Emerging roles of linker histones in regulating chromatin structure and function (original) (raw)
. Author manuscript; available in PMC: 2018 Apr 12.
Published in final edited form as: Nat Rev Mol Cell Biol. 2017 Oct 11;19(3):192–206. doi: 10.1038/nrm.2017.94
Abstract
Together with core histones, which make up the nucleosome, the linker histone (H1) is one of the five main histone protein families present in chromatin in eukaryotic cells. H1 binds to the nucleosome to form the next structural unit of metazoan chromatin, the chromatosome, which may help chromatin to fold into higher-order structures. Despite their important roles in regulating the structure and function of chromatin, linker histones have not been studied as extensively as core histones. Nevertheless, substantial progress has been made recently. The first near-atomic resolution crystal structure of a chromatosome core particle and an 11 Å resolution cryo-electron microscopy-derived structure of the 30 nm nucleosome array have been determined, revealing unprecedented details about how linker histones interact with the nucleosome and organize higher-order chromatin structures. Moreover, several new functions of linker histones have been discovered, including their roles in epigenetic regulation and the regulation of DNA replication, DNA repair and genome stability. Studies of the molecular mechanisms of H1 action in these processes suggest a new paradigm for linker histone function beyond its architectural roles in chromatin.
Genomic DNA in eukaryotic cells is packaged into chromatin (FIG. 1), the structure of which controls essentially all nuclear processes involving DNA, including transcription, DNA replication and DNA repair. The packaging of DNA into chromatin is primarily guided by two major types of small, positively charged proteins: the core histones (H2A, H2B, H3 and H4) and the linker histone (H1). The first level of DNA packaging involves the association of DNA with the core histones and the formation of the nucleosome core particle1–3 (FIG. 1), the recurring structural unit of chromatin. The nucleosome core particle contains an octamer of core histones (two copies each of H2A, H2B, H3 and H4), around which ~147 bp of DNA winds in a left-handed super-helical manner4,5 (FIG. 2a). Within the nucleosome core particle, each core histone forms a histone fold structure with a flexible amino-terminal tail (FIG. 2b). The nucleosome core particle with an additional variable length of DNA (linker DNA) is termed the nucleosome (FIG. 1). Further packaging of DNA involves the formation of the chromatosome core particle6–8 (FIG. 2a), the next recurring structural unit of chromatin, consisting of a linker histone bound to the nucleosome with ~10 bp of DNA at both the entry and the exit sites of the nucleosome core particle. The complex containing the nucleosome and a linker histone will be subsequently referred to as the chromatosome (FIG. 1). Linker histones in metazoans have a conserved tripartite structure9,10 (FIG. 2c) consisting of a short, flexible N-terminal tail, a central globular domain and a long, intrinsically disordered and highly basic carboxy-terminal tail. The globular domain has a structure with a winged helix fold11 and preference for recognition of the nucleosome12. Both core and linker histones mainly use positively charged Arg and Lys residues to interact with the backbone phosphates of DNA through electrostatic interactions in the nucleosome and chromatosome core particles (FIG. 2d,e).
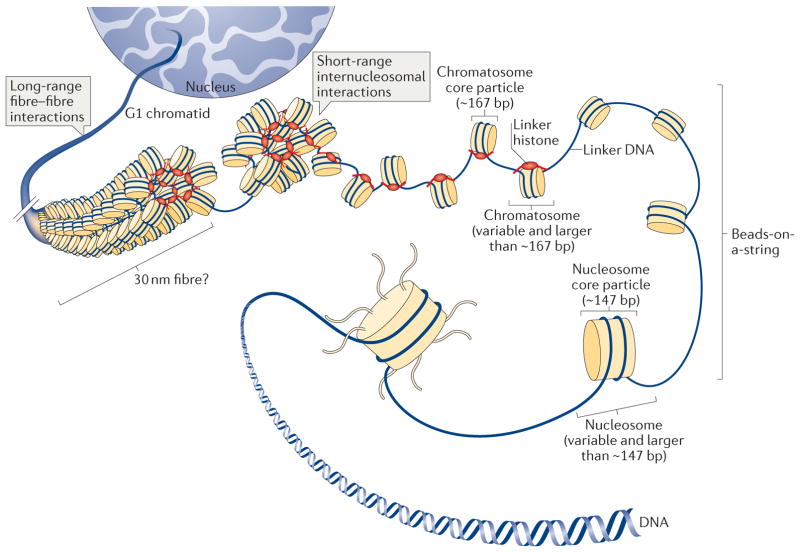
Figure 1. Multiple levels of chromatin folding.
DNA compaction within the interphase nucleus occurs through a hierarchy of histone-dependent interactions, including the formation of the nucleosome core particle, strings of nucleosomes (bead-on-a-string arrangement), the chromatosome core particle and 30 nm fibres (the existence of which is debatable in vivo and which may only be relevant over short lengths of chromatin) and the association of individual fibres, which eventually produces tertiary structures.
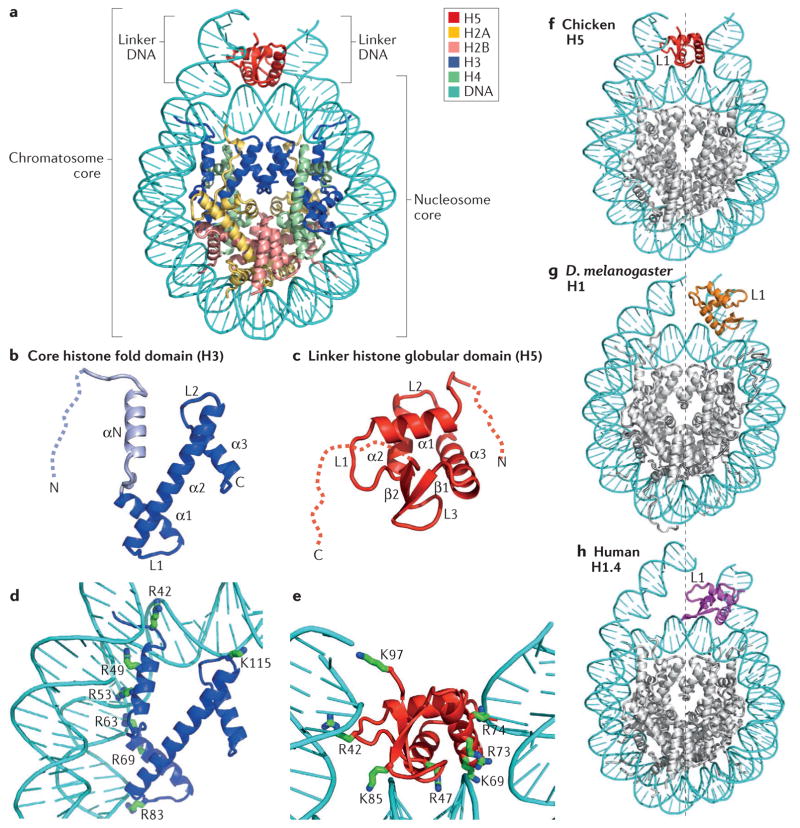
Figure 2. Structural illustration of the folded core regions of a chromatosome and representative interactions between histones and DNA.
a | The crystal structure of the chromatosome core containing the globular domain of chicken H5 (H1.0; shown in red) and fold regions of core histones (H2A, H2B, H3 and H4; all colour-coded) (Protein Data Bank identifier (PDB ID): 4QLC). The globular domain sits on the dyad of the nucleosome and interacts with both linker DNAs. b | The H3 structure from part a. The structural region from α1 to α3 (in blue) is termed the histone fold, which is shared by all core histones. The dashed line represents the intrinsically disordered histone tail. c | The structure of the folded globular domain of H5 from part a. The dashed line is used to illustrate the intrinsically disordered tails. In parts b and c, N and C indicate amino termini and carboxy termini, respectively. L indicates loop regions. d | Main interactions between DNA and the core histone H3 in the nucleosome (PDB ID: 4QLC). e | Main interactions between DNA and the globular domain of H5 (PDB ID: 4QLC). Lys (K) and Arg (R) residues that presumably form electrostatic interactions with the DNA phosphates are shown in sticks and are labelled with their residue numbers. f | The on-dyad binding mode observed in the crystal structure of the mono-nucleosome bound to the globular domain of H5 (H1.0), as in part a. g | The off-dyad binding mode observed in the NMR structural model of the mono-nucleosome bound to the Drosophila melanogaster linker histone H1 (REF. 46). h | The off-dyad binding mode observed in the cryo-electron microscopy structure of the nucleosome array containing human linker histone variant H1.4 (REF. 47). The L1 loop in the globular domain is labelled to highlight the difference in the orientation of the globular domain for each binding mode. The dashed line in parts f–h indicates the nucleosome dyad.
Core histones condense DNA and reduce its accessibility, leading to transcription inhibition in vitro13. However, in vivo functions of core histones are more elaborate, which is reflected by their important roles in the regulation of transcription14. The ability of core histones to regulate transcription is largely dependent on their post-translational modifications (PTMs)15,16, which regulate histone interactions with other proteins and thereby allow modification (either overcoming or solidifying) of the intrinsic histone barrier to transcription. Various experiments have identified specific and conserved PTMs of core histones that are linked to either transcription activation or repression17. Various proteins that add, recognize and remove these PTMs, termed writers, readers and erasers, respectively, have also been identified and structurally characterized18. In addition, numerous core histone chaperones, which facilitate core histone deposition or removal from chromatin, have also been characterized19.
In contrast to core histones, much less is known about the roles of linker histones in regulating the structure and function of chromatin. For example, in a recent review on the molecular hallmarks of epigenetic control16, the roles of linker histones were not even discussed. Moreover, even though it is known that linker histones are also subjected to PTMs (BOX 1), the respective writers, readers and erasers for any specific modification have not been fully established for these histones. Although several proteins have been suggested to have linker histone chaperone functions, such as nucleosome assembly protein 1 (Nap1) in Saccharomyces cerevisiae20 and nuclear autoantigenic sperm protein (NASP) in animals21, they have not been characterized in detail. Even more strikingly, no structures of linker histones in complex with any other proteins have been determined. This lack of understanding of the molecular biology of linker histones is partly attributed to technical difficulties: incomplete sequence coverage of linker histones in mass spectrometry approaches and a lack of high-quality antibodies. In addition, studies utilizing S. cerevisiae, which possesses low levels of a non-essential, non-canonical linker histone, have not been as fruitful as they have been for understanding the functions of core histones. Moreover, the existence of multiple redundant linker histone variants or subtypes in many metazoans has further limited progress in understanding their in vivo functional roles22. Nevertheless, these difficulties are being overcome, and the roles for linker histones in regulating many aspects of chromatin structure and function in vivo have begun to emerge. In this Review, we highlight some of the most recent experimental findings and provide some perspectives for future studies of linker histones and their biology.
Box 1. Linker histone post-translational modifications.
Like core histones, linker histones (H1) can be subjected to various post-translational modifications (PTMs), which are found in both the globular domain and the tails of H1. Even though H1 PTMs are much less understood than those of core histones, it is known that they have important roles in regulating chromatin structure and function. Studies show that PTMs in the globular domain of the somatic H1.2 to H1.4 variants are highly conserved but that those at the tails vary substantially, consistent with the sequence conservation features of these variants: the sequences in their globular domains are highly conserved (BOX 2), whereas the sequences in their tails vary substantially. These differences in PTMs between the different H1 variants are likely to correspond to the distinct functions of linker histone variants140. For a comprehensive review of H1 PTMs, the reader is directed to REF. 15. Below is a summary of well-studied H1 PTMs and their known functions or functional implications.
H1 phosphorylation
H1 phosphorylation is a highly complex and dynamic modification15. The level of phosphorylation is often cell cycle-regulated141. In both human and mouse cells, the levels of H1 phosphorylation are lowest in G1 phase, rise during S and G2, reach maximum levels at metaphase and sharply decrease thereafter. Phosphorylation of H1.4 at Ser27 inhibits the binding of heterochromatin protein 1 (HP1) to methylated Lys26 (see below)135, whereas Ser35 phosphorylation results in the dissociation of this H1 variant from mitotic chromatin142. Phosphorylation of H1.2 on Thr146 leads to dissociation of H1.2 from p53 and can activate the expression of p53 target genes, inducing apoptosis143. Phosphorylation of H1.2 and H1.5 at Ser172 localizes these variants to active DNA replication foci and to active transcription sites144. Phosphorylation of H1.4 at Ser187 facilitates gene activation by the glucocorticoid nuclear hormone receptor145.
H1 methylation
Methylation of H1.4 at Lys26 in human cells recruits HP1, leading to heterochromatin formation and gene silencing135. In Drosophila melanogaster, H1 methylated at Lys27 accumulates at pericentromeric heterochromatin in metaphase, potentially contributing to heterochromatin146.
H1 acetylation
Acetylation of H1.4 at Lys26 is related to the formation of facultative heterochromatin82, whereas Lys34 acetylation of H1.4 is associated with transcription activation147.
H1 citrullination
Citrullination of H1.2 to H1.4 at Arg54 has been shown to promote cell pluripotency and cell reprogramming to pluripotency. Mechanistically, it displaces H1 from chromatin, promoting an open chromatin state27.
H1 ubiquitylation
H1.5 mono-ubiquitylation might be important for the resistance against HIV-1 observed in mouse cells producing HIV-1 resistance factors148. Lys63 ubiquitylation of H1x serves as an important mark for recognition by factors involved in the maintenance of genome stability116.
Other H1 PTMs
H1 can also harbour various other PTMs, including formylation, denitration, ADP-ribosylation, crotonylation and lysine 2-hydroxyisobutyrylation, but their functions and cellular context remain to be elucidated15.
Linker histone variants
The linker histone family includes multiple variants23 (BOX 2). In humans and mice, 11 variants have been identified, including seven somatic subtypes (H1.0, H1.1 to H1.5, and H1x), three testis-specific subtypes (H1t, H1T2 and HILS1) and one oocyte-specific subtype (H1oo). H1.1 to H1.5 are expressed in a replication-dependent manner, whereas H1.0 and H1x are replication-independent and can be expressed in non-proliferating cells. Accordingly, H1.1 to H1.5, but also H1x, are ubiquitously expressed. By contrast, H1.0 accumulates in terminally differentiated cells. In line with this pattern of expression, in amphibian and avian organisms, H1.0 (known as H5 in birds) is associated with the formation of highly condensed and inert chromatin that is characteristic of terminally differentiated cells, such as nucleated erythrocytes, where it constitutes the majority of the H1 pool (for example, in chicken erythrocytes, H5 constitutes ~60% of the total amount of linker histones)24. Drosophila melanogaster larvae and adult flies express a single H1 variant. Flies also express an additional H1 variant during embryonic development. This H1 variant is termed BigH1 due to its longer N-terminal tail25, and because it is encoded by a single gene in the genome, it provides a convenient model system for studying the functions of linker histone variants.
Box 2. Linker histone variants and sequence alignment of their globular domains.
Linker histones typically comprise ~200 amino acids and exist as variants with varying sequences (see the figure; c denotes chicken, h denotes human and d denotes fly isoforms). The positively charged residues in the globular domains are important for nucleosome binding and are well conserved between species and individual variants (highlighted in blue in the figure)36,40. In contrast to the high sequence conservation among the globular domains of H1.0 to H1.5, the linker histone tails among the variants are much less conserved. For the nomenclature of linker histone variants, see REF. 149.

H1 variants are modified and regulated by PTMs26 (BOX 1), but these modifications and their functional differences and importance are understudied compared with PTMs of core histone. Nevertheless, some interesting functional data are now starting to emerge. For example, it has been recently shown that citrullination of a single residue (Arg54) within the globular domain of the linker histone variant H1.2 is important for its removal from chromatin, leading to de-condensation of chromatin during reprogramming to pluripotency27. Another recent study reported that silencing of the differentiation-associated H1.0 variant in tumour cells leads to self-renewal, promoting changes in chromatin organization and gene expression and resulting in a more malignant cancer phenotype28.
Roles in chromatin organization
To understand the function of chromatin, it is crucial to determine its structure at high resolution. Although the structure of the nucleosome core particle at atomic resolution was solved more than a decade ago29, determination of the structures of the folded chromatin containing linker histones, and accordingly the structure of the chromatosome core particle, has been very challenging. Nevertheless, important progress has been made in resolving higher-order chromatin structures, including chromatosomes, the 30 nm chromatin fibre and topologically associating domains (TADs), and in elucidating the interactions involved, as well as the role of linker histones in establishing these higher-order structures30.
Linker histone interactions with the nucleosome and the chromatosome structure
Both the nucleosome and the chromatosome were identified in the 1970s (REFS 1,2,7). Since then, substantial efforts have been made to solve their structures. A near-atomic resolution structure of the nucleosome core particle was solved in 1997 (REF. 5), which allowed the visualization of the side chains of amino acids of core histones and how they interact with DNA. Various structural models of the chromatosome core particle have been proposed over the years9,31–39, but it was only in 2015 that the first crystal structure of the nucleosome–linker histone complex at near-atomic resolution, which included 167 bp of DNA and the globular domain of chicken H5, was published40. In this crystal structure (FIG. 2a), the globular domain binds on the nucleosome dyad and interacts with the linker DNA at both the entry and the exit sites of the nucleosome. The interactions with the nucleosome mainly occur between positively charged Lys and Arg residues of the globular domain and the phosphates of the DNA backbone in the nucleosome, similar to those between core histones and DNA (FIG. 2d,e). This crystal structure is in agreement with previous in vitro experimental data investigating the binding of the H5 globular domain to nucleosomes41,42, as well as data showing the binding of the globular domain of mouse H1.0 and its mutants to chromatin in vivo36, suggesting that the linker histone binds to the nucleosome in the same way in vitro and in vivo. Using longer linker DNA and full-length linker histone H1.0, it was further shown by NMR and cryo-electron microscopy (cryo-EM) that the tails of the linker histones and the longer linker DNA in the chromatosome do not have a role in determining the binding mode between the nucleosome and the globular domain of the linker histone43,44. However, measurements of binding affinity and cryo-EM structural studies showed that within a single chromatosome, the C-terminal tail of H1.0 appears to be preferentially associated with only one of the two available linker DNAs44,45, which was speculated to have a role in influencing the assembly and properties of higher-order chromatin structures44.
A previous structural model of the chromatosome core particle was built from NMR studies in solution and contained the globular domain of H1 from D. melanogaster46. In this model, H1 binds off the dyad and appears to interact with one linker DNA, instead of both linker DNAs as in the crystal structure of H5 discussed above (FIG. 2f,g). The different binding modes were explained by a subsequent study, which identified five key residues in the globular domains of H5 and D. melanogaster H1 that determine the binding location of the globular domain in the chromatosome (on-dyad versus off-dyad binding)43, suggesting that a small number of residues in the globular domain of a linker histone variant can determine its binding mode.
Another structural model was generated for the human linker histone variant H1.4 in a nucleosome array using single-particle cryo-EM47. In this 11 Å resolution structural model, the globular domain of H1.4 binds off the nucleosome dyad (FIG. 2h). However, the orientation of the globular domain in this model is different from that described for D. melanogaster H1 discussed above40 (FIG. 2g,h). In addition, the tails of H1.4 were largely not observable in this cryo-EM study47, suggesting that they do not stably bind to linker DNA in any specific manner. Intriguingly, the globular domain of human H1.5, which has essentially the same amino acid sequence as the globular domain of H1.4 (BOX 2), was found to bind the dyad in single nucleosomes38,44. It has been suggested that the difference in the binding mode between the single chromatosome and the nucleosome array could be caused either by glutaraldehyde crosslinking used in the cryo-EM experiment or by the folded structure of the nucleosome array, in which re-orientation of the linker DNA disrupts the intrinsic on-dyad binding mode44.
These structural studies revealed detailed interactions between the linker histone and DNA in the nucleosome, which are essential for understanding the effects of mutations and PTMs in the globular domain of linker histones on chromatin structure and function. The observation that linker histone variants may bind to the nucleosome in different modes, which is associated with distinct structures of condensed nucleosome arrays — with on-dyad binding corresponding to more condensed chromatin architecture40,43 — is also likely to be related to chromatin function.
Contribution of linker histones to establishing 30 nm chromatin fibres
It has long been known that in the presence of linker histones, chromatin is prone to forming higher-order structures, including a fibre with a diameter of ~30 nm in vitro48. However, the structural nature of this 30 nm chromatin fibre and whether this structure is relevant in vivo have remained elusive8,49–60. The above-mentioned cryo-EM study of the nucleosome arrays associated with human H1.4 (REF. 47) has shed important light on this topic. This study revealed that in the presence of the linker histone, nucleosome arrays arrange into a twisted left-handed double helix with a zigzagged, two-start tetra-nucleosome as the repeating structural unit (FIG. 3a). The globular domains of the linker histone between these tetra-nucleosome units form a dimer, which appears to be responsible for the twisted feature of the double helix. This cryo-EM structure supports the previously proposed zigzag, two-start organization of the 30 nm chromatin fibre53,54 and contradicts the cryo-EM model obtained for the nucleosome array containing linker histone H5 (in the presence of additional 1 mM MgCl2), which suggested a single-start interdigitated nucleosome organization for the nucleosome array61. Intriguingly, an earlier cryo-EM study in the absence of salt and crosslinking fixation (commonly used in cryo-EM studies) showed that the nucleosome array condensed by H5 features loosely condensed nucleosomes with a zigzag arrangement62. These results suggest that nucleosome array structures are prone to environmental perturbations, and it is important to use non-invasive experimental methods such as NMR and cryo-EM in the absence of chemical crosslinking to investigate them.
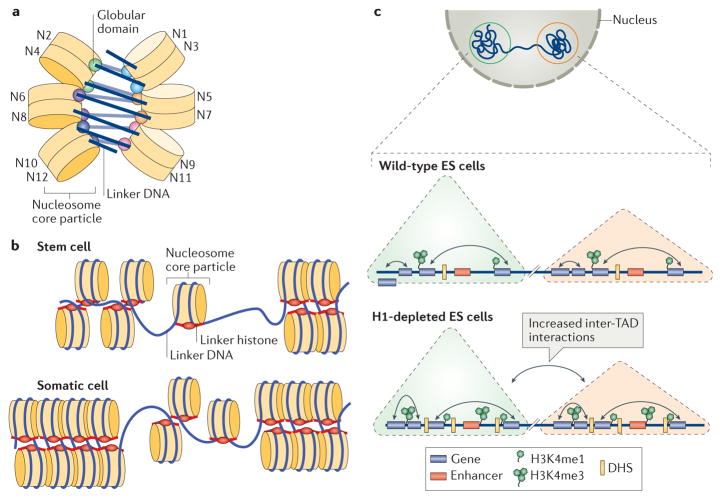
Figure 3. Roles of linker histones in chromatin folding.
a | A cartoon illustration of the cryo-electron microscopy structure of the nucleosome array condensed by human linker histone H1.4 (REF. 47). The nucleosome array structure is a twisted double helix with tetra-nucleosomes as the structural unit. The globular domain of the linker histone in each nucleosome is mainly associated with one linker DNA. The globular domains of the linker histones in the nucleosomes that interact between neighbouring tetra-nucleosome units form a dimer. The linker DNA connecting the two nucleosome core particles between neighbouring tetra-nucleosome units is not associated with the globular domain of the linker histones. The tails of the linker histones in the structure are not observed. b | Nucleosomes in the presence of linker histones are likely to form chromatosomes, which are arranged in heterogeneous groups, termed ‘nucleosome clutches’, along the chromatin fibre in a cell type-specific manner: in stem cells, smaller clutches are typically observed compared with differentiated cells. This different organization of chromatosomes corresponds to differences in chromatin compaction and heterochromatization: larger clutches are associated with heterochromatin formation. c | In interphase, the chromosome is structurally organized into distinct topologically associating domains (TADs; triangles). In H1-depleted mouse embryonic stem (ES) cells, the overall genome organization into TADs is not majorly affected. In addition, interactions (black double-headed arrows) within TADs do not change. However, within gene-dense TADs, long-range inter-TAD interactions increase. In addition, new DNase hypersensitive sites (DHSs) and sites of histone H3 Lys4 mono-methylation and trimethylation (H3K4me1 and H3K4me3, respectively) are established, indicating changes in the epigenetic landscape of the cell. Part c is adapted with permission from REF. 150, Macmillan Publishers Limited.
More recently, an investigation of chromatin structure in vivo was performed using stochastic optical reconstruction microscopy (STORM)63. The chromatin fibre was visualized at the single-cell level in interphase cells, with a resolution of ~20 nm. It was observed that nucleosomes are assembled in discrete heterogeneous groups of varying sizes, termed ‘nucleosome clutches’ (FIG. 3b). Notably, differentiated human fibroblasts contained larger clutches (~8 nucleosomes per clutch) compared with stem cells induced from fibroblasts (~4 nucleosomes per clutch). Increased levels of linker histones in larger and denser clutches were also well correlated with heterochromatin markers. The results from this study confirm the role of the linker histones in chromatin folding but also suggest that in vivo 30 nm chromatin structures only exist as short fragments rather than as continuously folded fibres. This observation is also consistent with the recent findings that no regular 30 nm chromatin fibre structures could be observed in native chromatin using electron-microscopy-assisted nucleosome interaction capture crosslinking experiments in combination with mesoscale chromatin modelling64 and small angle X-ray diffraction59. In addition, small clusters of nucleosome arrays primarily containing tetra-nucleosomes have also been observed in force-stretched nucleosome arrays in vitro65.
The role of linker histones in organizing TADs
Chromatin conformation capture studies have identified TADs as a conserved feature of higher-order chromatin structures66–69. TADs are defined as continuous regions of chromatin (100 kb to 10 Mb of DNA) within which DNA–DNA interactions are much more frequent than in other chromatin regions. To investigate the role of linker histones in TAD structures, high-throughput chromatin conformation capture was applied to wild-type and linker-histone-depleted (triple knockout of H1.2, H1.3 and H1.4, resulting in an ~50% decrease in total levels of linker histones) mouse embryonic stem (ES) cells22,30. It was found that the location and size of TADs were largely unaltered between wild-type and linker-histone-depleted cells, but the frequency of their inter-domain interactions increased, suggesting that structural changes within TADs occurred when the H1-to-nucleosome stoichiometry was reduced. Moreover, the increase in inter-TAD interactions was found to correlate with changes in the epigenetic landscape of cells, involving changes (increases at some loci and decreases at others) in activating histone marks — histone H3 Lys4 methylation (H3K4me1) and trimethylation (H3K4me3) — and an increase in the number of DNA hypersensitivity sites (DHSs), which are associated with decreased DNA methylation (suggesting a decrease in chromatin compaction) (FIG. 3c). These results suggest that linker histones are important for higher-order chromatin interactions and modulate local chromatin topology within the nucleus, thereby providing additional mechanisms regulating chromatin organization and function.
Biological functions
Linker histones have long been known to have important roles in the regulation of chromatin structure and gene expression70. Recent studies have provided new insights into the in vivo roles of linker histones in epigenetic regulation and regulation of DNA replication, DNA repair and genome stability. These topics are reviewed below, along with a discussion of the molecular mechanisms of H1 action.
Linker histones in regulating the epigenetic state of chromatin
The epigenetic landscape of eukaryotic genomes is maintained and dynamically regulated by PTMs of the core histones and DNA methylation, which are distributed in a highly regulated fashion in chromatin. Recent evidence suggests that the distribution of H1 is also not uniform but, rather, that it is modulated depending on the genomic context. This view is supported by studies using the DNA adenine methyltransferase identification (DamID) technique in both fly and human cells71,72 and by several studies involving chromatin immunoprecipitation followed by deep sequencing23,73–77. For instance, H1 is depleted in promoter regions of actively transcribed genes that are enriched for ‘active’ histone marks, such as H3K4me3, whereas H1 occupancy is increased within silenced chromatin domains enriched for repressive histone marks, including methylated H3K9 and H3K27.
The positive and negative correlations between the presence of H1 and that of certain histone marks make it plausible that H1 occupancy is governed through a distinctive recognition of the histone code and/or that, vice versa, H1 regulates the core histone PTMs in a locus-specific manner. As already hinted above, the presence of H1 has been implicated in altering the epigenetic code and in interfacing with particular modified core histone states (FIG. 4a). It has been known for over 35 years that H1 occupancy strongly correlates with hypoacetylation of core histones78,79. Apparently, H1 can repress histone acetylation by negatively regulating histone acetyltransferases (HATs), as was demonstrated for human p300/CBP-associated factor (PCAF; also known as KAT2B)80. It was proposed that the linker histone tails hinder the access of PCAF to H3, thereby preventing its modification (FIG. 4a). Furthermore, H1 is required for the maintenance of female germline stem cells in D. melanogaster, where its depletion selectively augments H4K16 acetylation (H4K16ac), causing premature differentiation81. In this case, H1 regulates H4K16ac by antagonizing the H3K16-specific HAT MOF (FIG. 4a), depletion of which rescued H1-knockdown phenotypes81. In addition, the interaction between H1 and histone deacetylases (HDACs) may contribute to the negative correlation between H1 and core histone acetylation. It has been shown that the human HDAC sirtuin 1 (SIRT1), which, apart from H4K16ac and H3K9ac, also uses H1K26ac as a substrate, is tethered to H1-containing chromatin and deacetylates core histones. These changes have been associated with a decrease in H3K79me2, which is a mark preventing spreading of transcriptionally silenced heterochromatin. Overall, it has been proposed that the SIRT1–H1-dependent mechanism contributes to spreading of hypomethylated H3K79 and heterochromatin formation82 (FIG. 4a).
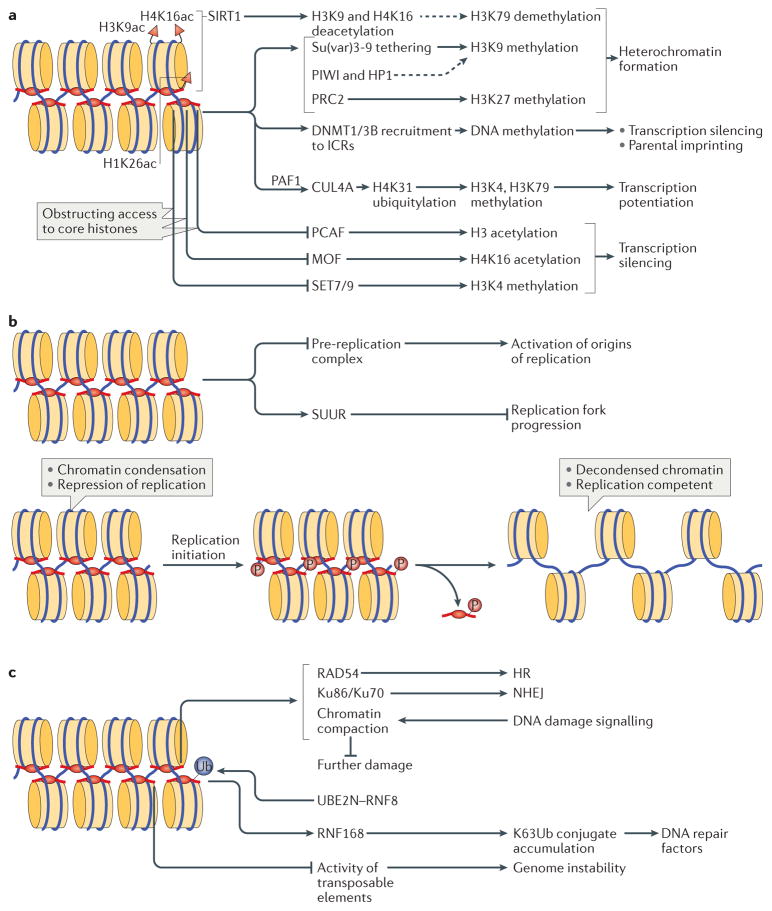
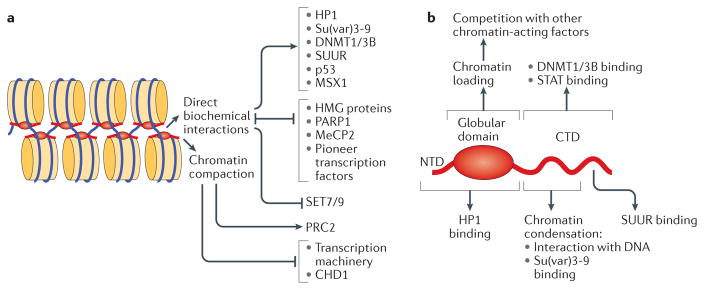
Figure 4. Biological functions of linker histones.
a | Linker histones (H1) are implicated in the regulation of the epigenetic landscape of the cell by interacting with several epigenetic modifiers and by regulating their recruitment to chromatin and/or activity, which affects chromatin organization. H1 acetylated at K26 (H1K26ac) is a binding partner for the deacetylase sirtuin 1 (SIRT1), which deacetylates both core histones (H3K9ac and H4K16ac) and H1; an undefined SIRT1–H1-dependent mechanism has been further linked to hypomethylation of H3 at Lys79 (H3K79). H1 is known to recruit the histone methyltransferase Su(var)3-9, as well as DNA methyltransferases DNMT1 and DNMT3B to chromatin. Furthermore, H1 interacts with PIWI proteins and heterochromatin protein 1 (HP1), modulating histone methylation and heterochromatin formation. H1 is also a substrate for methyltransferase Polycomb repressive complex 2 (PRC2) and promotes its activity. During transcription, H1 was also shown to recruit the E3 ubiquitin ligase cullin 4A (CUL4A) and RNA polymerase II-associated factor 1 (PAF1), which is necessary for CUL4A activity. By bringing CUL4A and PAF1 together, H1 promotes CUL4A-mediated ubiquitylation of H4, which further drives methylation of core histones. H1 also repels and/or interferes with the activity of several core histone modifying enzymes, including p300/CBP-associated factor (PCAF), MOF and SET7/9. b | H1-mediated mechanisms of DNA replication control. H1 represses DNA replication at the stage of replication initiation by inhibiting the assembly of the pre-replication complex and at the stage of replication fork progression by tethering the SNF2-like ATPase protein suppressor of underreplication (SUUR). In addition, H1 undergoes S phase-dependent phosphorylation (P), which results in H1 dissociation from chromatin, leading to large-scale chromatin decondensation and the activation of origins of replication. c | The roles of H1 in DNA repair, genomic stability and DNA damage signalling. H1 is involved in DNA repair via both homologous recombination (HR) and non-homologous end joining (NHEJ) through interactions with RAD54 and with Ku86 and Ku70, respectively. H1 also facilitates ubiquitin-dependent signalling at DNA double-strand breaks: H1 is ubiquitylated by the E2 ubiquitin-conjugating enzyme UBE2N and the E3 ubiquitin ligase RNF8, and recruits the E3 ubiquitin ligase RNF168 to promote accumulation of K63-linked ubiquitin conjugates, resulting in the binding of repair factors. Chromatin compaction promoted by H1 may help to limit further DNA damage. Suppression of transposable element activity by H1 also contributes to genome stability. ICRs, imprinting control regions.
Perhaps one of the better understood paradigms of the connection between H1 and the histone code is its role in marking pericentric heterochromatin of polytene chromosomes in D. melanogaster with methylated H3K9, the major core histone PTM associated with heterochromatin formation83. In more detail, H1 was shown to physically interact with the H3K9-specific histone methyltransferase (HMT) Su(var)3-9, assisting in tethering it to chromatin84 (FIG. 4a). Apart from regulating chromatin in pericentric regions, H1 was shown to facilitate methylation of H3K9 within transposable elements, resulting in their robust transcriptional repression84.
Interestingly, transposon repression in somatic cells in fly ovaries does not depend on H1-mediated methylation of H3K9 but still requires H1 and involves methylated H3K9. In this case, heterochromatization depends on the interaction of H1 with PIWI proteins and heterochromatin protein 1 (HP1; in particular, its isoform HP1α)85. Both transcriptional and post-transcriptional silencing of transposons can be mediated in female germline cells by PIWI-interacting RNAs (piRNAs), which can guide PIWI-dependent silencing machinery to deposit H3K9me3 at target genomic loci. It was demonstrated that in D. melanogaster, physical interaction between PIWI and H1 facilitates H1 recruitment to transposon sequences, which contributes to chromatin compaction and heterochromatin formation85. Accordingly, depletion of PIWI decreased H1 density at a subset of transposons, leading to their derepression. Interestingly, although the silencing requires H1, H3K9 methylation and HP1, the abrogation of H1 expression increases the target loci chromatin accessibility without affecting H3K9me3 density, whereas the loss of HP1 does not affect H1 occupancy, suggesting that these different components independently contribute to heterochromatin establishment. Thus, H1 can contribute to heterochromatic silencing through various biochemical pathways.
Similarly to its role in excluding PCAF from binding to chromatin (discussed above), H1 has the ability to prevent binding of HMTs that establish positive methylation marks, such as H3K4 methylation marks, thereby potentiating the repressed state of chromatin. It was demonstrated that in mouse ES cells, H1 selectively inhibits the binding of SET7/9 (also known as SETD7) to two regions, under maternal imprinting control: H19 and Meg3 (REF. 86), which results in decreased H3K4me2 levels and contributes to the silencing of these regions (FIG. 4a). Given the specific repressive action of H1 on this HMT, the molecular mechanism possibly requires a physical interaction between the proteins and/or specific masking of the H3K4 substrate within the H3 N-terminal tail by H1. An alternative mechanism based on H1-dependent restriction of H3 tail mobility was proposed87. The tail mobility constraining effects of H1 were observed in vitro and were shown to restrict the histone modifying activity of multiple H3 tail modifying enzymes, including the HMTs G9a (also known as EHMT2) and Polycomb repressive complex 2 (PRC2)87.
H1 has been biochemically linked to another global silencing mark, H3K27 methylation. In this case, it has been demonstrated that, at least in vitro, H1-containing oligonucleosomes present a favourable substrate for the human PRC2–EZH2 complex and stimulate its enzymatic activity to methylate H3K27 (REF. 88) (FIG. 4a). This observation can be explained by direct physical interactions between H1 and specific hPRC2 subunits — SUZ12, EED and AEBP2 (REF. 88). Additionally, H1-dependent structural changes in the chromatin fibre (see above) may enhance the affinity of the enzyme89. In a reciprocal mechanism, it has been shown that human H1.2 exhibits preferential binding to H3K27me3 nucleosomes in vitro. Furthermore, H3K27 methylation by EZH2 strongly and specifically stimulates H1.2 occupancy in cells90. Thus, H1 and the H3K27 methylation mark appear to establish a positive feedback loop that may mutually reinforce chromatin silencing effects.
H1.2 was also found to interact with the E3 ubiquitin ligase cullin 4A (CUL4A) and to promote H4K31 ubiquitylation, which is further associated with increased loading of the ‘positive’ H3K4me3 and H3K79me2 marks in target genes and potentiation of their transcription91. In this context, H1.2 selectively recognizes phosphorylated Ser2 of an elongating RNA polymerase II (Pol II) and recruits, in addition to CUL4A, RNA Pol II-associated factor 1 (PAF1), which is necessary for CUL4A activity. Overall, H1.2 serves as a bridge to couple CUL4A and PAF1 during transcription, thereby supporting the establishment of transcription-promoting histone marks (FIG. 4a). Therefore, linker histones may also have essential roles in maintaining active transcription states at certain loci.
Another important epigenetic mark in chromatin, present in some but not all eukaryotes, is DNA methylation92. A relationship between H1 and DNA methylation has been described in both mammals and plants. For example, even though overall it resulted in only a few gene expression changes, depletion of H1 in mouse ES cells prominently affected regions regulated by DNA methylation. These included the imprinted loci H19 and Meg3, which were hypomethylated in their imprinting control regions (ICRs) in H1-depleted cells22. At about the same time, a link between H1 and DNA methylation in plants was reported, which showed that knockdown of H1 in Arabidopsis thaliana reproduced the developmental abnormalities of DNA hypomethylation mutants93. Subsequent work with the H1-depleted mouse ES cells showed that, in addition to regulating methylation of the maternally imprinted loci H19 and Meg3, H1 is also involved in regulating an X-linked homeobox gene cluster94. Remarkably, in this case it was found that only the subset of genes that are paternally imprinted are affected by H1. Other studies revealed that H1-depleted ES cells exhibit impaired differentiation, which could be associated with a reduced ability to methylate and repress the expression of the Oct4 (also known as Pou5f1) pluripotency gene95. Collectively, these studies strongly suggest that H1 is able to promote DNA methylation at specific loci. Accordingly, re-expressing certain H1 subtypes in H1-depleted ES cells led to increased DNA methylation within the ICRs of H19 and Meg3 and repression of these loci. Furthermore, it was found that several H1 subtypes directly interact with the DNA methyltransferases DNMT1 and DNMT3B and help recruit them to the ICRs86 (FIG. 4a). Genome-wide studies of the H1-depleted ES cells support the view that H1 promotes DNA methylation at many regulatory regions, particularly at enhancer regions30. The role of H1 in regulating DNA methylation may be important in many processes, including disease pathogenesis. For example, very high rates of mutation in H1-encoding genes have been observed in B cell lymphomas, and many of these mutations abrogate the interaction of H1 with DNMT3B96.
Regulation of DNA replication by linker histones
DNA replication, perhaps even more so than transcription, requires a dramatic reorganization of chromatin structure in order to provide the replication machinery access to the DNA template. Therefore, it is expected that linker histones have important roles in the control of DNA replication97. Surprisingly, early experiments using an in vitro replication system based on viral (simian virus 40 (SV40)) DNA and HeLa cell cytosolic extracts indicated that purified native H1, when present at physiological ratios to nucleosomal histones, failed to repress SV40 replication98. Only at molecular ratios greater than 1 with respect to nucleosomes was H1 able to partially inhibit the reaction. However, the results were strikingly different when H1 was purified from cells synchronized at different phases of the cell cycle: H1 from G0-phase and M-phase cells, unlike H1 from S−phase cells, could strongly inhibit replication of SV40 DNA when present in equimolar amounts to nucleosomes99. This difference is likely to be attributable to the altered compaction of the chromatin template and hypothetically may depend on cell cycle-specific PTMs of H1 (REF. 99).
In addition, a strong reduction of the DNA replication rate by H1 was observed using frog egg extracts and recombinant mouse H1.2-permeabilized sperm chromatin100. The inhibition of replication occurred during pre-replication complex formation (FIG. 4b), whereas the fork progression was largely unaffected100,101. Notably, in this system, different H1 variants show varying capabilities to block replication; this has been associated with the different structures of their C-terminal domains, which correspond to the differential affinity of the individual H1 variants to chromatin102.
A role for H1 in the regulation of replication has been recently shown in vivo, in larval tissues of D. melanogaster undergoing endoreplication103. In this context, H1 was found to be an upstream effector of protein suppressor of underreplication (SUUR). Recruitment of SUUR to intercalary heterochromatin of polytene chromosomes causes retardation of replication fork progression, leading to less efficient endoreplication and, consequently, reduced copy numbers (underreplication) of loci within intercalary heterochromatin104. Localization of SUUR to intercalary heterochromatin was dependent on normal levels of H1, indicating that H1 is involved in the regulation of DNA endoreplication rates of late-replicating regions of chromatin by promoting the deposition of SUUR in these regions (FIG. 4b). Additional evidence suggested further SUUR-independent roles for H1 in replication103. Strikingly, H1 was shown to exhibit a very dynamic temporal distribution pattern in polytene chromosomes during the endocycle. At the earliest stage of the endocycle S (endo-S) phase, H1 was deposited specifically into late-replicating loci and then redistributed in the course of replication. The pre-replicative marking of intercalary heterochromatin with H1 precedes the subsequent deposition of SUUR, which occurs in these domains only after a delay of several hours. Thus, early in the endo-S phase, H1 may act directly, without SUUR, to suppress the activation of late origins of replication, thereby inhibiting DNA endoreplication of these loci until later in endo-S103. Although the mechanism of replication-independent H1 loading into intercalary heterochromatin is not understood, it is possible that the highly restricted distribution of H1 in early endo-S is established through epigenetic mechanisms, such as H1 tethering to the H3K27me3 mark90, which is highly abundant in intercalary heterochromatin105. The upstream signal or signals that guide H1 spatial distribution in early endo-S are likely to be installed during G1, when the temporal programme of replication is determined106.
Previous experiments in Physarum polycephalum led to a similar conclusion that H1 is an essential regulator of replication timing. However, in this case, the late replication programme was completely disrupted by H1 depletion107. Furthermore, phosphorylation of H1 facilitated its removal and late replication origin activation107. A negative correlation between H1 phosphorylation and association with chromatin has been established in other experimental systems, including Tetrahymena thermophila108 and mammals109,110. Importantly, H1 undergoes S phase-dependent phosphorylation that involves CDC45, cyclin A and cyclin-dependent kinase 2 and results in large-scale chromatin decondensation109 (FIG. 4b; see also BOX 1). Therefore, the dynamic pre-replicative association of H1 with chromatin and replication-dependent dispersal of H1 appear to be regulated by the phosphorylation state of H1 in the nucleus and can make important contributions to the control of DNA replication.
The role of linker histones in DNA repair and genome stability
Another major group of processes of nuclear DNA metabolism requiring substantial reorganization of nucleosome structure and chromatin decompaction is related to DNA damage and involves DNA repair reactions. Thus, chromatin structure defects produced by the elimination or reduction of linker histone content in chromatin may affect these reactions and thereby genome stability. In fact, HHO1, the non-essential S. cerevisiae homologue of H1, was demonstrated almost 15 years ago to be required for the suppression of DNA repair by homologous recombination (HR) and the recombination-dependent mechanism of telomere maintenance111. Furthermore, HHO1 represses HR of the ribosomal DNA (rDNA) locus, similar to, but independent of, the HDAC Sir2 (REF. 112). In D. melanogaster, H1 depletion is known to derepress transposable elements84,113, which may contribute to the genomic instability observed in H1-depleted flies (FIG. 4c). Interestingly, upon H1 knockdown, larval imaginal disc and salivary gland cells accumulate extrachromosomal circular DNA originating from the rDNA gene cluster due to hyper-recombination and genomic rearrangements113, a phenotype previously observed for _Su(var)3_-9 mutants. Consistently, H1 depletion causes a genome-wide increase in the incidence of double-stranded DNA breaks (DSBs) in fly cells113.
H1 also mediates genome stability-related processes in vertebrates. Chicken DT40 cells lacking one of the six H1 variants, H1R, exhibit elevated sensitivity to a DNA alkylating agent methyl methanesulfonate (MMS), and based on multiple lines of evidence, it was proposed that H1R is involved in the RAD54-mediated HR DSB repair pathway114 (FIG. 4c). By contrast, H1-depleted mouse ES cells were found to be hyper-resistant to DNA damage115, which was attributed to decompaction of chromatin upon H1 depletion resulting in increased DNA damage response signalling generated at DNA breaks. Additionally, depletion of H1 in mouse ES cells led to an increase in the frequency of telomeric sister chromatid exchange events and in telomere length by an as yet unknown mechanism115.
Importantly, linker histones have been implicated in physical interactions with multiple components of the DNA repair machinery and DNA damage response factors. Human H1, but not core histones, is a major chromatin-associated target of the E2 ubiquitin-conjugating enzyme UBE2N (also known as UBC13) that together with E3 ubiquitin ligase RNF8 act in the ubiquitin-dependent DSB signalling pathway116. Ubiquitylated H1 is recognized by the E3 ubiquitin ligase RNF168 and thus serves as a mark that promotes further accumulation of Lys63-linked ubiquitin conjugates at DSBs, which stimulates the recruitment of DNA repair factors (FIG. 4c). Human H1.0 interacts with Ku86 and Ku70 (REF. 117), which bind as a heterodimer to DSBs and are required for the non-homologous end joining DNA repair pathway and V(D)J recombination (FIG. 4c).
Finally, DNA damage signalling causes global chromatin fibre compaction, which may help to protect the genome from additional lesions. This compaction may be further stabilized by linker histones118 (FIG. 4c).
Molecular mechanisms of action
The conventional view of the biochemical activities of H1 in the regulation of nuclear functions limits its roles to altering chromatin architecture119. This paradigm primarily focuses on the inhibitory, DNA access-restricting properties of H1. However, in addition to the impact on chromatin structure per se, binding of H1 to linker DNA provides an obstacle to DNA-binding proteins, including histone modifiers (see above and FIG. 4a) and transcription factors120 (FIG. 5a). H1–nucleosome association may also inhibit the translocation of processive, DNA-tracking enzymes, such as RNA and DNA polymerases and chromatin remodelling factors. For instance, oligonucleosome-bound H1 impedes ATP-dependent remodelling by chromodomain-helicase-DNA-binding protein 1 (CHD1) in vitro121 (FIG. 5a). Interestingly, Chd1 and His1 genetically interact and share a large subset of regulatory targets in D. melanogaster in vivo122, which led to the speculation that CHD1 and H1 cooperate in the negative regulation of transcriptional elongation and/or repression of cryptic promoters in gene bodies.
Figure 5. Biochemical activities of linker histones.
a | Alternative molecular mechanisms used by linker histones (H1) to modulate the activity of chromatin. Direct biochemical interactions with H1 facilitate or inhibit chromatin binding of various structural proteins, enzymes and transcription factors; in addition, direct competition mechanisms control the mutually exclusive distribution patterns of H1 with various chromatin-interacting proteins. In general, H1-dependent chromatin compaction interferes with transcription initiation by preventing nucleosome remodelling and the binding of sequence-specific transcription factors, as well as the binding and translocation of general transcription factors. However, it has been shown that the compacted chromatin state established by H1 stimulates the activity of Polycomb repressive complex 2 (PRC2). b | Structural domains of the H1 polypeptide involved in H1 deposition, physical interactions and regulatory functions. Distinct regions within the globular domain and the carboxy-terminal domain (CTD) mediate the multiple biochemical activities of H1. CHD1, chromodomain-helicase-DNA-binding protein 1; HMG, high-mobility group; HP1, heterochromatin protein 1; MeCP2, methyl-CpG-binding protein 2; NTD, amino-terminal domain; PARP1, poly(ADP-ribose) polymerase 1; STAT, signal transducer and activator of transcription; SUUR, suppressor of underreplication.
Another canonical interpretation of the observed repressive activities of H1 in DNA metabolism processes is H1-dependent chromatin compaction. An added degree of three-dimensional folding is generally considered inhibitory due to partial internalization of linker DNA and the exterior surfaces of nucleosome core particles, causing steric hindrance of these elements (‘template obstruction model’)123,124 (FIG. 5a). However, quantitative biochemical experiments indicate that this hierarchical folding of chromatin need not be repressive. For example, assembly of ‘dense chromatin’ substrates that mimic the nucleosome repeat length of H1-containing chromatin (and are prone to folding into higher-order structures) can stimulate the activity of certain nuclear enzymes, such as PRC2 (REF. 89) (FIG. 5a).
Nevertheless, the inhibitory role of linker histones on chromatin transactions is valid in many contexts. A variation of the template obstruction model — involving competitive binding — was postulated for the interplay between linker histones and high-mobility group (HMG) proteins125, highly abundant, ubiquitous chromatin components that affect chromatin condensation and enhance DNA access for regulatory factors. Each distinct HMG family member associates with a separate set of chromatin binding sites, and it was proposed that H1 is able to compete with all of the HMG proteins, thereby regulating global chromosome organization (FIG. 5a). Similarly, a reciprocal, mutual exclusion mechanism guides the nucleosome binding properties of H1 and poly(ADP-ribose) polymerase 1 (PARP1), thus establishing specific transcriptional outcomes in mammalian cells126 (FIG. 5a). Competition mechanisms are also involved in chromatin binding of so-called ‘pioneer’ transcription factors. During cell differentiation, the pioneer factors are usually the first to bind enhancer elements, and their binding is necessary to alter the epigenetic landscape, trigger transcriptional competency of enhancers and implement cell type-specific expression programmes127. Importantly, the prototypical pioneer factor forkhead box protein A1 (FOXA1; also known as HNF3α) contains a winged helix motif that is highly homologous to that of H1 and can thus efficiently displace H1 from chromatin128,129. This displacement may constitute an obligatory first step of the transcriptional response to retinoic acid in ES cells130. Finally, strong evidence exists, both in vitro and in cell culture, for competition between human methyl-CpG-binding protein 2 (MeCP2) and H1 for common binding sites131, which may be important for their combinatorial regulation of gene expression (FIG. 5a). Interestingly, the association of MeCP2 with oligonucleosomes appears to impart the architecture (zigzag motif) characteristic of H1-containing chromatin, suggesting that MeCP2 interacts with nucleosomes in a similar fashion and is able to promote chromatin compaction131.
Surprising recent observations implicate H1 in active recruitment mechanisms that facilitate binding of a wide range of nuclear factors to chromatin via direct physical interactions (FIG. 5a). As described above, D. melanogaster H1 is required for tethering the H3K9 HMT Su(var)3-9 and the SNF2-like ATPase SUUR to their cognate loci in larval polytene chromosomes84,103. Binding of specific murine H1 subtypes to DNMT1 and DNMT3B helps to mediate their recruitment to ICRs, DNA methylation and gene repression86. In addition, in D. melanogaster, a specific association of H1 with signal transducer and activator of transcription (STAT) provides a molecular reservoir for STAT in chromatin, which promotes heterochromatin integrity and regulates the availability of this transcription factor in JAK–STAT signalling, thus conferring on H1 a tumour-suppressor function132. Along these lines, it was also reported that p53-mediated transcriptional repression and tumour suppression is dependent on interaction of p53 with human H1.2 (REF. 133). Similarly, cooperation between mouse H1.5 and the transcription repressor MSX1 has an essential role in myogenesis134. Mammalian and fly linker histones were also shown to bind HP1 proteins84,135,136 and may contribute to HP1 enrichment in heterochromatin and to chromatin silencing (see above). Importantly, these distinct modes of action (DNA binding versus tethering versus competition with chromatin effectors) are biochemically separable and map to distinct polypeptide segments of the H1 molecule (FIG. 5b). Whereas the globular domain of H1 is required for its deposition into chromatin and for physical interactions with HP1 (REFS 36,137), different regions of the C-terminal domain bind to linker DNA10, promote oligonucleosome condensation138,139 and mediate interactions of H1 with DNMTs, STAT, Su(var)3-9 and SUUR86,103,132,137.
The multiple molecular mechanisms by which H1 regulates genetic activity and other nuclear processes are underscored by the dynamic, locus-specific, activity-and cell cycle-dependent variations of H1 distribution in vivo71,72,103. It is conceivable that these alternative mechanisms are engaged in a context-specific fashion and depend on the local abundance of H1, its PTMs (BOX 1) and/or additional cofactors and interacting proteins. In summary, the biochemical functions of H1 in the regulation of nuclear DNA metabolism should not be limited to a single, one-size-fits-all DNA compaction paradigm. Rather, H1 appears to be an active biochemical player in chromatin and a potent effector of multiple aspects of chromosome structure and chromatin functions.
Conclusions and perspectives
Substantial progress in elucidating the roles of linker histones in chromatin structure and function has been made in the past several years. The recent determinations of the structure of the chromatosome core particle and the 30 nm nucleosome array provide breakthrough insights into two long-standing problems in the chromatin field, revealing that different linker histone variants can show different modes of binding to the nucleosome, which can be associated with distinct higher-order structures of chromatin. This progress should encourage future structural studies of chromatosomes and nucleosome arrays containing other linker histone variants (importantly, without crosslinking fixation of the sample, which may distort the linker histone–nucleosome interaction interface) to bring about a better understanding of how the diversity in the H1 protein family leads to distinct chromatin structures with different functional properfties. Progress at the structural level has been matched by equally exciting insights into the roles of H1 in several fundamental processes necessary for genome maintenance and function. A new paradigm has emerged in which H1 performs these various functions not simply as a chromatin architectural protein but also through intimate collaborations, direct interactions and competition with other protein factors, as well as with the epigenetic marks on DNA and core histones in chromatin. It is very likely that H1 carries out its functions in vivo as a part of multi-protein complexes. Defining the composition of these complexes, the specific H1 variants and H1 PTMs within them, and their locus-specific effects are exciting areas for future research.
Acknowledgments
The authors apologize to those colleagues whose works were not cited due to subjects not covered or limitations in the number of references permitted. The authors’ work is supported by grants from the U.S. National Institutes of Health/National Institute of General Medical Sciences to D.V.F. (GM074233) and A.I.S. (GM093190 and GM116143) and by the intramural research programme of the Center for Cancer Research, National Cancer Institute, National Institutes of Health (B.-R.Z. and Y.B.)
Glossary
Winged helix fold
A compact structural motif in proteins consisting of three α-helices and two or three β-strands and a loop (wing) between the last two β-strands.
Histone chaperones
A group of proteins that bind histones and regulate nucleosome assembly, initially coined to describe the function of nucleoplasmin in the prevention of histone–DNA aggregation during nucleosome assembly.
Citrullination
The conversion of the amino acid arginine in a protein into the amino acid citrulline by deimination.
Facultative heterochromatin
A chromatin region in which genes are silenced through a mechanism such as histone methylation or PIWI-interacting RNA (piRNA).
Nucleosome dyad
The middle point of the DNA that is on the two-fold symmetry axis in the nucleosome core particle structure.
Stochastic optical reconstruction microscopy
(STORM). A form of light microscopy where nearby fluorophores are excited individually, which allows reconstruction of images with resolution beyond the diffraction limit of light.
DNA hypersensitivity sites
(DHSs). Regions of chromatin that are sensitive to cleavage by DNase I enzymes.
DNA adenine methyltransferase identification
(DamID). Molecular protocol used to map binding sites of DNA-associated factors in eukaryotes. The DNA-binding factor is fused with a prokaryotic DNA methyltransferase and expressed in vivo; the fusion protein labels DNA in the vicinity of binding sites with a non-naturally occurring methyl-A.
Histone code
A hypothesis that certain functions of the genome are governed by recognition of combinatorial chemical modifications of histones.
Polytene chromosomes
Oversized chromosomes resulting from chromatin expansion due to polyploidization found in the salivary glands of Drosophila melanogaster.
Transposable elements
Also known as transposons. Segments of DNA that can change their location in the genome.
PIWI proteins
A class of regulatory proteins responsible for maintaining incomplete differentiation in stem cells and maintaining the stability of cell division rates in germline cells.
Heterochromatin protein 1
(HP1). A family of proteins associated with heterochromatin structure formation and maintenance. The HP1 family includes several isoforms (HP1α, HP1β, etc.).
Imprinting
An epigenetic phenomenon in which specific genes are expressed only from DNA inherited from one of the parents, owing to silencing of the chromatin in the other parental genome.
Homeobox gene cluster
A cluster of a large family of similar genes that direct the formation of many body structures during early embryonic development.
Endoreplication
Replication of the nuclear genome in the absence of cell division, which leads to elevated nuclear gene content and polyploidy.
Intercalary heterochromatin
Heterochromatin, other than centromeric heterochromatin, dispersed throughout eukaryotic chromosomes.
Imaginal disc
One of the parts of a holometabolous insect larva that will become a portion of the outside of the adult insect (for example, a wing) during the larval to pupal transition (metamorphosis).
V(D)J recombination
Unique mechanism of somatic DNA recombination that occurs in developing B and T cells to give rise to a diverse repertoire of immunoglobulins.
Cryptic promoters
Genomic sequences in eukaryotes that may be intermittently utilized for transcription initiation; core promoter elements of cryptic promoters are poorly defined (do not strongly interact with general transcription factors) and are obstructed by nucleosome structure and thus silenced in normal cells.
Poly(ADP-ribose) polymerase 1
(PARP1). A nuclear enzyme in eukaryotes that post-translationally modifies proteins by poly ADP-ribosylation.
Retinoic acid
Metabolite of vitamin A (retinol) and a ligand for the retinoic acid receptor transcription factor, an essential intercellular signalling molecule that guides anterior–posterior patterning during early development of chordate animals.
JAK–STAT signalling
Extracellular chemical signalling pathway that regulates the transcription of multiple genes involved in proliferation, differentiation, immunity, and others. The signalling cascade involves Janus kinase (JAK) cell surface receptors and signal transducers and activators of transcription (STATs).
Footnotes
Author contributions
All authors contributed equally to researching data for the article, discussion of content and writing and reviewing of the manuscript before submission.
Competing interests statement
The authors declare no competing interests.
References
- 1.Olins AL, Olins DE. Spheroid chromatin units (v bodies) Science. 1974;183:330–332. doi: 10.1126/science.183.4122.330. [DOI] [PubMed] [Google Scholar]
- 2.Kornberg RD. Chromatin structure: a repeating unit of histones and DNA. Science. 1974;184:868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- 3.Kornberg RD, Thomas JO. Chromatin structure; oligomers of the histones. Science. 1974;184:865–868. doi: 10.1126/science.184.4139.865. [DOI] [PubMed] [Google Scholar]
- 4.Arents G, Moudrianakis EN. Topography of the histone octamer surface: repeating structural motifs utilized in the docking of nucleosomal DNA. Proc Natl Acad Sci USA. 1993;90:10489–10493. doi: 10.1073/pnas.90.22.10489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 6.Noll M, Kornberg RD. Action of micrococcal nuclease on chromatin and the location of histone H1. J Mol Biol. 1977;109:393–404. doi: 10.1016/s0022-2836(77)80019-3. [DOI] [PubMed] [Google Scholar]
- 7.Simpson RT. Structure of the chromatosome, a chromatin particle containing 160 base pairs of DNA and all the histones. Biochemistry. 1978;17:5524–5531. doi: 10.1021/bi00618a030. This study defines the chromatosome core particle for the first time. [DOI] [PubMed] [Google Scholar]
- 8.Thoma F, Koller T, Klug A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J Cell Biol. 1979;83:403–427. doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allan J, Hartman PG, Crane-Robinson C, Aviles FX. The structure of histone H1 and its location in chromatin. Nature. 1980;288:675–679. doi: 10.1038/288675a0. [DOI] [PubMed] [Google Scholar]
- 10.Allan J, Mitchell T, Harborne N, Bohm L, Crane-Robinson C. Roles of H1 domains in determining higher order chromatin structure and H1 location. J Mol Biol. 1986;187:591–601. doi: 10.1016/0022-2836(86)90337-2. [DOI] [PubMed] [Google Scholar]
- 11.Ramakrishnan V, Finch JT, Graziano V, Lee PL, Sweet RM. Crystal structure of globular domain of histone H5 and its implications for nucleosome binding. Nature. 1993;362:219–223. doi: 10.1038/362219a0. [DOI] [PubMed] [Google Scholar]
- 12.Singer DS, Singer MF. Studies on the interaction of H1 histone with superhelical DNA: characterization of the recognition and binding regions of H1 histones. Nucleic Acids Res. 1976;3:2531–2547. doi: 10.1093/nar/3.10.2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lorch Y, LaPointe JW, Kornberg RD. Nucleosomes inhibit the initiation of transcription but allow chain elongation with the displacement of histones. Cell. 1987;49:203–210. doi: 10.1016/0092-8674(87)90561-7. [DOI] [PubMed] [Google Scholar]
- 14.Nagai S, Davis RE, Mattei PJ, Eagen KP, Kornberg RD. Chromatin potentiates transcription. Proc Natl Acad Sci USA. 2017;114:1536–1541. doi: 10.1073/pnas.1620312114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Izzo A, Schneider R. The role of linker histone H1 modifications in the regulation of gene expression and chromatin dynamics. Biochim Biophys Acta. 2016;1859:486–495. doi: 10.1016/j.bbagrm.2015.09.003. [DOI] [PubMed] [Google Scholar]
- 16.Allis CD, Jenuwein T. The molecular hallmarks of epigenetic control. Nat Rev Genet. 2016;17:487–500. doi: 10.1038/nrg.2016.59. [DOI] [PubMed] [Google Scholar]
- 17.Eberharter A, Becker PB. Histone acetylation: a switch between repressive and permissive chromatin. Second in review series on chromatin dynamics. EMBO Rep. 2002;3:224–229. doi: 10.1093/embo-reports/kvf053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Torres IO, Fujimori DG. Functional coupling between writers, erasers and readers of histone and DNA methylation. Curr Opin Struct Biol. 2015;35:68–75. doi: 10.1016/j.sbi.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hammond CM, Stromme CB, Huang H, Patel DJ, Groth A. Histone chaperone networks shaping chromatin function. Nat Rev Mol Cell Biol. 2017;18:141–158. doi: 10.1038/nrm.2016.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shintomi K, et al. Nucleosome assembly protein-1 is a linker histone chaperone in Xenopus eggs. Proc Natl Acad Sci USA. 2005;102:8210–8215. doi: 10.1073/pnas.0500822102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richardson RT, et al. Nuclear autoantigenic sperm protein (NASP), a linker histone chaperone that is required for cell proliferation. J Biol Chem. 2006;281:21526–21534. doi: 10.1074/jbc.M603816200. [DOI] [PubMed] [Google Scholar]
- 22.Fan Y, et al. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell. 2005;123:1199–1212. doi: 10.1016/j.cell.2005.10.028. This study shows for the first time that H1 participates in epigenetic regulation in vivo by affecting DNA methylation at specific loci. [DOI] [PubMed] [Google Scholar]
- 23.Hergeth SP, Schneider R. The H1 linker histones: multifunctional proteins beyond the nucleosomal core particle. EMBO Rep. 2015;16:1439–1453. doi: 10.15252/embr.201540749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bates DL, Thomas JO. Histones H1 and H5: one or two molecules per nucleosome? Nucleic Acids Res. 1981;9:5883–5894. doi: 10.1093/nar/9.22.5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perez-Montero S, Carbonell A, Moran T, Vaquero A, Azorin F. The embryonic linker histone H1 variant of Drosophila, dBigH1, regulates zygotic genome activation. Dev Cell. 2013;26:578–590. doi: 10.1016/j.devcel.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 26.Kowalski A, Palyga J. Modulation of chromatin function through linker histone H1 variants. Biol Cell. 2016;108:339–356. doi: 10.1111/boc.201600007. [DOI] [PubMed] [Google Scholar]
- 27.Christophorou MA, et al. Citrullination regulates pluripotency and histone H1 binding to chromatin. Nature. 2014;507:104–108. doi: 10.1038/nature12942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Torres CM, et al. The linker histone H1.0 generates epigenetic and functional intratumor heterogeneity. Science. 2016;353:aaf1644. doi: 10.1126/science.aaf1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richmond TJ, Davey CA. The structure of DNA in the nucleosome core. Nature. 2003;423:145–150. doi: 10.1038/nature01595. [DOI] [PubMed] [Google Scholar]
- 30.Geeven G, et al. Local compartment changes and regulatory landscape alterations in histone H1-depleted cells. Genome Biol. 2015;16:289. doi: 10.1186/s13059-015-0857-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thoma F, Koller T. Influence of histone H1 on chromatin structure. Cell. 1977;12:101–107. doi: 10.1016/0092-8674(77)90188-x. [DOI] [PubMed] [Google Scholar]
- 32.Pruss D, et al. An asymmetric model for the nucleosome: a binding site for linker histones inside the DNA gyres. Science. 1996;274:614–617. doi: 10.1126/science.274.5287.614. [DOI] [PubMed] [Google Scholar]
- 33.Zhou YB, Gerchman SE, Ramakrishnan V, Travers A, Muyldermans S. Position and orientation of the globular domain of linker histone H5 on the nucleosome. Nature. 1998;395:402–405. doi: 10.1038/26521. [DOI] [PubMed] [Google Scholar]
- 34.An W, Leuba SH, van Holde K, Zlatanova J. Linker histone protects linker DNA on only one side of the core particle and in a sequence-dependent manner. Proc Natl Acad Sci USA. 1998;95:3396–3401. doi: 10.1073/pnas.95.7.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fan L, Roberts VA. Complex of linker histone H5 with the nucleosome and its implications for chromatin packing. Proc Natl Acad Sci USA. 2006;103:8384–8389. doi: 10.1073/pnas.0508951103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brown DT, Izard T, Misteli T. Mapping the interaction surface of linker histone H1(0) with the nucleosome of native chromatin in vivo. Nat Struct Mol Biol. 2006;13:250–255. doi: 10.1038/nsmb1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cui F, Zhurkin VB. Distinctive sequence patterns in metazoan and yeast nucleosomes: implications for linker histone binding to AT-rich and methylated DNA. Nucleic Acids Res. 2009;37:2818–2829. doi: 10.1093/nar/gkp113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Syed SH, et al. Single-base resolution mapping of H1-nucleosome interactions and 3D organization of the nucleosome. Proc Natl Acad Sci USA. 2010;107:9620–9625. doi: 10.1073/pnas.1000309107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pachov GV, Gabdoulline RR, Wade RC. On the structure and dynamics of the complex of the nucleosome and the linker histone. Nucleic Acids Res. 2011;39:5255–5263. doi: 10.1093/nar/gkr101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou BR, et al. Structural mechanisms of nucleosome recognition by linker histones. Mol Cell. 2015;59:628–638. doi: 10.1016/j.molcel.2015.06.025. This paper presents the first near-atomic resolution crystal structure of the nucleosome bound to the globular domain of chicken H5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goytisolo FA, et al. Identification of two DNA-binding sites on the globular domain of histone H5. EMBO J. 1996;15:3421–3429. [PMC free article] [PubMed] [Google Scholar]
- 42.Duggan MM, Thomas JO. Two DNA-binding sites on the globular domain of histone H5 are required for binding to both bulk and 5 S reconstituted nucleosomes. J Mol Biol. 2000;304:21–33. doi: 10.1006/jmbi.2000.4205. [DOI] [PubMed] [Google Scholar]
- 43.Zhou BR, et al. A small number of residues can determine if linker histones are bound on or off dyad in the chromatosome. J Mol Biol. 2016;428:3948–3959. doi: 10.1016/j.jmb.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bednar J, et al. Structure and dynamics of a 197 bp nucleosome in complex with linker histone H1. Mol Cell. 2017;66:384–397.e8. doi: 10.1016/j.molcel.2017.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.White AE, Hieb AR, Luger K. A quantitative investigation of linker histone interactions with nucleosomes and chromatin. Sci Rep. 2016;6:19122. doi: 10.1038/srep19122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou BR, et al. Structural insights into the histone H1-nucleosome complex. Proc Natl Acad Sci USA. 2013;110:19390–19395. doi: 10.1073/pnas.1314905110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Song F, et al. Cryo-EM study of the chromatin fiber reveals a double helix twisted by tetranucleosomal units. Science. 2014;344:376–380. doi: 10.1126/science.1251413. This paper presents the first cryo-EM structure of the 30 nm nucleosome array containing human linker histone variant H1.4, in which the linker histone is visible and binds off the nucleosome dyad. [DOI] [PubMed] [Google Scholar]
- 48.Finch JT, Klug A. Solenoidal model for superstructure in chromatin. Proc Natl Acad Sci USA. 1976;73:1897–1901. doi: 10.1073/pnas.73.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McGhee JD, Nickol JM, Felsenfeld G, Rau DC. Higher order structure of chromatin: orientation of nucleosomes within the 30 nm chromatin solenoid is independent of species and spacer length. Cell. 1983;33:831–841. doi: 10.1016/0092-8674(83)90025-9. [DOI] [PubMed] [Google Scholar]
- 50.Widom J, Klug A. Structure of the 300Å chromatin filament: x-ray diffraction from oriented samples. Cell. 1985;43:207–213. doi: 10.1016/0092-8674(85)90025-x. [DOI] [PubMed] [Google Scholar]
- 51.Ghirlando R, Felsenfeld G. Hydrodynamic studies on defined heterochromatin fragments support a 30 nm fiber having six nucleosomes per turn. J Mol Biol. 2008;376:1417–1425. doi: 10.1016/j.jmb.2007.12.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Worcel A, Strogatz S, Riley D. Structure of chromatin and the linking number of DNA. Proc Natl Acad Sci USA. 1981;78:1461–1465. doi: 10.1073/pnas.78.3.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Williams SP, et al. Chromatin fibers are left-handed double helices with diameter and mass per unit length that depend on linker length. Biophys J. 1986;49:233–248. doi: 10.1016/S0006-3495(86)83637-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Woodcock CL, Frado LL, Rattner JB. The higher-order structure of chromatin: evidence for a helical ribbon arrangement. J Cell Biol. 1984;99:42–52. doi: 10.1083/jcb.99.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dorigo B, et al. Nucleosome arrays reveal the two-start organization of the chromatin fiber. Science. 2004;306:1571–1573. doi: 10.1126/science.1103124. [DOI] [PubMed] [Google Scholar]
- 56.Schalch T, Duda S, Sargent DF, Richmond TJ. X-ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature. 2005;436:138–141. doi: 10.1038/nature03686. [DOI] [PubMed] [Google Scholar]
- 57.Routh A, Sandin S, Rhodes D. Nucleosome repeat length and linker histone stoichiometry determine chromatin fiber structure. Proc Natl Acad Sci USA. 2008;105:8872–8877. doi: 10.1073/pnas.0802336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Robinson PJ, Rhodes D. Structure of the ‘30 nm’ chromatin fibre: a key role for the linker histone. Curr Opin Struct Biol. 2006;16:336–343. doi: 10.1016/j.sbi.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 59.Maeshima K, Imai R, Tamura S, Nozaki T. Chromatin as dynamic 10-nm fibers. Chromosoma. 2014;123:225–237. doi: 10.1007/s00412-014-0460-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Scheffer MP, Eltsov M, Frangakis AS. Evidence for short-range helical order in the 30-nm chromatin fibers of erythrocyte nuclei. Proc Natl Acad Sci USA. 2011;108:16992–16997. doi: 10.1073/pnas.1108268108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Robinson PJ, Fairall L, Huynh VA, Rhodes D. EM measurements define the dimensions of the “30-nm” chromatin fiber: evidence for a compact, interdigitated structure. Proc Natl Acad Sci USA. 2006;103:6506–6511. doi: 10.1073/pnas.0601212103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bednar J, et al. Nucleosomes, linker DNA, and linker histone form a unique structural motif that directs the higher-order folding and compaction of chromatin. Proc Natl Acad Sci USA. 1998;95:14173–14178. doi: 10.1073/pnas.95.24.14173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ricci MA, Manzo C, Garcia-Parajo MF, Lakadamyali M, Cosma MP. Chromatin fibers are formed by heterogeneous groups of nucleosomes in vivo. Cell. 2015;160:1145–1158. doi: 10.1016/j.cell.2015.01.054. [DOI] [PubMed] [Google Scholar]
- 64.Grigoryev SA, et al. Hierarchical looping of zigzag nucleosome chains in metaphase chromosomes. Proc Natl Acad Sci USA. 2016;113:1238–1243. doi: 10.1073/pnas.1518280113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li W, et al. FACT remodels the tetranucleosomal unit of chromatin fibers for gene transcription. Mol Cell. 2016;64:120–133. doi: 10.1016/j.molcel.2016.08.024. [DOI] [PubMed] [Google Scholar]
- 66.Nora EP, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dixon JR, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hou C, Li L, Qin ZS, Corces VG. Gene density, transcription, and insulators contribute to the partition of the Drosophila genome into physical domains. Mol Cell. 2012;48:471–484. doi: 10.1016/j.molcel.2012.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sexton T, et al. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012;148:458–472. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 70.Wolffe AP. Histone H1. Int J Biochem Cell Biol. 1997;29:1463–1466. doi: 10.1016/s1357-2725(97)00026-5. [DOI] [PubMed] [Google Scholar]
- 71.Braunschweig U, Hogan GJ, Pagie L, van Steensel B. Histone H1 binding is inhibited by histone variant H3.3. EMBO J. 2009;28:3635–3645. doi: 10.1038/emboj.2009.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Izzo A, et al. The genomic landscape of the somatic linker histone subtypes H1.1 to H1.5 in human cells. Cell Rep. 2013;3:2142–2154. doi: 10.1016/j.celrep.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 73.Li JY, Patterson M, Mikkola HK, Lowry WE, Kurdistani SK. Dynamic distribution of linker histone H1.5 in cellular differentiation. PLoS Genet. 2012;8:e1002879. doi: 10.1371/journal.pgen.1002879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cao K, et al. High-resolution mapping of h1 linker histone variants in embryonic stem cells. PLoS Genet. 2013;9:e1003417. doi: 10.1371/journal.pgen.1003417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Millan-Arino L, et al. Mapping of six somatic linker histone H1 variants in human breast cancer cells uncovers specific features of H1.2. Nucleic Acids Res. 2014;42:4474–4493. doi: 10.1093/nar/gku079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nalabothula N, et al. The chromatin architectural proteins HMGD1 and H1 bind reciprocally and have opposite effects on chromatin structure and gene regulation. BMC Genomics. 2014;15:92. doi: 10.1186/1471-2164-15-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mayor R, et al. Genome distribution of replication-independent histone H1 variants shows H1.0 associated with nucleolar domains and H1X associated with RNA polymerase II-enriched regions. J Biol Chem. 2015;290:7474–7491. doi: 10.1074/jbc.M114.617324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Reczek PR, Weissman D, Huvos PE, Fasman GD. Sodium butyrate induced structural changes in HeLa cell chromatin. Biochemistry. 1982;21:993–1002. doi: 10.1021/bi00534a026. [DOI] [PubMed] [Google Scholar]
- 79.Schroter H, Gomez-Lira MM, Plank KH, Bode J. The extent of histone acetylation induced by butyrate and the turnover of acetyl groups depend on the nature of the cell line. Eur J Biochem. 1981;120:21–28. doi: 10.1111/j.1432-1033.1981.tb05664.x. [DOI] [PubMed] [Google Scholar]
- 80.Herrera JE, West KL, Schiltz RL, Nakatani Y, Bustin M. Histone H1 is a specific repressor of core histone acetylation in chromatin. Mol Cell Biol. 2000;20:523–529. doi: 10.1128/mcb.20.2.523-529.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sun J, et al. Histone H1-mediated epigenetic regulation controls germline stem cell self-renewal by modulating H4K16 acetylation. Nat Commun. 2015;6:8856. doi: 10.1038/ncomms9856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vaquero A, et al. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol Cell. 2004;16:93–105. doi: 10.1016/j.molcel.2004.08.031. [DOI] [PubMed] [Google Scholar]
- 83.Lu X, et al. Linker histone H1 is essential for Drosophila development, the establishment of pericentric heterochromatin, and a normal polytene chromosome structure. Genes Dev. 2009;23:452–465. doi: 10.1101/gad.1749309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lu X, et al. Drosophila H1 regulates the genetic activity of heterochromatin by recruitment of Su(var)3-9. Science. 2013;340:78–81. doi: 10.1126/science.1234654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Iwasaki YW, et al. Piwi modulates chromatin accessibility by regulating multiple factors including histone H1 to repress transposons. Mol Cell. 2016;63:408–419. doi: 10.1016/j.molcel.2016.06.008. References 84 and 85 elucidate two different mechanisms by which H1 is involved in repression of genetic activity in heterochromatin. [DOI] [PubMed] [Google Scholar]
- 86.Yang SM, Kim BJ, Norwood Toro L, Skoultchi AI. H1 linker histone promotes epigenetic silencing by regulating both DNA methylation and histone H3 methylation. Proc Natl Acad Sci USA. 2013;110:1708–1713. doi: 10.1073/pnas.1213266110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Stutzer A, et al. Modulations of DNA contacts by linker histones and post-translational modifications determine the mobility and modifiability of nucleosomal H3 tails. Mol Cell. 2016;61:247–259. doi: 10.1016/j.molcel.2015.12.015. [DOI] [PubMed] [Google Scholar]
- 88.Martin C, Cao R, Zhang Y. Substrate preferences of the EZH2 histone methyltransferase complex. J Biol Chem. 2006;281:8365–8370. doi: 10.1074/jbc.M513425200. [DOI] [PubMed] [Google Scholar]
- 89.Yuan W, et al. Dense chromatin activates Polycomb repressive complex 2 to regulate H3 lysine 27 methylation. Science. 2012;337:971–975. doi: 10.1126/science.1225237. [DOI] [PubMed] [Google Scholar]
- 90.Kim JM, et al. Linker histone H1.2 establishes chromatin compaction and gene silencing through recognition of H3K27me3. Scientif Rep. 2015;5:16714. doi: 10.1038/srep16714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kim K, et al. Linker histone H1.2 cooperates with Cul4A and PAF1 to drive H4K31 ubiquitylation-mediated transactivation. Cell Rep. 2013;5:1690–1703. doi: 10.1016/j.celrep.2013.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Du J, Johnson LM, Jacobsen SE, Patel DJ. DNA methylation pathways and their crosstalk with histone methylation. Nat Rev Mol Cell Biol. 2015;16:519–532. doi: 10.1038/nrm4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wierzbicki AT, Jerzmanowski A. Suppression of histone H1 genes in Arabidopsis results in heritable developmental defects and stochastic changes in DNA methylation. Genetics. 2005;169:997–1008. doi: 10.1534/genetics.104.031997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Maclean JA, et al. The rhox homeobox gene cluster is imprinted and selectively targeted for regulation by histone H1 and DNA methylation. Mol Cell Biol. 2011;31:1275–1287. doi: 10.1128/MCB.00734-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang Y, et al. Histone H1 depletion impairs embryonic stem cell differentiation. PLoS Genet. 2012;8:e1002691. doi: 10.1371/journal.pgen.1002691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Li H, et al. Mutations in linker histone genes HIST1H1 B, C, D, and E; OCT2 (POU2F2); IRF8; and ARID1A underlying the pathogenesis of follicular lymphoma. Blood. 2014;123:1487–1498. doi: 10.1182/blood-2013-05-500264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Flickinger RA. Possible role of H1 histone in replication timing. Dev Growth Differ. 2015;57:1–9. doi: 10.1111/dgd.12190. [DOI] [PubMed] [Google Scholar]
- 98.Halmer L, Gruss C. Influence of histone H1 on the in vitro replication of DNA and chromatin. Nucleic Acids Res. 1995;23:773–778. doi: 10.1093/nar/23.5.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Halmer L, Gruss C. Effects of cell cycle dependent histone H1 phosphorylation on chromatin structure and chromatin replication. Nucleic Acids Res. 1996;24:1420–1427. doi: 10.1093/nar/24.8.1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lu ZH, Sittman DB, Brown DT, Munshi R, Leno GH. Histone H1 modulates DNA replication through multiple pathways in Xenopus egg extract. J Cell Sci. 1997;110:2745–2758. doi: 10.1242/jcs.110.21.2745. [DOI] [PubMed] [Google Scholar]
- 101.Lu ZH, Sittman DB, Romanowski P, Leno GH. Histone H1 reduces the frequency of initiation in Xenopus egg extract by limiting the assembly of prereplication complexes on sperm chromatin. Mol Biol Cell. 1998;9:1163–1176. doi: 10.1091/mbc.9.5.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.De S, et al. Histone H1 variants differentially inhibit DNA replication through an affinity for chromatin mediated by their carboxyl-terminal domains. Gene. 2002;292:173–181. doi: 10.1016/s0378-1119(02)00675-3. [DOI] [PubMed] [Google Scholar]
- 103.Andreyeva EN, et al. Regulatory functions and chromatin loading dynamics of linker histone H1 during endoreplication in Drosophila. Genes Dev. 2017;31:603–616. doi: 10.1101/gad.295717.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nordman JT, et al. DNA copy-number control through inhibition of replication fork progression. Cell Rep. 2014;9:841–849. doi: 10.1016/j.celrep.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sher N, et al. Developmental control of gene copy number by repression of replication initiation and fork progression. Genome Res. 2012;22:64–75. doi: 10.1101/gr.126003.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dimitrova DS, Gilbert DM. The spatial position and replication timing of chromosomal domains are both established in early G1 phase. Mol Cell. 1999;4:983–993. doi: 10.1016/s1097-2765(00)80227-0. [DOI] [PubMed] [Google Scholar]
- 107.Thiriet C, Hayes JJ. Linker histone phosphorylation regulates global timing of replication origin firing. J Biol Chem. 2009;284:2823–2829. doi: 10.1074/jbc.M805617200. References 103 and 107 implicate H1 in the regulation of replication timing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Dou Y, Mizzen CA, Abrams M, Allis CD, Gorovsky MA. Phosphorylation of linker histone H1 regulates gene expression in vivo by mimicking H1 removal. Mol Cell. 1999;4:641–647. doi: 10.1016/s1097-2765(00)80215-4. [DOI] [PubMed] [Google Scholar]
- 109.Alexandrow MG, Hamlin JL. Chromatin decondensation in S-phase involves recruitment of Cdk2 by Cdc45 and histone H1 phosphorylation. J Cell Biol. 2005;168:875–886. doi: 10.1083/jcb.200409055. References 107, 108 and 109 show the importance of phosphorylation of H1 for its association with chromatin and its effects on gene expression and DNA replication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lever MA, Th’ng JP, Sun X, Hendzel MJ. Rapid exchange of histone H1.1 on chromatin in living human cells. Nature. 2000;408:873–876. doi: 10.1038/35048603. [DOI] [PubMed] [Google Scholar]
- 111.Downs JA, Kosmidou E, Morgan A, Jackson SP. Suppression of homologous recombination by the Saccharomyces cerevisiae linker histone. Mol Cell. 2003;11:1685–1692. doi: 10.1016/s1097-2765(03)00197-7. [DOI] [PubMed] [Google Scholar]
- 112.Li C, Mueller JE, Elfline M, Bryk M. Linker histone H1 represses recombination at the ribosomal DNA locus in the budding yeast Saccharomyces cerevisiae. Mol Microbiol. 2008;67:906–919. doi: 10.1111/j.1365-2958.2007.06101.x. [DOI] [PubMed] [Google Scholar]
- 113.Vujatovic O, et al. Drosophila melanogaster linker histone dH1 is required for transposon silencing and to preserve genome integrity. Nucleic Acids Res. 2012;40:5402–5414. doi: 10.1093/nar/gks224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hashimoto H, et al. Histone H1 variant, H1R is involved in DNA damage response. DNA Repair (Amst ) 2007;6:1584–1595. doi: 10.1016/j.dnarep.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 115.Murga M, et al. Global chromatin compaction limits the strength of the DNA damage response. J Cell Biol. 2007;178:1101–1108. doi: 10.1083/jcb.200704140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Thorslund T, et al. Histone H1 couples initiation and amplification of ubiquitin signalling after DNA damage. Nature. 2015;527:389–393. doi: 10.1038/nature15401. [DOI] [PubMed] [Google Scholar]
- 117.Kalashnikova AA, et al. Linker histone H1.0 interacts with an extensive network of proteins found in the nucleolus. Nucleic Acids Res. 2013;41:4026–4035. doi: 10.1093/nar/gkt104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hamilton C, Hayward RL, Gilbert N. Global chromatin fibre compaction in response to DNA damage. Biochem Biophys Res Commun. 2011;414:820–825. doi: 10.1016/j.bbrc.2011.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sera T, Wolffe AP. Role of histone H1 as an architectural determinant of chromatin structure and as a specific repressor of transcription on Xenopus oocyte 5S rRNA genes. Mol Cell Biol. 1998;18:3668–3680. doi: 10.1128/mcb.18.7.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Laybourn PJ, Kadonaga JT. Role of nucleosomal cores and histone H1 in regulation of transcription by RNA polymerase II. Science. 1991;254:238–245. doi: 10.1126/science.254.5029.238. [DOI] [PubMed] [Google Scholar]
- 121.Maier VK, Chioda M, Rhodes D, Becker PB. ACF catalyses chromatosome movements in chromatin fibres. EMBO J. 2008;27:817–826. doi: 10.1038/sj.emboj.7601902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kavi H, et al. A genetic screen and transcript profiling reveal a shared regulatory program for Drosophila linker histone H1 and chromatin remodeler CHD1. G3 (Bethesda) 2015;5:677–687. doi: 10.1534/g3.115.016709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Leuba SH, Zlatanova J, van Holde K. On the location of linker DNA in the chromatin fiber. Studies with immobilized and soluble micrococcal nuclease. J Mol Biol. 1994;235:871–880. doi: 10.1006/jmbi.1994.1045. [DOI] [PubMed] [Google Scholar]
- 124.Tremethick DJ. Higher-order structures of chromatin: the elusive 30 nm fiber. Cell. 2007;128:651–654. doi: 10.1016/j.cell.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 125.Postnikov YV, Bustin M. Functional interplay between histone H1 and HMG proteins in chromatin. Biochim Biophys Acta. 2016;1859:462–467. doi: 10.1016/j.bbagrm.2015.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Krishnakumar R, et al. Reciprocal binding of PARP-1 and histone H1 at promoters specifies transcriptional outcomes. Science. 2008;319:819–821. doi: 10.1126/science.1149250. Reference 126 demonstrates a genome-wide, reciprocal binding pattern of H1 and PARP1 and their opposite effects on gene activity, and reference 125 reviews similar competition between H1 and HMG proteins. [DOI] [PubMed] [Google Scholar]
- 127.Magnani L, Eeckhoute J, Lupien M. Pioneer factors: directing transcriptional regulators within the chromatin environment. Trends Genet. 2011;27:465–474. doi: 10.1016/j.tig.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 128.Cirillo LA, et al. Binding of the winged-helix transcription factor HNF3 to a linker histone site on the nucleosome. EMBO J. 1998;17:244–254. doi: 10.1093/emboj/17.1.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Cirillo LA, et al. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell. 2002;9:279–289. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- 130.Taube JH, Allton K, Duncan SA, Shen L, Barton MC. Foxa1 functions as a pioneer transcription factor at transposable elements to activate Afp during differentiation of embryonic stem cells. J Biol Chem. 2010;285:16135–16144. doi: 10.1074/jbc.M109.088096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ghosh RP, Horowitz-Scherer RA, Nikitina T, Shlyakhtenko LS, Woodcock CL. MeCP2 binds cooperatively to its substrate and competes with histone H1 for chromatin binding sites. Mol Cell Biol. 2010;30:4656–4670. doi: 10.1128/MCB.00379-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Xu N, Emelyanov AV, Fyodorov DV, Skoultchi AI. Drosophila linker histone H1 coordinates STAT-dependent organization of heterochromatin and suppresses tumorigenesis caused by hyperactive JAK-STAT signaling. Epigenetics Chromatin. 2014;7:16. doi: 10.1186/1756-8935-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kim K, et al. Isolation and characterization of a novel H1.2 complex that acts as a repressor of p53-mediated transcription. J Biol Chem. 2008;283:9113–9126. doi: 10.1074/jbc.M708205200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lee H, Habas R, Abate-Shen C. MSX1 cooperates with histone H1b for inhibition of transcription and myogenesis. Science. 2004;304:1675–1678. doi: 10.1126/science.1098096. [DOI] [PubMed] [Google Scholar]
- 135.Daujat S, Zeissler U, Waldmann T, Happel N, Schneider R. HP1 binds specifically to Lys26-methylated histone H1.4, whereas simultaneous Ser27 phosphorylation blocks HP1 binding. J Biol Chem. 2005;280:38090–38095. doi: 10.1074/jbc.C500229200. [DOI] [PubMed] [Google Scholar]
- 136.Nielsen AL, et al. Heterochromatin formation in mammalian cells: interaction between histones and HP1 proteins. Mol Cell. 2001;7:729–739. doi: 10.1016/s1097-2765(01)00218-0. [DOI] [PubMed] [Google Scholar]
- 137.Kavi H, Emelyanov AV, Fyodorov DV, Skoultchi AI. Independent biological and biochemical functions for individual structural domains of Drosophila linker histone H1. J Biol Chem. 2016;291:15143–15155. doi: 10.1074/jbc.M116.730705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lu X, Hamkalo B, Parseghian MH, Hansen JC. Chromatin condensing functions of the linker histone C-terminal domain are mediated by specific amino acid composition and intrinsic protein disorder. Biochemistry. 2009;48:164–172. doi: 10.1021/bi801636y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.McBryant SJ, Lu X, Hansen JC. Multifunctionality of the linker histones: an emerging role for protein-protein interactions. Cell Res. 2010;20:519–528. doi: 10.1038/cr.2010.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Starkova TY, et al. Post-translational modifications of linker histone H1 variants in mammals. Phys Biol. 2017;14:016005. doi: 10.1088/1478-3975/aa551a. [DOI] [PubMed] [Google Scholar]
- 141.Talasz H, Helliger W, Puschendorf B, Lindner H. In vivo phosphorylation of histone H1 variants during the cell cycle. Biochemistry. 1996;35:1761–1767. doi: 10.1021/bi951914e. [DOI] [PubMed] [Google Scholar]
- 142.Chu CS, et al. Protein kinase A-mediated serine 35 phosphorylation dissociates histone H1.4 from mitotic chromosome. J Biol Chem. 2011;286:35843–35851. doi: 10.1074/jbc.M111.228064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kim K, et al. Functional interplay between p53 acetylation and H1.2 phosphorylation in p53-regulated transcription. Oncogene. 2012;31:4290–4301. doi: 10.1038/onc.2011.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Talasz H, Sarg B, Lindner HH. Site-specifically phosphorylated forms of H1.5 and H1.2 localized at distinct regions of the nucleus are related to different processes during the cell cycle. Chromosoma. 2009;118:693–709. doi: 10.1007/s00412-009-0228-2. [DOI] [PubMed] [Google Scholar]
- 145.Zheng Y, et al. Histone H1 phosphorylation is associated with transcription by RNA polymerases I and II. J Cell Biol. 2010;189:407–415. doi: 10.1083/jcb.201001148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Bonet-Costa C, et al. Combined bottom-up and top-down mass spectrometry analyses of the pattern of post-translational modifications of Drosophila melanogaster linker histone H1. J Proteomics. 2012;75:4124–4138. doi: 10.1016/j.jprot.2012.05.034. [DOI] [PubMed] [Google Scholar]
- 147.Kamieniarz K, et al. A dual role of linker histone H1.4 Lys 34 acetylation in transcriptional activation. Genes Dev. 2012;26:797–802. doi: 10.1101/gad.182014.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Lesner A, et al. Monoubiquitinated histone H1B is required for antiviral protection in CD4+T cells resistant to HIV-1. Biochemistry. 2004;43:16203–16211. doi: 10.1021/bi0492758. [DOI] [PubMed] [Google Scholar]
- 149.Talbert PB, et al. A unified phylogeny-based nomenclature for histone variants. Epigenetics Chromatin. 2012;5:7. doi: 10.1186/1756-8935-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Izzo A, Schneider R. H1 gets the genome in shape. Genome Biol. 2016;17:8. doi: 10.1186/s13059-016-0872-9. [DOI] [PMC free article] [PubMed] [Google Scholar]