Pathogenesis of taste impairment and salivary dysfunction in COVID-19 patients (original) (raw)
Abstract
Coronavirus disease 2019 (COVID-19) is a highly transmissible pandemic disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The characteristics of the disease include a broad range of symptoms from mild to serious to death, with mild pneumonia to acute respiratory distress syndrome and complications in extrapulmonary organs. Taste impairment and salivary dysfunction are common early symptoms in COVID-19 patients. The mouth is a significant entry route for SARS-COV-2, similar to the nose and eyes. The cells of the oral epithelium, taste buds, and minor and major salivary glands express cell entry factors for SARS-COV-2, such as ACE2, TMPRSS2, and Furin. We describe the occurrence of taste impairment and salivary dysfunction in COVID-19 patients and show immunohistochemical findings regarding the cell entry factors in the oral tissue. We review and describe the pathogeneses of taste impairment and salivary dysfunction. Treatment for the oral disease is also described. Recently, it was reported that some people experience persistent and prolonged taste impairment and salivary dysfunction, described as post-COVID-19 syndrome or long COVID-19, after the acute illness of the infection has healed. To resolve these problems, it is important to understand the pathogenesis of oral complications. Recently, important advances have been reported in the understanding of gustatory impairment and salivary dysfunction. Although some progress has been made, considerable effort is still required for in-depth elucidation of the pathogenesis.
Keywords: COVID-19, SARS-CoV-2, Taste impairment, Salivary gland disorder, Gustatory impairment, Xerostomia, Long COVID, Pathogenesis
1. Introduction
Coronavirus disease 2019 (COVID-19) emerged in Wuhan, China, in December 2019 and then rapidly spread worldwide. This pandemic disease is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which is an enveloped virus with a positive-sense single-stranded RNA genome. The disease varies in severity, ranging from no symptoms to critical illness. Symptoms of COVID-19 at illness onset are variable and commonly include fever or chills, cough, shortness of breath or difficulty breathing, fatigue, muscle or body ache, headache, new loss of taste or smell, sore throat, congestion or runny nose, nausea or vomiting, and diarrhea [1]. The illness severity can range from mild to critical. Mild symptoms include mild pneumonia (81%); severe symptoms include dyspnea, hypoxia, or more than 50% lung involvement on imaging (14%); and critical symptoms include respiratory failure, shock, or multiorgan system dysfunction (5%) [1,2]. The global death-to-case ratio, which reflects the number of deaths divided by the number of diagnosed cases within a given time interval, was 2.2% (2,974,830/138,340,920) as of 15 April 2021 [3].
The mouth is a significant entry route for SARS-CoV-2, similar to the nose and eyes. The cells of the oral epithelium, taste buds, and minor and major salivary glands express cell entry factors for SARS-CoV-2, such as angiotensin-converting enzyme 2 (ACE2), transmembrane protease serine 2 (TMPRSS2), and Furin [[4], [5], [6], [7], [8], [9], [10], [11], [12], [13], [14], [15], [16]]. Recently, an international multidisciplinary team conducted research to comprehend the role of the oral cavity in SARS-CoV-2 infection [16]. The study results show that SARS-CoV-2 can infect cells in the mouth and salivary glands and that infected cells harbor replicating viruses. Infected cells can be a source of pathogens to spread to other organs.
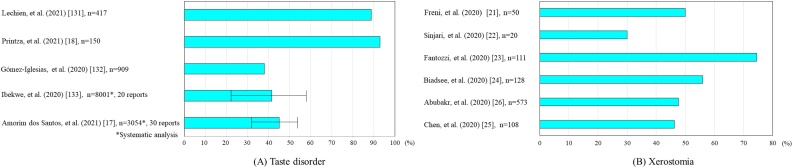
Taste impairment same olfactory deficiency, both disturbances of neurosensory perception, are early common symptoms in COVID-19 patients [[17], [18], [19], [20]]. A systematic review reported that the loss of taste occurred in 41.47% of COVID-19 patients (20 studies, 8001 COVID-19 patients). Xerostomia is another highly prevalent symptom for nearly half of patients [[21], [22], [23], [24], [25], [26]].
Although most COVID-19 patients recover completely within a few weeks, some people experience persistent and prolonged symptoms, described as post-COVID-19 syndrome or long COVID-19, after the acute illness of the infection has healed [[27], [28], [29]]. The symptoms include fatigue, dyspnea, cough, fever, headache, heart palpitation, chest pain, joint or muscle pain, depression, deteriorated mental activity, etc. Impairment of taste is also highly frequent among those long-lasting symptoms. A question-based survey at 6 months after COVID-19 reported that loss of smell or taste was the second most common persistent symptom after fatigue [29]. Some individuals also suffer from long-lasting xerostomia [18,[30], [31], [32]]. Understanding the pathogeneses of these symptoms will provide new insights for managing patients with these symptoms. However, the pathogeneses of these symptoms are not fully understood. The aim of this review is to describe hypotheses regarding their pathogeneses.
2. Taste impairment and salivary dysfunction in COVID-19 patients
2.1. Taste impairment
The taste impairment in COVID-19 patients is reported as hypogeusia, a diminished sense of taste; dysgeusia, an alteration or distortion of taste sense; and ageusia, a complete loss of taste. A systematic review by Amorim dos Santos et al. (analyzed 33 cross-sectional studies, 10,220 patients) provided a large and comprehensive analysis of the prevalence, onset, and duration of symptoms of each of the previously reported gustatory disorders (dysgeusia, hypogeusia, and ageusia) [17]. They reported that the overall prevalence of gustatory disorders was 45%, including 38% for dysgeusia, 35% for hypogeusia, 24% for ageusia, and a mean duration of 15 days, after which the patients fully recovered. With regard to COVID-19 severity, an association was observed between dysgeusia and mild and/or moderate COVID-19 cases, with an odds ratio of 2.09 compared to severe cases. Regarding regional differences in the prevalence of gustatory disorders in COVID-19 patients, 53% of North American patients, 50% of European patients, and 27% of Asian patients were affected. Furthermore, the odds ratio for males and females with gustatory disorders was 1.64. However, in this review, diagnostic methods were mixed, including telephone, online, and in-person questionnaires and medical record reviews. Limitations may exist in aggregating and analyzing these reports as a source. According to a telephone interview survey of patients in Greece by Printza et al., the median time from the onset of COVID-19 to the survey in 150 patients was 61 days. Thirty-eight percent had gustatory disorders, of which 6% were mild, 21% moderate, 5% severe, and 61% very severe. Forty-two patients (79%) regained their sense of taste by 61 days [18].
On the other hand, a cross-sectional study by Nouchi et al. reported that 29% of patients with olfactory or gustatory disorders still had persistent disease [19]. In a letter by Nguyen et al., 200 patients were randomly selected and surveyed to evaluate their recovery rate at least 6 months after onset [20]. The results showed that of 125 patients with taste and smell disturbances during the acute phase of COVID-19, 38.5% reported only partial recovery of taste, and 11.5% had no recovery at all. In addition, female patients (73.3%) were more likely to report persistent symptoms than male patients (26.7%), according to the study. This finding suggests that taste deprivation may be a possible sequelae of COVID-19 (Fig. 1A).
Fig. 1.
Patients with symptoms % [[131], [132], [133]].
(A) Taste disorder. (B) Xerostomia.
2.2. Xerostomia
The incidence of xerostomia in various reports was 50% [21], 30% [22], 74.5% [23], 56% [24], 47.6% [26], and 46.3% [25], with an average of approximately 50%. Although it is less common than olfactory dysfunction, nearly half of patients show this symptom, indicating that xerostomia is a trigger for deterioration of oral health status. There are few reports on the duration of xerostomia, but Eghbali Zarch et al. reported it to be 14 days [33]. SARS-CoV-2 is detected in saliva from the earliest stages, ACE2-positive salivary gland epithelial cells are the initial target of SARS-CoV-2, and salivary gland function may be affected in the early stages of infection [34]. In a cross-sectional study of 108 patients in Wuhan with confirmed COVID-19, 46% of patients complained of xerostomia as one of their symptoms [25].
Oral symptoms may be the initial manifestation of COVID-19 before typical symptoms, such as fever, dry cough, fatigue, and shortness of breath, occur. There are few reports of xerostomia as a sequela after recovery from COVID-19 infection; Gherlone et al. reported that dry mouth was seen in 30% of COVID-19 patients at follow-up after recovery and was significantly associated with diabetes and chronic obstructive pulmonary disease (COPD) [32]. Although there are a variety of therapeutic medicines for COVID-19, these drugs cause oral mucosal symptoms and xerostomia. For example, chloroquine/hydroxychloroquine, which is used for the treatment of malaria and certain autoimmune diseases, can cause allergic lichenoid reactions and xerostomia. The combination of lopinavir and ritonavir, both approved for the treatment of HIV, may be associated with side effects such as xerostomia, stomatitis, and oral ulcers. Interferon-β, a drug for multiple sclerosis, also can cause xerostomia as a side effect [35] (Fig. 1B).
3. Physiological aspects of taste buds and salivary gland
3.1. Physiological aspects of taste buds
The human tongue has approximately 3000 taste buds, with a vertical length of 50−80 μm and a horizontal length of 30−50 μm. Taste buds are composed of 50–90 cells in clusters, and the base is slightly wider than the apex [36]. Four cell types are present: dark-toned cells (type I cells) occupy 50–70% of taste buds. Light-toned cells (type II cells) occupy 15–30 % of the taste buds. Taste cells (type III) occupy 5–15 % of the taste buds. Type IV cells (support cells) are present at the base of the taste buds [37]. The five basic tastes currently recognized are sweet, salty, sour, bitter, and umami. In particular, sweet, bitter, and umami are transmitted by G protein-coupled (GPC) receptors in taste buds [38]. A typical sweet stimulus is sucrose, a disaccharide. In 1999, Hoon et al. identified two taste-specific GPC receptors, TR1 and TR2 (later referred to as Tas1r1 and Tas1r2) [39]. Tas1rs and Tas2rs are expressed separately in type II cells in the taste buds [40].
Bitter taste receptors are diverse: according to a review by Meyerhof, approximately 30 families of TAS2R bitter taste receptors have been identified in mammals [41]. They have evolved through adaptive diversification and are linked to chromosomal loci known to influence bitterness in mice and humans. TAS2R widely detects several bitter substances, including amides and alkaloids such as strychnine, caffeine, and quinine, and certain amino acids, urea, fatty acids, phenols, amines, esters, and other compounds that can also induce bitterness. Furthermore, some mineral salts, such as potassium, magnesium, and calcium salts, can also taste bitter. Therefore, the persistence of bitterness in the loss of taste in COVID-19 may be explained by the fact that it is sensitive to a variety of substances.
The acceptance of umami taste is based on umami receptors, which are heterodimers composed of TAS1R1 (taste receptor type 1, member 1) and TAS1R3 [42], which collectively detect several amino acids, such as umami-associated l-glutamate and 5′-ribonucleotides [38].
As for the reception of salty taste, it was found to be the passage of sodium ions through specific transport pathways in the apical region of taste buds [43]. Sodium chloride (NaCl) evokes salty taste via amiloride-sensitive and amiloride-insensitive mechanisms in taste cells [40], and ENaC alpha (also known as SCNN1A) encodes a putative amiloride-sensitive salt taste receptor. However, the specific molecular features of cells mediating sodium-induced taste have not yet been well defined, although there are reports [44] that taste is mediated by type III cells in the taste buds. Similarly, acid taste perception is also mediated by type III cells in the taste buds. The involvement of amiloride-sensitive epithelial sodium channels (ENaC), GPR4, a proton-sensitive GPCR, and the transient receptor potential (TRP) family channel PKD2L1 and its related partner PKD1L3 has been shown, but the details are still unclear [38].
The renin-angiotensin system (RAS) is a signaling pathway involved in the regulation of blood volume, natriuresis, blood pressure, blood flow, and homeostasis in response to various stimuli. It is a signaling pathway involved in regulation (homeostasis) [45]. Updated RAS pathways, which include ACE2, are available in the following reference [46]. Generally, ACE2 metabolizes angiotensin I to angiotensin (1–9) or angiotensin II (Ang II) to angiotensin (1–7) (Ang (1–7)), which contribute to vasodilation. Moreover, ACE2 is known to convert angiotensin A to alamandine, an angiotensin-related peptide that contributes to aortic relaxation. Shigemura et al. [47] reported that ACE2, which metabolizes Ang II to Ang (1–7) and is encoded by the GPC receptor Mas and counteracts the effects of Ang II, was found not only in filiform and vallate papillae but also in the lingual epithelium [48]. These results suggest that ACE2 may be localized in taste buds. These results further suggest that Ang II produced locally in taste buds is rapidly degraded by ACE2, supporting the hypothesis of short-term regulation of taste sensitivity (e.g., GPC receptors that are responsible for bitterness) by the local RAS.
3.2. Physiology of salivary gland
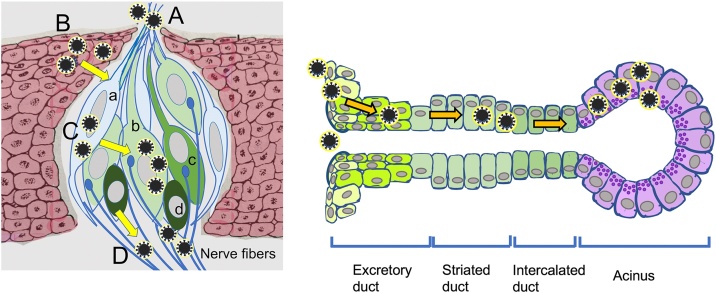
The glands that secrete saliva consist of the parotid, submandibular, and sublingual glands, which are called the three major salivary glands. In addition, there are minor salivary glands that are distributed throughout the submucosa of the oral cavity, excluding the gingiva and the anterior part of the hard palate [49,50]. The parotid gland secretes serous saliva, the submandibular gland secretes a mixture of serous and mucous saliva, and the sublingual gland secretes predominantly mucous saliva. The minor salivary glands (with the exception of the lingual serous gland, known as Ebner's gland) principally secrete mucous saliva [49,50]. The salivary glands are controlled under the autonomous nervous system. Stimulation of either the parasympathetic or sympathetic nervous system stimulates salivary gland secretion, but the effect of parasympathetic stimulation is stronger and longer lasting. Parasympathetic innervation of the parotid gland originates from the preganglionic fibers of the glossopharyngeal nerve synapsing from the inferior salivary nucleus and relays into the otic ganglion. Postganglionic fibers of the glossopharyngeal nerve convey to the parotid gland, passing via the auriculotemporal nerve. The parasympathetic innervation of the submandibular and sublingual glands originates in the superior salivary nucleus, where the preganglionic fibers of the facial nerve synapse into the submandibular ganglion and relay as postganglionic fibers to the submandibular and sublingual glands. The role of these autonomic nerves may lead to the development of future treatments for chronic xerostomia and salivary gland atrophy [49,50]. Saliva has several mucosal protectant, buffering, tooth remineralization, antibacterial, tissue repair, digestive, and gustatory aid functions. When saliva production is reduced, oral mucosa and tooth protection, buffering, antibacterial, taste alteration, digestion, and tissue repair activities are diminished. Due to the lack of innate antimicrobial defenses, xerostomia increases the risk of tooth decay and bacterial and fungal infections [[49], [50], [51]].
4. SARS-CoV-2 cell entry
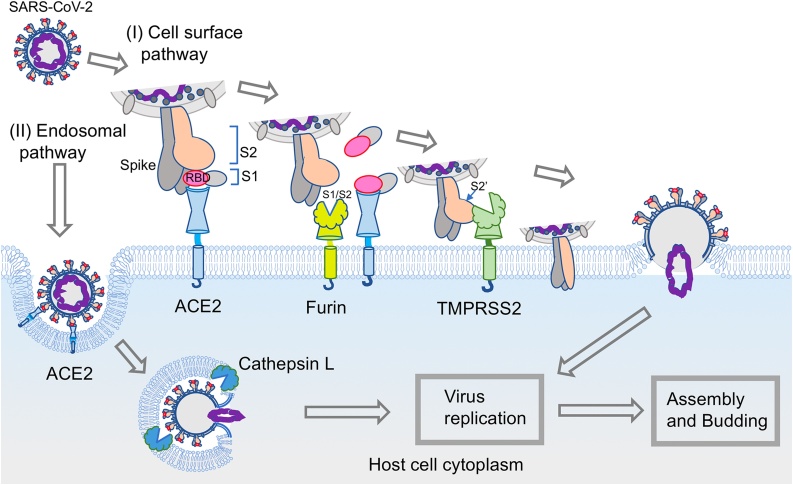
SARS-CoV-2 enters cells through host membrane-bound peptidases, ACE2 and TMPRSS2 (pathway I, Fig. 2). This entry mechanism is similar to that of SARS-CoV-1, which caused an outbreak of severe acute respiratory syndrome in 2003. Spike proteins of SARS-CoV-2 bind to ACE2 first as the receptor. The spike of the virus is a trimer of glycoproteins that consist of an S1 subunit and an S2 subunit. The S1 subunit contains an ACE2 recognition motif in the receptor-binding domain (RBD) [52]. After viral attachment to the host ACE2, the spike protein subunit S1 is proteolytically cleaved between the S1 and S2 domains (S1/S2) and disassociated by Furin, which is a proprotein convertase localized to the host cell membrane. Subsequently, TMPRSS2 cleaves the S2′ site of the S2 subunit. These cleavages cause a series of conformational changes in the spike protein, and the structural change in the S2 subunit has an important role in membrane fusion, which results in transfer of the viral RNA into the host cell cytoplasm [[53], [54], [55], [56]].
Fig. 2.
Model of the SARS-CoV-2 entry mechanism. SARS-CoV-2 utilizes two host entry routes. One is a surface membrane pathway (I), and the other route is the endosomal pathway (II). (I) In the surface membrane pathway, the RBD of the viral spike protein (S1 subunit) initially binds to the host receptor ACE2. Subsequently, Furin cleaves the spike protein at the S1/S2 site, and then the S1 subunit is dissociated from the rest of the spike protein. Another membrane-bound protease, TMPRSS2, then cleaves at the S2′ site of the S2 subunit to expose the fusion peptide for host cell membrane fusion. These events lead to a series of conformational changes that result in fusion between the viral envelope and the host cell membrane. The viral genome is released into the host cell cytoplasm. (II) Following interaction of the spike protein with the host cell receptor ACE2 on the cell membrane, the virus is endocytosed. The spike protein is processed by cathepsin L for cleavage to S1 and S2 in the endosome, which allows fusion of the viral membrane with the endosomal membrane. After that, the virus genome is released.
SARS-CoV-2 can utilize another entry route for infection (pathway II, Fig. 2), in which S1/S2 site cleavage of the spike protein prior to invading the host cell membrane is not required [57]. This endosomal route, clathrin- and non-clathrin-mediated endocytosis, is accomplished without plasma membrane proteases, such as TMPRSS2 [58,59]. After severe acute respiratory syndrome coronavirus (SARS-CoV) binds to ACE2 and internalizes into endosomes, cathepsin L is essential for fusion between the virus envelope and the endosome vesicular membrane [58]. SARS-CoV-2 is also able to internalize and release its viral genome via this endosomal route.
Coronaviruses, including SARS-CoV-2, can enter host cells via both routes, with a preference for the cell surface pathway over the endosomal pathway [60,61]. Many cell line-based studies show that the viral tropism of SARS-CoV-2 for various cell types likely reflects the differential expression of key host proteins involved in viral attachment and entry [61,62]. The varying expression of the SARS-CoV-2 cellular receptor ACE2 and proteases, such as Furin, TMPRSS2, cathepsin L, elastase, and trypsin, affects the infectivity and clinical symptoms of the disease.
Coronaviruses have fewer mutations than most RNA viruses because they employ genetic proofreading machinery during replication. However, since the pandemic began in 2019 and was caused by the original Wuhan-Hu-1 strain, many mutational changes in SARS-CoV-2 have been reported. Most are single-nucleotide mutations occurring in the gene encoding the spike glycoprotein. The first major variant, called D614G, an aspartic acid-to-glycine substitution at position 614 of the spike protein, appeared in the spring of 2020, and thereafter, the original strain was replaced by the variant. After that, other variants, B.1.1.7 (Alpha variant, the so-called UK strain, N501Y), B.1.351 (Beta variant, the so-called South African strain, N501Y.V2), P.1 (the so-called Brazil strain), B.1.617.2 (Delta variant, the so-cold Indian strain) and miscellaneous variants (20E [EU1], 20A [EU2], etc.), have been identified and led to concerns about increased infectivity and transmission [63,64]. The D614G mutation has become common to nearly all variants. This mutation is likely to make spike proteins more or less able to bind ACE2. The resulting conformational changes in the variants cause cell entry enhancement [65] (Fig. 2).
5. Cell entry factors for SARS-CoV-2 in oral and maxillofacial tissues
5.1. Angiotensin-converting enzyme 2 (ACE2) expression in oral and maxillofacial tissues
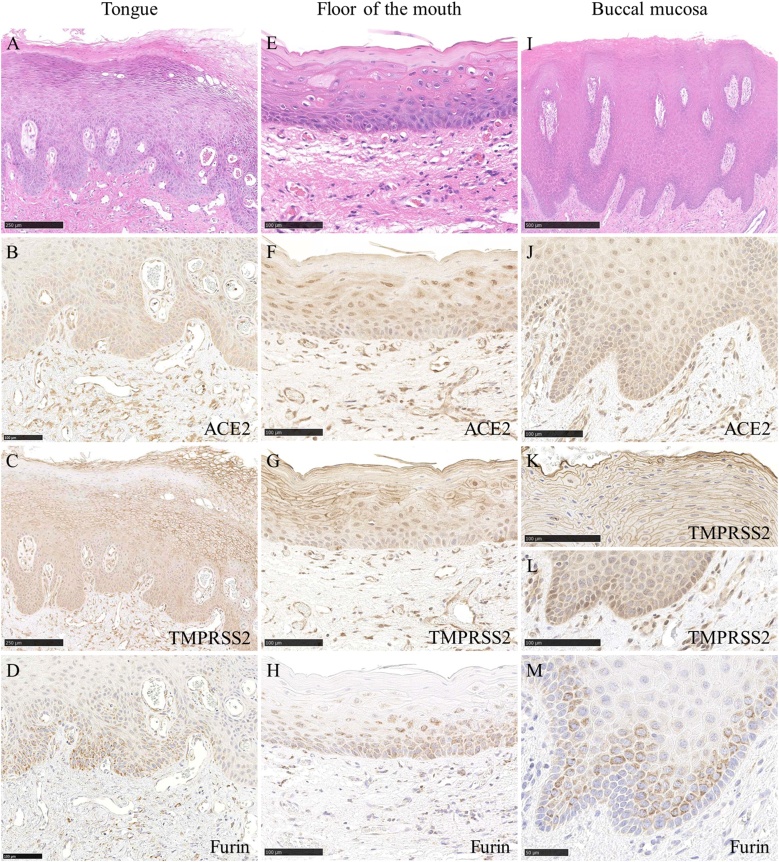
The SARS-CoV-2 receptor on human cells is ACE2, which is the same as the SARS-CoV receptor [55,[66], [67], [68], [69], [70]]. ACE2 is abundantly expressed in a variety of cells residing in many different human organs, such as the lung, small intestine, kidney, and skin [71,72]. Xu et al. reported the expression of ACE2 in oral tissue using a public RNA-sequencing database [8]. Since then, the expression of ACE2 in the oral mucosal epithelium has been reported, including in our report (31 of 36 cases, 86.1%) [[9], [10], [11],[13], [14], [15]] (Fig. 3B, F, J).
Fig. 3.
Expression of ACE2. Expression of ACE2 was observed in the nuclei and cytoplasm of the spinous and basal cell layers of the epithelium and endothelial cells of the tongue (B), oral floor (F) and buccal mucosa (J). Expression of TMPRSS2 was observed in the cell membrane of the spinous cell layers of the epithelium and endothelial cells of the tongue (C), cell membrane of the spinous cell layers and nuclei of the spinous and basal cell layers, endothelial cells of the oral floor (G), cell membrane of the spinous cell layers and nuclei of the basal cell layers, and endothelial cells of the buccal mucosa (K, L). Expression of Furin was observed in a dotted pattern and the cytoplasm of the spinous and basal cell layers of the epithelium of the tongue (D), oral floor (H) and buccal mucosa (M). Scale bars: A; 250 μm, B; 100 μm, C; 250 μm, D-H; 100 μm, I; 500 μm, J–L; 100 μm, M; 50 μm.
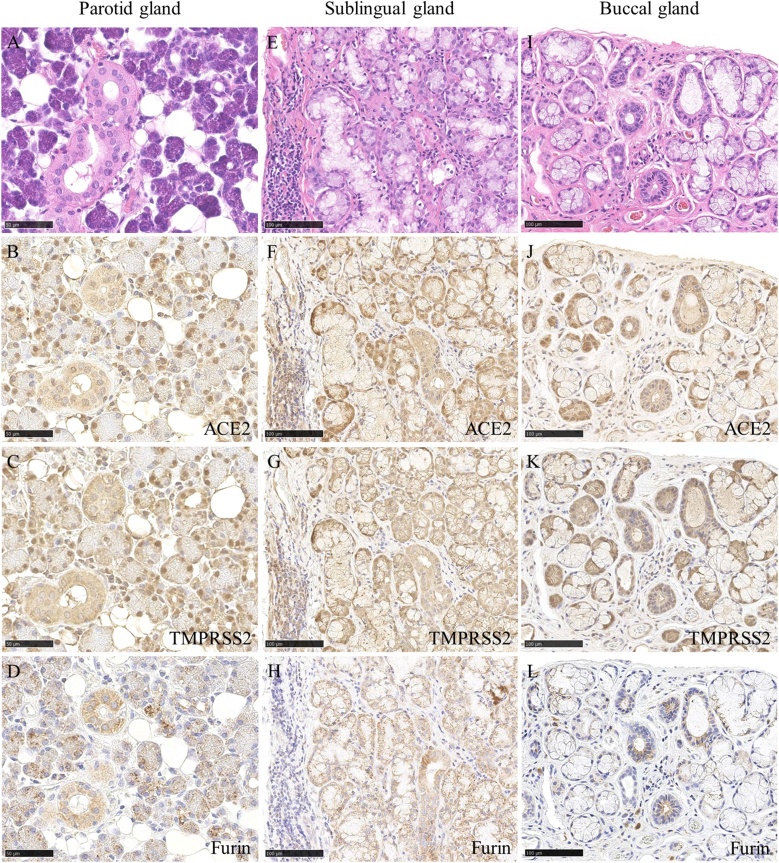
It has been reported that saliva is the cause of SARS-CoV-2 infection. This hypothesis suggests that oral tissue, including salivary glands, plays a role as an organ for viral growth in subclinical SARS-CoV-2 infections [73,74]. Accordingly, the expression of ACE2 in the salivary glands (22 of 25 cases, 88.0%) and ducts (26 of 27 cases, 96.3%) has been reported, including in our report [[9], [10], [11],[13], [14], [15]]. We reported that ACE2 expression occurs in most of the serous cells but very few mucous cells [15] (Fig. 4B, F, J).
Fig. 4.
Expression of ACE2 was observed in the serous cells, ductal epithelium and adipocytes of the parotid gland (B), serous demilunes, ductal epithelium and endothelial cells of the sublingual gland (F) and the buccal gland (J). Expression of TMPRSS2 was observed in the serous cells, ductal epithelium and adipocytes of the parotid gland (C), serous demilunes, ductal epithelium and endothelial cells of the sublingual gland (G) and the buccal gland (K). Expression of Furin was observed in the serous cells and ductal epithelium of the parotid gland (D), serous demilunes, ductal epithelium of the sublingual gland (H) and the buccal gland (L). Scale bars: A–D; 50 μm, E–L; 100 μm.
Expression of ACE2 in the taste buds of the tongue has been reported [9]. Han et al. reported the single-cell profiles of tongue tissues with typical gene markers of taste buds, such as TAS1R3, TAS2R4, TAS2R14, SNAP25, and NCAM1. They reported that the distribution of ACE2-positive cells is correlated with that of taste-related gene-marked cells [12]. Type II taste cell marker genes include TAS1R2 and TAS1R3 for sweet taste, TAS1R1 and TAS1R3 for umami taste, and TAS2Rs for bitter taste, and type III taste cell marker genes include NCAM1 and SNAP25. The distribution of type II and type III marker genes in the tongue suggests the coexistence of ACE2 and individual taste cells [38,75]. Wang reported that ACE2 is enriched in a subpopulation of epithelial cells in nongustatory filiform papillae but not in taste papillae or taste buds of adult mice [5].
The expression of ACE2 in vascular endothelial cells and adipocytes has been reported [66,71,76]. We reported that ACE2 expression was found in capillary endothelial cells (55 of 63 cases, 87.3%) and adipocytes (36 of 44 cases, 81.8%) that nourish the parotid and minor salivary glands and oral mucosal lamina propria [15] (Figs. 3B, F, J, 4 B, F, J). Adipocytes secrete various types of adipokines and are potential targets and reservoirs for SARS-CoV-2. Although the amount of adipose tissue in the parotid stromal and oral lamina propria is limited, it may contribute to the proinflammatory changes that underlie COVID-19 progression.
5.2. TMPRSS2 expression in oral and maxillofacial tissues
The expression of TMPRSS2 in the esophagus, jejunum and ileum has been reported [71]. It has been reported that TMPRSS2 is expressed in the oral mucosal epithelium and the salivary glands [6,9,13,14]. The expression of TMPRSS2 was shown to be stronger in serous acini than in mucous acini [14]. TMPRSS2 expression has been reported to be positive in the salivary gland duct [9]. However, it has been reported that ACE2 expression is absent in the striated ducts [14]. The expression of TMPRSS2 in the taste buds of the tongue has been reported [9]. Our immunostaining revealed TMPRSS2 expression in the tongue, buccal and oral floor mucosal epithelium, serous glands, salivary gland ducts, vascular endothelial cells, and adipocytes (Figs. 3C, G, K, L, 4 C, G, K).
5.3. Furin expression in oral and maxillofacial tissues
The expression of Furin in the lachrymal glands, colon, liver, and kidneys has been reported [77]. It has been reported that Furin is expressed in the oral mucosal epithelium [7,9]. The expression of Furin in the taste buds of the tongue and the salivary glands (serous glands) has been reported [9]. It has been reported that Furin is expressed in the endothelial cells of oral mucosal tissues [7]. Our immunohistochemical staining revealed Furin expression in the tongue, buccal and oral floor mucosal epithelium, serous gland, salivary gland duct, and vascular endothelial cells but not in adipocytes (Figs. 3D, H, M, 4 D, H, L).
5.4. Cathepsin expression in oral and maxillofacial tissues
Cathepsin B and L are abundantly expressed in many different human organs, such as the minor salivary glands, lung, small intestine, liver, kidney, and heart [72]. The expression of Cathepsin L between the epithelium and the adjacent subepithelial connective tissue in periodontal disease has been reported [78].
6. Pathogenesis of taste impairment and salivary gland disturbances
6.1. SARS-CoV-2 damages infected cells
Ultrastructural studies show that SARS-CoV-2 damages cells with viral distribution within the cytoplasm, and many virus particles are encapsulated in cytoplasmic vesicles. Autophagic vacuoles and fragmented mitochondria were detected in a coculture experiment using human pulmonary alveolar epithelial cells [79]. SARS-CoV-2 infection experiments in organotypic human airway epithelial culture showed findings similar to those of the cell culture study, with extensive apoptosis observation [80]. Ultrastructural autopsy findings in endothelial cells and alveolar cells in SARS-CoV-2-infected patients have been reported [81]. SARS-CoV-2 replication triggers the release of proinflammatory cytokines and chemokines, and cell death is induced in airway epithelial cells, alveolar epithelial cells, and vascular endothelial cells, in which high ACE2 expression is observed [82,83].
6.2. How does SARS-CoV-2 cause damage to gustatory sensory cells?
ACE2 is expressed in the taste buds and the peripheral oral mucosa, stratified squamous epithelium of the dorsal tongue, and gingiva in humans [9]. In a mouse study, ACE2 mRNA was expressed in fungiform and circumvallate papillae [47]. Replication of SARS-CoV-2 in infected gustatory cells in the taste bud can generate inflammation and eventually destroy the cells. This direct cell damage may cause malfunction of the gustatory system. However, a study showed that gustatory cells in taste buds do not express ACE2. Wang et al. performed a single-cell RNA-sequencing analysis of adult mouse tongue epithelial cells, and ACE2 mRNA was enriched in the basal region of nongustatory filiform papillae but not in the taste papillae or taste buds. The authors hypothesized that taste loss in COVID-19 patients is unlikely to be caused by an initial direct infection of SARS-CoV-2 in taste bud cells [5]. The virus could invade through ACE2-expressing stratified squamous epithelium of the dorsal tongue and filiform papillae in the vicinity of the taste bud and eventually progress to gustatory cell infection.
Interestingly, a similar indirect infection mechanism is reported in the olfactory system. Nasal mucosa biopsy specimens of chronic rhinosinusitis patients were used for an ACE2 immunohistochemical study. High levels of ACE2 were found in nasal cells, which are called sustentacular cells. These nonneural cells are located in the olfactory neuroepithelium. The cells in the olfactory neuroepithelium had a 200-fold to 700-fold increase in ACE2 protein levels compared to those in other nose and tracheal cells. However, olfactory sensory neurons in the olfactory neuroepithelium showed an absence of ACE2 protein expression [84]. In a study of postmortem tissue from COVID-19 patients, quantitative PCR with reverse transcription, immunohistochemistry, and electron microscopy analyses revealed that viral RNA was most common in the olfactory mucosa, which is composed of the olfactory epithelium, olfactory neurons, sustentacular cells, Bowman's glands, and blood vessels. Electron microscopic analysis of the olfactory mucosa from one individual with a high viral RNA load revealed intact viral particles in the olfactory mucosa. Furthermore, SARS-CoV spike protein has been detected in olfactory sensory neurons, which do not express ACE2 [85,86]. SARS-CoV-2 therefore infects the olfactory mucosa and possibly infects olfactory sensory neurons in an indirect manner (Fig. 5).
Fig. 5.
Hypothetical SARS-CoV-2 cell entry mechanism in the taste bud and salivary gland.
Taste bud (left). A; Microvilli of taste sensory cells allow SARS-CoV-2 entry into the cells. B; Non-ACE2-expressing gustatory cells are infected through ACE2-positive neighboring cells. C; SARS-CoV-2 directly invades taste receptor cells via cell surface ACE2 and TMPRSS2 expression. D; SARS-CoV-2 neuroinvasion can occur at the neural-mucosal interface by transmucosal entry via regional nervous structures. (a), Type I cell; (b), Type II cell; (c), Type III cell; (d) basal cell.
Salivary gland (right). SARS-CoV-2 initially enters epithelial cells close to the salivary duct orifice and/or lining salivary gland ducts through ACE2 binding. TMPRSS2 and Furin are also expressed in salivary gland ducts. Secretary cells in acinus are eventually infected with the virus.
6.3. How does indirect infection occur in gustatory cells?
One possible mechanism is direct cell-to-cell viral transmission. The actin–myosin cytoskeletal system, through tunneling nanotubes (TNTs), consists of filamentous cellular projections that facilitate intracellular exchange of cellular organelles, proteins, and substances. Many viruses can use TNTs for transmission from infected cells to neighboring cells [87]. A cell culture infection model of SARS-CoV-2 using the Vero cell line (kidney epithelial cells, African green monkey) was used for scanning electron microscopic study. Viral particles were found around cell surface membrane ruffles and filopodium-like structures. Other viral particles were wrapped with thin cellular projections that resembled nanotubes. Membrane bridges that connected two cells showed the presence of virus particles on their surface [88]. This finding may explain why non-ACE2-expressing gustatory cells are infected through ACE2-positive neighboring cells (Fig. 5).
6.4. Is SARS-CoV-2 neuroinvasive?
It is known that human coronaviruses are neuroinvasive and neurotropic and can induce overactivation of the immune system [89,90]. Additionally, similar to pathogens of two preceding epidemics (SARS-CoV-1 and Middle East respiratory syndrome coronavirus (MERS-CoV)), SARS-CoV-2 also exhibits neuroinvasion [91]. Neurological manifestations, ranging from anosmia, dysgeusia, headache, encephalitis, etc., in patients with COVID-19 are one of the main characteristics of the disease [92,93]. SARS-CoV-2 infection may directly damage the nervous system in a cytopathic manner [94]. SARS-CoV-2 neuroinvasion can occur at the neural-mucosal interface by transmucosal entry via regional nervous structures [85,95]. This mechanism may be the case for infection of gustatory neurons in the taste buds and subsequent alterations of taste perception. However, it remains to be confirmed whether this concept is applicable for direct neural invasion of SARS-CoV-2 [96,97] (Fig. 5).
6.5. Does cellular morphology affect SARS-CoV-2 infection?
Viruses, including coronaviruses, can attach to the cilia of epithelial cells [98]. An ultrastructural study regarding the nasal mucosal changes in common cold patients was reported. The loss of cilia and ciliated cells was found in a previous study [99]. Another transmission electron microscopic study in a human subject showed that coronaviruses adhered to the microvilli and cilia in nasal epithelial cells [100]. In a SARS-CoV-2-inoculated macaque experiment, the virus antigen was present in ciliated nasal epithelial cells [101]. Murine leukemia virus, with a single-stranded RNA genome, can induce lateral movement along microvilli of polarized epithelial cells before cell entry [102]. Similar to nasal epithelial cells, taste cells possess microvilli at the apical end of the cells. The microvilli protrude from the pores of taste buds toward the oral cavity to capture tastants [103,104]. It is presumed that microvilli of taste sensory cells allow SARS-CoV-2 entry into the cells (Fig. 5).
6.6. Short recovery time of taste impairment
Many patients complain of disturbance of taste as an initial symptom accompanied by disturbance of smell in their COVID-19 course [105]. The recovery time of most patients is short, varying from 4 to 17 days [[106], [107], [108], [109]]. Sensory impairments are primed at initial entry sites for the virus, for example, the oral and nasal regions. The taste buds on the dorsum surface of the tongue afford virus entry, and susceptible taste sensory cells are directly or indirectly infected by the virus. Taste buds contain tightly packed taste receptor cells with life spans of 5–20 days in mammals [110]. Thus, the damaged taste sensory cells tend to recover quickly, indicating that taste stem cells may not be completely destroyed. The most likely explanation for the characteristic symptoms and the time course is that the virus infection triggers inflammation in the taste sensory structures and taste buds and that the subsequent damage to taste receptor cells is reparable.
6.7. Long-lasting taste impairment
Some patients suffer from a long-lasting impairment of smell and taste [18,30,31]. Approximately 10% of patients in a two-month follow-up survey [18] and 11% of patients in a 6-month survey of infected individuals did not show any recovery, and only partial recovery was present in 30% of the cases [31]. It is interesting that for these chemosensory systems, smell and taste, deficiency occurs simultaneously in COVID-19; furthermore, similar long-lasting symptom rates are reported for both. An inflammatory collapse of the taste receptor apparatus may not be cause for the delay of recovery or long-lasting deficiency. The neuroinvasive or neurotropic properties of SARS-CoV-2 may be relevant to sensory sequelae. A postmortem study of COVID-19 patients suggested one possibility of virus entry in the nervous system via the neural-mucosal interface in the olfactory mucosa [85]. Taste buds can also be another virus entry point to neurons. Taste buds are packed with 50–100 taste cells, which are anatomically classified into four types: type I, supporting cells; type II, cells for sweet, bitter, and umami sensation; type III, cells for sour sensation; and type IV, stem cells. Cooper et al. reviewed the relationship between SARS-CoV-2 infection and taste sensory cells. In their review, studies and reports indicated that type III taste receptor cells in mice express ACE2, whereas type II taste receptor cells express less ACE2. Type II and type III cells express little to no TMPRSS2. Cathepsins, which are proteases that cleave the SARS-CoV-2 spike protein, are abundant in these two cell types [96]. Although type I and type IV taste sensory cells have not been investigated, it is interesting that SARS-CoV-2 may have a cytotropic nature for gustatory sensing disturbances. Forty-one taste-deficient COVID-19 patients were prospectively surveyed, and it was found that a significant loss of sour (33.3%), salty (17.9%), and sweet and bitter tastes (10.3%) were recognized [111]. These results may reflect virus neurotropic invasion and possible nervous system damage. Each different gustatory sensory cell (for sweet, bitter, umami, and sour sensors) connects each appropriate taste partner ganglion neuron in a taste bud. SARS-CoV-2 may invade these afferent neurons, and the colonization by the virus interferes with sensory nerve conduction for a long time.
There are many examples of long-term sequelae after viral infection. Measles, Ebola virus, Chikungunya virus, and Epstein-Barr virus have the potential to lead to autoimmune reactions [112,113]. Although more research is required to elucidate the pathogenesis of long COVID, another hypothesis for the persistence of the long-lasting impairment may include a disturbed immune response in neuron compartments [28,114].
6.8. How does SARS-CoV-2 affect xerostomia?
The expression of ACE2 in minor salivary glands was shown to be higher than that in lungs [115]. In an animal study using Chinese macaques, ACE2 was found to be expressed in epithelial cells lining salivary gland ducts. After intranasal inoculations of a pseudovirus for SARS-CoV-1 that targets the same entry factor, ACE2, as SARS-CoV-2, epithelial cells lining salivary gland ducts were productively infected by the pseudovirus [116]. The results suggested that SARS-CoV-2 could infect salivary gland cells. Salivary gland specimens from COVID-19 patient autopsies were studied with ACE2 and SARS-CoV-2 spike in situ hybridization (ISH) and immunohistochemistry [16]. Chronic sialadenitis, including focal lymphocytic sialadenitis, was the most common feature. SARS-CoV-2 mRNA was detected in ACE2-expressing ducts and acini of the minor and parotid salivary glands. Viral transcript expression was regionally dependent on the acinar unit. Additionally, in an acutely infected individual, the replication of SARS-CoV-2 was confirmed. Sialadenitis was correlated with the T cell response, and mild-to-moderate sialadenitis was demonstrated with focal lymphocytic inflammation and epithelial injury. These study results may explain the cause of xerostomia. SARS-CoV-1 RNA can be detected in saliva before disease-associated lung lesions appear [115]. Salivary gland infection with SARS-CoV-2 may initiate in an early stage of the disease. A similar phenomenon has been found in clinical courses of xerostomia among COVID-19 patients. The reduced salivary flow may partially affect taste impairment (Fig. 5).
6.9. Varying severity and time course of impairment of taste and disturbed salivary gland function
The SARS-CoV-2 viral load of nasopharyngeal swab samples has been shown to correlate with the severity of COVID-19 [117]. Plasma levels of interferons and cytokines have been observed to correlate with nasopharyngeal viral load. The viral load correlated significantly with the levels of IFNα, IFNγ, TNF and tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) in COVID-19 patients [83]. Viral load may affect the severity and time course of taste impairment and disturbed salivary gland function.
Cytotropic infection with SARS-CoV-2 is largely dependent on the expression of ACE2 and other entry factors, as shown in Fig. 2. Type 2 diabetes is one of the major risks for more severe COVID-19. Notably, ACE2 expression is elevated in patients with type 2 diabetes for metabolic adaptation [118,119]. This aspect implies that the amount of ACE2 expression in taste buds and salivary glands may affect the severity of their impairment and malfunction, though it is not clearly understood what pathological mechanisms cause the severity differences including a variety of taste impairment types such as hypogeusia, dysgeusia, and ageusia.
In general, cellular invasion and replication of SARS-CoV-2 cause pyroptosis that triggers secretion of proinflammatory cytokines and chemokines, including IL-6 and IP-10, macrophage inflammatory proteins from neighboring epithelial cells, fibroblasts, endothelial cells, and macrophages [120]. These proteins attract monocytes, macrophages and T cells to the site of infection and promote further inflammation. T cell-based cellular immunity plays an important role in the early phase of SARS-CoV-2 infection to clear the virus and resolve the disease, and one of the functions is the production of IFNγ [114]. In a study of autopsy and outpatient samples of COVID-19 salivary glands, T lymphocytic inflammation (CD3) with increased proportions of B lymphocytes (CD20) and infiltration were predominant in focal lymphocytic sialadenitis [16]. These inflammatory and immune responses to SARS-CoV-2 in taste buds and salivary gland tissues may vary in individuals, including based on sex, age, and genetic variation.
7. Treatment for oral problems in COVID-19 patients
7.1. Treatment recommendation for taste disorders
The duration of dysgeusia varies from 4 to 17 days [[106], [107], [108], [109]]. Seventy-nine percent of patients reportedly improve and do not require immediate treatment. However, it has been reported that 11.5% of patients do not recover at all and have residual disability. In particular, SARS-CoV-2 exhibits neural invasion [91]. Therefore, neurostimulants (steroids, B vitamins, and ATP) may be expected to treat taste disorders in COVID-19 patients. Levy [121] provided a commentary on the recommended treatment for the sequelae of olfactory and gustatory abnormalities, stating that at present there are few interventions for the treatment of olfactory and gustatory dysfunction after infection but that oral and topical corticosteroids, phosphodiesterase inhibitors, nasal calcium buffers, nasal sodium citrate, nasal vitamin A, which may act to promote olfactory neurogenesis, and nerve regeneration or antiinflammatory agents are effective [122,123], which may act via regeneration or antiinflammation activity, but that there is little quality evidence to justify their routine use in clinical practice.
7.2. Treatment of the sequelae of xerostomia
There are no reports on the response of xerostomia to the sequelae of COVID-19, but infection treatment and oral hygiene have been reported to reduce the severity of the disease [124]. There are also various therapeutic agents for COVID-19, but these medications can cause oral mucosal symptoms as well as xerostomia. For example, chloroquine/hydroxychloroquine, used to treat malaria and certain autoimmune diseases, can cause allergic lichenoid reactions and xerostomia as oral symptoms. The combination of lopinavir and ritonavir, both approved for the treatment of HIV, may be associated with side effects such as xerostomia, stomatitis, and oral ulcers. Interferon-β, a medication for multiple sclerosis, also can cause xerostomia as a side effect [35]. Existing saliva secretagogues, such as cevimeline hydrochloride hydrate, pilocarpine hydrochloride, and Chinese herbal medicine, may also be promising [[125], [126], [127], [128]]. Zinc supplementation can also be a potential treatment for xerostomia as well as taste dysfunction. The reason for this is that human salivary glands express metabotropic zinc receptor/GPC receptor 39 (ZnR/GPR39), which regulates saliva secretion from the submandibular gland. However, many patients with COVID-19 show decreased serum zinc levels [129,130], and zinc deficiency is thus believed to be a contributing factor to xerostomia associated with COVID-19. However, to evaluate the treatment of xerostomia, it is necessary to perform objective evaluations, such as measuring saliva volume and analyzing salivary components, before and after COVID-19 diagnosis to show a close correlation with the virus infection [22].
8. Conclusion
Taste impairment and salivary dysfunction are high-incidence symptoms in COVID-19 patients. This symptom lasts long after the infection disappears in some people and leads to deterioration of their quality of life (QOL). This discomfort can be a serious problem for those patients. To resolve these problems, it is important to understand the pathogenesis of the disease. Much important work has been carried out to understand COVID-19 pathogenesis and develop treatments: the latest studies include immunohistochemical examination, RT-PCR, quantitative PCR, ISH, electron microscopic studies, single-cell RNA-sequencing analysis, neutralization studies, cell culture studies, organotypic tissue culture studies, animal studies, transgenic animal studies, and so on. Recently, important advances have been reported in gustatory and salivary impairment. Although some progress has been made, considerable effort is still required for the in-depth elucidation of the pathogenesis.
Conflict of interest
The authors declare that they have no conflicts of interest.
Author contributions
M.T. and Y.O. contributed to the conceptualization and design of the article. M.T. wrote introduction (Section 1), SARS-CoV-2 cell entry (Section 4), pathogenesis of taste impairment and salivary gland disturbances section (Section 6), and conclusion. Y.O. performed immunohistochemical examination and reviewed cell entry factors for SARS-CoV-2 (Section 5). K.Y. wrote taste impairment in COVID-19 patients (Section 2.1), physiological aspects of taste buds (Section 3.1), and treatment for oral problems in COVID-19 patients (Section 7). S.T. wrote salivary dysfunction in COVID-19 patients (Section 2.2), physiology of salivary gland (Section 3.2), and treatment for oral problems in COVID-19 patients (7). All authors reviewed and contributed to preparation of the final manuscript.
References
- 1.Centers for Disease Control and Prevention; 2021. Clinical presentation, summary of recent changes, interim clinical guidance for management of patients with confirmed coronavirus disease (COVID-19) updated Feb. 16, 2021, COVID-19. https://www.cdc.gov/coronavirus/2019-ncov/hcp/clinical-guidance-managementpatients.html [Accessed 30 April] [Google Scholar]
- 2.Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the chinese center for disease control and prevention. JAMA. 2020;323:1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 3.Johns Hopkins University & Medicine . 2021. Coronavirus resource center, critical trends, mortality analyses.https://coronavirus.jhu.edu/data/mortality April 15. [Google Scholar]
- 4.Roper S.D., Chaudhari N. Taste buds: cells, signals and synapses. Nat Rev Neurosci. 2017;18:485–497. doi: 10.1038/nrn.2017.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Z., Zhou J., Marshall B., Rekaya R., Ye K., Liu H.X. SARS-CoV-2 receptor ACE2 is enriched in a subpopulation of mouse tongue epithelial cells in nongustatory papillae but not in taste buds or embryonic oral epithelium. ACS Pharmacol Transl Sci. 2020;3:749–758. doi: 10.1021/acsptsci.0c00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song J., Li Y., Huang X., Chen Z., Li Y., Liu C. Systematic analysis of ACE2 and TMPRSS2 expression in salivary glands reveals underlying transmission mechanism caused by SARS-CoV-2. J Med Virol. 2020;92:2556–2566. doi: 10.1002/jmv.26045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhong M., Lin B., Pathak J.L., Gao H., Young A.J., Wang X. ACE2 and furin expressions in oral epithelial cells possibly facilitate COVID-19 infection via respiratory and fecal-oral routes. Front Med (Lausanne) 2020;7 doi: 10.3389/fmed.2020.580796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu H., Zhong L., Deng J., Peng J., Dan H., Zeng X. High expression of ACE2 receptor of 2019-nCoV on the epithelial cells of oral mucosa. Int J Oral Sci. 2020;12:8. doi: 10.1038/s41368-020-0074-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sakaguchi W., Kubota N., Shimizu T., Saruta J., Fuchida S., Kawata A. Existence of SARS-CoV-2 entry molecules in the oral cavity. Int J Mol Sci. 2020;21:6000. doi: 10.3390/ijms21176000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Descamps G., Verset L., Trelcat A., Hopkins C., Lechien J.R., Journe F. ACE2 protein landscape in the head and neck region: the conundrum of SARS-CoV-2 infection. Biology. 2020;9:235. doi: 10.3390/biology9080235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Usami Y., Hirose K., Okumura M., Toyosawa S., Sakai T. Brief communication: immunohistochemical detection of ACE2 in human salivary gland. Psychol Behav Sci Int J. 2020 doi: 10.1002/osi2.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han Q., Peng J., Xu H., Chen Q. Taste cell is abundant in the expression of ACE2 receptor of 2019-nCoV. Preprints. 2020 doi: 10.20944/preprints202004.0424.v1. https://www.preprints.org/manuscript/202004.0424/v1 . [Accessed 14 December 2020] [DOI] [Google Scholar]
- 13.Lechien J.R., Radulesco T., Calvo-Henriquez C., Chiesa-Estomba C.M., Hans S., Barillari M.R. ACE2 & TMPRSS2 expressions in head & neck tissues: a systematic review. Head Neck Pathol. 2021;15:225–235. doi: 10.1007/s12105-020-01212-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sawa Y., Ibaragi S., Okui T., Yamashita J., Ikebe T., Harada H. Expression of SARS-CoV-2 entry factors in human oral tissue. J Anat. 2021;238:1341–1354. doi: 10.1111/joa.13391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoshimura K., Toya S., Okada Y. Morphological analysis of angiotensin-converting enzyme 2 expression in the salivary glands and associated tissues. J Hard Tissue Biol. 2021;30:265–272. [Google Scholar]
- 16.Huang N., Pérez P., Kato T., Mikami Y., Okuda K., Gilmore R.C. SARS-CoV-2 infection of the oral cavity and saliva. Nat Med. 2021;27:892–903. doi: 10.1038/s41591-021-01296-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amorim Dos Santos J., Normando A.G.C., Carvalho da Silva R.L., Acevedo A.C., De Luca Canto G., Sugaya N. Oral manifestations in patients with COVID-19: a living systematic review. J Dent Res. 2021;100:141–154. doi: 10.1177/0022034520957289. [DOI] [PubMed] [Google Scholar]
- 18.Printza A., Katotomichelakis M., Valsamidis K., Metallidis S., Panagopoulos P., Panopoulou M. Smell and taste loss recovery time in COVID-19 patients and disease severity. J Clin Med. 2021;10:966. doi: 10.3390/jcm10050966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nouchi A., Chastang J., Miyara M., Lejeune J., Soares A., Ibanez G. Prevalence of hyposmia and hypogeusia in 390 COVID-19 hospitalized patients and outpatients: a cross-sectional study. Eur J Clin Microbiol Infect Dis. 2021;40:691–697. doi: 10.1007/s10096-020-04056-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nguyen N.N., Hoang V.T., Lagier J.C., Raoult D., Gautret P. Long-term persistence of olfactory and gustatory disorders in COVID-19 patients. Clin Microbiol Infect. 2021 doi: 10.1016/j.cmi.2020.12.021. S1198-743X(20)30781-30783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Freni F., Meduri A., Gazia F., Nicastro V., Galletti C., Aragona P. Symptomatology in head and neck district in coronavirus disease (COVID-19): a possible neuroinvasive action of SARS-CoV-2. Am J Otolaryngol. 2020;41 doi: 10.1016/j.amjoto.2020.102612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sinjari B., D’Ardes D., Santilli M., Rexhepi I., D’Addazio G., Di Carlo P. SARS-CoV-2 and oral manifestation: an observational, human study. J Clin Med. 2020;9:3218. doi: 10.3390/jcm9103218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fantozzi P.J., Pampena E., Di Vanna D., Pellegrino E., Corbi D., Mammucari S. Xerostomia, gustatory and olfactory dysfunctions in patients with COVID-19. Am J Otolaryngol. 2020;41 doi: 10.1016/j.amjoto.2020.102721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Biadsee A., Biadsee A., Kassem F., Dagan O., Masarwa S., Ormianer Z. Olfactory and oral manifestations of COVID-19: sex-related symptoms—a potential pathway to early diagnosis. Otolaryngol Head Neck Surg. 2020;163:722–728. doi: 10.1177/0194599820934380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen L., Zhao J., Peng J., Li X., Deng X., Geng Z. Detection of SARS-CoV-2 in saliva and characterization of oral symptoms in COVID-19 patients. Cell Prolif. 2020;53 doi: 10.1111/cpr.12923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abubakr N., Salem Z.A., Kamel A.H.M. Oral manifestations in mild-to-moderate cases of COVID-19 viral infection in the adult population. Dent Med Probl. 2021;58:7–15. doi: 10.17219/dmp/130814. [DOI] [PubMed] [Google Scholar]
- 27.Nalbandian A., Sehgal K., Gupta A., Madhavan M.V., McGroder C., Stevens J.S. Post-acute COVID-19 syndrome. Nat Med. 2021;27:601–615. doi: 10.1038/s41591-021-01283-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brodin P. Immune determinants of COVID-19 disease presentation and severity. Nat Med. 2021;27:28–33. doi: 10.1038/s41591-020-01202-8. [DOI] [PubMed] [Google Scholar]
- 29.Logue J.K., Franko N.M., McCulloch D.J., McDonald D., Magedson A., Wolf C.R. Sequelae in adults at 6 months after COVID-19 infection. JAMA Netw Open. 2021;4 doi: 10.1001/jamanetworkopen.2021.0830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Horvath L., Lim J.W.J., Taylor J.W., Saief T., Stuart R., Rimmer J. Smell and taste loss in COVID-19 patients: assessment outcomes in a Victorian population. Acta Otolaryngol. 2021;141:299–302. doi: 10.1080/00016489.2020.1855366. [DOI] [PubMed] [Google Scholar]
- 31.Riestra-Ayora J., Yanes-Diaz J., Esteban-Sanchez J., Vaduva C., Molina-Quiros C., Larran-Jimenez A. Long-term follow-up of olfactory and gustatory dysfunction in COVID-19: 6 months case-control study of health workers. Eur Arch Otorhinolaryngol. 2021:1–7. doi: 10.1007/s00405-021-06764-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gherlone E.F., Polizzi E., Tetè G., De Lorenzo R., Magnaghi C., Rovere Querini P. Frequent and persistent salivary gland ectasia and oral disease after COVID-19. J Dent Res. 2021;100:464–471. doi: 10.1177/0022034521997112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eghbali Zarch R., Hosseinzadeh P. COVID-19 from the perspective of dentists: a case report and brief review of more than 170 cases. Dermatol Ther. 2021;34 doi: 10.1111/dth.14717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shamsoddin E. Saliva: a diagnostic option and a transmission route for 2019-nCoV. Evid Based Dent. 2020;21:68–70. doi: 10.1038/s41432-020-0104-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Farook F.F., Mohamed Nuzaim M.N., Taha Ababneh K., Alshammari A., Alkadi L. COVID-19 pandemic: oral health challenges and recommendations. Eur J Dent. 2020;14:S165–S170. doi: 10.1055/s-0040-1718641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fawcett D.W. Oral cavity and associated glands; Taste buds. In: Fawcett D.W., Bloom W., editors. Bloom and Fawcett, a textbook of histology. 12th ed. Chapman & Hall; New York: 1994. p. 562. [Google Scholar]
- 37.Farbman A.I. Fine structure of the taste bud. J Ultrastruct Res. 1965;12:328–350. doi: 10.1016/s0022-5320(65)80103-4. [DOI] [PubMed] [Google Scholar]
- 38.Roper S.D. Signal transduction and information processing in mammalian taste buds. Pflugers Arch. 2007;454:759–776. doi: 10.1007/s00424-007-0247-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoon M.A., Adler E., Lindemeier J., Battey J.F., Ryba N.J., Zuker C.S. Putative mammalian taste receptors: a class of taste-specific GPCRs with distinct topographic selectivity. Cell. 1999;96:541–551. doi: 10.1016/s0092-8674(00)80658-3. [DOI] [PubMed] [Google Scholar]
- 40.Chandrashekar J., Hoon M.A., Ryba N.J., Zuker C.S. The receptors and cells for mammalian taste. Nature. 2006;444:288–294. doi: 10.1038/nature05401. [DOI] [PubMed] [Google Scholar]
- 41.Meyerhof W. Elucidation of mammalian bitter taste. Rev Physiol Biochem Pharmacol. 2005;154:37–72. doi: 10.1007/s10254-005-0041-0. [DOI] [PubMed] [Google Scholar]
- 42.Nelson G., Chandrashekar J., Hoon M.A., Feng L., Zhao G., Ryba N.J. An amino-acid taste receptor. Nature. 2002;416(March(6877)):199–202. doi: 10.1038/nature726. Epub 2002 Feb 24. PMID: 11894099. [DOI] [PubMed] [Google Scholar]
- 43.Heck G.L., Mierson S., DeSimone J.A. Salt taste transduction occurs through an amiloride-sensitive sodium transport pathway. Science. 1984;223(Jan (4634)):403–405. doi: 10.1126/science.6691151. PMID: 6691151. [DOI] [PubMed] [Google Scholar]
- 44.Ohmoto M., Jyotaki M., Foskett J.K., Matsumoto I. Sodium-Taste Cells RequireSkn-1a for Generation and Share Molecular Features with Sweet, Umami, and Bitter Taste Cells. eNeuro. 2020;7(December (6)) doi: 10.1523/ENEURO.0385-20.2020. ENEURO. 0385-20.2020.PMID: 33219051; PMCID: PMC7729297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Williams V.R., Scholey J.W. Angiotensin-converting enzyme 2 and renal disease. Curr Opin Nephrol Hypertens. 2018;27:35–41. doi: 10.1097/MNH.0000000000000378. [DOI] [PubMed] [Google Scholar]
- 46.2021. Renin-angiotensin system — Homo sapiens (human) KEGG PATHWAY database KEGG: Kyoto encyclopedia of genes and genomes. https://www.genome.jp/keggbin/show_pathway?hsa04614 [Accessed 30 April] [Google Scholar]
- 47.Shigemura N., Takai S., Hirose F., Yoshida R., Sanematsu K., Ninomiya Y. Expression of renin–angiotensin system components in the taste organ of mice. Nutrients. 2019;11:2251. doi: 10.3390/nu11092251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mario E.G., Santos S.H., Ferreira A.V., Bader M., Santos R.A., Botion L.M. Angiotensin-(1-7) Mas-receptor deficiency decreases peroxisome proliferator-activated receptor gamma expression in adipocytes. Peptides. 2012;33:174–177. doi: 10.1016/j.peptides.2011.11.014. [DOI] [PubMed] [Google Scholar]
- 49.Ghannam M.G., Singh P. StatPearls Publishing, StatPearls Publishing LLC; Treasure Island (FL): 2021. Anatomy, head and neck, salivary glands. StatPearls.https://www.ncbi.nlm.nih.gov/books/NBK538325/ [PubMed] [Google Scholar]
- 50.Bordoni B., Varacallo M. StatPearls Publishing, StatPearls Publishing LLC; Treasure Island (FL): 2021. Anatomy, head and neck, temporomandibular joint. StatPearls.https://www.ncbi.nlm.nih.gov/books/NBK538486/ [PubMed] [Google Scholar]
- 51.Pedersen A.M.L., Sørensen C.E., Proctor G.B., Carpenter G.H., Ekström J. Salivary secretion in health and disease. J Oral Rehabil. 2018;45:730–746. doi: 10.1111/joor.12664. [DOI] [PubMed] [Google Scholar]
- 52.Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thomas G. Furin at the cutting edge: from protein traffic to embryogenesis and disease. Nat Rev Mol Cell Biol. 2002;3:753–766. doi: 10.1038/nrm934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A. Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci U S A. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Qing E., Gallagher T. SARS coronavirus redux. Trends Immunol. 2020;41:271–273. doi: 10.1016/j.it.2020.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tang T., Bidon M., Jaimes J.A., Whittaker G.R., Daniel S. Coronavirus membrane fusion mechanism offers a potential target for antiviral development. Antiviral Res. 2020;178 doi: 10.1016/j.antiviral.2020.104792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Inoue Y., Tanaka N., Tanaka Y., Inoue S., Morita K., Zhuang M. Clathrin-dependent entry of severe acute respiratory syndrome coronavirus into target cells expressing ACE2 with the cytoplasmic tail deleted. J Virol. 2007;81:8722–8729. doi: 10.1128/JVI.00253-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Iwata-Yoshikawa N., Okamura T., Shimizu Y., Hasegawa H., Takeda M., Nagata N. TMPRSS2 contributes to virus spread and immunopathology in the airways of murine models after coronavirus infection. J Virol. 2019;93:e01815–18. doi: 10.1128/JVI.01815-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shirato K., Kawase M., Matsuyama S. Wild-type human coronaviruses prefer cell-surface TMPRSS2 to endosomal cathepsins for cell entry. Virology. 2018;517:9–15. doi: 10.1016/j.virol.2017.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181 doi: 10.1016/j.cell.2020.02.052. 271–280.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Murgolo N., Therien A.G., Howell B., Klein D., Koeplinger K., Lieberman L.A. SARS-CoV-2 tropism, entry, replication, and propagation: considerations for drug discovery and development. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lauring A.S., Hodcroft E.B. Genetic variants of SARS-CoV-2—what do they mean? JAMA. 2021;325:529–531. doi: 10.1001/jama.2020.27124. [DOI] [PubMed] [Google Scholar]
- 64.Moore J.P., Offit P.A. SARS-CoV-2 vaccines and the growing threat of viral variants. JAMA. 2021;325:821–822. doi: 10.1001/jama.2021.1114. [DOI] [PubMed] [Google Scholar]
- 65.Callaway E. The coronavirus is mutating — does it matter? Nature. 2020;585:174–177. doi: 10.1038/d41586-020-02544-6. [DOI] [PubMed] [Google Scholar]
- 66.Hamming I., Timens W., Bulthuis M.L., Lely A.T., Navis G., van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ou X., Liu Y., Lei X., Li P., Mi D., Ren L. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol. 2020;94:e00127–20. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Saha P., Banerjee A.K., Tripathi P.P., Srivastava A.K., Ray U. A virus that has gone viral: amino acid mutation in S protein of Indian isolate of Coronavirus COVID-19 might impact receptor binding, and thus, infectivity. Biosci Rep. 2020;40 doi: 10.1042/BSR20201312. BSR20201312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bourgonje A.R., Abdulle A.E., Timens W., Hillebrands J.L., Navis G.J., Gordijn S.J. Angiotensin-converting enzyme 2 (ACE2), SARS-CoV-2 and the pathophysiology of coronavirus disease 2019 (COVID-19) J Pathol. 2020;251:228–248. doi: 10.1002/path.5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Darbani B. The expression and polymorphism of entry machinery for COVID-19 in human: juxtaposing population groups, gender, and different tissues. Int J Environ Res Public Health. 2020;17:3433. doi: 10.3390/ijerph17103433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Azzi L., Carcano G., Gianfagna F., Grossi P., Gasperina D.D., Genoni A. Saliva is a reliable tool to detect SARS-CoV-2. J Infect. 2020;81:e45–e50. doi: 10.1016/j.jinf.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xu R., Cui B., Duan X., Zhang P., Zhou X., Yuan Q. Saliva: potential diagnostic value and transmission of 2019-nCoV. Int J Oral Sci. 2020;12:11. doi: 10.1038/s41368-020-0080-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tsuchiya H. Oral symptoms associated with COVID-19 and their pathogenic mechanisms: a literature review. Dent J (Basel) 2021;9:32. doi: 10.3390/dj9030032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Amraei R., Rahimi N. COVID-19, renin-angiotensin system and endothelial dysfunction. Cells. 2020;9:1652. doi: 10.3390/cells9071652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jin X., Xu K., Jiang P., Lian J., Hao S., Yao H. Virus strain from a mild COVID-19 patient in Hangzhou represents a new trend in SARS-CoV-2 evolution potentially related to Furin cleavage site. Emerg Microbes Infect. 2020;9:1474–1488. doi: 10.1080/22221751.2020.1781551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Trabandt A., Müller-Ladner U., Kriegsmann J., Gay R.E., Gay S. Expression of proteolytic cathepsins B, D, and L in periodontal gingival fibroblasts and tissues. Lab Invest. 1995;73:205–212. [PubMed] [Google Scholar]
- 79.Wang P., Luo R., Zhang M., Wang Y., Song T., Tao T. A cross-talk between epithelium and endothelium mediates human alveolar-capillary injury during SARS-CoV-2 infection. Cell Death Dis. 2020;11:1042. doi: 10.1038/s41419-020-03252-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhu N., Wang W., Liu Z., Liang C., Wang W., Ye F. Morphogenesis and cytopathic effect of SARS-CoV-2 infection in human airway epithelial cells. Nat Commun. 2020;11:3910. doi: 10.1038/s41467-020-17796-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Borczuk A.C., Salvatore S.P., Seshan S.V., Patel S.S., Bussel J.B., Mostyka M. COVID-19 pulmonary pathology: a multi-institutional autopsy cohort from Italy and New York City. Mod Pathol. 2020;33:2156–2168. doi: 10.1038/s41379-020-00661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yap J.K.Y., Moriyama M., Iwasaki A. Inflammasomes and Pyroptosis as therapeutic targets for COVID-19. J Immunol. 2020;205:307–312. doi: 10.4049/jimmunol.2000513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lucas C., Wong P., Klein J., Castro T.B.R., Silva J., Sundaram M. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584:463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen M., Shen W., Rowan N.R., Kulaga H., Hillel A., Ramanathan M., Jr. Elevated ACE-2 expression in the olfactory neuroepithelium: implications for anosmia and upper respiratory SARS-CoV-2 entry and replication. Eur Respir J. 2020;56 doi: 10.1183/13993003.01948-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Meinhardt J., Radke J., Dittmayer C., Franz J., Thomas C., Mothes R. Olfactory transmucosal SARS-CoV-2 invasion as a port of central nervous system entry in individuals with COVID-19. Nat Neurosci. 2021;24:168–175. doi: 10.1038/s41593-020-00758-5. [DOI] [PubMed] [Google Scholar]
- 86.Lemprière S. SARS-CoV-2 detected in olfactory neurons. Nat Rev Neurol. 2021;17:63. doi: 10.1038/s41582-020-00449-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jones J.E., Le Sage V., Lakdawala S.S. Viral and host heterogeneity and their effects on the viral life cycle. Nat Rev Microbiol. 2021;19:272–282. doi: 10.1038/s41579-020-00449-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Caldas L.A., Carneiro F.A., Higa L.M., Monteiro F.L., da Silva G.P., da Costa L.J. Ultrastructural analysis of SARS-CoV-2 interactions with the host cell via high resolution scanning electron microscopy. Sci Rep. 2020;10:16099. doi: 10.1038/s41598-020-73162-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Netland J., Meyerholz D.K., Moore S., Cassell M., Perlman S. Severe acute respiratory syndrome coronavirus infection causes neuronal death in the absence of encephalitis in mice transgenic for human ACE2. J Viol. 2008;82:7264–7275. doi: 10.1128/JVI.00737-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Desforges M., Le Coupanec A., Brison E., Meessen-Pinard M., Talbot P.J. Neuroinvasive and neurotropic human respiratory coronaviruses: potential neurovirulent agents in humans. Adv Exp Med Biol. 2014;807:75–96. doi: 10.1007/978-81-322-1777-0_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hu J., Jolkkonen J., Zhao C. Neurotropism of SARS-CoV-2 and its neuropathological alterations: similarities with other coronaviruses. Neurosci Biobehav Rev. 2020;119:184–193. doi: 10.1016/j.neubiorev.2020.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mao L., Jin H., Wang M., Hu Y., Chen S., He Q. Neurologic manifestations of hospitalized patients with coronavirus disease 2019 in Wuhan, China. JAMA Neurol. 2020;77:683–690. doi: 10.1001/jamaneurol.2020.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Paterson R.W., Brown R.L., Benjamin L., Nortley R., Wiethoff S., Bharucha T. The emerging spectrum of COVID-19 neurology: clinical, radiological and laboratory findings. Brain. 2020;143:3104–3120. doi: 10.1093/brain/awaa240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.van Eijk L.E., Binkhorst M., Bourgonje A.R., Offringa A.K., Mulder D.J., Bos E.M. COVID-19: immunopathology, pathophysiological mechanisms, and treatment options. J Pathol. 2021 doi: 10.1002/path.5642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Keyhanian K., Umeton R.P., Mohit B., Davoudi V., Hajighasemi F., Ghasemi M. SARS-CoV-2 and nervous system: from pathogenesis to clinical manifestation. J Neuroimmunol. 2020;350 doi: 10.1016/j.jneuroim.2020.577436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cooper K.W., Brann D.H., Farruggia M.C., Bhutani S., Pellegrino R., Tsukahara T. COVID-19 and the chemical senses: supporting players take center stage. Neuron. 2020;107:219–233. doi: 10.1016/j.neuron.2020.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Meunier N., Briand L., Jacquin-Piques A., Brondel L., Pénicaud L. COVID 19-Induced smell and taste impairments: putative impact on physiology. Front Physiol. 2020;11 doi: 10.3389/fphys.2020.625110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Li W., Li M., Ou G. COVID-19, cilia, and smell. FEBS J. 2020;287:3672–3676. doi: 10.1111/febs.15491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rautiainen M., Nuutinen J., Kiukaanniemi H., Collan Y. Ultrastructural changes in human nasal cilia caused by the common cold and recovery of ciliated epithelium. Ann Otol Rhinol Raryngol. 1992;101:982–987. doi: 10.1177/000348949210101204. [DOI] [PubMed] [Google Scholar]
- 100.Afzelius B.A. Ultrastructure of human nasal epithelium during an episode of coronavirus infection. Virchows Arch. 1994;424:295–300. doi: 10.1007/BF00194614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rockx B., Kuiken T., Herfst S., Bestebroer T., Lamers M.M., Oude Munnink B.B. Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science. 2020;368:1012–1015. doi: 10.1126/science.abb7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lehmann M.J., Sherer N.M., Marks C.B., Pypaert M., Mothes W. Actin- and myosin-driven movement of viruses along filopodia precedes their entry into cells. J Cell Biol. 2005;170:317–325. doi: 10.1083/jcb.200503059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chaudhari N., Roper S.D. The cell biology of taste. J Cell Biol. 2010;190:285–296. doi: 10.1083/jcb.201003144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Behrens M., Born S., Redel U., Voigt N., Schuh V., Raguse J.D. Immunohistochemical detection of TAS2R38 protein in human taste cells. PLoS One. 2012;7 doi: 10.1371/journal.pone.0040304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mastrangelo A., Bonato M., Cinque P. Smell and taste disorders in COVID-19: from pathogenesis to clinical features and outcomes. Neurosci Lett. 2021;748 doi: 10.1016/j.neulet.2021.135694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Levinson R., Elbaz M., Ben-Ami R., Shasha D., Levinson T., Choshen G. Time course of anosmia and dysgeusia in patients with mild SARS-CoV-2 infection. Infect Dis (Lond) 2020;52:600–602. doi: 10.1080/23744235.2020.1772992. [DOI] [PubMed] [Google Scholar]
- 107.Sedaghat A.R., Gengler I., Speth M.M. Olfactory dysfunction: a highly prevalent symptom of COVID-19 with public health significance. Otolaryngol Head Neck Surg. 2020;163:12–15. doi: 10.1177/0194599820926464. [DOI] [PubMed] [Google Scholar]
- 108.Reiter E.R., Coelho D.H., Kons Z.A., Costanzo R.M. Subjective smell and taste changes during the COVID-19 pandemic: short term recovery. Am J Otolaryngol. 2020;41 doi: 10.1016/j.amjoto.2020.102639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sakalli E., Temirbekov D., Bayri E., Alis E.E., Erdurak S.C., Bayraktaroglu M. Ear nose throat-related symptoms with a focus on loss of smell and/or taste in COVID-19 patients. Am J Otolaryngol. 2020;41(November–December) doi: 10.1016/j.amjoto.2020.102622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lee H., Macpherson L.J., Parada C.A., Zuker C.S., Ryba N.J.P. Rewiring the taste system. Nature. 2017;548:330–333. doi: 10.1038/nature23299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Singer-Cornelius T., Cornelius J., Oberle M., Metternich F.U., Brockmeier S.J. Objective gustatory and olfactory dysfunction in COVID-19 patients: a prospective cross-sectional study. Eur Arch Otorhinolaryngol. 2021:1–8. doi: 10.1007/s00405-020-06590-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bellini W.J., Rota J.S., Lowe L.E., Katz R.S., Dyken P.R., Zaki S.R. Subacute sclerosing panencephalitis: more cases of this fatal disease are prevented by measles immunization than was previously recognized. J Infect Dis. 2005;192:1686–1693. doi: 10.1086/497169. [DOI] [PubMed] [Google Scholar]
- 113.Altmann D.M., Boyton J.R. Confronting the pathophysiology of long covid. BMJ Opin. 2020 https://blogs.bmj.com/bmj/2020/12/09/confronting-the-pathophysiology-of-long-covid/ [Google Scholar]
- 114.Karlsson A.C., Humbert M., Buggert M. The known unknowns of T cell immunity to COVID-19. Sci Immunol. 2020 doi: 10.1126/sciimmunol.abe8063. [DOI] [PubMed] [Google Scholar]
- 115.Xu J., Li Y., Gan F., Du Y., Yao Y. Salivary glands: potential reservoirs for COVID-19 asymptomatic infection. J Dent Res. 2020;989 doi: 10.1177/0022034520918518. Epub 2020 Apr 9. [DOI] [PubMed] [Google Scholar]
- 116.Liu L., Wei Q., Alvarez X., Wang H., Du Y., Zhu H. Epithelial cells lining salivary gland ducts are early target cells of severe acute respiratory syndrome coronavirus infection in the upper respiratory tracts of rhesus macaques. J Virol. 2011;85:4025–4030. doi: 10.1128/JVI.02292-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Liu Y., Yan L.M., Wan L., Xiang T.X., Le A., Liu J.M. Viral dynamics in mild and severe cases of COVID-19. Lancet Infect Dis. 2020;20:656–657. doi: 10.1016/S1473-3099(20)30232-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Batlle D., Jose Soler M., Ye M. ACE2 and diabetes: ACE of ACEs? Diabetes. 2010;59:2994–2996. doi: 10.2337/db10-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ayres J.S. A metabolic handbook for the COVID-19 pandemic. Nat Metab. 2020;2:572–585. doi: 10.1038/s42255-020-0237-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tay M.Z., Poh C.M., Rénia L., MacAry P.A., Ng L.F.P. The trinity of COVID-19: immunity, inflammation and intervention. Nat Rev Immunol. 2020;20:363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Levy J.M. Treatment recommendations for persistent smell and taste dysfunction following COVID-19—the coming deluge. JAMA Otolaryngol Head Neck Surg. 2020;146:733. doi: 10.1001/jamaoto.2020.1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Whitcroft K.L., Hummel T. Clinical diagnosis and current management strategies for olfactory dysfunction: a review. JAMA Otolaryngol Head Neck Surg. 2019;145:846–853. doi: 10.1001/jamaoto.2019.1728. [DOI] [PubMed] [Google Scholar]
- 123.Yan C.H., Rathor A., Krook K., Ma Y., Rotella M.R., Dodd R.L. Effect of Omega-3 supplementation in patients with smell dysfunction following endoscopic sellar and parasellar tumor resection: a multicenter prospective randomized controlled trial. Neurosurgery. 2020;87(August (2)):E91–E98. doi: 10.1093/neuros/nyz559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Coke C.J., Davison B., Fields N., Fletcher J., Rollings J., Roberson L. SARS-CoV-2 infection and oral health: therapeutic opportunities and challenges. J Clin Med. 2021;10:156. doi: 10.3390/jcm10010156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Gil-Montoya J.A., Silvestre F.J., Barrios R., Silvestre-Rangil J. Treatment of xerostomia and hyposalivation in the elderly: a systematic review. Med Oral Patol Oral Cir Bucal. 2016;21:e355–66. doi: 10.4317/medoral.20969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Tanasiewicz M., Hildebrandt T., Obersztyn I. Xerostomia of various etiologies: a review of the literature. Adv Clin Exp Med. 2016;25:199–206. doi: 10.17219/acem/29375. [DOI] [PubMed] [Google Scholar]
- 127.Veilleux M.P., Moriyama S., Yoshioka M., Hinode D., Grenier D. A review of evidence for a therapeutic application of traditional japanese kampo medicine for oral Diseases/Disorders. Medicines (Basel) 2018;5:35. doi: 10.3390/medicines5020035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Nakagawa Y. Management of dry mouth in Sjogren’s syndrome. Jpn Dent Sci Rev. 2011;47:115–123. [Google Scholar]
- 129.Jothimani D., Kailasam E., Danielraj S., Nallathambi B., Ramachandran H., Sekar P. COVID-19: poor outcomes in patients with zinc deficiency. Int J Infect Dis. 2020;100:343–349. doi: 10.1016/j.ijid.2020.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Vogel-González M., Talló-Parra M., Herrera-Fernández V., Pérez-Vilaró G., Chillón M., Nogués X. Low zinc levels at admission associates with poor clinical outcomes in SARS-CoV-2 infection. Nutrients. 2021;13:562. doi: 10.3390/nu13020562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lechien J.R., Chiesa-Estomba C.M., De Siati D.R., Horoi M., Le Bon S.D., Rodriguez A. Olfactory and gustatory dysfunctions as a clinical presentation of mild-to-moderate forms of the coronavirus disease (COVID-19): a multicenter European study. Eur Arch Otorhinolaryngol. 2020;277:2251–2261. doi: 10.1007/s00405-020-05965-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gómez-Iglesias P., Porta-Etessam J., Montalvo T., Valls-Carbó A., Gajate V., Matías-Guiu J.A. An online observational study of patients with olfactory and gustory alterations secondary to SARS-CoV-2 infection. Front Public Health. 2020;8(May):243. doi: 10.3389/fpubh.2020.00243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ibekwe T.S., Fasunla A.J., Orimadegun A.E. Systematic review and meta-analysis of smell and taste disorders in COVID-19. OTO Open. 2020;4 doi: 10.1177/2473974X20957975. 2473974X20957975. [DOI] [PMC free article] [PubMed] [Google Scholar]