Structural basis of West Nile virus neutralization by a therapeutic antibody - PubMed (original) (raw)
Structural basis of West Nile virus neutralization by a therapeutic antibody
Grant E Nybakken et al. Nature. 2005.
Abstract
West Nile virus is a mosquito-borne flavivirus closely related to the human epidemic-causing dengue, yellow fever and Japanese encephalitis viruses. In establishing infection these icosahedral viruses undergo endosomal membrane fusion catalysed by envelope glycoprotein rearrangement of the putative receptor-binding domain III (DIII) and exposure of the hydrophobic fusion loop. Humoral immunity has an essential protective function early in the course of West Nile virus infection. Here, we investigate the mechanism of neutralization by the E16 monoclonal antibody that specifically binds DIII. Structurally, the E16 antibody Fab fragment engages 16 residues positioned on four loops of DIII, a consensus neutralizing epitope sequence conserved in West Nile virus and distinct in other flaviviruses. The E16 epitope protrudes from the surface of mature virions in three distinct environments, and docking studies predict Fab binding will leave five-fold clustered epitopes exposed. We also show that E16 inhibits infection primarily at a step after viral attachment, potentially by blocking envelope glycoprotein conformational changes. Collectively, our results suggest that a vaccine strategy targeting the dominant DIII epitope may elicit safe and effective immune responses against flaviviral diseases.
Conflict of interest statement
The authors declare competing financial interests: details accompany the paper on www.nature.com/nature.G.E.N., T.O., M.S.D. and D.H.F. have filed provisional patents based on this and other studies. S.J. and S.B. are employed by and M.S.D. consults for MacroGenics.
Figures
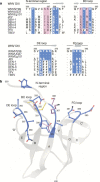
Figure 1. Crystal structure of the E16 Fab in complex with DIII of West Nile virus envelope glycoprotein.
a, Ribbon diagram of the complex, with DIII depicted in dark blue, the antibody heavy chain in green and the light chain in cyan. b, Molecular surface representation of the E16–DIII interface highlighting E16 VH (green) and VL (cyan) contact residues (left), as well as DIII contact residues (blue) and the residues defined by yeast surface display (magenta) (right). c, Flow cytometry of yeast cells expressing wild-type or mutant versions of West Nile virus DIII. d, Detailed interactions of DIII residues Ser 306 and Lys 307 with E16, with interfacial waters (red) evident in the composite electron density omit map. e, Interactions of Thr 330 and Thr 332 at the E16 interface.
Figure 2. The E16 binding site defines a West Nile virus dominant neutralizing epitope that is divergent in other flaviviruses.
a, Sequence of the four segments of West Nile virus (WNV) DIII contacted by E16 aligned with the analogous residues of other flaviviruses. The DIII residues contacted by E16 are highlighted in blue and magenta, and deletions are indicated with a hash symbol. DEN, dengue virus; JEV, Japanese encephalitis virus; TBEV, tick-borne encephalitis virus; YFV, yellow fever virus. b, Structure of the West Nile virus dominant neutralizing epitope as defined by the E16–DIII complex.
Figure 3. E16 decoration of envelope glycoprotein assemblies.
a, E16 docked onto the DEN-2 envelope glycoprotein dimer (1OAN) through DIII (blue). Super-positioning indicates that binding probably occurs without DI (red) or DII (yellow) contacts. The distances between DIII contact sites (141 Å) and the Fab termini (239 Å) preclude bivalent antibody engagement. b, E16 docked onto the post-fusion DEN-2 envelope glycoprotein trimer (1OK8) indicates accessibility of the binding site. c, DIII from the E16 complex (blue), the pre-fusion DEN-2 (1OKE, light grey) and post-fusion DEN-2 (1OK8, dark grey) reveal the conserved structure of the domains. The interaction of E16 with the flavivirus conserved Tyr 302 in the N-terminal region of West Nile virus DIII is highlighted. d, The E16 structural epitope is mapped in magenta onto the cryo-electron microscopic reconstruction of the West Nile virus virion7 presented as 2.0-Å-radius Cα atoms. e, E16–DIII complexes docked around the icosahedral three-fold axis. f, DIII situated around the outer ring of the five-fold axis is permissive to E16 binding, but the inner ring appears to exclude E16 engagement. g, Saturation binding by E16 is predicted to entail the binding of 120 out of 180 West Nile virus epitopes with exclusion of binding to the inner-five-fold DIIIs (magenta). Panels e and f are magnified views of the boxed regions in g.
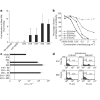
Figure 4. Mechanism of E16-mediated neutralization of West Nile virus.
a, Two DI/DII-specific neutralizing monoclonal antibodies (E53 and E60) block cellular attachment significantly more than the DIII-specific neutralizing antibodies (E16 and E24) or controls (no antibody, non-neutralizing monoclonal antibody E22 or anti-SARS ORF7a). Fold-reductions are reported, with standard deviations, as the average of four to seven independent experiments performed in triplicate. b, Dose-dependent blockade of West Nile virus infection by E16 and E53 in pre- and post-adsorption assays. The data are one of three representative experiments performed in duplicate. c, The DIII-specific monoclonal antibodies effectively inhibit West Nile virus infection of macrophages, whereas DI/DII-specific E5 and E60 monoclonal antibodies enhance infection. The data are one of three representative experiments performed in duplicate, with the dotted line representing the limit of sensitivity of the assay. Error bars represent the standard deviation. d, Pre-incubation with unlabelled monoclonal antibodies followed by addition of APC conjugates reveals that both E16 and E60 are self-competitive but not cross-competitive for envelope glycoprotein binding.
Similar articles
- Crystal structure of west nile virus envelope glycoprotein reveals viral surface epitopes.
Kanai R, Kar K, Anthony K, Gould LH, Ledizet M, Fikrig E, Marasco WA, Koski RA, Modis Y. Kanai R, et al. J Virol. 2006 Nov;80(22):11000-8. doi: 10.1128/JVI.01735-06. Epub 2006 Aug 30. J Virol. 2006. PMID: 16943291 Free PMC article. - The Fc region of an antibody impacts the neutralization of West Nile viruses in different maturation states.
Lee PD, Mukherjee S, Edeling MA, Dowd KA, Austin SK, Manhart CJ, Diamond MS, Fremont DH, Pierson TC. Lee PD, et al. J Virol. 2013 Dec;87(24):13729-40. doi: 10.1128/JVI.02340-13. Epub 2013 Oct 9. J Virol. 2013. PMID: 24109224 Free PMC article. - A plant-produced vaccine protects mice against lethal West Nile virus infection without enhancing Zika or dengue virus infectivity.
Lai H, Paul AM, Sun H, He J, Yang M, Bai F, Chen Q. Lai H, et al. Vaccine. 2018 Mar 27;36(14):1846-1852. doi: 10.1016/j.vaccine.2018.02.073. Epub 2018 Feb 26. Vaccine. 2018. PMID: 29490880 Free PMC article. - The molecular basis of antibody-mediated neutralization of West Nile virus.
Oliphant T, Diamond MS. Oliphant T, et al. Expert Opin Biol Ther. 2007 Jun;7(6):885-92. doi: 10.1517/14712598.7.6.885. Expert Opin Biol Ther. 2007. PMID: 17555373 Review. - Mapping and analysis of West Nile virus-specific monoclonal antibodies: prospects for vaccine development.
Throsby M, Ter Meulen J, Geuijen C, Goudsmit J, de Kruif J. Throsby M, et al. Expert Rev Vaccines. 2007 Apr;6(2):183-91. doi: 10.1586/14760584.6.2.183. Expert Rev Vaccines. 2007. PMID: 17408368 Review.
Cited by
- Development of Antibody-Based Therapeutics Against West Nile Virus in Plants.
Sun H, Lesio J, Chen Q. Sun H, et al. Methods Mol Biol. 2023;2585:211-225. doi: 10.1007/978-1-0716-2760-0_19. Methods Mol Biol. 2023. PMID: 36331777 - Plasticity of a critical antigenic determinant in the West Nile virus NY99 envelope protein domain III.
Plante JA, Torres M, Huang CY, Beasley DWC. Plante JA, et al. Virology. 2016 Sep;496:97-105. doi: 10.1016/j.virol.2016.05.024. Epub 2016 Jun 7. Virology. 2016. PMID: 27284640 Free PMC article. - The Japanese Encephalitis Antigenic Complex Viruses: From Structure to Immunity.
Khare B, Kuhn RJ. Khare B, et al. Viruses. 2022 Oct 8;14(10):2213. doi: 10.3390/v14102213. Viruses. 2022. PMID: 36298768 Free PMC article. Review. - Deep Mutational Scanning Comprehensively Maps How Zika Envelope Protein Mutations Affect Viral Growth and Antibody Escape.
Sourisseau M, Lawrence DJP, Schwarz MC, Storrs CH, Veit EC, Bloom JD, Evans MJ. Sourisseau M, et al. J Virol. 2019 Nov 13;93(23):e01291-19. doi: 10.1128/JVI.01291-19. Print 2019 Dec 1. J Virol. 2019. PMID: 31511387 Free PMC article. - Identification of novel small-molecule inhibitors of West Nile virus infection.
Noueiry AO, Olivo PD, Slomczynska U, Zhou Y, Buscher B, Geiss B, Engle M, Roth RM, Chung KM, Samuel M, Diamond MS. Noueiry AO, et al. J Virol. 2007 Nov;81(21):11992-2004. doi: 10.1128/JVI.01358-07. Epub 2007 Aug 22. J Virol. 2007. PMID: 17715228 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases