. 1993;12:595–600. doi: 10.1002/j.1460-2075.1993.tb05691.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lemon B, Tjian R. Orchestrated response: a symphony of transcription factors for gene control. Genes & development. 2000;14:2551–2569. doi: 10.1101/gad.831000. [DOI] [PubMed] [Google Scholar]
- 17.van Essen D, Engist B, Natoli G, Saccani S. Two modes of transcriptional activation at native promoters by NF-kappaB p65. PLoS biology. 2009;7:e73. doi: 10.1371/journal.pbio.1000073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kretzschmar M, Kaiser K, Lottspeich F, Meisterernst M. A novel mediator of class II gene transcription with homology to viral immediate-early transcriptional regulators. Cell. 1994;78:525–534. doi: 10.1016/0092-8674(94)90429-4. [DOI] [PubMed] [Google Scholar]
- 19.Ikeda K, Stuehler T, Meisterernst M. The H1 and H2 regions of the activation domain of herpes simplex virion protein 16 stimulate transcription through distinct molecular mechanisms. Genes to cells : devoted to molecular & cellular mechanisms. 2002;7:49–58. doi: 10.1046/j.1356-9597.2001.00492.x. [DOI] [PubMed] [Google Scholar]
- 20.Neely KE, et al. Activation domain-mediated targeting of the SWI/SNF complex to promoters stimulates transcription from nucleosome arrays. Molecular cell. 1999;4:649–655. doi: 10.1016/s1097-2765(00)80216-6. [DOI] [PubMed] [Google Scholar]
- 21.Marinho HS, Real C, Cyrne L, Soares H, Antunes F. Hydrogen peroxide sensing, signaling and regulation of transcription factors. Redox biology. 2014;2:535–562. doi: 10.1016/j.redox.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu X, et al. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nature biotechnology. 2014;32:670–676. doi: 10.1038/nbt.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johannessen CM, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468:968–972. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nazarian R, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–977. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Musgrove EA, Sutherland RL. Biological determinants of endocrine resistance in breast cancer. Nature reviews. Cancer. 2009;9:631–643. doi: 10.1038/nrc2713. [DOI] [PubMed] [Google Scholar]
- 26.Prahallad A, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 2012;483:100–103. doi: 10.1038/nature10868. [DOI] [PubMed] [Google Scholar]
- 27.Corcoran RB, et al. EGFR-mediated re-activation of MAPK signaling contributes to insensitivity of BRAF mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer discovery. 2012;2:227–235. doi: 10.1158/2159-8290.CD-11-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Villanueva J, et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer cell. 2010;18:683–695. doi: 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi H, Kong X, Ribas A, Lo RS. Combinatorial treatments that overcome PDGFRbeta-driven resistance of melanoma cells to V600EB-RAF inhibition. Cancer research. 2011;71:5067–5074. doi: 10.1158/0008-5472.CAN-11-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johannessen CM, et al. A melanocyte lineage program confers resistance to MAP kinase pathway inhibition. Nature. 2013;504:138–142. doi: 10.1038/nature12688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dorsam RT, Gutkind JS. G-protein-coupled receptors and cancer. Nature reviews. Cancer. 2007;7:79–94. doi: 10.1038/nrc2069. [DOI] [PubMed] [Google Scholar]
- 32.Lappano R, Maggiolini M. G protein-coupled receptors: novel targets for drug discovery in cancer. Nature reviews. Drug discovery. 2011;10:47–60. doi: 10.1038/nrd3320. [DOI] [PubMed] [Google Scholar]
- 33.Franke TF. PI3K/Akt: getting it right matters. Oncogene. 2008;27:6473–6488. doi: 10.1038/onc.2008.313. [DOI] [PubMed] [Google Scholar]
- 34.Desgrosellier JS, Cheresh DA. Integrins in cancer: biological implications and therapeutic opportunities. Nature reviews. Cancer. 2010;10:9–22. doi: 10.1038/nrc2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barretina J, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin WM, et al. Modeling genomic diversity and tumor dependency in malignant melanoma. Cancer research. 2008;68:664–673. doi: 10.1158/0008-5472.CAN-07-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilks C, et al. The Cancer Genomics Hub (CGHub): overcoming cancer through the power of torrential data. Database : the journal of biological databases and curation. 2014 doi: 10.1093/database/bau093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rizos H, et al. BRAF inhibitor resistance mechanisms in metastatic melanoma: spectrum and clinical impact. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014;20:1965–1977. doi: 10.1158/1078-0432.CCR-13-3122. [DOI] [PubMed] [Google Scholar]
- 39.Konieczkowski DJ, et al. A melanoma cell state distinction influences sensitivity to MAPK pathway inhibitors. Cancer discovery. 2014;4:816–827. doi: 10.1158/2159-8290.CD-13-0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anders C, Niewoehner O, Duerst A, Jinek M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature. 2014;513:569–573. doi: 10.1038/nature13579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gilbert LA, et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell. 2014 doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hsu PD, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luo B, et al. Highly parallel identification of essential genes in cancer cells. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:20380–20385. doi: 10.1073/pnas.0810485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barbie DA, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liberzon A, et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27:1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smalley KS. Understanding melanoma signaling networks as the basis for molecular targeted therapy. The Journal of investigative dermatology. 2010;130:28–37. doi: 10.1038/jid.2009.177. [DOI] [PubMed] [Google Scholar]
- 50.Wong PP, et al. Histone demethylase KDM5B collaborates with TFAP2C and Myc to repress the cell cycle inhibitor p21(cip) (CDKN1A) Molecular and cellular biology. 2012;32:1633–1644. doi: 10.1128/MCB.06373-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hart MJ, et al. Direct stimulation of the guanine nucleotide exchange activity of p115 RhoGEF by Galpha13. Science. 1998;280:2112–2114. doi: 10.1126/science.280.5372.2112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SuppInfo
SuppTable6
SuppTable7