Understanding human fetal pancreas development using subpopulation sorting, RNA sequencing and single-cell profiling (original) (raw)
ABSTRACT
To decipher the populations of cells present in the human fetal pancreas and their lineage relationships, we developed strategies to isolate pancreatic progenitors, endocrine progenitors and endocrine cells. Transcriptome analysis of the individual populations revealed a large degree of conservation among vertebrates in the drivers of gene expression changes that occur at different steps of differentiation, although notably, sometimes, different members of the same gene family are expressed. The transcriptome analysis establishes a resource to identify novel genes and pathways involved in human pancreas development. Single-cell profiling further captured intermediate stages of differentiation and enabled us to decipher the sequence of transcriptional events occurring during human endocrine differentiation. Furthermore, we evaluate how well individual pancreatic cells derived in vitro from human pluripotent stem cells mirror the natural process occurring in human fetuses. This comparison uncovers a few differences at the progenitor steps, a convergence at the steps of endocrine induction, and the current inability to fully resolve endocrine cell subtypes in vitro.
KEY WORDS: Diabetes, Endocrine, Human, Islets, Pancreas, Stem cells
Summary: This study develops sorting strategies to isolate specific pancreatic populations and use single-cell profiling to evaluate whether pancreatic cells derived from hPSCs mirror the process occurring in human fetuses.
INTRODUCTION
Because of the limited availability of primary human tissue, much of our knowledge of human organ development is extrapolated from animal models. It is remarkable that the mechanisms of organ formation are conserved enough between vertebrate species so as to enable biologists to control the differentiation of organ-specific cells from human pluripotent stem cells. In the pancreas, directed differentiation informed by the knowledge of development has enabled the field to progress to the production of beta cells, with the ambition of transplanting these cells to treat diabetic patients who, depending on subtype, have reduced β-cell function, or are without them entirely (D'Amour et al., 2006). Over the years, the protocols have become more efficient, adaptable to multiple pluripotent stem cell lines and lead to the production of ever-more representative functional cells (Pagliuca et al., 2014; Rezania et al., 2014; Russ et al., 2015; Nostro et al., 2015; Ameri et al., 2017; Cogger et al., 2017). Yet, _in vitro_-generated cells do not fully recapitulate the function of bona fide β cells, notably lacking a tight control of insulin secretion upon glucose stimulation (Johnson, 2016). Accordingly, it is very important at this point to compare the cells we produce in vitro with endogenous cell types. Moreover, comparing progenitors with intermediates in the differentiation process may help to pinpoint where the processes diverge, and how we can improve them. Some divergences may originate from previously underappreciated differences between human pancreas development and those model organ vertebrates such as mouse, which are much easier to study.
The pancreas is both a digestive and an endocrine organ. The digestive function is ensured by the acinar cells that secrete digestive enzymes into the pancreatic ducts. The ductal cells also participate in the process, notably by neutralizing stomach acidity. Pancreatic endocrine cells are clustered into islets of Langerhans that are composed of five different types of endocrine cells, α, β, δ, ε and PP, secreting glucagon, insulin, somatostatin, ghrelin and pancreatic polypeptide, respectively.
Pancreas development begins with the invagination of the foregut into dorsal and ventral buds at embryonic day (E) 8 in the mouse and at ∼4 weeks of development (WD) in humans (Jennings et al., 2013; Larsen and Grapin-Botton, 2017). In both species, pancreatic buds contain multipotent progenitors that are characterized by the expression of several transcription factors, such as PDX1, NKX6-1 and SOX9 (Jonsson et al., 1994; Stoffers et al., 1997; Piper et al., 2004; Seymour et al., 2007; Jennings et al., 2013; Cebola et al., 2015). They proliferate and differentiate into all pancreatic lineages (acinar, ductal and endocrine). In the mouse, proliferation is dependent on signals from the mesenchyme and also from cell to cell interactions, notably via the NOTCH pathway, which activates the transcription factor HES1 (Bhushan et al., 2001; Pan and Wright, 2011; Jensen et al., 2000). The function of the NOTCH pathway appears to be conserved in humans (Jeon et al., 2009; Zhu et al., 2016; Jennings et al., 2017).
In mice, endocrine differentiation occurs from multipotent or bipotent endocrine-ductal progenitors and is marked by the expression of the transcriptional factor NEUROG3 (Solar et al., 2009). Many of these mechanisms appear to be conserved in humans, though we know little about the existence of multipotent versus bipotent progenitors (Zhu et al., 2016). Pancreatic endocrine cell differentiation starts at E9 in the mouse and at 8 WD in humans, with the expression of the transcription factor NEUROG3 (Gu et al., 2002; Jennings et al., 2013; Salisbury et al., 2014). NEUROG3 deficiency leads to an important reduction in, or absence of, pancreatic endocrine cell development, in both mouse and human, and in models of human embryonic stem cell (hESC) differentiation towards endocrine cells (Gradwohl et al., 2000; Rubio-Cabezas et al., 2011; McGrath et al., 2015; Zhu et al., 2016).
There are many similarities, but also differences, in pancreatic development between rodent and human. Whereas pancreatic endocrine cell development occurs in two waves in rodents, a single wave of endocrine cell differentiation was described in humans (Pictet et al., 1972; Jennings et al., 2013; Salisbury et al., 2014). Another example is represented by the transcription factor NKX2-2, which is expressed in rodents by early pancreatic progenitors upstream of NEUROG3, whereas its onset is downstream of NEUROG3 in humans (Jennings et al., 2013). Many genes acting downstream of NEUROG3, some of which are direct targets, have been identified in the mouse (Dassaye et al., 2016). Some control endocrine differentiation in all endocrine cell types, whereas others are specific to one or to several subtypes. Important endocrine genes are also expressed in the human fetal pancreas, including NEUROD1, NKX2-2, PAX4, PAX6 and ISL1 (Lyttle et al., 2008; Jeon et al., 2009). Their sequence of activation and their function have been studied in stem cell models of pancreatic differentiation (Liu et al., 2014; Zhu et al., 2016; Petersen et al., 2017). The degree of conservation of gene function and developmental mechanisms between human and other vertebrates require more extensive investigation.
In the field of hematopoiesis, the identification of cell surface markers using flow cytometry has been instrumental in understanding lineage relationships and differentiation mechanisms (Eaves, 2015), an approach we initiated to study human fetal pancreatic cell differentiation. Using a combination of the cell surface markers GP2, ECAD (also known as CDH1), CD142 (also known as F3) and SUSD2 on human fetal pancreatic epithelial cells, we have previously identified distinct endocrine populations at different stages of their development (Ramond et al., 2017). In the present study, we provide in-depth transcriptional profiling of these populations using RNA sequencing (RNA-seq). In addition, we improve the purity of the sorted populations by expanding the sorting strategy to include an additional surface marker, CD133 (also known as PROM1), to exclude ductal cells, along with using granularity to distinguish endocrine cells. Using the improved sorting scheme, we isolate highly pure populations from human fetal pancreata and study their composition at the single-cell level using qPCR. Our analysis focuses on 9 WD pancreata and shows that, at a single time point, different steps of the endocrine differentiation path can be captured. Our findings furthermore enable us to benchmark the pancreatic cell types produced in vitro from human pluripotent stem cells (hPSCs) using a well-established endocrine-biased differentiation protocol (Rezania et al., 2014).
RESULTS
RNA-seq profiling of human fetal pancreatic populations
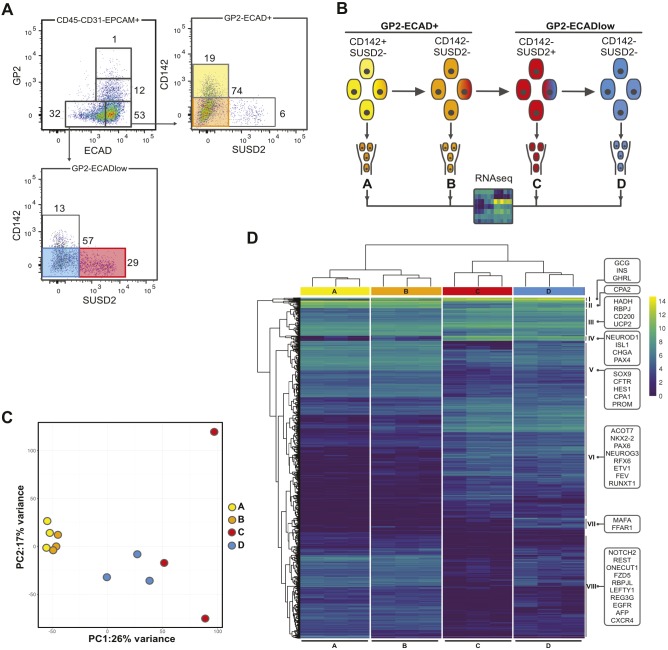
We have previously reported that flow cytometry with specific cell surface markers can be used to purify distinct populations that sequentially differentiate during human fetal pancreas development. Following gating on epithelial cells (CD45−CD31−EPCAM+GP2−), four distinct populations were sorted from human fetal pancreata at 9 WD: ECAD+CD142+ (population A), ECAD+CD142− (population B), ECADlowCD142−SUSD2+ (population C) and ECADlowCD142−SUSD2− (population D). We found that NEUROG3 is first detected in population B but culminates in population C. Endocrine markers, especially INS, are first detected in population C and later (from 10 WD) in population D (Ramond et al., 2017). Here, we used this sorting strategy to isolate similar populations from three individual human fetal pancreata at 9 WD and analyzed them by RNA sequencing (Fig. 1A,B; Fig. S1 for details on the gating strategy). Transcripts encoding the markers that were used for cell sorting, namely CDH1, EPCAM, F3, GP2, PECAM1 (CD31), PTPRC (CD45) and SUSD2 followed the expected expression pattern, validating the experimental procedure (Fig. S2A).
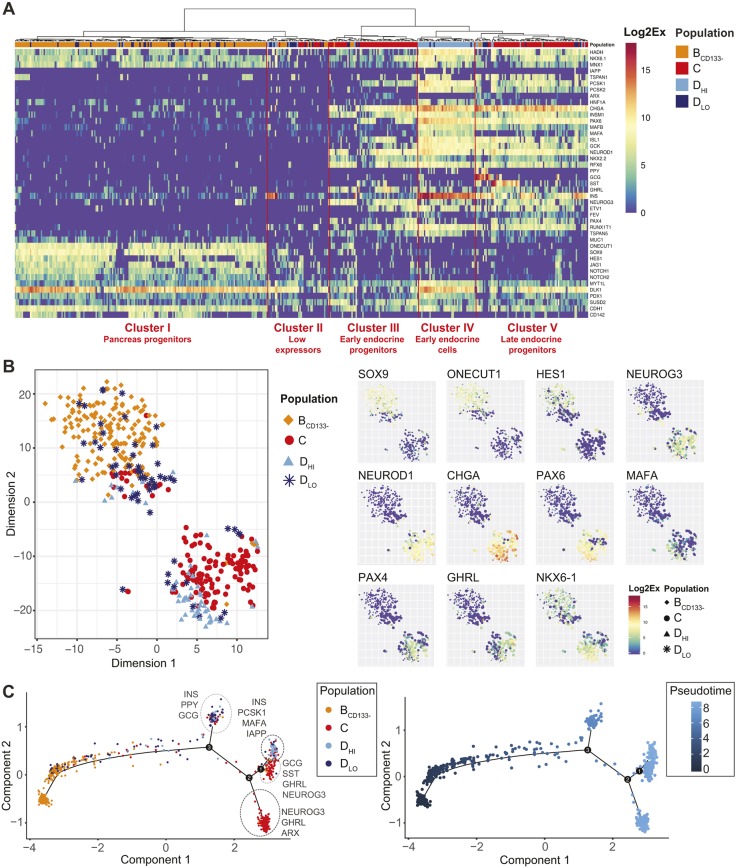
Fig. 1.
Characterization of sorted cell populations from human fetal pancreata by RNA-seq at 9 WD. (A) Flow cytometry analysis of the expression of GP2 and ECAD on CD45−CD31−EPCAM+ cells at 9 WD. CD142 and SUSD2 expression was analyzed in GP2−ECAD+ and GP2−ECAD− cells. FACS plots are representative of three independent pancreata at 9 WD. (B) Scheme of the experimental setup: GP2−ECAD+CD142+SUSD2− (population A, yellow), GP2−ECAD+CD142−SUSD2− (population B, orange), GP2−ECADlowCD142−SUSD2+ (population C, red) and GP2−ECADlowCD142−SUSD2− (population D, blue) were cell sorted and analyzed by RNA-seq. All populations were CD45−CD31−EPCAM+ and derived from three independent pancreata at 9 WD. The population color code is re-used throughout the article. (C) Principal component analysis map on the RNA-seq from all four sorted populations in triplicate at 9 WD. (D) Heatmap displaying the expression of the 1007 differentially expressed genes in triplicate in all four populations in RPKM, along with the hierarchical clustering. Clusters are named on the right side and some gene examples are indicated in boxes. For more extensive examples, Table S1 highlights genes enriched in the four sorted cell populations.
Principal component analysis on the RNA-seq from the four populations revealed that populations A and B clustered together, whereas populations D and C clustered independently (Fig. 1C). Hierarchical clustering on 1007 differentially expressed genes revealed that the first branching occurred between populations A and B and populations C and D (Fig. 1D). Moreover, eight clusters of genes with similar expression patterns were identified. Clusters I and II mark genes which were highly expressed in at least three of the populations and had a diversity of expression patterns. Cluster III includes genes expressed at high levels, with minor changes in A versus B and C versus D. Clusters IV and VI contain known endocrine progenitor and endocrine genes, which were expressed at higher levels in C and D, with some increase already in B for cluster IV. Several of these genes are highlighted in Fig. 1D. In contrast, clusters V and VIII, which contained pancreas progenitor markers (HES1 and REST for example), were enriched in A and B and, for VIII, the decrease was particularly strong in C (Fig. 1D). Cluster VII was characterized by genes expressed at higher levels in population D compared with the other populations, some of which are known mature endocrine cell markers (Fig. 1D and Table S1). In addition, we identified differential expression of several genes associated with different signaling pathways between the populations, such as EGFR, CXCR4 and RARG, enriched in populations A and B (Table S1).
We then performed a gene set enrichment analysis on populations A to D using the Gene Ontology Consortium database (www.geneontology.org). Biological processes related to hormone regulation and secretion appeared specifically enriched in populations C and D compared with populations A and B (Fig. S3 and Table S2). Moreover, biological processes, such as regulation of insulin secretion, regulation of hormone secretion, regulation of hormone levels, insulin secretion, hormone transport and hormone secretion, were further enriched in population D (Fig. S3 and Table S2). Taken together, our data indicate that populations C and D contain endocrine cells.
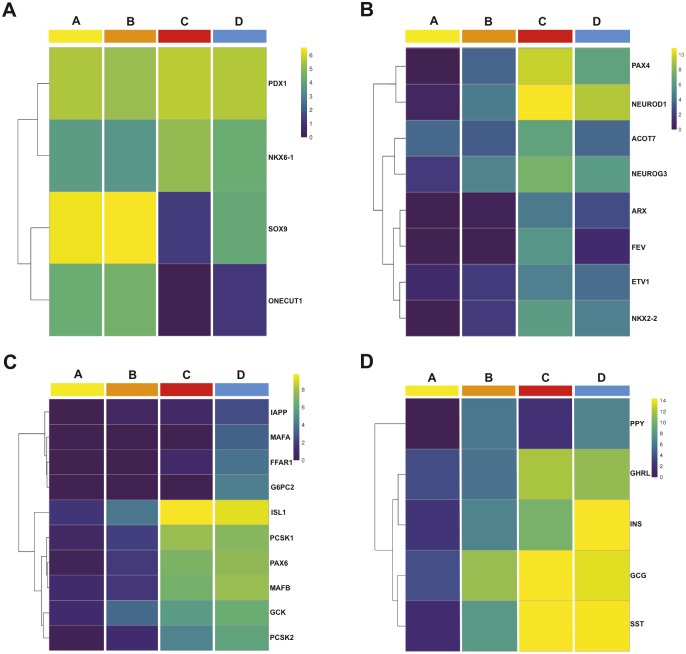
To understand the composition of each cell population with respect to multipotent pancreatic progenitors, endocrine progenitors and endocrine cells, we generated heatmaps. PDX1, a factor expressed in humans by pancreatic progenitors at early time points and later on in β, δ and duct cells, was equally detected in all four populations (Fig. 2A) (Jennings et al., 2015). It was also the case for NKX6-1, a marker of early pancreatic progenitors, which is later restricted to β-cells (Fig. 2A) (Jennings et al., 2013). The high expression of SOX9 and ONECUT1 in populations A and B suggested that they contained pancreatic progenitors (Fig. 2A) (Larsen and Grapin-Botton, 2017). With regards to endocrine progenitors, NEUROG3 was expressed at low levels in population A, at increased levels in population B, at the highest levels in population C and at lower levels in population D (Fig. 2B). The NEUROG3 targets FEV, ETV1, PAX4, ARX, ACOT7, NKX2-2 and NEUROD1 followed similar patterns (Fig. 2B). These data thus indicate that population A is enriched in multipotent pancreatic progenitors, whereas population C is enriched in endocrine progenitors. Populations C and D also expressed the highest level of the endocrine cell marker genes ISL1, CHGA, MAFB, PAX6 and PCSK1 (Fig. 2C and Fig. 1D, cluster IV and VI) in addition to the genes encoding the hormones GCG, SST, GHRL and INS (Fig. 2D). PPY expression levels appeared enriched in populations B and D. INS expression levels were 13 times higher in population D compared with population C (Fig. 2D and Fig. S2B). In addition, a set of β-cell-specific genes (MAFA, PCSK1, IAPP, G6PC2, FFAR1, SLC30A8) was enriched in population D compared with population C, suggesting that the endocrine cells in population D were more mature than those in population C (Fig. 1D cluster VII, Fig. 2D). We also observed upregulation of genes with unknown function in pancreas development, such as ELAVL4 and EYA2 in population C. Additional examples of novel genes marking each population can be found in Table S1.
Fig. 2.
Characterization of populations A, B, C and D at 9 WD. Heatmaps displaying the expression of multipotent genes (PDX1, NKX6-1, SOX9 and ONECUT1) (A); progenitor genes and early endocrine markers (PAX6, PAX4, NEUROD1, ACOT7, NEUROG3, ARX, FEV, ETV1 and NKX2-2) enriched in population C (B); Endocrine genes (IAPP, MAFA, FFAR1, G6PC2, ISL1, PCSK1, MAFB, GCK and PCSK2) enriched in population D (C); and pancreatic hormones (PPY, GHRL, INS, GCG and SST) in sorted populations A, B, C and D (D). Heatmaps are displayed using a log2 expression scale.
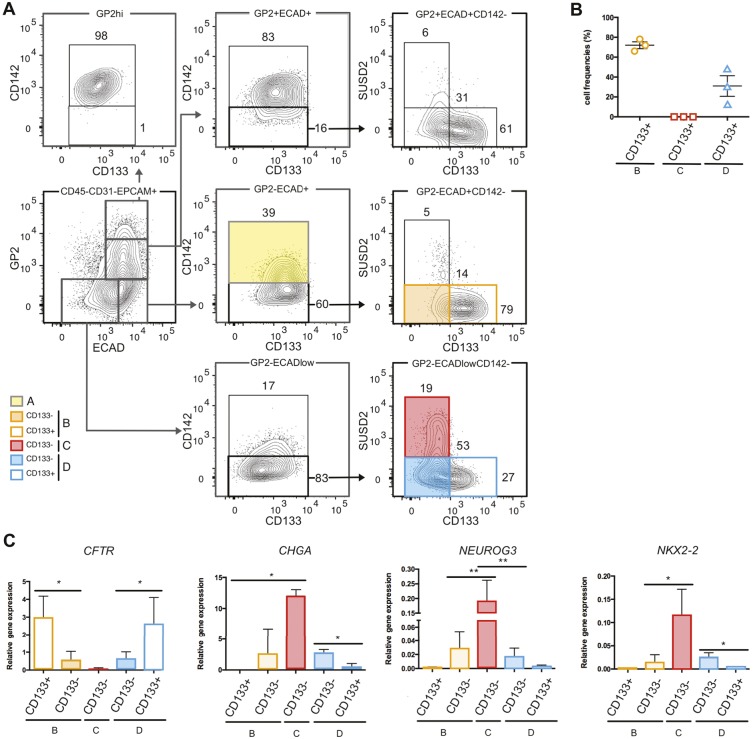
CD133 marks ductal cells
We have previously shown, by single-cell qPCR, that population B contained a majority of duct-like cells (78% positive for CFTR) and 16% NEUROG3+ endocrine progenitors, whereas population C was highly enriched in NEUROG3+ cells (74%) with rare CFTR+ cells (2%) (Ramond et al., 2017). Our RNA-seq data supported this claim (Fig. S2B), but also showed that, compared with the other populations, population D expressed the highest level of INS, with CFTR levels similar to population A. Therefore, we searched for additional markers to deplete CFTR+ duct-like cells from populations B and D. In the pancreas, ductal cells express the transmembrane protein CD133 (Sugiyama et al., 2007; Lardon et al., 2008). Fluorescence-activated cell sorting (FACS) analyses indicated that the C population was CD133−, which correlates with the low frequency of duct-like cells in this population (Fig. 3A,B), whereas populations B and D contained cells with different levels of CD133 (Fig. 3A,B). The frequency of CD133+ cells was highest in population B compared with population D (72±6% versus 31±18%) (Fig. 3B). We next sorted cells from populations B to D based on their CD133 levels. We performed RT-qPCR for the expression of CFTR, marking ductal cells, and CHGA, NEUROG3 and NKX2-2, marking endocrine cells. We found that the CD133− populations expressed lower levels of CFTR and were enriched in CHGA, NEUROG3 and NKX2-2 (Fig. 3C). We concluded that in the human fetal pancreas, CD133 marks duct-like cells and can be used for their depletion from endocrine cell-containing populations.
Fig. 3.
CD133 marks human fetal pancreatic ductal cells. (A) Flow cytometry analysis displaying the expression of CD133 in populations B, C and D at 11 WD. FACS plots are representative of three independent pancreata at 11 WD. (B) Cell frequencies of population B CD133+, population C CD133+ and population D CD133+ from three independent pancreata at 10 WD. (C) Expression of CFTR, CHGA, NEUROG3 and NKX2-2 (9-11 WD) by RT-qPCR in population B CD133+, population B CD133−, population C CD133−, population D CD133− and population D CD133+. *P<0.05, **P<0.001 (_t_-test; data are mean±s.e.m. of three independent pancreata).
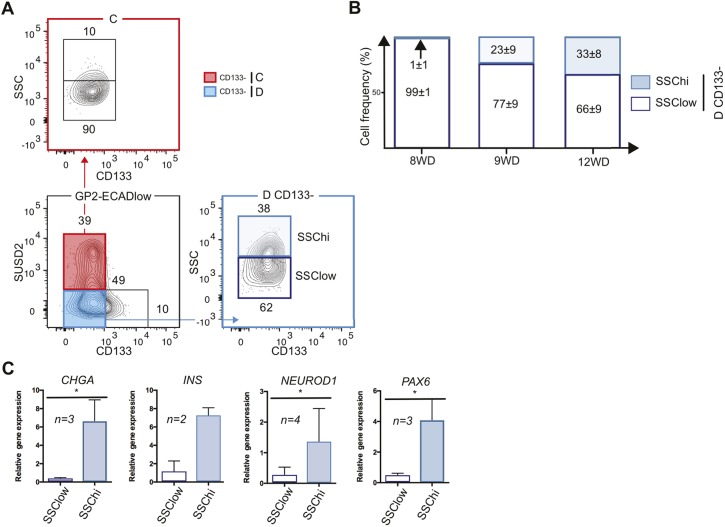
Granularity parameter isolates fetal endocrine cells in population D CD133− (DCD133−)
Pancreatic endocrine cells contain hormone-rich secretory granules, in contrast to other pancreatic cell types. Cell granularity can be assessed by flow cytometry in the side scatter (SSC) parameter. This method has previously been used to enrich for endocrine cells from the adult pancreas (Pipeleers, 1987; Rui et al., 2017). We therefore investigated whether granularity could be used to further enrich for hormone-positive cells. In the human fetal pancreas at 10 WD, population C (which is CD133−) contained a low frequency (15±7%) of SSChi cells, whereas population D depleted from duct cells (CD133−) contained a higher frequency (33±14%) of SSChi cells (Fig. 4A) (percentages are mean±s.e.m. from three independent pancreata at 10 WD). The frequency of SSChi cells in population D was age dependent, as SSChi cells first appeared at 9 WD and their frequency increased thereafter (Fig. 4B). We sorted SSChi and SSClow cells from population DCD133− (named as DHI and DLO, respectively) and performed RT-qPCR for the expression of endocrine genes (CHGA, INS, NEUROD1 and PAX6). RT-qPCR results demonstrated that endocrine markers were enriched in population DHI (Fig. 4C). We concluded that, as in the adult, endocrine cells in the human fetal pancreas were the most granular.
Fig. 4.
Endocrine cells in population D are the most granular. (A) Flow cytometry displaying the granulometry (SSC) in population C and population DCD133− at 10 WD. FACS plots are representative of three independent pancreata at 10 WD. (B) Frequency of SSChi and SSClow in population DCD133− at 8, 9 and 12 WD (data are mean±s.e.m. of three independent pancreata at each stage). (C) Expression of CHGA, INS, NEUROD1 and PAX6 at (10-12 WD) by RT-qPCR in population DCD133−, SSClow or SSChi. *P<0.05 (_t_-test; data are mean±s.e.m. of three independent pancreata).
Single-cell profiling reveals heterogeneity in the sorted human fetal pancreatic populations
To address the heterogeneity within populations BCD133−, C, DLO and DHI described earlier, we analyzed sorted cells by single-cell qPCR for the expression of 91 genes (see Materials and Methods for details and Table S3 for information on primers). Unsupervised hierarchical clustering of the data showed that the major differences in gene expression were found between cells from population BCD133− (Cluster I) versus cells from populations C and DHI (Clusters III-V) (Fig. 5A and Fig. S4A). Associated with Cluster I, we found a mixed population consisting of cells that, in general, had low levels of gene expression, most likely for technical reasons (Cluster II). Most cells of Cluster I expressed ECAD and key pancreatic progenitor markers such as ONECUT1, SOX9, MNX1 and NKX6-1, whereas HES1 and JAG1 were more heterogeneously expressed (Fig. 5A and Fig. S5). Furthermore, a few cells had initiated NEUROG3 expression (Fig. 5A,B). The YAP/TAZ target gene CTGF also displayed a heterogeneous profile, activated in ∼50% of the cells (Fig. S4A and Fig. S5). Though the cells were sorted based on their lack of the surface marker CD142, a subpopulation of the cells expressed the transcript encoding CD142 (Fig. 5A).
Fig. 5.
Single-cell profiling of sorted human fetal pancreatic cells. (A) Heatmap showing gene expression for a selected set of genes in individual cells sorted from human fetal pancreas based on cell-surface markers and granularity discriminating populations BCD133−, C, DHI and DLO. The genes with greatest variance are shown. A heatmap with all genes is provided in Fig. S4A. Cells of populations BCD133− and C are derived from three individual pancreata at 9 WD; cells from populations DHI and DLO are derived from two individual pancreata at 9 WD. (B) t-SNE plot of single-cell qPCR data from human fetal pancreas colored by population (left panel) or according to expression level of selected genes (right panel). See also Fig. S5. (C) Developmental trajectory for endocrine differentiation in human fetal pancreas using pseudotemporal ordering with Monocle. Cells on the trajectory are colored according to population (left panel) or pseudotime (right panel). Specific genes characteristic for each branch are indicated on the left panel. See also Fig. S6.
The second group of clusters (III-V) comprised endocrine progenitors (C) and early endocrine cells (DHI). All clusters shared the expression of NEUROG3, RFX6, CHGA, NEUROD1, NKX2-2, INSM1 and SUSD2, with some noise, as commonly seen for low abundance genes when starting from small amounts of RNA. Cells from Cluster III appear to be early progenitors that do not express PCSK2 or MAFB transcripts and largely exhibit no hormone expression (except for GHRL in a subpopulation) (Fig. 5A,B). Furthermore, PDX1, NKX6-1 and MNX1 transcripts were downregulated compared with pancreatic progenitors. Cluster III was intrinsically heterogeneous, with at least four subpopulations that may represent different levels of maturity or distinct subtypes. These included a group of cells likely representing a precursor to α-cells, expressing ARX, PCSK1, ISL1 and ETV1. Another group expressed robust NKX6-1, in addition to PCSK1, PAX6, ETV1 and PAX4. Cluster V likely contains more mature endocrine progenitor cells, with low levels of PCSK1 and PCSK2, high CHGA and PAX6 levels, and heterogeneous hormone expression. Cells from population C were the main contributors to these two clusters. Finally, Cluster IV was composed of early endocrine cells (mainly DHI cells) with high levels of endocrine markers, with most cells expressing high levels of INS. Cells from population DLO did not appear to represent a homogenous population, but were intermingled with the clusters described above (Fig. 5A,B).
Visualizing the data using dimensionality reduction via t-distributed stochastic neighbor embedding (t-SNE, Van Der Maaten and Hinton, 2008) identified similar populations as hierarchical clustering, with population B largely forming a separate cluster from populations C and DHI (Fig. 5B and Fig. S5). The DHI cluster overlapped, to some extent, a cluster representing cells of population C, highlighting a high degree of similarity between these populations that are both committed towards endocrine differentiation. t-SNE further confirmed that population DLO is heterogeneous, encompassing cells with profiles similar to BCD133− or DHI. Violin plots highlighted the subpopulation of cells within the B population that had activated NEUROG3, and showed that NEUROD1 was expressed at similar levels in populations C and DHI, whereas DHI expressed higher levels of CHGA, PAX6 and MAFA in a subset of cells (Fig. S4B).
As the data indicated that cells of different maturation stages were present in the pancreas at 9 WD, we next used the algorithm Monocle (Trapnell et al., 2014) to infer a developmental trajectory for the differentiating cells and pinpoint branching points to different cell fates. Through the construction of a minimal spanning tree, cells were linked in a pseudotemporal order, with population B as the starting population, giving rise to four branches (Fig. 5C and Fig. S6). The pseudotime reflected downregulation of pancreatic progenitor-associated genes (e.g. SOX9 and HES1) and upregulation of endocrine-biased genes (e.g. NEUROG3, CHGA and NKX2-2) (Fig. S6B). The first branch off of the main trunk represented polyhormonal endocrine cells (Fig. 5C, Fig. S6A-C), the second branch further diverged into two endocrine populations (likely on the path to β- and α/δ-cells, respectively), and the third branch represented endocrine progenitors (Fig. 5C, Fig. S6A-C). Heat map visualization of temporal gene expression distinguished cells on the β-cell versus α/δ-cell track (Fig. S6D, cells diverging from branching point 1).
Taken together, based on what is known about each marker in mouse pancreas development, the single-cell analysis shows that the FACS strategy enables discrimination between distinct populations of human pancreas progenitors, endocrine progenitors and endocrine cells. In addition, it shows that subpopulations are found within these three groups, which likely represent maturation intermediates co-existing at the same time point because of heterochrony of differentiation in individual cells.
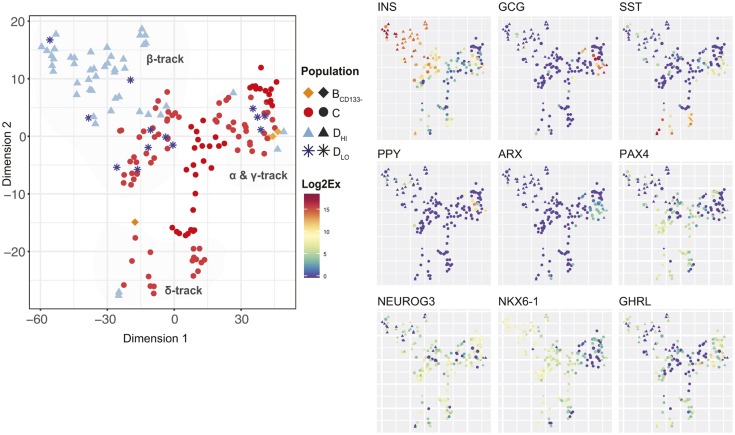
Endocrine subtype selection is initiated in endocrine progenitors
We next investigated whether cells differentiating towards different endocrine subtypes expressing different hormones could be captured, and when they diverged. To gain power, we computationally excluded cells of Clusters I and II (pancreatic progenitors and low expressors) and performed t-SNE analysis on the remaining endocrine-biased cells (Fig. 6). With this data visualization, the cells distribute into a three-branch star, each branch ending with different hormonal expression (Fig. 6). One of the branches contains cells with a gradient of INS expression. Another contains cells expressing GCG and the α-cell marker ARX. Some cells in this branch also express PPY and low levels of INS and SST. The third branch contains cells with the highest level of SST in cells at the tip of the branch. To investigate whether the co-expression of multiple hormones was also evident at the level of protein expression, we performed immunofluorescence stainings on human fetal pancreas sections at 10 WD. We assessed the combination of INS with GCG or SST, and GCG with GHRL, and found rare cells co-expressing each of these three combinations of hormones (Fig. S7). We did not evaluate the expression of PPY.
Fig. 6.
Gene expression profile for human fetal endocrine progenitors and early endocrine cells. t-SNE plot of single-cell gene expression data for the endocrine-biased cluster (Cluster III-V) formed by the hierarchical clustering shown in Fig. 5A (left panel). Cells are colored according to population (left panel) or gene expression level (right panel) for selected genes. Colours reflect the Log2Ex scale.
At the transcriptional level, we furthermore observed co-expression of NEUROG3 with hormones. In the t-SNE analysis, most endocrine progenitors (C population) express NEUROG3 transcripts, arguing that endocrine subtype selection is initiated in endocrine progenitors and continues in population D (Fig. 6). The α-cell specifier ARX and the β-cell specifier NKX6-1 are already expressed in subpopulations of C cells in different branches. Even though PAX4 is commonly described as a β-cell specifier in mice and humans, it is restricted to endocrine progenitors of population C, as is GHRL (Fig. 6). PAX4 and GHRL were absent from most cells in population DHI, suggesting that they are endocrine progenitor markers (Fig. 6). In conclusion, the single-cell gene expression profiling shows the initiation of endocrine subtype specification in endocrine progenitors in humans, and reveals unexpected expression patterns for PAX4 and GHRL.
_In vitro_-produced endocrine cells follow paths similar to those of their in vivo counterparts, but do not resolve hormonal subtypes
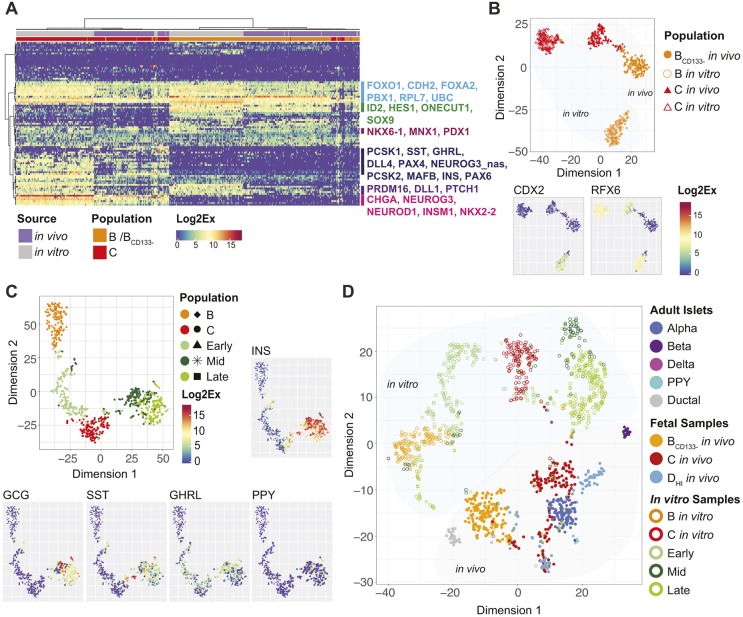
We have previously shown that ECAD, CD142 and SUSD2 markers also discriminate B and C populations generated in vitro from hPSCs (Ramond et al., 2017). To evaluate the degree of similarity between in vivo and in vitro populations, we furthermore performed single-cell qPCR on _in vitro_-differentiated cells. To simulate the biological variability between individual pancreata, we used three different hPSC lines [SA121 hESCs and induced pluripotent stem cells (iPSCs) AD2.1 and AD3.1] differentiated until stage 5 day 1 (S5D1) using a protocol adapted from Rezania et al. (2014) (with minor modifications – see Materials and Methods for details). All three lines efficiently generated NKX6-1+ cells and initiated NEUROG3 expression as assessed by flow cytometry, indicating differentiation towards the pancreatic endocrine lineage (Fig. S8). The hPSC-derived cells were then sorted based on ECAD, CD142 and SUSD2 as population B or C and analyzed by single-cell qPCR. Hierarchical clustering of the data showed that cells segregated first by population (B and C) regardless of their in vivo or in vitro origin, suggesting that similar populations are generated both in vivo and in vitro (Fig. 7A and Fig. S9). The second clustering distinguished cells from in vitro origin from the fetal cells, indicating that there are source-based differences in gene expression. Some of this difference might be driven by a lower level of gene expression in cells extracted from the fetal pancreata; however, normalizing gene expression to the housekeeping gene RPL7 did not significantly alter the clustering (data not shown). Likewise, the four clusters were also generated by t-SNE analysis (Fig. 7B), discriminating populations B and C, and cells generated in vivo from those produced in vitro. The decreased distance between populations B and C in vivo suggests that they are more similar than their in vitro counterparts.
Fig. 7.
Comparison of the single-cell expression profile of pancreatic cells generated in vitro with in vivo fetal and adult pancreata. (A) Heatmap showing the gene expression level in single cells of populations B and C isolated from human fetal pancreas (in vivo) or from hPSC-derived cultures (in vitro). Selected genes are indicated, colours reflect the Log2Ex scale. A heatmap with all gene annotations is provided in Fig. S9. (B) t-SNE plot of single-cell qPCR data for populations B and C isolated from human fetal pancreas (in vivo) or from hPSC-derived cultures (in vitro). Gene expression level in individual cells for RFX6 and CDX2 is indicated in the bottom plots. (C) t-SNE plot combining data from the present study on sorted hPSC-derived cells (populations C and D) with data from a previously published dataset comprising endocrine-biased hPSC-derived cells collected at different time points (early, stage 4 day 1 and 3; mid, stage 5 day 3+stage 6 day 2; late, stage 6 day 7+stage 7 day 7). Expression level of hormonal genes are mapped onto the t-SNE plot as indicated. (D) t-SNE plot combining the data from C with data on the sorted cell populations from human fetal pancreas (populations B, C and DHI) from the present study and data on adult human islet cells (from the same previously published dataset as used in C). See also Fig. S10.
_In vitro_-produced population B was distinct from the in vivo reference as the former expressed both RFX6 and CDX2 (Fig. 7B), which may indicate that this population retains a mixed pancreas-duodenum fate or are equivalent to earlier pancreas progenitors. Fetal pancreatic progenitors had robust NKX6-1 and DLK1 expression; however, only a subpopulation of the hPSC-derived cells was NKX6-1+ and DLK1+ (Fig. S9). Sporadic expression of GPM6A, CDKN1A and BMP5 was detected among _in vitro_-generated cells, whereas these transcripts were absent from fetal cells. Finally, ZNF453 was robustly expressed in most _in vitro_-derived cells, but only in a small subpopulation in vivo (Fig. S9).
To further extend the comparison between in vivo and _in vitro_-derived cells, and to assess their maturity, we combined the single-cell qPCR datasets of the human fetal cells (populations BCD133−, C and DHI) and the _in vitro_-differentiated hPSC-derived cells (populations B and C) (this study), with a previously published single-cell qPCR dataset covering several stages of in vitro differentiation, along with endocrine cells from human adult islets analyzed with the same set of primers (Petersen et al., 2017). The hPSC-derived cells from this dataset were generated using the same protocol as in the present study, and were isolated from stage 4 (early), end of stage 5 and early stage 6 (mid), and end of stage 6 and 7 (late) of the differentiation protocol using a NEUROG3-GFP reporter hESC line. First, we evaluated how the sorted hPSC-derived B and C cells aligned with cells from the additional differentiation stages (Fig. 7C). The B population formed a separate cluster close to the stage 4/early population (Fig. 7C), whereas population C was positioned between cells from the early and mid stages. We next visualized the complete combined dataset comprising all in vivo and in vitro samples (Fig. 7D). On the t-SNE plot, the _x_-axis separated cells from immature pancreas progenitors (left) to most differentiated endocrine (right) in both in vivo and in vitro conditions. The _y_-axis separated cells produced in vitro (up) from in vivo differentiation (down) (Fig. 7D). The different maturation stages are largely inferred from known sequences of gene activation during mouse pancreas differentiation. The _x_-axis organizes cells at the different stages of differentiation that co-exist at stage 5 in the differentiation protocol, and also reflects a temporal progression of differentiation/days of in vitro differentiation (Fig. 7C,D and Fig. S10). We validated some of the progression at the protein level, including SOX9-only cells, rare cells co-expressing SOX9 and NEUROG3 (Fig. S11A), a vast majority of NEUROG3 cells expressing NKX2-2 (Fig. S11B,C) and a few cells co-expressing NEUROG3 and hormones (Fig. S12A-C). Although hPSC-derived cells initiate the expression of β-cell maturity markers MAFA and IAPP, they do not reach the expression levels seen in the adult β-cells (Fig. S10). Finally, we found that _in vitro_-produced cells do not resolve hormonal subtypes and continue to co-express multiple hormones at the transcriptional level (Fig. 7C). Although a subset of cells generated with this differentiation protocol is also polyhormonal at the protein level, the majority express only INS or GCG (Rezania et al., 2014; Petersen et al., 2017). As PCSK1 was often co-expressed with INS and GCG transcripts, we speculated that these cells might produce C peptide and GLP1, a peptide processed by PCSK1 from the GCG transcript. By immunofluorescence staining, we could indeed detect GLP1, with some cells co-expressing GLP1 and C peptide/proinsulin (Fig. S12D). In conclusion, our single-cell data, combined with the sorting strategy, provide a platform for future studies to further delineate lineage allocation of individual endocrine cell types of the human pancreas.
DISCUSSION
The developmental mechanisms of the mouse and zebrafish pancreas have been characterized in depth. Numerous studies have uncovered markers for different cell types, the lineage hierarchies between cells that are present at different time points, the role of numerous genes, notably transcription factors, and extracellular signals and their regulatory mechanisms (Larsen and Grapin-Botton, 2017). These studies, and limited comparisons with human development, have shown a large conservation of developmental mechanisms between species, but have also revealed notable differences in the ratio between different endocrine cell types, and in the precise gene expressed in a family of transcription factors endowed with similar properties (Flasse et al., 2013; Jennings et al., 2015). The limited information we have in human development most likely leads us to underestimate differences. In our previous work, we identified markers that enable the enrichment of human fetal pancreatic progenitors (ECAD+CD142+/population A), acinar cells (GP2hi) and cells at different steps on the endocrine differentiation pathway initiating NEUROG3 expression (ECAD+CD142−/population B), activating downstream genes (ECADlowCD142−SUSD2+/population C) and endocrine cells (ECADlowCD142−SUSD2−/population D) (Ramond et al., 2017). Extending our initial analysis, which was limited to a few transcripts and previous transcriptome analysis of the whole human fetal pancreas, our current transcriptome analysis of these populations enables the assessment of the nature and purity of the isolated populations, the conservation of stage-specific regulators of pancreas development with other vertebrates, and the identification of potentially interesting new players. It also documents the cell types in which genes that have variants predisposed to monogenic primary or syndromic diabetes (HADH, GCK, HNF1A, HNF1B, INS, ISL1, NEUROD1, NEUROG3, PAX4, PAX6, PCSK1, PDX1, RFX6, SPINK1, UCP2) or to type 2 diabetes (NOTCH2, GCK, PAX4, HNF1B, HNF1A) are expressed (Fuchsberger et al., 2016). Their expression at this stage may contribute to a predisposition to diabetes.
Population A contains mostly pancreas progenitors, but the expression of several acinar cell markers (CPA1, CPA2, NR5A2 and RBPJL) suggests that this population either contains early acinar cells (notably, there is no mature marker such as amylase), or corresponds to the early multipotent progenitors observed in mice, a population that has the potential to differentiate into endocrine or acinar cells (Zhou et al., 2007). These are not detected in population B, most of which is initiating endocrine differentiation while retaining pancreas progenitor markers. These two progenitor populations are enriched in receptors for multiple signaling pathways. The presence of EGFR, ERBB3, FGFR2 and SPROUTY2 in populations A and B suggests that, as in mouse, the EGF and FGF pathways are used in human pancreas progenitors, possibly for their maintenance and proliferation, as was shown in mice (Miralles et al., 1999; Bhushan et al., 2001; Miettinen et al., 2000). Interestingly, the pathway may be conserved between species, but differences in the ligands that activate the pathway were observed. As an example, FGF10, which activates FGFR2b in the mouse, is expressed at low levels in the human pancreatic mesenchyme, in which it is replaced by FGF7, another activator of the same pathway (Ramond et al., 2017). The receptors for these pathways may be used as alternative sorting markers, as could CXCR4, which is enriched in this population and has previously been used to sort endoderm. The presence of the RARG receptor is also suggestive of retinoic acid signaling. This pathway has been shown in mice to initially promote the formation of the pancreatic primordium, which takes place at earlier stages than those studied here, and endocrine progenitor formation, though RARA is the most highly expressed retinoic acid receptor in the mouse epithelium (Tulachan et al., 2003; Molotkov et al., 2005; Martín et al., 2005). FZD5 enrichment is in agreement with the observation that Wnt signaling promotes pancreas progenitor expansion in mice (Baumgartner et al., 2014; Afelik et al., 2015). TCF7L2, a transcription factor that mediates canonical Wnt pathway activity, is also enriched in populations A and B. It is a major susceptibility locus for type 2 diabetes, and it is therefore possible that its variant affects pancreas development, in addition to roles in β-cell function that have previously been reported (Liu and Habener, 2010). The progenitors in populations A and B are also enriched in signaling molecules such as GDNF, which promotes pancreatic innervation and is also secreted by multipotent and bipotent progenitors in mice, and PDGFC, of unknown activity in the mouse pancreas (Muñoz-Bravo et al., 2013). The enrichment of LEFTY1 is enigmatic, and is possibly a remnant of left-right asymmetry determination at this late stage or an antagonism to NODAL, and has not been reported in mice.
Our profiling also detects numerous transcription factors that have been reported to maintain progenitors in the mouse (SOX9, HNF1B, OC1, HES1, REST), and that have previously been detected in the human pancreatic bud at 6-7 WD (Larsen and Grapin-Botton, 2017). It also suggests new transcription factors that are active in pancreas progenitors such as HEYL, a Notch target possibly acting redundantly to HES1, GRHL2 and OVOL2. With regard to genes involved in morphogenesis, the expression of the RhoGAP STARD13 in these populations suggests a conservation of its role in tip cell morphogenesis (Petzold et al., 2013). Only a few markers were specific to the B population, notably the proneural gene ATOH8, which represses the _NEUROG3_-induced differentiation program in the mouse (Ejarque et al., 2016). Its absence in population C suggests an expression at the early phase of NEUROG3 expression, or even earlier. Population B also exhibits enrichment in several gene ontology (GO) terms linked to cell migration, suggesting that these cells have initiated delamination.
Population C is enriched with NEUROG3, and also with many of its targets that have previously been described in rodent models, such as PAX6, PAX4, NEUROD1, NKX2-2, ACOT7, CELSR3, ARX, FEV and ETV1. In agreement with the observations in mouse, CDKN1C and RIPPLY3 upregulation may play a role in the cell cycle block observed in endocrine progenitors (Georgia et al., 2006; Osipovich et al., 2014). Several genes with reported activity in neurons are also upregulated in this population, some of which have an unknown function in pancreas development, such as ELAVL4, EYA2, FGF12 or FGF14.
The profile of population D shows that many components of the secretory machinery of endocrine cells are activated during development. Moreover, the activation of ACVR1C suggests the involvement of activin signaling during the late stages of endocrine differentiation, possibly recapitulating the importance of ACVR1C in modulating insulin secretion in mice (Bertolino et al., 2008).
Taken together, our study reveals a large degree of conservation in the expression of genes involved in mouse pancreas development in humans. Our RNA-seq data also reveal new genes that are expressed in specific subpopulations during human pancreatic development, which will be useful for dissecting endocrinogenesis in this organ.
Single-cell qPCR conducted for NEUROG3, NEUROD1, NKX2-2 and CFTR on populations B and C has previously shown that these populations were not pure (Ramond et al., 2017). In the present study, we applied an extra step to the sorting scheme to increase purity of the populations, as we also performed a more comprehensive single-cell qPCR analysis. Although cells in population BCD133− were largely homogeneous in regard to the expression hallmarks of pancreas progenitors (SOX9, ONECUT1 and HES1), we also observed heterogeneity. The YAP/TAZ target gene CTFG was expressed in only a subpopulation of the pancreatic progenitors, indicating that Hippo signaling is active in these cells (Zhao et al., 2008). Notably, the Hippo signaling pathway has recently been identified as a key regulator of the expansion of human pancreatic progenitors (Cebola et al., 2015). Our data may therefore suggest that, at 9 WD, there are discrete populations of pancreatic progenitors in the human pancreas with differential proliferative capacities. We further capture the initiation of NEUROG3 at different levels among pancreatic progenitors and, in a few cells, the initiation of target genes NEUROD1, PAX6 and CHGA.
Based on the pseudotemporal ordering of cells proposed by Monocle, combined with knowledge on mouse pancreas development and previous studies of human fetal pancreatic tissue, we can infer a sequence of gene activation during differentiation of endocrine cells in the human pancreas. Population C showed a general downregulation of pancreas progenitor markers, as NEUROG3 and its targets NEUROD1, RUNX1T1, NKX2-2, INSM1, RFX6, PAX4 and CHGA were upregulated and the hormones were initiated at low levels, followed shortly after by PCSK1, GCK, PCSK2, PAX6 and MAFB. The onset of hormone transcripts in NEUROG3+ endocrine progenitors that we have previously observed in vitro (Petersen et al., 2017) thus appears to recapitulate development in vivo. This was not expected from studying hormones and the NEUROG3 protein in mice, but would be worth assessing transcriptionally. It would be interesting to know when these proteins are expressed during human fetal development and the degree to which they are co-expressed, but the availability of antibodies against human proteins and the number of combinations that can be used on the same sample is a limit to such studies. However, as a proof of concept, we show that we can detect different steps of progression at the protein level, including SOX9-only cells, a few cells co-expressing SOX9 and NEUROG3, a vast majority of NEUROG3 cells expressing NKX2-2 and a few cells co-expressing NEUROG3 and hormones. Although NKX2-2 is known to be expressed in human α- and β-cells (Riedel et al., 2012), we report here its expression at an earlier stage in endocrine progenitors. The elimination of cells with low granularity from population D enabled the isolation of more mature endocrine progenitors, along with what we assume to be the most mature endocrine cells present in the human pancreas at 9 WD. Comparison with adult islets shows that the fetal cells need to mature further to reach the adult state. For example, although they activate MAFA, which is detected from E13.5 in the mouse pancreas, they express little IAPP, a highly expressed β-cell-specific gene (Nishi et al., 1990; Artner et al., 2010).
Our previous observations in hESC-derived cells showed that GHRL was activated early in endocrine progenitors, as soon as NEUROG3 was activated (Petersen et al., 2017). This is now confirmed in vivo. GHRL is later repressed, as the cells acquire the expression of other hormonal genes. The function of GRHL at this early stage is worth further investigation. We have also previously reported a similarly early activation of PAX4, seemingly in all cells initiating NEUROG3 expression (Petersen et al., 2017). The early expression of PAX4 has been reported in the mouse to mark differentiated β-cells, and to be repressed by ARX in α-cells. However, the early stage of PAX4 expression in humans, and how it becomes repressed in β-cells that do not express ARX, suggests that in humans it may function, and be regulated, differently. As we have previously reported (Petersen et al., 2017), we confirmed in this study that hPSC-derived endocrine cells co-express transcripts of several hormones. We can now further conclude that this is also the case for a subset of endocrine cells formed during human fetal pancreas development, although it is much less prevalent. Whereas the fetal α-cells (marked by ARX expression) co-express transcripts for GCG, INS (at low level) and PPY, the cells on the β-cell path only express INS transcripts. The cells expressing SST, though not numerous, also co-expressed other hormone transcripts such as INS and GCG. Though multiple hormonal transcripts are detected above background in all _in vitro-_derived cells, we have previously shown that the majority do not express multiple hormonal peptides (Petersen et al., 2017). Different processing of prohormones is likely to be involved, as the levels of PCSK1 and PCSK2 are variable in these cells and many have detectable GLP1, an alternative peptide processed from the GCG transcript in the presence of PCSK1. It is also possible that the level of transcripts is too low to detect the INS peptide, or they may not be translated (Riedel et al., 2012).
The resource we provide, and the discovery of the sequence of molecular events, sheds light on how cell types are specified in humans and, combined with genome editing in stem cell models of human pancreas development, will be of further use for understanding the mechanisms by which an ever expanding number of genes predispose to diabetes. It also provides a reference to assess the quality of protocols and pancreatic cells produced in vitro from hPSCs.
MATERIALS AND METHODS
Human fetal pancreas tissue
Human fetal pancreata were isolated from surgical abortion carried out by suction aspiration between 9 and 12 weeks of development post conception in compliance with the French bioethics legislation and the guidelines of Inserm (Castaing et al., 2001; Capito et al., 2013; Scharfmann et al., 2014). Approval was obtained from Agence de Biomédecine, the French competent authority, along with maternal written consent. Fetal ages are displayed as weeks of development post conception, confirmed by hand and foot morphology.
Maintenance and differentiation of human pluripotent stem cell lines
The study was carried out with one hESC line (Takara Bio, SA121) and two iPSC lines (StemBANCC, SB AD2.1 and SB AD3.1) (Heins et al., 2004; van de Bunt et al., 2016). These cell lines have been confirmed to be pluripotent by the evaluation of pluripotency marker expression, trilineage differentiation and karyotyping, and tested negative for mycoplasma contamination. All hPSC lines were cultured in mTeSR1 medium (STEMCELL Technologies) on hESC-qualified Matrigel (Corning) and passaged every 3-4 days, or when confluent, by dissociating to a single-cell suspension using TrypLE Select (Thermo Fisher Scientific). Cells were seeded onto freshly coated Matrigel in mTeSR1 with 5 µM Tiger (ROCK inhibitor, Sigma-Aldrich), and medium was replenished daily. Differentiation to the pancreatic lineage was conducted essentially as previously described (Petersen et al., 2017; Ramond et al., 2017) based on a protocol published by Rezania et al. (2014) with minor modifications. Briefly, cells were dissociated to single cells using TrypLE Select and resuspended in mTeSR1 with 5 µM Tiger. Cells were seeded at a concentration of 0.3-0.35×106 cells/cm2 onto growth-factor reduced Matrigel on CellBIND surfaces (Corning). Cells were incubated for 24 h before starting the differentiation. Modifications from the original protocol (Rezania et al., 2014) comprise: MCDB131 medium (Life Technologies) was used as basal medium throughout the differentiation in place of BLAR medium; activin A (PeproTech) was used at 100 ng/ml during stage 1 instead of GDF8. CHIR99201 (Axon Medchem) was used at 3 µM and 0.3 µM for the first and second day of stage 1, respectively, instead of MCX-928; cells were kept in 2D culture throughout the differentiation. The differentiation efficiency with this protocol was reported in figure 1 of Petersen et al. (2017) and is further documented in Fig. S8. Experiments with stem cell lines were approved by the scientific committee of ‘Region Hovedstaden’.
Flow cytometry
Before flow cytometry analysis and/or sorting, fetal pancreata and hPSC-derived cultures were dissociated into single-cell suspensions as follows: fetal pancreata were rinsed with Hanks Balanced Salt Solution (HBSS) (Gibco) to remove contaminating blood cells, and gently dissected with forceps under a binocular magnifying lens (Leica MZ12.5). Pancreata were then incubated for 5 min in collagenase V (0.5 mg/ml; Sigma-Aldrich) in HBSS with Ca2+ and Mg2+. Cells were rinsed in HBSS and then incubated for 5 min in trypsin (0.05%) (Gibco). Finally, cells were rinsed in HBSS supplemented with 20% fetal calf serum (FCS) (Eurobio). Differentiated hPSCs were washed with PBS without Ca2+ and Mg2+ (Invitrogen) and incubated with TrypLE select for 1-3 min.
For staining with cell-surface markers, cells were incubated with antibodies for 20 min in FACS medium (HBSS+2% FCS), then rinsed in FACS medium and resuspended in FACS medium with propidium iodide (1/4000) (Sigma-Aldrich). For intracellular staining of hPSC-derived cultures, dissociated cells were fixed for 20 min in 4% paraformaldehyde (PFA) and then rinsed with PBS with 1% bovine serum albumin (BSA) (Miltenyi Biotec, MACS Buffer). Cells were permeabilized for 30 min at 4°C in PBS with 5% donkey serum (Millipore) and 0.2% Triton X-100 (Sigma-Aldrich) and then incubated with primary antibodies in blocking solution (PBS with 0.1% Triton X-100 and 5% donkey serum) overnight at 4°C, rinsed and resuspended in PBS with 1% BSA. For secondary antibody staining, cells were further incubated with secondary antibodies in blocking buffer for 1 h at room temperature. Finally, cells were washed twice in PBS with 1% BSA and resuspended in PBS with 1% BSA for analysis.
The following antibodies were used: anti-CD45-PerCP/Cy5.5 (1/20, clone 2D1, BioLegend), anti-CD31-PerCP/Cy5.5 (1/20, clone WM59, BioLegend), anti-CD235a-PerCP/Cy5.5 (1/20, clone HI264, BioLegend), anti-EPCAM-Brillant Violet 605 (1/20, clone 9C4, BioLegend), anti-ECAD-PE-Cy7 (1/20, clone 67A4, BioLegend), anti-CD133-APC (1/20, clone 315-2C11, BioLegend), anti-GP2-PE (1/5, clone 3G7H9, MBL International), anti-SUSD2-VioBrightFITC (1/20, W5C5, Miltenyi Biotec), anti-CD142-BV711 (1/20, clone HTF-1, BD Biosciences), anti-NKX6-1-PE (1/40, clone R11-560, BD Biosciences), sheep anti-NEUROG3 (1/300, AF3444, R&D Systems), mouse anti-NKX2-2 (1/200, F4.5A5, Developmental Studies Hybridoma Bank), C-peptide-Alexa Fluor 647 (1/200, clone U8-424, BD Biosciences) and Glucagon-BV421 (1/80, clone U16-850, BD Biosciences). Secondary antibodies were from Jackson ImmunoResearch: anti-sheep Alexa Fluor 647 (1/500, 313-605-003) and anti-mouse Alexa Fluor 647 (1/500, 615-605-214). For each antibody, optimal dilution was determined by titration. Cell sorting was carried out using an ARIA III and a BD FACS Fusion (BD Biosciences) and cell analysis was carried out using a BD LSRFortessa (BD Biosciences). Data were analyzed using FlowJo 10.4.1 software. Dead cells were excluded from analyses.
Immunohistochemistry
Human fetal pancreatic sections (4-5 μm thick) were prepared and processed as previously described (Castaing et al., 2005). The following primary antibodies were used: mouse anti-glucagon (1:2000, G2654, Sigma-Aldrich); mouse anti-insulin (1:1000, I2018, Sigma-Aldrich), mouse anti-ghrelin (1:500, clone 1ML-1D7, Millipore) and rabbit anti-somatostatin (1:500, 10566, DAKO). The secondary antibodies used were: anti-rabbit Alexa Fluor 488 (1:400, A11034, Life Technologies) and anti-mouse Alexa Fluor 594 (1:400, 115 585 003, Jackson ImmunoResearch). Nuclei were stained using the Hoechst 33342 fluorescent stain (0.3 mg/ml, Invitrogen).
For staining of hPSC-derived cultures, cells were washed with PBS and fixed with 4% PFA at RT for 30 min. Cells were then washed with PBS and permeabilized for 10 min with 0.5% Triton X-100 in PBS. Cells were incubated in blocking buffer [0.1 M Tris-HCl (pH 7.5), 0.15 M NaCl, 0.5% TSA Blocking Reagent (PerkinElmer)] for 30 min, followed by overnight incubation at 4°C with primary antibodies diluted in 0.1% Triton X-100 in PBS. The following day, cells were washed 3×5 min with PBS and incubated with secondary antibodies in 0.1% Triton X-100 in PBS for 1 h at room temperature. Primary antibodies were: sheep anti-NEUROG3 (1/1000, AF3444, R&D Systems), mouse anti-NKX2-2 (1/200, F4.5A5, Developmental Studies Hybridoma Bank), rabbit anti-SOX9 (1/2000, AB5535, Millipore), rat anti-C-peptide/proinsulin (1/200, GN_ID4, Developmental Studies Hybridoma Bank), mouse anti-glucagon (1/2000, G2654, Sigma-Aldrich), rabbit anti-GLP1 (1/500, ab22625, Abcam).
Bulk RT-qPCR on human fetal pancreatic cell populations
For human fetal pancreas, 50 to 100 cells were sorted in 9 μl of RT/pre-amp mix from the One-Step qRT-PCR Kit (Invitrogen). Pre-amplified (20 cycles) cDNAs were obtained according to the manufacturer's protocol and used for qPCR reaction. The pre-amplified cDNAs were then used for qPCR using the TaqMan protocol. RT, pre-amplification and qPCR were performed using TaqMan primers from Applied Biosystems. The following TaqMan primers were used: PPIA (Hs04194521_s1), CHGA (Hs00900370_m1), NEUROD1 (Hs01922995_s1), INS (Hs02741908), PAX6 (Hs01088114_m1), CFTR (Hs00357011_m1), NKX2-2 (Hs00159616_m1) and NEUROG3 (Hs01875204_s1). RT-qPCR results are presented in arbitrary units (AU) relative to expression of the control gene PPIA. qPCR was performed on QuantStudio (Thermo Fisher Scientific) following the manufacturer's instructions.
RNA extraction and sequencing of human fetal pancreatic cell populations
RNA-seq was performed on cells from three independent pancreata at 9 WD. Cells were FACS sorted into TRIzol Reagent (Life Technologies, 15596018) according to the expression of surface markers EPCAM, CD45, CD31, GP2, CD142, ECAD and SUSD2 (see Fig. S1 for gating strategy). For each population, A, B, C and D, we purified 1228, 2300, 1916 and 5244 cells from pancreas 1; 892, 1600, 993 and 1000 cells from pancreas 2, and 331, 1021, 433 and 548 cells from pancreas 3, respectively. RNA was extracted from each population using TRIzol Reagent according to manufacturer's guidelines. RNA was quantified and assessed for quality using the 2100 Bioanalyzer (Agilent). Library preparation and sequencing was performed at the Oxford Genomics Centre (Wellcome Trust Centre for Human Genetics, University of Oxford). Libraries were prepared using the SMARTer Ultra Low Input HV kit (634820, Clontech). All libraries were multiplexed and sequenced over multiple lanes of Illumina HiSeq4000 as 75-nucleotide paired-end reads to a minimum depth of 20 million read pairs.
Transcript quantification
Raw sequencing reads were aligned to the primary assemblies and 1000 Genomes phase 2 decoy sequences for human genome reference GRCh37.p13 with STAR v2.5.1, using GENCODE release 19 as the transcriptome reference (Dobin et al., 2013). Gene level read counts for differential expression analysis were quantified for all protein-coding and long non-coding transcripts present in GENCODE release 19 using featureCounts (Liao et al., 2014). For plotting and filtering purposed, we also normalized the gene counts to transcripts per million (TPM).
Differential expression analysis
Differential expression was performed on all autosomal protein-coding and long non-coding RNA genes. Only genes demonstrating consistent expression (>1 count per million) in all samples of at least one population were included in the analysis. Gene counts were normalized using voom, and genes that demonstrated significant expression differences between the populations were identified with the moderated _F_-test implemented in limma (R/Bioconductor) (Ritchie et al., 2015). Significant genes were assigned to the population corresponding to their maximum _F_-statistic. Results were corrected for multiple testing using the Benjamini–Hochberg procedure, with a false discovery rate of <1% the significance threshold.
GO term enrichment
To assess differentially expressed genes of each population for overrepresentation of GO terms, we used the g:Profiler web server with all ‘Biological Process’ (‘BP’) terms (Reimand et al., 2016). GO terms were required to contain a minimum of three and maximum of 1000 genes, with a minimum overlap of three genes between the GO term and gene sets from each population needed for inclusion. All genes studied in the differential expression analysis were used as the background set. _P_-values were adjusted for multiple testing according to Benjamini–Hochberg.
Single-cell qPCR
Live, dissociated cells were stained for and sorted according to expression of the surface markers CD142, ECAD and SUSD2 as described above. Cells derived from human fetal pancreas were additionally stained for CD45, CD31 and CD235a to exclude hematopoietic, endothelial and erythrocyte cells. EPCAM was used to exclude mesenchymal cells (CD45-CD31-EPCAM-), CD133 to exclude ductal cells (CD45−CD31−EPCAM+CD133+) and GP2 to exclude acinar cells (CD45−CD31−EPCAM+GP2hi), respectively. Single cells were sorted by FACS into individual wells of 96-well PCR plates containing NP40 Detergent Surfact-Amps Solution (Thermo Fisher Scientific), SUPERase-In RNase Inhibitor (Ambion), Superscript VILO-cDNA Synthesis reaction mix (Invitrogen) and nuclease-free water. Samples were stored at −80°C until downstream processing. Reverse transcription, specific target amplification and qPCR was performed according to the manufacturer's instructions (Fluidigm, protocol 68000088_G1, appendix B). Briefly, RNA was denatured at 65°C for 90 s, followed by the addition of Superscript VILO cDNA Synthesis enzyme mix (Invitrogen) and T4 Gene 32 Protein (New England Biolabs) for reverse transcription. cDNA was then pre-amplified for 20 cycles with TaqMan PreAmp Master Mix (Invitrogen) using a pool of all primers, followed by exonuclease I (New England Biolabs) treatment. All reactions were performed on a OneStepPlus system (Applied Biosystems). Pre-amplified cDNA was subsequently diluted 5× and mixed with 2× SSo Fast EvaGreen Supermix (Bio-Rad) and DNA Binding Dye Sample Loading Reagent (Fluidigm) before loading onto a primed 96.96 chip (Fluidigm). Primers were mixed with Assay Loading Reagent (Fluidigm) and DNA suspension buffer (Teknova) to a concentration of 5 μM. The concentration of each primer was 500 nM in the final reaction. qPCR was performed on a Biomark HD system (Fluidigm) for 30 cycles followed by melt curve generation. Primer sequences can be found in Table S3. A total of 91 primers were used, of which 86 had previously been validated (Petersen et al., 2017).
Initial processing of single-cell qPCR data and quality control
Ct values that produced melting curves beyond the validated temperature range and/or Ct values that were greater than or equal to the Limit of Detection (LOD) Ct value were treated as a non-detectable transcript/non-specific amplicon and were set to LOD Ct (LOD Ct=22 for this primer set). Cells that expressed the housekeeping genes UBC and RPL7 above the LOD Ct value or at very low or high level (greater than the mean Ct value±3× s.d.) were excluded from further analysis. Ct values were transformed into Log2Ex values (Log2Ex=LOD−Raw Ct). No-template control (NTC) and control bulk cDNA samples were included on each chip for technical quality assessment. Single-cell cDNA from three individual human fetal pancreata at 9 WD and hPSC-derived cells from populations B/BCD133− and C was equally distributed between six 96.96 chips. cDNA from three additional individual human fetal pancreata from populations DHI and DLO was equally distributed among three 96.96 chips, along with some samples representing B and C cells to enable data merging. In total, 864 samples were analyzed by single-cell qPCR distributed on nine 96.96 chips. Among these, 36 samples were controls that were excluded from downstream computational analysis. Based on the expression level for UBC and RPL7 as described above, 181 cells were excluded. Among these were all DHI and DLO samples from one pancreas that all had low/undetectable gene expression. Consequently, 683 cells were included in the final analysis. A detailed table that shows how many cells were processed from each population and pancreata can be found in Table S3.
Single-cell qPCR data analysis
Single-cell qPCR data were analyzed with a custom script in R. As the sorted populations were run on different 96.96 chips and in two different batches, chip-chip variance was analyzed using the control bulk cDNA samples included on each chip. Because of low technical variance between chips, data from all nine chips were combined without normalization. Processed single-cell qPCR data for additional hPSC-differentiation stages and human mature islet cells were obtained from a previously published dataset (Petersen et al., 2017). These data were merged with data from the present study, after checking for variation based on the expression of housekeeping genes and using ComBat to check for batch effects. The 86 primers in common for the two datasets were used for analysis (see Table S3 for the list of primers). For statistical analysis, each cell was treated as a biological replicate. Error bars indicate s.d. or s.e.m. as indicated in the figure legends. Pseudotemporal ordering of cells from human fetal pancreas was carried out using Monocle (‘monocle’ package, R), an unsupervised algorithm with a high temporal resolution that enables the visualization of bifurcation points or cell fates in pseudotime during developmental progression (Trapnell et al., 2014). A differential expression test was performed on all cells based on the population (BCD133−, C, DHI and DLO) and the top 50 significant genes were used in the Monocle ordering algorithm.
Supplementary Material
Supplementary information
Footnotes
Competing interests
M.V.D.B., M.B.K.P., N.L.B., M.H. and C.H. are or have been employees of Novo Nordisk and may hold shares in the company.
Author contributions
Conceptualization: C.R., B.S.B.-T., A.A., M.V.D.B., M.B.K.P., N.L.B., A.L.G., M.I.M., C.H., A.G.-B., R.S.; Methodology: C.R., B.S.B.-T., A.A., M.V.D.B., M.B.K.P., N.G., C.B., C.H., R.S.; Formal analysis: A.A., M.V.D.B., R.S.; Investigation: C.R., B.S.B.-T., M.B.K.P., N.G., C.B., C.H.; Resources: N.L.B., C.H., A.G.-B.; Data curation: A.A., M.V.D.B.; Writing - original draft: C.R., B.S.B.-T., A.A., A.G.-B., R.S.; Writing - review & editing: C.R., B.S.B.-T., A.A., M.V.D.B., M.B.K.P., N.L.B., N.G., C.B., A.L.G., M.H., M.I.M., C.H., R.S.; Visualization: C.R., B.S.B.-T., A.A., M.V.D.B., A.G.-B.; Supervision: A.L.G., M.H., M.I.M., C.H., A.G.-B., R.S.; Project administration: A.L.G., M.H., M.I.M., C.H., A.G.-B., R.S.; Funding acquisition: A.L.G., M.H., M.I.M., C.H., A.G.-B., R.S.
Funding
This work was supported by the HumEn project funded by the European Union's Seventh Framework Programme for Research, (602587 to R.S. and A.G.-B.), the Fondation Bettencourt Schueller (R.S.) and the Fondation Francophone pour la Recherche sur le Diabete (FFRD) (R.S.). The R.S. laboratory belongs to the Laboratoire d'Excellence consortium Revive and to the Departement Hospitalo-Universitaire (DHU) Department of Autoimmune and Hormonal Disease. M.H., M.I.M., C.H. and R.S. have also received support from the Innovative Medicines Initiative Joint Undertaking (IMI JU) (115439), resources of which are composed of financial contributions from the European Union's Seventh Framework Programme (FP7/2007-2013) and companies of The European Federation of Pharmaceutical Industries and Associations (EFPIA). This publication reflects only the authors' views and neither the IMI JU, EFPIA nor the European Commission is liable for any use that may be made of the information contained herein. C.R. is supported by the Aide aux Jeunes Diabétiques. B.S.B.-T. is supported by the Copenhagen Bioscience PhD program financed by the Novo Nordisk Fonden (NNF16CC0020994). A.A. is supported by the Danmarks Grundforskningsfond (DNRF 116 to the StemPhys project). A.G.-B.’s lab received funding from the Novo Nordisk Fonden (NNF17CC0027852). M.V.D.B. was supported by a Novo Nordisk postdoctoral fellowship run in partnership with the University of Oxford. A.L.G. is a Wellcome Trust Senior Fellow in Basic Biomedical Science (0951010, 200837). Deposited in PMC for release after 6 months.
Data availability
Sequence data for this study has been deposited at the European Genome-phenome Archive (EGA), under accession number EGAS00001003127.
References
- Afelik S., Pool B., Schmerr M., Penton C. and Jensen J. (2015). Wnt7b is required for epithelial progenitor growth and operates during epithelial-to-mesenchymal signaling in pancreatic development. _Dev. Biol._399, 204-217. 10.1016/j.ydbio.2014.12.031 [DOI] [PubMed] [Google Scholar]
- Ameri J., Borup R., Prawiro C., Ramond C., Schachter K. A., Scharfmann R. and Semb H. (2017). Efficient generation of glucose-responsive beta cells from isolated GP2(+) human pancreatic progenitors. _Cell Rep._19, 36-49. 10.1016/j.celrep.2017.03.032 [DOI] [PubMed] [Google Scholar]
- Artner I., Hang Y., Mazur M., Yamamoto T., Guo M., Lindner J., Magnuson M. A. and Stein R. (2010). MafA and MafB regulate genes critical to beta-cells in a unique temporal manner. _Diabetes_59, 2530-2539. 10.2337/db10-0190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgartner B. K., Cash G., Hansen H., Ostler S. and Murtaugh L. C. (2014). Distinct requirements for beta-catenin in pancreatic epithelial growth and patterning. _Dev. Biol._391, 89-98. 10.1016/j.ydbio.2014.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolino P., Holmberg R., Reissmann E., Andersson O., Berggren P.-O. and Ibáñez C. F. (2008). Activin B receptor ALK7 is a negative regulator of pancreatic beta-cell function. _Proc. Natl. Acad. Sci. USA_105, 7246-7251. 10.1073/pnas.0801285105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhushan A., Itoh N., Kato S., Thiery J. P., Czernichow P., Bellusci S. and Scharfmann R. (2001). Fgf10 is essential for maintaining the proliferative capacity of epithelial progenitor cells during early pancreatic organogenesis. _Development_128, 5109-5117. [DOI] [PubMed] [Google Scholar]
- Capito C., Simon M.-T., Aiello V., Clark A., Aigrain Y., Ravassard P. and Scharfmann R. (2013). Mouse muscle as an ectopic permissive site for human pancreatic development. _Diabetes_62, 3479-3487. 10.2337/db13-0554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaing M., Péault B., Basmaciogullari A., Casal I., Czernichow P. and Scharfmann R. (2001). Blood glucose normalization upon transplantation of human embryonic pancreas into beta-cell-deficient SCID mice. _Diabetologia_44, 2066-2076. 10.1007/s001250100012 [DOI] [PubMed] [Google Scholar]
- Castaing M., Guerci A., Mallet J., Czernichow P., Ravassard P. and Scharfmann R. (2005). Efficient restricted gene expression in beta cells by lentivirus-mediated gene transfer into pancreatic stem/progenitor cells. _Diabetologia_48, 709-719. 10.1007/s00125-005-1694-6 [DOI] [PubMed] [Google Scholar]
- Cebola I., Rodríguez-Seguí S. A., Cho C. H.-H., Bessa J., Rovira M., Luengo M., Chhatriwala M., Berry A., Ponsa-Cobas J., Maestro M. A. et al. (2015). TEAD and YAP regulate the enhancer network of human embryonic pancreatic progenitors. _Nat. Cell Biol._17, 615-626. 10.1038/ncb3160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogger K. F., Sinha A., Sarangi F., McGaugh E. C., Saunders D., Dorrell C., Mejia-Guerrero S., Aghazadeh Y., Rourke J. L., Screaton R. A. et al. (2017). Glycoprotein 2 is a specific cell surface marker of human pancreatic progenitors. _Nat. Commun._8, 331 10.1038/s41467-017-00561-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amour K. A., Bang A. G., Eliazer S., Kelly O. G., Agulnick A. D., Smart N. G., Moorman M. A., Kroon E., Carpenter M. K. and Baetge E. E. (2006). Production of pancreatic hormone-expressing endocrine cells from human embryonic stem cells. _Nat. Biotechnol._24, 1392-1401. 10.1038/nbt1259 [DOI] [PubMed] [Google Scholar]
- Dassaye R., Naidoo S. and Cerf M. E. (2016). Transcription factor regulation of pancreatic organogenesis, differentiation and maturation. _Islets_8, 13-34. 10.1080/19382014.2015.1075687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M. and Gingeras T. R. (2013). STAR: ultrafast universal RNA-seq aligner. _Bioinformatics_29, 15-21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaves C. J. (2015). Hematopoietic stem cells: concepts, definitions, and the new reality. _Blood_125, 2605-2613. 10.1182/blood-2014-12-570200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejarque M., Mir-Coll J., Gomis R., German M. S., Lynn F. C. and Gasa R. (2016). Generation of a conditional allele of the transcription factor atonal homolog 8 (Atoh8). _PLoS ONE_11, e0146273 10.1371/journal.pone.0146273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flasse L. C., Pirson J. L., Stern D. G., Von Berg V., Manfroid I., Peers B. and Voz M. L. (2013). Ascl1b and Neurod1, instead of Neurog3, control pancreatic endocrine cell fate in zebrafish. _BMC Biol._11, 78 10.1186/1741-7007-11-78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchsberger C., Flannick J., Teslovich T. M., Mahajan A., Agarwala V., Gaulton K. J., Ma C., Fontanillas P., Moutsianas L., McCarthy D. J. et al. (2016). The genetic architecture of type 2 diabetes. _Nature_536, 41-47. 10.1038/nature18642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgia S., Soliz R., Li M., Zhang P. and Bhushan A. (2006). p57 and Hes1 coordinate cell cycle exit with self-renewal of pancreatic progenitors. _Dev. Biol._298, 22-31. 10.1016/j.ydbio.2006.05.036 [DOI] [PubMed] [Google Scholar]
- Gradwohl G., Dierich A., LeMeur M. and Guillemot F. (2000). neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. _Proc. Natl. Acad. Sci. USA_97, 1607-1611. 10.1073/pnas.97.4.1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu G., Dubauskaite J. and Melton D. A. (2002). Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from duct progenitors. _Development_129, 2447-2457. [DOI] [PubMed] [Google Scholar]
- Heins N., Englund M. C., Sjöblom C., Dahl U., Tonning A., Bergh C., Lindahl A., Hanson C. and Semb H. (2004). Derivation, characterization, and differentiation of human embryonic stem cells. _Stem Cells_22, 367-376. 10.1634/stemcells.22-3-367 [DOI] [PubMed] [Google Scholar]
- Jennings R. E., Berry A. A., Kirkwood-Wilson R., Roberts N. A., Hearn T., Salisbury R. J., Blaylock J., Piper Hanley K. and Hanley N. A. (2013). Development of the human pancreas from foregut to endocrine commitment. _Diabetes_62, 3514-3522. 10.2337/db12-1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings R. E., Berry A. A., Strutt J. P., Gerrard D. T. and Hanley N. A. (2015). Human pancreas development. _Development_142, 3126-3137. 10.1242/dev.120063 [DOI] [PubMed] [Google Scholar]
- Jennings R. E., Berry A. A., Gerrard D. T., Wearne S. J., Strutt J., Withey S., Chhatriwala M., Piper Hanley K., Vallier L., Bobola N. et al. (2017). Laser capture and deep sequencing reveals the transcriptomic programmes regulating the onset of pancreas and liver differentiation in human embryos. _Stem Cell Rep._9, 1387-1394. 10.1016/j.stemcr.2017.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen J., Pedersen E. E., Galante P., Hald J., Heller R. S., Ishibashi M., Kageyama R., Guillemot F., Serup P. and Madsen O. D. (2000). Control of endodermal endocrine development by Hes-1. _Nat. Genet._24, 36-44. 10.1038/71657 [DOI] [PubMed] [Google Scholar]
- Jeon J., Correa-Medina M., Ricordi C., Edlund H. and Diez J. A. (2009). Endocrine cell clustering during human pancreas development. _J. Histochem. Cytochem._57, 811-824. 10.1369/jhc.2009.953307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. D. (2016). The quest to make fully functional human pancreatic beta cells from embryonic stem cells: climbing a mountain in the clouds. _Diabetologia_59, 2047-2057. 10.1007/s00125-016-4059-4 [DOI] [PubMed] [Google Scholar]
- Jonsson J., Carlsson L., Edlund T. and Edlund H. (1994). Insulin-promoter-factor 1 is required for pancreas development in mice. _Nature_371, 606-609. 10.1038/371606a0 [DOI] [PubMed] [Google Scholar]
- Lardon J., Corbeil D., Huttner W. B., Ling Z. and Bouwens L. (2008). Stem cell marker prominin-1/AC133 is expressed in duct cells of the adult human pancreas. _Pancreas_36, e1-e6. 10.1097/mpa.0b013e318149f2dc [DOI] [PubMed] [Google Scholar]
- Larsen H. L. and Grapin-Botton A. (2017). The molecular and morphogenetic basis of pancreas organogenesis. _Semin. Cell Dev. Biol._66, 51-68. 10.1016/j.semcdb.2017.01.005 [DOI] [PubMed] [Google Scholar]
- Liao Y., Smyth G. K. and Shi W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. _Bioinformatics_30, 923-930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Liu Z. and Habener J. F. (2010). Wnt signaling in pancreatic islets. _Adv. Exp. Med. Biol._654, 391-419. 10.1007/978-90-481-3271-3_17 [DOI] [PubMed] [Google Scholar]
- Liu H., Yang H., Zhu D., Sui X., Li J., Liang Z., Xu L., Chen Z., Yao A., Zhang L. et al. (2014). Systematically labeling developmental stage-specific genes for the study of pancreatic β-cell differentiation from human embryonic stem cells. _Cell Res._24, 1181-1200. 10.1038/cr.2014.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyttle B. M., Li J., Krishnamurthy M., Fellows F., Wheeler M. B., Goodyer C. G. and Wang R. (2008). Transcription factor expression in the developing human fetal endocrine pancreas. _Diabetologia_51, 1169-1180. 10.1007/s00125-008-1006-z [DOI] [PubMed] [Google Scholar]
- Martín M., Gallego-Llamas J., Ribes V., Kedinger M., Niederreither K., Chambon P., Dollé P. and Gradwohl G. (2005). Dorsal pancreas agenesis in retinoic acid-deficient Raldh2 mutant mice. _Dev. Biol._284, 399-411. 10.1016/j.ydbio.2005.05.035 [DOI] [PubMed] [Google Scholar]
- McGrath P. S., Watson C. L., Ingram C., Helmrath M. A. and Wells J. M. (2015). The basic helix-loop-helix transcription factor NEUROG3 is required for development of the human endocrine pancreas. _Diabetes_64, 2497-2505. 10.2337/db14-1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miettinen P. J., Huotari M., Koivisto T., Ustinov J., Palgi J., Rasilainen S., Lehtonen E., Keski-Oja J. and Otonkoski T. (2000). Impaired migration and delayed differentiation of pancreatic islet cells in mice lacking EGF-receptors. _Development_127, 2617-2627. [DOI] [PubMed] [Google Scholar]
- Miralles F., Czernichow P., Ozaki K., Itoh N. and Scharfmann R. (1999). Signaling through fibroblast growth factor receptor 2b plays a key role in the development of the exocrine pancreas. _Proc. Natl. Acad. Sci. USA_96, 6267-6272. 10.1073/pnas.96.11.6267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molotkov A., Molotkova N. and Duester G. (2005). Retinoic acid generated by Raldh2 in mesoderm is required for mouse dorsal endodermal pancreas development. _Dev. Dyn._232, 950-957. 10.1002/dvdy.20256 [DOI] [PubMed] [Google Scholar]
- Muñoz-Bravo J. L., Hidalgo-Figueroa M., Pascual A., López-Barneo J., Leal-Cerro A. and Cano D. A. (2013). GDNF is required for neural colonization of the pancreas. _Development_140, 3669-3679. 10.1242/dev.091256 [DOI] [PubMed] [Google Scholar]
- Nishi M., Sanke T., Nagamatsu S., Bell G. I. and Steiner D. F. (1990). Islet amyloid polypeptide. A new beta cell secretory product related to islet amyloid deposits. _J. Biol. Chem._265, 4173-4176. [PubMed] [Google Scholar]
- Nostro M. C., Sarangi F., Yang C., Holland A., Elefanty A. G., Stanley E. G., Greiner D. L. and Keller G. (2015). Efficient generation of NKX6-1+ pancreatic progenitors from multiple human pluripotent stem cell lines. _Stem Cell Rep._4, 591-604. 10.1016/j.stemcr.2015.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osipovich A. B., Long Q., Manduchi E., Gangula R., Hipkens S. B., Schneider J., Okubo T., Stoeckert C. J., Takada S. and Magnuson M. A. (2014). Insm1 promotes endocrine cell differentiation by modulating the expression of a network of genes that includes Neurog3 and Ripply3. _Development_141, 2939-2949. 10.1242/dev.104810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliuca F. W., Millman J. R., Gürtler M., Segel M., Van Dervort A., Ryu J. H., Peterson Q. P., Greiner D. and Melton D. A. (2014). Generation of functional human pancreatic β cells in vitro. _Cell_159, 428-439. 10.1016/j.cell.2014.09.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan F. C. and Wright C. (2011). Pancreas organogenesis: from bud to plexus to gland. _Dev. Dyn._240, 530-565. 10.1002/dvdy.22584 [DOI] [PubMed] [Google Scholar]
- Petersen M. B. K., Azad A., Ingvorsen C., Hess K., Hansson M., Grapin-Botton A. and Honoré C. (2017). Single-cell gene expression analysis of a human ESC model of pancreatic endocrine development reveals different paths to β-cell differentiation. _Stem Cell Rep._9, 1246-1261. 10.1016/j.stemcr.2017.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petzold K. M., Naumann H. and Spagnoli F. M. (2013). Rho signalling restriction by the RhoGAP Stard13 integrates growth and morphogenesis in the pancreas. _Development_140, 126-135. 10.1242/dev.082701 [DOI] [PubMed] [Google Scholar]
- Pictet R. L., Clark W. R., Williams R. H. and Rutter W. J. (1972). An ultrastructural analysis of the developing embryonic pancreas. _Dev. Biol._29, 436-467. 10.1016/0012-1606(72)90083-8 [DOI] [PubMed] [Google Scholar]
- Pipeleers D. (1987). The biosociology of pancreatic B cells. _Diabetologia_30, 277-291. 10.1007/BF00299019 [DOI] [PubMed] [Google Scholar]
- Piper K., Brickwood S., Turnpenny L. W., Cameron I. T., Ball S. G., Wilson D. I. and Hanley N. A. (2004). Beta cell differentiation during early human pancreas development. _J. Endocrinol._181, 11-23. 10.1677/joe.0.1810011 [DOI] [PubMed] [Google Scholar]
- Ramond C., Glaser N., Berthault C., Ameri J., Kirkegaard J. S., Hansson M., Honoré C., Semb H. and Scharfmann R. (2017). Reconstructing human pancreatic differentiation by mapping specific cell populations during development. _eLife_6, e27564 10.7554/eLife.27564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimand J., Arak T., Adler P., Kolberg L., Reisberg S., Peterson H. and Vilo J. (2016). g:Profiler-a web server for functional interpretation of gene lists (2016 update). _Nucleic Acids Res._44, W83-W89. 10.1093/nar/gkw199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezania A., Bruin J. E., Arora P., Rubin A., Batushansky I., Asadi A., O'Dwyer S., Quiskamp N., Mojibian M., Albrecht T. et al. (2014). Reversal of diabetes with insulin-producing cells derived in vitro from human pluripotent stem cells. _Nat. Biotechnol._32, 1121-1133. 10.1038/nbt.3033 [DOI] [PubMed] [Google Scholar]
- Riedel M. J., Asadi A., Wang R., Ao Z., Warnock G. L. and Kieffer T. J. (2012). Immunohistochemical characterisation of cells co-producing insulin and glucagon in the developing human pancreas. _Diabetologia_55, 372-381. 10.1007/s00125-011-2344-9 [DOI] [PubMed] [Google Scholar]
- Ritchie M. E., Phipson B., Wu D., Hu Y., Law C. W., Shi W. and Smyth G. K. (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. _Nucleic Acids Res._43, e47 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio-Cabezas O., Jensen J. N., Hodgson M. I., Codner E., Ellard S., Serup P. and Hattersley A. T. (2011). Permanent neonatal diabetes and enteric anendocrinosis associated with biallelic mutations in NEUROG3. _Diabetes_60, 1349-1353. 10.2337/db10-1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rui J., Deng S., Arazi A., Perdigoto A. L., Liu Z. and Herold K. C. (2017). β cells that resist immunological attack develop during progression of autoimmune diabetes in NOD mice. _Cell Metab._25, 727-738. 10.1016/j.cmet.2017.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russ H. A., Parent A. V., Ringler J. J., Hennings T. G., Nair G. G., Shveygert M., Guo T., Puri S., Haataja L., Cirulli V. et al. (2015). Controlled induction of human pancreatic progenitors produces functional beta-like cells in vitro. _EMBO J._34, 1759-1772. 10.15252/embj.201591058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salisbury R. J., Blaylock J., Berry A. A., Jennings R. E., De Krijger R., Piper Hanley K. and Hanley N. A. (2014). The window period of NEUROGENIN3 during human gestation. _Islets_6, e954436 10.4161/19382014.2014.954436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharfmann R., Pechberty S., Hazhouz Y., von Bülow M., Bricout-Neveu E., Grenier-Godard M., Guez F., Rachdi L., Lohmann M., Czernichow P. et al. (2014). Development of a conditionally immortalized human pancreatic β cell line. _J. Clin. Invest._124, 2087-2098. 10.1172/JCI72674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seymour P. A., Freude K. K., Tran M. N., Mayes E. E., Jensen J., Kist R., Scherer G. and Sander M. (2007). SOX9 is required for maintenance of the pancreatic progenitor cell pool. _Proc. Natl. Acad. Sci. USA_104, 1865-1870. 10.1073/pnas.0609217104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solar M., Cardalda C., Houbracken I., Martín M., Maestro M. A., De Medts N., Xu X., Grau V., Heimberg H., Bouwens L. et al. (2009). Pancreatic exocrine duct cells give rise to insulin-producing beta cells during embryogenesis but not after birth. _Dev. Cell_17, 849-860. 10.1016/j.devcel.2009.11.003 [DOI] [PubMed] [Google Scholar]
- Stoffers D. A., Zinkin N. T., Stanojevic V., Clarke W. L. and Habener J. F. (1997). Pancreatic agenesis attributable to a single nucleotide deletion in the human IPF1 gene coding sequence. _Nat. Genet._15, 106-110. 10.1038/ng0197-106 [DOI] [PubMed] [Google Scholar]
- Sugiyama T., Rodriguez R. T., McLean G. W. and Kim S. K. (2007). Conserved markers of fetal pancreatic epithelium permit prospective isolation of islet progenitor cells by FACS. _Proc. Natl. Acad. Sci. USA_104, 175-180. 10.1073/pnas.0609490104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Cacchiarelli D., Grimsby J., Pokharel P., Li S., Morse M., Lennon N. J., Livak K. J., Mikkelsen T. S. and Rinn J. L. (2014). The dynamics and regulators of cell fate decisions are revealed by pseudo-temporal ordering of single cells. _Nat. Biotechnol._32, 381-386. 10.1038/nbt.2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulachan S. S., Doi R., Kawaguchi Y., Tsuji S., Nakajima S., Masui T., Koizumi M., Toyoda E., Mori T., Ito D. et al. (2003). All-trans retinoic acid induces differentiation of ducts and endocrine cells by mesenchymal/epithelial interactions in embryonic pancreas. _Diabetes_52, 76-84. 10.2337/diabetes.52.1.76 [DOI] [PubMed] [Google Scholar]
- van de Bunt M., Lako M., Barrett A., Gloyn A. L., Hansson M., McCarthy M. I., Beer N. L. and Honoré C. (2016). Insights into islet development and biology through characterization of a human iPSC-derived endocrine pancreas model. _Islets_8, 83-95. 10.1080/19382014.2016.1182276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Maaten L. J. P. and Hinton G. E. (2008). Visualizing high- dimensional data using t-SNE. _J. Mach. Learn. Res._9, 2579-2605. [Google Scholar]
- Zhao B., Ye X., Yu J., Li L., Li W., Li S., Yu J., Lin J. D., Wang C.-Y., Chinnaiyan A. M. et al. (2008). TEAD mediates YAP-dependent gene induction and growth control. _Genes Dev._22, 1962-1971. 10.1101/gad.1664408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Law A. C., Rajagopal J., Anderson W. J., Gray P. A. and Melton D. A. (2007). A multipotent progenitor domain guides pancreatic organogenesis. _Dev. Cell_13, 103-114. 10.1016/j.devcel.2007.06.001 [DOI] [PubMed] [Google Scholar]
- Zhu Z., Li Q. V., Lee K., Rosen B. P., González F., Soh C. L. and Huangfu D. (2016). Genome editing of lineage determinants in human pluripotent stem cells reveals mechanisms of pancreatic development and diabetes. _Cell Stem Cell_18, 755-768. 10.1016/j.stem.2016.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information