The Herpes Simplex Virus US11 Protein Effectively Compensates for the γ134.5 Gene if Present before Activation of Protein Kinase R by Precluding Its Phosphorylation and That of the α Subunit of Eukaryotic Translation Initiation Factor 2 (original) (raw)
Abstract
In herpes simplex virus-infected cells, viral γ134.5 protein blocks the shutoff of protein synthesis by activated protein kinase R (PKR) by directing the protein phosphatase 1α to dephosphorylate the α subunit of eukaryotic translation initiation factor 2 (eIF-2α). The amino acid sequence of the γ134.5 protein which interacts with the phosphatase has high homology to a domain of the eukaryotic protein GADD34. A class of compensatory mutants characterized by a deletion which results in the juxtaposition of the α47 promoter next to US11, a γ2 (late) gene in wild-type virus-infected cells, has been described. In cells infected with these mutants, protein synthesis continues even in the absence of the γ134.5 gene. In these cells, PKR is activated but eIF-2α is not phosphorylated, and the phosphatase is not redirected to dephosphorylate eIF-2α. We report the following: (i) in cells infected with these mutants, US11 protein was made early in infection; (ii) US11 protein bound PKR and was phosphorylated; (iii) in in vitro assays, US11 blocked the phosphorylation of eIF-2α by PKR activated by poly(I-C); and (iv) US11 was more effective if present in the reaction mixture during the activation of PKR than if added after PKR had been activated by poly(I-C). We conclude the following: (i) in cells infected with the compensatory mutants, US11 made early in infection binds to PKR and precludes the phosphorylation of eIF-2α, whereas US11 driven by its natural promoter and expressed late in infection is ineffective; and (ii) activation of PKR by double-stranded RNA is a common impediment countered by most viruses by different mechanisms. The γ134.5 gene is not highly conserved among herpesviruses. A likely scenario is that acquisition by a progenitor of herpes simplex virus of a portion of the cellular GADD34 gene resulted in a more potent and reliable means of curbing the effects of activated PKR. US11 was retained as a γ2 gene because, like many viral proteins, it has multiple functions.
The herpes simplex virus 1 (HSV-1) genome encodes two sets of functions. The first and paramount are functions related to viral gene expression, replication of viral DNA, synthesis of virion proteins, assembly, packaging, and egress of the virus from the infected cell. The second set of functions, no less important in the survival of the virus in the human population, is creation of the environment necessary to maximize the yield and spread of virus from cell to cell and from infected to uninfected individuals (reviewed in reference 38). Of these known genes, several play a significant role in abating or delaying a host response to infection. The earliest to be expressed is the UL41 gene which encodes a protein that is introduced into the cell in virions during infection (26, 27). This protein reduces the synthesis of host proteins by causing the destruction of mRNA in a rather nonspecific manner and therefore could be expected to reduce the synthesis of cellular proteins deleterious to viral replication (26, 27, 44).
A second and very different approach to blocking host defense mechanisms is exemplified by infected cell protein 47 (ICP47). Proteosomal degradation of viral proteins could be expected to produce antigenic peptides which, if presented on the cell surface, could provoke a cytotoxic cell response early in infection and thus reduce viral yield. ICP47, an α protein made immediately after infection, blocks the presentation of antigenic peptides on the surface of the infected cells (20).
The focus of this laboratory has been on a third viral pathway designed to block cellular response to infection. In cells infected with most viruses, the synthesis of complementary mRNA leads to activation of double-stranded RNA-dependent protein kinase R (PKR). This enzyme phosphorylates the α subunit of eukaryotic translation initiation factor 2 (eIF-2α) (23). A consequence of this phosphorylation is total shutoff of protein synthesis. This would be an example of a noble sacrifice of the infected cell for the sake of survival of the organism were it not for the fact that viruses, while activating the PKR kinase pathway by making double-stranded RNA, also express functions which block this host defense system (2–4, 6, 7, 10, 28, 30, 34). In the case of HSV-1, more than 50% of the viral DNA is represented late in infection in the form of cRNA (21, 25), and the gene whose product blocks the consequences of activation of PKR is γ134.5 (7). In the absence of the gene, eIF-2α is phosphorylated and protein synthesis is impaired beginning approximately 5 h after infection (7, 9). In its presence, protein synthesis continues unabated even though PKR is activated (9). Recent studies have shown that the carboxyl terminus of the γ134.5 gene binds to the protein phosphatase 1α (PP1) and redirects it to dephosphorylate eIF-2α (19). The effectiveness of the γ134.5-PP1 complex is apparent from the observation that the rate of dephosphorylation of eIF-2α in cells infected with wild-type virus is more than 1000 times that of uninfected cells or cells infected with the γ134.5− virus (5, 19).
The studies described in this report concern another aspect of virus-induced block of the consequence of activation of PKR. Briefly, Mohr and Gluzman reported that serial passage of a γ134.5− mutant resulted in the selection of a compensatory mutation capable of sustained protein synthesis (35). A characteristic of the compensatory mutants isolated by Mohr and Gluzman is a deletion in the α47 gene resulting in the juxtaposition of the promoter of the α47 gene next to the 5′ end of US11, a late (γ2) viral gene. Preliminary studies of those mutants revealed that PKR was activated in cells infected with either the wild-type parent or the γ134.5− virus, but protein synthesis was unaffected in cells infected with wild-type virus or the mutant carrying the compensatory mutations (5, 18).
In an attempt to define the phenotype of the virus carrying the compensatory mutation, we constructed a mutant lacking the γ134.5 and the US8 to -12 genes. This mutant, designated R5103, activated PKR and caused a shutoff of protein synthesis (5). We then inserted into the R5103 genome a DNA fragment consisting of the intact US10 gene and the US11 open reading frame fused to the α47 promoter. This virus, designated R5104, activated PKR but did not induce the shutoff of protein synthesis. Consistent with the conclusion of Mohr and Gluzman (35), the mutation maps in the domain inserted into the R5104 virus (5). Further studies yielded two significant observations. First, in stark contrast to lysates of cells infected with R5103 and other γ134.5− mutants, the lysates of R5104 virus failed to phosphorylate the α subunit of eIF-2 (5). Second, in striking contrast to lysates of wild-type virus-infected cells, the phosphatase activity of lysates of R5104 virus-infected cells specific for eIF-2α could not be differentiated from that of mock-infected cells or those of cells infected with other γ134.5− mutants (5). These results indicated that the compensatory mutation blocks PKR from phosphorylating eIF-2α.
The studies summarized in this report focused on US11 protein. We report that in cells infected with the R5104 recombinant the US11 protein is made early in infection, that US11 protein interacts with PKR and blocks the phosphorylation of eIF-2α by activated PKR in in vitro assays, and that the effectiveness of the US11 protein is greater if the protein is present in the reaction before activation of PKR than if it is after PKR has been activated by the addition of poly(I-C). We also found that US11 is phosphorylated in the presence of activated PKR but not in its absence. We conclude that US11 may have been an ancient mechanism for blocking the effects of activated PKR and that it has been supplanted by acquisition of the carboxyl-terminal domain of the γ134.5 protein from a cellular gene. We also note that US11 protein made late in infection, after PKR has been activated, is ineffective.
Relevant to this report are some of the properties of the US11 protein. US11 is one of the most abundant viral proteins expressed at late times in viral infection (22, 31). It binds mRNA in a sequence- and conformation-specific fashion (39–41). In HSV-1-infected cells, US11 suppresses the synthesis of a truncated RNA colinear with the 5′ domain of the UL34 mRNA (40). The protein accumulates in nucleoli, in the cytoplasm in association with the 60S ribosomal subunit, and it is also packaged in virions (31, 37, 41). In newly infected cells, the US11 protein has been found associated with ribosomes (41).
Recently a plethora of reports suggested that US11 may have novel functions not readily apparent from its localization in the infected cell. Thus, US11 protein has been reported to have functions similar to those of human immunodeficiency Tat and Rev proteins and has also been reported to complement Rev function in a Rev− human immunodeficiency virus mutant (11). The US11 protein has been reported to confer thermotolerance and help restore protein synthesis in HeLa cells subjected to thermal injury (12).
MATERIALS AND METHODS
Cells and viruses.
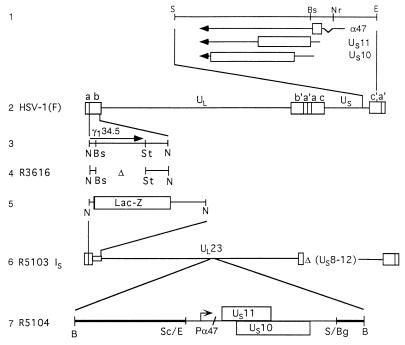
All cell lines were obtained from the American Type Culture Collection. The properties of HSV-1(F), the prototype HSV-1 strain used in these studies, and the recombinant viruses R3616, R5103, and R5104 derived from it have been described elsewhere (5, 6, 13) and are represented in Fig. 1. The stocks of HSV-1(F) were prepared in HEp-2 cells. All recombinant virus stocks and all virus titrations were done in Vero cells. SK-N-SH and HeLa cells were used for protein radiolabeling studies and for the preparation of virus-infected cell lysates. Cells were grown and maintained in Dulbecco’s modified Eagle’s medium supplemented with 5% newborn calf serum (Vero and HeLa) or with 5% fetal bovine serum (SK-N-SH).
FIG. 1.
Schematic representation of the HSV-1 genome and of genome domains relevant to this report. Line 1, sequence arrangement of the domains of the genes US10, US11, and α47 (US12) located at the terminus of unique short (US) sequence shown in the prototype (P) orientation. The coding sequences are shown as rectangular boxes; thin lines and arrows represent the transcriptional unit and polarity of the genes. Line 2, sequence arrangement of HSV-1 genome. The rectangular boxes represent the inverted repeat sequences ab and _b′a_′ flanking the unique long (UL) sequence and inverted repeats c′a′ and ca flanking the US sequence. Line 3, map location of the Δ134.5 gene. In wild-type virus, the Δ134.5 gene is present in both copies of the inverted repeats. Line 4, representation of the structure of R3616 DNA in which both copies of the Δ134.5 had been deleted. Line 5, structure of the single ab sequence of the R5103 genome in which the Δ134.5 gene had been replaced with the E. coli lacZ gene. Line 6, sequence arrangement of the R5103 genome. In both the R5103 and parent R7023 genomes (29), the genes US8 to -12 as well as most of the internal inverted repeats had been deleted. The US sequence is in an inverted (IS) arrangement. Line 7, sequence arrangement of DNA fragment inserted into the _Bgl_II site of the _Bam_HI Q fragment, resulting in construction of the R5104 recombinant virus. The thick line represents the _Bam_HI Q fragment disrupted by the insertion of the fragment consisting of the α47 gene fused to the coding domain of the US11 gene. Abbreviations: B, _Bam_HI; Bg, _Bgl_II; Bs, _Bst_EII; E, _Eco_RI; N, _Nco_I; Nr, _Nru_I; S, _Sal_I; Sc, _Sac_II; St, _Stu_I.
Plasmids.
Plasmid pRB4508, described previously (1), encodes the final 93 codons of the UL10 open reading frame fused to the glutathione _S_-transferase (GST) coding sequence in plasmid pGEX-3x (Pharmacia). The UL10 gene of HSV-1 encodes the viral glycoprotein gM, which is incorporated into the virion and cellular membranes of infected cells. Plasmid pRB4766 was prepared by inserting the US11 open reading frame contained within a Klenow-blunted _Eco_RI-_Xho_I fragment into blunted, _Hin_dIII- and _Xho_I-digested plasmid pGEX-KG such that the coding sequence of GST was fused in frame to the entire coding domain of the US11 gene (33, 41a). Plasmid pGEX-PKR(wt), kindly provided by Bryan R. G. Williams, encodes full-length PKR fused to GST and has been described previously (32). In addition, the control plasmid pGEX-3x (Pharmacia) was used to generate GST protein to serve as a control in appropriate experiments.
Labeling of proteins with [35S]methionine and electrophoretic separation in denaturing gels.
Protein labeling experiments were done as previously described (7, 36). Two hours before harvest, infected SK-N-SH cells in 25-cm2 flasks were incubated in 1 ml of medium 199V (medium 199 supplemented with 1% calf serum) lacking methionine but supplemented with 50 μCi of [35S]methionine (1,000 Ci/mmol; Amersham). The cells were then rinsed twice with PBS-A (phosphate-buffered saline [PBS]), scraped in 1 ml of ice-cold PBS-A, pelleted, solubilized in disruption buffer, boiled, electrophoretically separated on a denaturing 12.5% (vol/vol) polyacrylamide gel cross-linked with N,_N_′-diallyltartardiamide, electrically transferred to a nitrocellulose sheet, and subjected to autoradiography on Kodak XAR5 film or immunoblotting.
Immunoblotting.
The nitrocellulose sheet containing the electrophoretically separated proteins was blocked with 5% skim milk in PBS-A (blocking solution) for at least 1 h, reacted with different antibodies diluted in blocking solution for at least 4 h, and rinsed five times in PBS-A–1% Tween for 15 min. The nitrocellulose filter was then reacted with an appropriate alkaline phosphatase-conjugated antibody diluted in blocking solution for approximately 90 min. The filter was then washed once in large volumes of PBS-A–1% Tween, washed four times in PBS-A, and developed by using 1× 5-bromo-4-chloro-3-indolylphosphate–nitroblue tetrazolium in AP buffer (100 mM Tris-HCl [pH 9.5], 5 mM MgCl2, 100 mM NaCl) (43).
The monoclonal antibodies against ICP0 (24) and US11 (41) have been described elsewhere.
GST protein affinity assay and immunoblotting.
HeLa cells were mock infected or infected with HSV-1(F) for 14 h. During the last 2 h of infection, the infected cell proteins were labeled by incubation in medium 199V containing [35S]methionine as described above. The cells were then rinsed twice with ice-cold PBS-A and resuspended in lysis solution (PBS-A containing 1% deoxycholate [DOC], 1% Nonidet P-40 [NP-40], and 5 mM sodium benzamidine) at a concentration of 4 × 106 cells/400 μl. The labeled infected cell lysates were then incubated with 20 μg of RNase A and 2 μg of RNase T1 for 90 min at 37°C. The labeled infected cell lysates were sonicated by using a 3-mm probe for 10 s, and the insoluble material was removed by centrifugation. The soluble infected cell proteins were transferred to a new tube and used in the experiments described below.
GST and GST-PKR fusion proteins were expressed in Escherichia coli BL21 cells and induced by addition of 0.1 mM isopropyl-β-d-thiogalactopyranoside. After a 90-min incubation, the bacteria were harvested. A 25-μl sample was then boiled, electrophoretically separated on a 10% (vol/vol) polyacrylamide minigel, and stained with Coomassie blue to quantify the relative amounts of GST and GST-PKR protein bound to glutathione-agarose beads (G-beads). Equivalent amounts of bound GST and GST-PKR protein were each reacted with 200 μl of the RNase-treated, labeled infected cell protein mixture described above for 12 h at 4°C. The beads were then harvested and washed seven times with PBS containing 1% DOC and 1% NP-40. The proteins bound to the G-beads were released by boiling in disruption buffer and electrophoretically separated on a sodium dodecyl sulfate (SDS)–denaturing 12.5% (vol/vol) polyacrylamide gel. The proteins were then electrically transferred to a nitrocellulose filter for autoradiography and immunoblotting.
In vitro phosphorylation of eIF-2α by PKR kinase.
Phosphorylation reaction mixtures were incubated at 34°C in 10 mM Tris-HCl (pH 7.5)–20 mM KCl–2 mM MgCl2 (TKM buffer) with the additions and for the times indicated in Results. The specific activity of the added [γ-32P]ATP (purchased from ICN) was 2 to 10 Ci/mmol. Reactions were stopped by adding an SDS-containing, denaturing solution (16); the products were analyzed by electrophoresis for 20 h at 44 V (3 V/cm) on denaturing 7% polyacrylamide gels as described elsewhere (16), followed by silver staining, drying, and autoradiography.
Preparation of protein components and source of materials.
Rabbit reticulocyte eIF-2 was highly purified from the ribosomal fraction as previously described (14). The nonactive form of PKR was partially purified free of eIF-2 from the same ribosome fraction by chromatography on DEAE-cellulose, ammonium sulfate fractionation, and then chromatography on phosphocellulose as described elsewhere (15). This preparation was added to phosphorylation reactions at a final concentration of 0.4 mg/ml. Recombinant chimeric proteins consisting of GST fused to either HSV-1 US11 or UL10 (1) were prepared as described above. Partially purified nonactivated PKR from rabbit reticulocyte lysate was activated by incubation with 0.1 μg of poly(I-C) (P-L Biochemicals) per ml, 0.10 mM ATP, and 2.5 mM MgCl2 as described previously (15).
RESULTS
R5104-infected cells produce the US11 protein earlier than HSV-1(F)-infected cells.
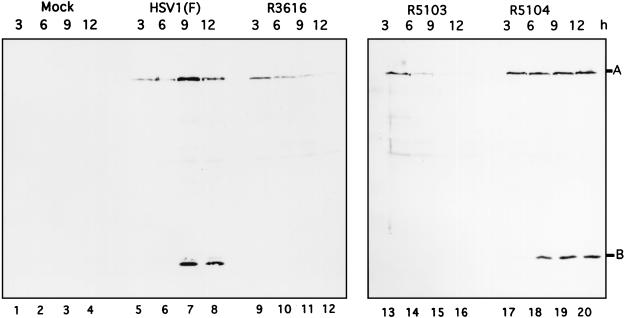
In the context of the wild-type genome, US11 is expressed as a γ2 gene. Since in R5104 the open reading frame of US11 was juxtaposed to the promoter of the α47 gene, it was necessary to verify that the US11 protein was made early rather than late in infection. Replicate 25-cm2 flask cultures of SK-N-SH cells were mock infected or infected with 10 PFU of HSV-1(F), R3616, R5103, and R5104. At 3, 6, 9, and 12 h postinfection, the cells were harvested, rinsed twice with PBS-A, and resuspended and boiled in disruption buffer (50 mM Tris-HCl [pH 7.0], 2% SDS, 700 mM β-mercaptoethanol, 2.75% sucrose); the infected cell proteins were subjected to electrophoresis on a denaturing 12.5% polyacrylamide gel and transferred to a nitrocellulose sheet as described in Materials and Methods. Immunoblotting of nitrocellulose filters with the US11 and α0 antibodies (Fig. 2) revealed that the US11 protein, labeled B, is easily detectable within 6 h in the R5104-infected cells (Fig. 2, lane 18), whereas in the HSV-1(F) infected cells, equivalent amounts of US11 are not produced until at least 9 h postinfection (lane 7). In cells infected with R3616, protein synthesis shutoff occurs at the onset of viral DNA synthesis; consequently the US11 protein, a true late γ2 protein, is not synthesized (lanes 9 to 12). The US11 protein is also not produced in the R5103-infected cells, as R5103 lacks the domain encoding this protein (lanes 13 to 16). In addition to being probed for the US11 protein, the nitrocellulose filters were probed with antiserum directed against the α0 protein, labeled A (24), showing equivalent loading of protein in each lane of the gel (Fig. 2).
FIG. 2.
Photograph of an immunoblot of lysates of wild-type and mutant viruses probed with US11 and ICP0 antibodies. Mock-infected SK-N-SH cells or SK-N-SH cells infected with HSV-1(F), R3616, R5103, or R5104 were harvested at 3, 6, 9, or 12 h after infection as described in Materials and Methods, lysed in disruption buffer, subjected to electrophoresis on an SDS–12.5% polyacrylamide gel, electrically transferred to a nitrocellulose sheet, and reacted with the anti-US11 or ICP0 monoclonal antibody as described in Materials and Methods. Positions of the US11 (labeled B) and ICP0 (labeled A) protein bands are indicated.
PKR binds the US11 protein in vitro.
In an earlier publication, we reported that a viral or virus-induced factor blocked the phosphorylation of eIF-2α by activated PKR (5). The purpose of this series of experiments was to determine whether the US11 protein could be involved in this process by specifically interacting with PKR.
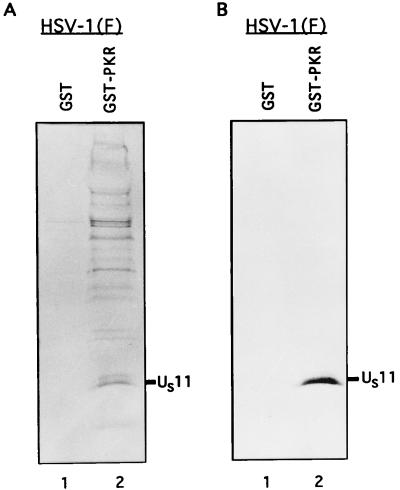
To test this hypothesis, we induced the expression of a GST-PKR fusion protein from a clone encoding the full-length PKR sequence fused to GST (32). The protein was bound to G-beads and rinsed four times with PBS-A–1% Triton X-100. The bound GST-PKR protein was then incubated for 12 h at 4°C with the [35S]methionine-labeled cell lysates, either RNase digested or untreated as described in Materials and Methods. Experiments using untreated and RNase-treated samples yielded similar results. The photograph shown in Fig. 3 is from the experiments using RNase-treated lysates. An equivalent amount of GST protein bound to beads was also incubated with the labeled mock- and HSV-1(F)-infected cell lysates prepared from 2 × 106 HeLa cells to test the specificity of the PKR protein binding. Following the incubation, the GST and GST-PKR protein samples were washed nine times with wash buffer (PBS-A, 1% NP-40, 1% DOC), resuspended in disruption buffer, and boiled for 3 min. The proteins were then separated on a denaturing 12.5% polyacrylamide gel, transferred to a nitrocellulose sheet for autoradiography (Fig. 3A), then blocked with 5% skim milk, and probed with an antibody to the US11 protein (Fig. 3B).
FIG. 3.
Autoradiogram and photograph of proteins from infected cell lysates that were bound to GST or GST-PKR, subjected to electrophoretic separation and autoradiography, and reacted with the anti-US11 antibody. Equivalent quantities of GST or GST-PKR bound to beads were reacted overnight with the radiolabeled, RNase-treated, and sonicated lysates from 2 × 106 HeLa cells infected with HSV-1(F) as described in Materials and Methods. The beads were then washed seven times with PBS containing 1% NP-40 and 1% DOC, denatured in disruption buffer, boiled, subjected to electrophoresis in a denaturing 12.5% polyacrylamide gel, electrically transferred to a nitrocellulose sheet, reacted with the anti-US11 antibody, and subjected to autoradiography. The position of the US11 protein band is indicated. (A) Autoradiographic image of the electrophoretically separated proteins bound to beads; (B) photograph of the immunoblot.
As shown in Fig. 3, lanes 2, the GST-PKR protein bound the HSV-1 US11 protein. The results suggest that the chimeric protein preferentially bound the faster-migrating form of the protein (Fig. 3B, lane 2). This interaction was mediated by the PKR protein inasmuch as GST by itself failed to bind the US11 protein (Fig. 3, lanes 1).
US11 protein precludes the phosphorylation of eIF-2α by activated PKR in vitro.
As reported in previously, the lysates of cells infected with the γ134.5− mutant R3616 lack the specific eIF-2α phosphatase activity characteristic of wild-type virus-infected cells (5, 19). Since the γ134.5− compensatory mutants do not phosphorylate eIF-2α (5), it was of interest to determine whether US11 played a role in this process. Two series of experiments were done to determine whether the US11 protein can suppress the phosphorylation of exogenously added eIF-2α by HSV-1-infected HeLa cell lysate in which PKR had been activated.
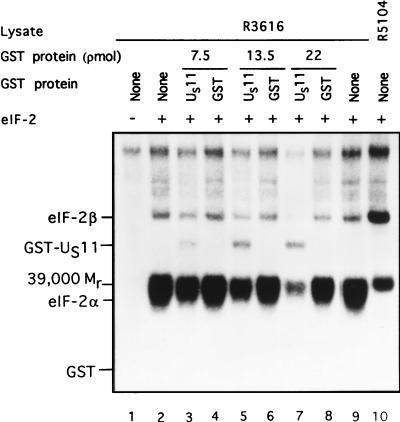
In the first series, limiting amounts of lysates from HeLa cells infected with the γ134.5− mutant R3616 were mixed with 5 pmol of purified eIF-2, [γ-32P]ATP (2 to 10 Ci/mmol) and increasing amounts of either purified GST or the chimeric protein GST-US11 in 10 μl of TKM buffer as previously described (5). The results (Fig. 4) demonstrated a dose-dependent inhibition of the phosphorylation of eIF-2α by GST-US11 but not by GST, indicating that inhibition was mediated by the US11 component. No phosphorylation of eIF-2α occurred in the absence of added eIF-2 (Fig. 4, lane 1) or in reaction mixtures containing lysates of HeLa cells infected with the R5104 mutant. Two observations are particularly noteworthy. First, the GST-US11 chimeric protein precluded the phosphorylation of all eIF-2-associated proteins (e.g., eIF-2β and the _M_r-39,000 protein) but had less effect on the cellular proteins which migrated near the top of the gel. Second, GST-US11 but not GST was phosphorylated in this reaction. Unlike the incremental reduction in phosphorylation of eIF-2α, the phosphorylation of GST-US11 was not dose dependent and saturated at approximately 13.5 pmol of added GST-US11.
FIG. 4.
Autoradiographic images of electrophoretically separated proteins from reaction mixtures containing lysates of cells infected with R3616 (γ134.5−) as a source of activated PKR, eIF-2, and either GST or GST-US11. Phosphorylation reactions contained the equivalent of 0.19 μl of lysates of R3616-infected cells (lanes 1 to 9) or R5104-infected cell lysate (lane 10), 5 pmol of eIF-2 (lanes 2 to 10), and GST-US11 (lanes 3, 5, and 7) or GST (lanes 4, 6, and 8) in concentrations shown in a final volume of 10 μl of TKM buffer. The mixtures were preincubated at 34°C for 30 s; the reaction was started by the addition of [γ-32P]ATP (final concentration, 0.04 mM) and terminated by the addition of an SDS denaturing solution after 1 min of further incubation at 34°C. The mixtures were subjected to electrophoresis in denaturing polyacrylamide gels and autoradiography. The positions of eIF-2α, eIF-2β, the _M_r-39,000 phosphoprotein, GST-US11, and GST are shown.
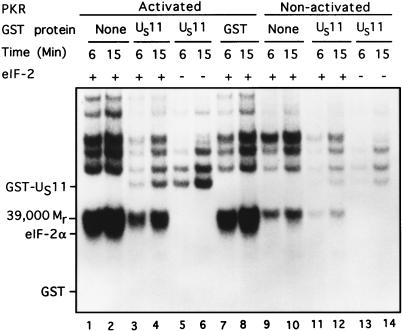
In the second series, we determined the effect of GST-US11 on the activity of PKR partially purified from rabbit reticulocyte ribosomes. Specifically, the reaction mixtures consisted of activated and nonactivated PKR, [γ-32P]ATP (2 to 10 Ci/mmol), 0.09 mM ATP, 2.5 mM MgCl2, and, where added, 5 pmol of eIF-2 and 22 pmol of GST-US11 or GST. On the basis of previous experiments involving infected cell lysates depicted in Fig. 4 and kinase reactions using serial dilutions of GST-US11 (not shown), we calculated the concentration of US11 protein which effectively blocked the phosphorylation of eIF-2α by the activated PKR. The results (Fig. 5) were as follows.
FIG. 5.
Autoradiographic images of electrophoretically separated proteins from reaction mixtures containing activated or nonactivated purified PKR, eIF-2, and either GST or GST-US11. PKR, partially purified from rabbit reticulocyte ribosomes as indicated in Material and Methods, was preincubated with 0.10 mM ATP and 2.5 mM MgCl2, either without (nonactivated PKR) or with poly(I-C) (0.1 μg/ml), for 20 min at 34°C. The contents of the reaction mixtures are given above the autoradiogram. The additions were 5 pmol of eIF-2, 22 pmol of GST-US11 or GST, and a final concentration of 0.09 mM [γ-32P]ATP in a final volume of 10.5 μl of TKM buffer. The reaction was begun by addition of activated (lanes 1 to 8) or nonactivated (lanes 9 to 14) PKR and incubation at 34°C. Aliquots (5.0 μl) were removed from each reaction mixture and placed into disruption buffer after 6 and 15 min, boiled, and subjected to electrophoresis in denaturing gels and autoradiography. Positions of the US11 protein, _M_r-39,000 phosphoprotein, and eIF-2α bands are shown. The position of GST is indicated, although it was not labeled.
(i) PKR activated by preincubation with poly(I-C) phosphorylated eIF-2α (Fig. 5, lanes 1 and 2), whereas nonactivated PKR did not (lanes 9 and 10). (ii) GST-US11, added at a level found to be just saturating in Fig. 4 and on the basis on serial dilution experiments (not shown), blocked the phosphorylation of eIF-2α by activated PKR (lanes 3 and 4), whereas GST had no effect (lanes 7 and 8). (iii) GST-US11 more effectively inhibited the phosphorylation of eIF-2α than that of the _M_r-39,000 protein and that of other proteins present in the reaction mixture, suggesting that under the conditions used, the inhibitory effects of US11 were more specific than those shown in Fig. 4. (iv) GST-US11 (lanes 3 to 6) but not GST (lanes 7 and 8) was phosphorylated in the presence of activated PKR. The levels of phosphorylated US11 were significantly reduced and barely detectable in the presence of nonactivated PKR (lanes 11 to 14). Moreover, the phosphorylation of GST-US11 by PKR was considerably greater in the absence (lanes 5 and 6) than in the presence (lanes 3 and 4) of eIF-2. This observation suggests that US11 protein and eIF-2α may be competing substrates for the activated PKR.
US11 is more effective in blocking the phosphorylation of PKR and of eIF-2α if present in the reaction mixture before rather than after the activation of PKR by poly(I-C).
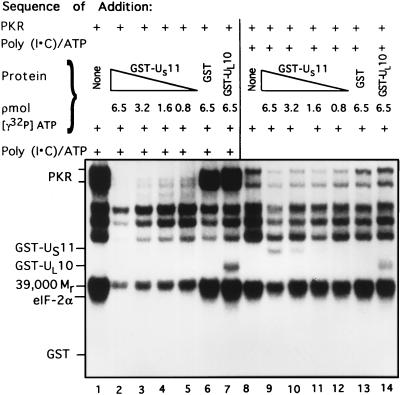
The purpose of this series of experiments was to test the effectiveness of GST-US11 in blocking the phosphorylation of eIF-2α before and after activation of PKR. In these experiments, the reaction mixtures contained identical amounts of reactants but the order of addition differed.
In the first part of experiment, the rabbit reticulocyte PKR was reacted with poly(I-C) after addition of GST or GST fusion proteins, and purified eIF-2. In the second part of the experiment, poly(I-C) was added first. The results (Fig. 6) indicate that GST-US11 was far more effective in blocking the phosphorylation of eIF-2α when added prior to activation of PKR than when added after PKR had already been exposed to poly(I-C). Specifically, the phosphorylation of eIF-2α was completely suppressed by 0.8 pmol of GST-US11 added before activation of PKR (Fig. 6, lane 5). In contrast, significant amounts of eIF-2α were phosphorylated in the presence of much higher concentrations of GST-US11 (e.g., 3.2 pmol [lane 10]) added after activation of PKR. In these experiments, GST alone and the GST-UL10 fusion protein had no effect on the phosphorylation of eIF-2α (lanes 6, 7, 13, and 14).
FIG. 6.
Autoradiographic images of electrophoretically separated proteins from reaction mixtures in which PKR was activated before or after addition of the GST-US11 chimeric protein. The order of addition of the reactants in mixtures 1 through 14 is shown at the top. Phosphorylation reaction mixtures contained of eIF-2 (2.5 pmol) and PKR; poly(I-C) (0.1 μg/ml) and 0.09 mM [γ-32P]ATP were added after (lanes 1 to 7) or before (lanes 8 to 14) addition of GST, GST-US11, or GST-UL10 in the concentrations shown. All components were in TKM buffer, and the final volume was 5.25 μl. The reaction mixtures were incubated at 34°C for 15 min. The reaction was stopped by the addition of disruption buffer, and the proteins were separated on denaturing gels and subjected to autoradiography. Positions of the reacting proteins are shown.
DISCUSSION
In an earlier report, we showed that a DNA fragment containing the US10 gene in its natural environment and the US11 gene driven by the α47 promoter imparted on the recipient γ134.5− virus (R5104) the phenotype of sustained protein synthesis, whereas in the absence of the inserted DNA fragment the parent virus (R5103) lacked this capacity. We reported also that the phenotype of R5104 was different from that of the wild-type virus in two important respects even though both viruses were capable of sustained protein synthesis. Specifically, unlike the wild-type virus, R5104 did not induce the phosphorylation of eIF-2α in infected cells by activated PKR. Moreover, the lysates of cells infected with the R5104 virus lacked the eIF-2α-specific phosphatase activity which is highly elevated and readily demonstrable in wild-type virus-infected cells. We concluded that the gene affected by the second site mutation which compensates for the absence of the γ134.5 gene operates by a mechanism different from that of the γ134.5 gene. The suspicion also focused on the US11 gene as the possible bearer of the compensatory mutation. But in fact, the only mutation that we could identify with certainty is the apparent replacement of the γ2 promoter with that of the α47 promoter.
In this article, we report the results of studies designed to test the hypothesis that the compensatory mutation is in fact the expression of the US11 as an early protein. The report consists of three parts. In the first, we showed that in cells infected with R5104, US11 is made earlier in infection. In the second series of experiments, we showed a physical interaction between PKR and US11 protein. In the third, we showed that in in vitro assays, in the presence of US11 the phosphorylation of eIF-2α is impaired. The key experiment in this series dealt with the order of addition of reagents. If US11 was added to the reaction mixture before PKR was activated, both phosphorylation of PKR and the phosphorylation of eIF-2α were blocked at lower concentrations of US11 protein than if US11 was added after PKR had been activated. The effect of US11 protein was dose dependent.
In essence, the conclusion to be reached from these studies is that US11 can substitute for the γ134.5 protein if it is present in appropriate amounts or form early in infection and that US11 protein blocks the shutoff of protein synthesis by binding to PKR and preventing it from phosphorylating eIF-2α. The results also raise three very interesting questions.
First and foremost, the mechanism by which US11 protein blocks PKR is not known. There are two clues, however. The first is that US11 is phosphorylated in the presence of PKR. There are, however, no data to unambiguously determine whether US11 irreversibly blocks PKR or whether it merely competes with eIF-2α for PKR. The second clue stems from the observation that US11 is less effective if added to the reaction mixture after activation of PKR. This observation suggests that activated PKR has a much lower affinity for US11 than the preactivated protein. If US11 binds to the activation site, it may be expected that bound US11 interferes with access to the PKR activation site by a third molecule.
The second question relates to the fact that US11 is packaged into the virion and upon entry of the virus into infected cells becomes associated with ribosomes. The question is why the US11 brought into the cell by the virus does not preclude the shutoff of protein synthesis by activated PKR. Indeed, in cells infected with R3616, which contain virion-associated US11, protein synthesis is shut off as early as in cells infected with the R5103 recombinant, which lacks the US11 gene (data not shown). These results suggest that US11 introduced into cells by the infecting virus may have a function other than that of precluding the shutoff of protein synthesis.
Finally, the question arises as to why γ134.5 protein evolved and supplanted US11 in precluding the shutoff of protein synthesis. The γ134.5 protein expresses at least two functions. The entire gene is required for viral replication in the central nervous system and only the 3′-terminal 70 codons are required to block the shutoff of protein synthesis by activated PKR. This domain of the gene is homologous to the corresponding domain of GADD34, a conserved mammalian gene expressed during growth arrest, during differentiation, or after DNA damage (8, 17).
We may speculate that the progenitor of HSV-1, HSV-2, and the simian B virus acquired this domain from the corresponding cellular gene and that in time, US11 acquired additional functions and may have evolved a late promoter. The selective pressure is not difficult to appreciate: whereas the modern US11 even under the best circumstances precludes the phosphorylation of eIF-2α, the virus carrying the γ134.5 gene renders the activated PKR totally impotent by dephosphorylating eIF-2α as rapidly as it is formed, since none is detected.
Viruses appear to have evolved myriad mechanisms designed to defeat the shutoff of proteins synthesis resulting from the activation of PKR by double-stranded RNA. These range from selective destruction or inactivation of PKR to precluding double-stranded RNA from activating PKR (2–4, 6, 7, 10, 28, 30, 34). In the case of poxviruses, two viral protein employing different pathways are dedicated to the task of nullifying PKR as a threat to viral replication (4, 10). HSV is unique in two respects. First, it has sequestered a cellular function to perform this task, and second, it may have retained the vestiges of an older, now cryptic mechanism to block this host response to infection.
ACKNOWLEDGMENTS
We thank Suzanne Hessefort for technical assistance.
This study was aided by grants from the National Cancer Institute (CA47451) to B.R. and from the National Heart, Lung, and Blood Institute (HL30121) to M.G. Grant support was also provided to K.A.C. by the Pediatric Scientist Development Program of the National Institute of Child Health and Human Development administered by the Association of Medical School Pediatric Department Chairmen Inc.
REFERENCES
- 1.Baines J D, Roizman B. The UL10 gene of herpes simplex virus 1 encodes a novel glycoprotein, gM, which is present in the virion and in the plasma membrane of infected cells. J Virol. 1993;67:1441–1452. doi: 10.1128/jvi.67.3.1441-1452.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Black T L, Barber G N, Katze M G. Degradation of the interferon-induced 68,000-Mr protein kinase by poliovirus requires RNA. J Virol. 1993;67:791–800. doi: 10.1128/jvi.67.2.791-800.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brand S R, Kobayashi R, Mathews M B. The tat protein in human immunodeficiency virus type 1 is a substrate and inhibitor of the inhibitor of the interferon-induced, virally activated protein kinase, PKR. J Biol Chem. 1997;272:8388–8395. doi: 10.1074/jbc.272.13.8388. [DOI] [PubMed] [Google Scholar]
- 4.Carroll K, Elroy-Stein O, Moss B, Jagus R. Recombinant vaccinia virus K3L gene product prevents activation of double-stranded RNA-dependent, initiation factor 2α-specific protein kinase. J Biol Chem. 1993;268:12837–12842. [PubMed] [Google Scholar]
- 5.Cassady K A, Gross M, Roizman B. The second-site mutation in the herpes simplex virus recombinants lacking the γ134.5 genes preclude shutoff of protein synthesis by blocking the phosphorylation of eIF-2α. J Virol. 1998;72:7005–7011. doi: 10.1128/jvi.72.9.7005-7011.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chou J, Kern E R, Whitley R J, Roizman B. Mapping of herpes simplex virus-1 neurovirulence to γ134.5, a gene nonessential for growth in cell culture. Science. 1990;250:1262–1266. doi: 10.1126/science.2173860. [DOI] [PubMed] [Google Scholar]
- 7.Chou J, Roizman B. The γ134.5 gene of herpes simplex virus 1 precludes neuroblastoma cells from triggering the total shutoff of protein synthesis characteristic of programmed cell death in neuronal cells. Proc Natl Acad Sci USA. 1992;89:3266–3270. doi: 10.1073/pnas.89.8.3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chou J, Roizman B. Herpes simplex virus 1 γ134.5 gene function, which blocks the host response to infection maps in the homologous domain of the genes expressed during growth arrest and DNA damage. Proc Natl Acad Sci USA. 1994;91:5247–5251. doi: 10.1073/pnas.91.12.5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chou J, Chen J-J, Gross M, Roizman B. Association of a Mr 90,000 phosphoprotein with protein kinase PKR in cells exhibiting enhanced phosphorylation of translation initiation factor eIF-2α and premature shutoff of protein synthesis after infection with γ134.5− mutants of herpes simplex virus 1. Proc Natl Acad Sci USA. 1995;92:10516–10520. doi: 10.1073/pnas.92.23.10516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davies M V, Chang H W, Jacobs B L, Kaufman R J. The E3L and K3L vaccinia virus gene products stimulate translation through inhibition of the double-stranded RNA-dependent protein kinase by different mechanisms. J Virol. 1993;67:1688–1692. doi: 10.1128/jvi.67.3.1688-1692.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diaz J J, Duc-Dodon M, Schaerer-Uthurralt N, Simonin D, Kindbeiter K, Gazzolo L, Madjar J J. Post-transcriptional transactivation of human retroviral envelope glycoprotein expression by herpes simplex virus US11 protein. Nature (London) 1996;379:273–277. doi: 10.1038/379273a0. [DOI] [PubMed] [Google Scholar]
- 12.Diaz-Latoud C, Diaz J J, Fabre-Jonca N, Kindbeiter K, Madjar J J, Arrigo A P. Herpes simplex virus US11 protein enhances recovery of protein synthesis and survival in heat shock treated HeLa cells. Cell Stress Chaperones. 1997;2:119–131. doi: 10.1379/1466-1268(1997)002<0119:hsvupe>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ejercito P M, Kieff E D, Roizman B. Characterization of herpes simplex virus strains differing in their effect on social behavior of cells. J Gen Virol. 1968;2:357–364. doi: 10.1099/0022-1317-2-3-357. [DOI] [PubMed] [Google Scholar]
- 14.Gross M, Kaplansky D A. Identification of a Mr = 39,000 phosphoprotein in highly purified preparations of rabbit reticulocyte eIF-2 that is distinct from the Mr = 35,000 subunit phosphorylated by the hemin-controlled translational repressor. J Biol Chem. 1980;255:6270–6275. [PubMed] [Google Scholar]
- 15.Gross M, Knish W M, Kwan A. Rabbit reticulocyte double-stranded RNA-activated protein kinase and the hemin-controlled translational repressor phosphorylate the same Mr 1500 peptide of eukaryotic initiation factor 2 alpha. FEBS Lett. 1981;125:223–226. doi: 10.1016/0014-5793(81)80724-7. [DOI] [PubMed] [Google Scholar]
- 16.Gross M, Kaplansky D A. Differential effect of Mn2+ on the hemin-controlled translational repressor and the double stranded RNA-activated inhibitor. Biochim Biophys Acta. 1983;740:255–263. doi: 10.1016/0167-4781(83)90134-3. [DOI] [PubMed] [Google Scholar]
- 17.He B, Chou J, Lieberman D A, Hoffman B, Roizman B. The carboxy terminus of murine MyD116 gene substitutes for the corresponding domain of the γ134.5 gene of herpes simplex virus to preclude the premature shutoff of total protein synthesis in infected human cells. J Virol. 1996;70:84–90. doi: 10.1128/jvi.70.1.84-90.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He B, Chou J, Brandimarti R, Mohr I, Gluzman Y, Roizman B. Suppression of the phenotype of γ134.5− herpes simplex virus 1: failure of activated RNA-dependent protein kinase to shut off protein synthesis is associated with a deletion in the domain of the α47 gene. J Virol. 1997;71:6049–6054. doi: 10.1128/jvi.71.8.6049-6054.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He B, Gross M, Roizman M. The γ134.5 protein of herpes simplex virus 1 complexes with protein phosphatase 1α to dephosphorylate the α subunit of eukaryotic translation initiation factor 2 and preclude the shutoff of protein synthesis by double-stranded RNA-activated protein kinase. Proc Natl Acad Sci USA. 1997;94:843–848. doi: 10.1073/pnas.94.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hill A, Jugovic P, York I, Russ G, Bennink J, Yewdell J, Ploegh H, Johnson D. Herpes simplex virus turns off the TAP to evade host immunity. Nature (London) 1995;375:411–415. doi: 10.1038/375411a0. [DOI] [PubMed] [Google Scholar]
- 21.Jacquemont B, Roizman B. RNA synthesis in cells infected with herpes simplex virus. X. Properties of viral symmetrical transcripts and double-stranded RNA prepared from them. J Virol. 1975;15:707–713. doi: 10.1128/jvi.15.4.707-713.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson P A, MacLean C, Marsden H S, Dalziel R G, Everett R D. The product of gene Us11 of herpes simplex virus type 1 is expressed as a true late gene. J Gen Virol. 1986;67:871–883. doi: 10.1099/0022-1317-67-5-871. [DOI] [PubMed] [Google Scholar]
- 23.Katze M. Regulation of interferon-induced PKR: can viruses cope? Trends Microbiol. 1995;3:75–78. doi: 10.1016/s0966-842x(00)88880-0. [DOI] [PubMed] [Google Scholar]
- 24.Kawaguchi Y, Van Sant C, Roizman B. Herpes simplex virus 1 α regulatory protein ICP0 interacts with and stabilizes the cell cycle regulator cyclin D3. J Virol. 1997;71:7328–7336. doi: 10.1128/jvi.71.10.7328-7336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kozak M, Roizman B. RNA synthesis in cells infected with herpes simplex virus. IX. Evidence for accumulation of abundant symmetrical transcripts in nuclei. J Virol. 1975;15:36–40. doi: 10.1128/jvi.15.1.36-40.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kwong A D, Frenkel N. Herpes simplex virus-infected cells contain a function(s) that destabilizes both host and viral mRNAs. Proc Natl Acad Sci USA. 1987;84:1926–1930. doi: 10.1073/pnas.84.7.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwong A D, Frenkel N. The herpes simplex virus virion host shutoff function. J Virol. 1989;63:4834–4839. doi: 10.1128/jvi.63.11.4834-4839.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee T G, Tang N, Thompson S, Miller J, Katze M G. The 58,000-dalton cellular inhibitor of the interferon-inducible double-stranded RNA-activated protein kinase (PKR) is a member of the tetratricopeptide repeat. Mol Cell Biol. 1994;14:2331–2342. doi: 10.1128/mcb.14.4.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Longnecker R, Roizman B. Generation of an inverting herpes simplex virus 1 mutant lacking the L-S junction a sequences, an origin of DNA synthesis and several genes including those specifying glycoprotein E and the α47 gene. J Virol. 1986;58:583–591. doi: 10.1128/jvi.58.2.583-591.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu Y, Wambach M, Katze M G, Krug R M. Binding of the influenza virus NS1 protein to the protein kinase that phosphorylates the eIF-2 translation initiation factor. Virology. 1995;214:222–228. doi: 10.1006/viro.1995.9937. [DOI] [PubMed] [Google Scholar]
- 31.Maclean C A, Rixon F J, Marsden H S. The products of gene US11 of herpes simplex virus type 1 are DNA-binding and localize to the nucleoli of infected cells. J Gen Virol. 1987;43:1921–1937. doi: 10.1099/0022-1317-68-7-1921. [DOI] [PubMed] [Google Scholar]
- 32.McMillan N A, Chun R F, Siderovski D P, Galabru J, Toone W M, Samuel C E, Mak T W, Hovanessian A G, Jeang K T, Williams B R. HIV-1 Tat directly interacts with the interferon-induced, double-stranded RNA-dependent kinase, PKR. Virology. 1995;213:413–424. doi: 10.1006/viro.1995.0014. [DOI] [PubMed] [Google Scholar]
- 33.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 34.Mathews M B, Schenk T. Adenovirus virus-associated RNA and translation control. J Virol. 1991;65:5657–5662. doi: 10.1128/jvi.65.11.5657-5662.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mohr I, Gluzman Y. A herpesvirus genetic element which affects translation in the absence of viral GADD34 function. EMBO J. 1996;15:4759–4766. [PMC free article] [PubMed] [Google Scholar]
- 36.Purves F C, Spector D, Roizman B. UL34, the target of the herpes simplex virus US3 protein kinase, is a membrane protein which in its unphosphorylated state associates with novel phosphoproteins. J Virol. 1992;66:4295–4303. doi: 10.1128/jvi.66.7.4295-4303.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Puvion-Dutilleul F, Pichard E, Shedrick P, Arratric F, Puvion E. Appearance of host-specific nucleolar proteins in intranuclear ‘dense bodies’ following herpes simplex infection. Eur J Cell Biol. 1985;39:458–468. [PubMed] [Google Scholar]
- 38.Roizman B R, Sears A E. Herpes simplex viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Virology. 3rd ed. New York, N.Y: Lippincott-Raven Press; 1996. pp. 2231–2295. [Google Scholar]
- 39.Roller R, Roizman B. The herpes simplex virus US11 open reading frame encodes a sequence-specific RNA-binding protein. J Virol. 1990;64:3463–3470. doi: 10.1128/jvi.64.7.3463-3470.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roller R, Roizman B. Herpes simplex virus 1 RNA-binding protein US11 negatively regulates the accumulation of a truncated viral mRNA. J Virol. 1991;65:5873–5879. doi: 10.1128/jvi.65.11.5873-5879.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roller R, Roizman B. The herpes simplex virus 1 RNA binding protein US11 is a virion component and associates with ribosomal 60S subunit. J Virol. 1992;66:3624–3632. doi: 10.1128/jvi.66.6.3624-3632.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41a.Roller, R., and B. Roizman. Unpublished data.
- 42.Smith D B, Corcoron L M. Expression and purification of glutathione-S-transferase fusion proteins. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Short protocols in molecular biology. 3rd ed. New York, N.Y: John Wiley & Sons, Inc.; 1995. pp. 16-28–16-31. [Google Scholar]
- 43.Ward P L, Campadelli-Fiume G, Avitabile E, Roizman B. Localization and putative function of the UL20 membrane protein in cells infected with herpes simplex virus 1. J Virol. 1994;68:7406–7417. doi: 10.1128/jvi.68.11.7406-7417.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zelus B D, Stewart R S, Ross J. The virion host shutoff protein of herpes simplex virus type 1: messenger ribonucleolytic activity in vitro. J Virol. 1996;70:2411–2419. doi: 10.1128/jvi.70.4.2411-2419.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]