Mitochondrial p32 is a critical mediator of ARF-induced apoptosis - PubMed (original) (raw)
Mitochondrial p32 is a critical mediator of ARF-induced apoptosis
Koji Itahana et al. Cancer Cell. 2008 Jun.
Abstract
The shared exon 2 of the p14ARF-p16INK4a locus is frequently mutated in human cancers. However, in contrast to the exon 1beta-encoded N-terminal half of ARF, the function of the exon 2-encoded C-terminal half of ARF has been elusive. Here, we report that the mitochondrial protein p32/C1QBP binds the ARF C terminus. We show that p32 is required for ARF to localize to mitochondria and induce apoptosis, and that ARF mutations specifically disrupting p32 binding can impair both of these functions. Wild-type ARF, but not a p32-binding-deficient ARF mutant, localizes to mitochondria, reduces mitochondrial membrane potential, and sensitizes cells to p53-induced apoptosis. These findings provide a potential explanation for the frequent human cancer mutations targeting the ARF C terminus.
Figures
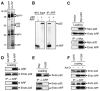
Figure 1. ARF Interacts with p32
(A) Identification of ARF binding proteins by mass spectrometry. Extracts from U2OS cells infected with adenovirus expressing GFP or ARF were immunoprecipitated with an anti-ARF antibody, resolved by SDS-PAGE, and visualized by silver staining. Peptide bands unique to samples infected with Ad-ARF were subjected to mass spectrometry. ARF, p32, B23 (a known ARF-binding protein), and IgG heavy and light chains (IgG-H and IgG-L) are indicated. (B) Direct interaction between p32 and ARF. Full-length p32 and ARF were in vitro translated in the presence of 35S-methionine. Equal amounts of p32 protein were incubated with (+) or without (−) ARF followed by immunoprecipitation with an anti-ARF antibody. The resulting ARF immunoprecipitates were separated by SDS-PAGE and visualized by autoradiography. (C) Coimmunoprecipitation (co-IP) of endogenous ARF with ectopically expressed p32. H1299 cells (p53-negative, ARF-positive) were transfected with Myc-p32 plasmid for 2 days, and cell extracts were immunoprecipitated with anti-Myc antibody. Western blotting was performed with anti-Myc and anti-ARF antibodies. Loading control represents 10% of cell lysate used for co-IP. (D) Co-IP of endogenous p32 with ectopically expressed ARF. Extracts from U2OS cells infected with Ad-ARF were immunoprecipitated with anti-ARF antibody. Western blotting was performed with anti-ARF and anti-p32 antibodies. Loading control represents approximately 10% of cell lysate used for co-IP. (E) Co-IP of ARF with p32from Raji cells. Cell extracts from Raji (Burkitt's lymphoma, expressing endogenous ARF and p32) cells were immunoprecipitated with anti-ARF antibody and blotted with anti-p32 and anti-ARF antibodies. HL60 (leukemia, ARF-negative) cells served as a negative control. (F) Increased ARF-p32 interaction following actinomycin D treatment. H1299 cells were treated with (+) or without (−) 5 nM actinomycin D for 36 hr. Cell extracts were immunoprecipitated with anti-ARF antibody and blotted with anti-p32 and anti-ARF antibodies. Extracts from U2OS (ARF-negative) cells served as a negative control. Loading control represents approximately 10% of cell lysate used for co-IP.
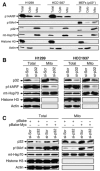
Figure 2. p32 Is Critical for a Subset of ARF to Localize to Mitochondria
(A) Mitochondrial localization of endogenous ARF. p53-negative H1299 and HCC1937 cells, and _p53_−/− MEFs were either prepared as SDS lysates (Total) or separated into cytoplasmic (Cyto) and mitochondrial (Mito) fractions. Equal amounts of protein were loaded in each lane. The membrane was blotted with antibodies to p14ARF, p19Arf, p32, mt-Hsp70 (mitochondrial heat shock protein 70, a mitochondrial protein marker), Histone H3 (a nuclear protein marker), and Actin (a cytoplasmic protein marker). (B) Downregulation of p32 by siRNA inhibits ARF mitochondrial localization. H1299 and HCC1937 cells were transiently transfected with a scrambled siRNA (si-Scr) or a p32 siRNA (si-p32) for 3 days. The cells were then isolated for mitochondrial fractions and analyzed by western blotting with indicated antibodies. (C) Downregulation of p32 by siRNA inhibits localization of oncogenic Myc-induced mouse p19Arf to mitochondria. Freshly prepared wild-type MEFs were infected with retrovirus carrying either no insert (pBabe) or oncogenic c-Myc (pBabe-Myc) and selected with puromycin for 3 days. The cells were then transiently transfected with scrambled siRNA (si-Scr) or p32 siRNA (si-p32) for 3 days. Total cell lysates and mitochondrial fractions were prepared and analyzed by western blotting with indicated antibodies.
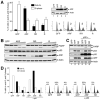
Figure 3. p32 Is Critical for ARF-Induced Apoptosis
(A) p32 depletion causes resistance of cells to ARF-induced apoptosis. U2OS cells were transfected for 24 hr with indicated siRNA and then infected for 2 days with adenovirus expressing either GFP or ARF. Cells that survived ARF overexpression were counted, and the percentages of surviving cells relative to GFP-expressing control cells were plotted. Error bars represent the mean ± SD of four independent experiments. (B) p32 depletion attenuates ARF-induced apoptosis. U2OS cells were treated as described in (A) and collected for flow cytometry analysis of cell-cycle profiles. The percentages of cells in S and sub-G1 phases are shown. (C) p32 depletion causes resistance of cells to ARF-induced PARP cleavage. U2OS cells were transfected with siRNA and infected with adenoviruses as described above. Cell extracts were analyzed by western blotting with antibodies to indicated proteins. (D) p32 depletion causes resistance of wild-type MEFs to c-Myc-induced apoptosis. Freshly prepared MEFs were infected with retrovirus carrying either no insert (pBabe) or oncogenic c-Myc (pBabe-Myc) and were selected with puromycin for 3 days. The cells were then transfected with siRNA for 2 days and subsequently cultured with (+) or without (−) serum for 24 hr. The percentages of cells surviving the treatment were quantified, and the results were plotted. Error bars represent the mean ± SD of four independent experiments. (E) A portion of the MEFs described in (D) was analyzed for p32 knockdown, p19Arf expression, and PARP cleavage by western blotting. Note that a second p32 siRNA oligos (si-p32-2) gives rise to identical results as the first p32 siRNA (si-p32).
Figure 4. Cancer-Derived ARF C-Terminal Mutations Impair ARF-p32 Binding
(A) Mapping of the p32 binding domain on human ARF. Extracts from U2OS cells transfected with plasmids encoding various ARF deletion mutants were immunoprecipitated with anti-Myc antibody and blotted with anti-Myc and anti-p32 antibodies. A diagram of human ARF is shown with the indicated p32-binding site (82–101). (B) Cancer-derived mutations in the p14ARF/p16INK4a gene locus are clustered at the 5′ portion of exon 2. The reported single missense mutations that affect the p16 sequence alone (gray bar) or both p16 and ARF (black bar) are shown in regard to the amino acid sequence of p16 (Greenblatt, 2006). Hotspot mutations at Arg98 and Arg99 of human ARF, overlapping with His83 and Asp84 in p16, are indicated with asterisks. (C) Amino acid sequence of the exon 2-encoded human ARF C terminus. Individual mutations analyzed in (D) are indicated with downward arrows. (D) Cancer-derived ARF C-terminal Arg mutations impair p32 binding. U2OS cells were transiently transfected with indicated plasmids expressing various Myc-tagged ARF proteins. Cell extracts were immunoprecipitated with anti-Myc antibody and blotted with the indicated antibodies.
Figure 5. p32-Binding-Deficient ARF Mutants Are Attenuated in Apoptotic Function
(A) ARF C-terminal Arg mutations impair ARF's apoptotic function. U2OS cells were infected with adenoviruses expressing either wild-type ARF (ARF) or p32-binding-deficient ARF RR98,99LS (RR) and ARF R98Q (R) mutants for 2 days. Cells (both floating and attached) were collected for flow cytometry analysis of cell-cycle distribution. The fraction of cells in sub-G1 phase (filled bar) and S phase (open bar) and the level of ARF protein expression (inset) are shown. Cell-cycle profiles are also shown to the right. Error bars represent the mean ± SD of three independent experiments. p < 0.01 for pairwise comparisons of cells in sub-G1 infected by wild-type ARF and each of the mutants. (B) ARF C-terminal Arg mutations impair ARF-mediated PARP cleavage, but not p53 induction. U2OS cells treated as described in (A) were analyzed for PARP cleavage and p53 induction by western blotting. (C) p32 enhances ARF-induced PARP cleavage. Cells were infected with adenoviruses expressing the indicated proteins for 2 days and then harvested to be analyzed for PARP cleavage. Note that p32-enhanced PARP cleavage was seen only with wild-type ARF, but not the p32-binding-deficient mutant ARF. (D) p32 enhances ARF-induced apoptosis but not ARF-induced cell-cycle arrest. U2OS cells were infected with adenoviruses expressing the indicated proteins for 2 days and harvested to determine the percentages of cells in sub-G1 and S phase fractions by flow cytometry analysis.
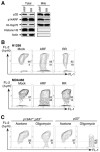
Figure 6. Wild-type ARF, but Not a p32-Binding-Deficient ARF Mutant, Localizes to Mitochondria and Reduces Mitochondrial Membrane Potential
(A) A p32-binding-deficient ARF mutant is impaired in mitochondrial localization. p53-negative H1299 cells were infected with adenovirus expressing either wild-type ARF (ARF) or a p32-binding-deficient ARF mutant (RR). Total SDS lysates and mitochondrial fractions were prepared 2 days after infection and analyzed by western blotting with antibodies to p32, ARF, mt-Hsp70, HistoneH3, and Actin. (B) Wild-type ARF, but not a p32-binding-deficient ARF (RR) mutant, reduces mitochondrial membrane potential (Δψm). p53-negative H1299 and MDA468 cells were either uninfected (Mock) or infected with adenoviruses expressing wild-type ARF (ARF) or a p32-binding-deficient ARF mutant (RR) for 2 days. Δψm was measured by incubating the cells with JC-1 dye followed by flow cytometry analysis. In nonapoptotic cells (high Δψm), JC-1 exists as a monomer in the cytosol (green, FL-1) and also accumulates as aggregates in the mitochondria, which stains red (FL-2). In apoptotic cells (low Δψm), JC-1 exists in monomeric form and stains the cytosol green. Cells with preserved Δψm are high in both FL-1 and FL-2 (top right of graph). Cells with loss of Δψm are high in FL-1 and low in FL-2 (bottom of graph). Flow cytometry data plots are shown with percentage of total events indicated. Data are representative of three independent experiments. (C) Endogenous ARF sensitizes cells to mitochondrial stress, leading to reduction of Δψm. Freshly prepared _p19Arf_−/−;_p53_−/− and _p53_−/− MEFs were incubated with acetone (as a control) or 2 μg/ml of oligomycin, an inhibitor of mitochondrial ATP synthase widely used to generate mitochondrial stress in cultured cells. After 48 hr of oligomycin treatment, the cells were collected and analyzed for Δψm by JC-1 dye.
Figure 7. ARF Synergizes with p53 to Induce Cytochrome c Release
(A) ARF expression did not induce cytochrome c release in H1299 cells. p53-negative H1299 and p53-positive U2OS cells were infected with increasing amounts of adenovirus expressing ARF for 2 days. Total SDS lysates and cytoplasmic fractions were prepared and analyzed by western blotting with indicated antibodies. (B) ARF synergizes with p53 to induce PARP cleavage. Saos2 (p53-negative) cells were infected with adenovirus expressing GFP, ARF, or p53 as indicated. Cells were analyzed by western blotting with antibodies against the indicated proteins. Note that the level of ectopically expressed ARF in the Saos2 cells is comparable to that of endogenous ARF in the H1299 cells. (C) A low level of p53 induces cytochrome c release in ARF-positive cells. _p19Arf_−/−;_p53_−/− and _p53_−/− MEFs were infected with adenovirus expressing p53 at a low moi. Two days after infection, total cell lysates and cytoplasmic fractions were prepared and analyzed by western blotting.
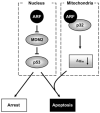
Figure 8. A Model for the Function of Mitochondrial ARF
Nuclear ARF stabilizes and activates p53 through its N-terminal interaction with and inhibition of MDM2 (left). A subset of ARF also localizes to the mitochondria through its C-terminal interaction with mitochondrial p32 (right). The mitochondria-localized ARF reduces Δψm and sensitizes cells to p53-induced apoptosis.
Similar articles
- Amino terminal hydrophobic import signals target the p14(ARF) tumor suppressor to the mitochondria.
Irvine M, Philipsz S, Frausto M, Mijatov B, Gallagher SJ, Fung C, Becker TM, Kefford RF, Rizos H. Irvine M, et al. Cell Cycle. 2010 Feb 15;9(4):829-39. doi: 10.4161/cc.9.4.10785. Epub 2010 Mar 2. Cell Cycle. 2010. PMID: 20107316 - Systematic genetic dissection of p14ARF-mediated mitochondrial cell death signaling reveals a key role for p21CDKN1 and the BH3-only protein Puma/bbc3.
Hemmati PG, Müer A, Gillissen B, Overkamp T, Milojkovic A, Wendt J, Dörken B, Daniel PT. Hemmati PG, et al. J Mol Med (Berl). 2010 Jun;88(6):609-22. doi: 10.1007/s00109-010-0606-5. Epub 2010 Apr 25. J Mol Med (Berl). 2010. PMID: 20419447 - ARF in the mitochondria: the last frontier?
Itahana K, Clegg HV, Zhang Y. Itahana K, et al. Cell Cycle. 2008 Dec;7(23):3641-6. doi: 10.4161/cc.7.23.7105. Epub 2008 Dec 16. Cell Cycle. 2008. PMID: 19033735 - The ARF-p53 senescence pathway in mouse and human cells.
Wadhwa R, Sugihara T, Taira K, Kaul SC. Wadhwa R, et al. Histol Histopathol. 2004 Jan;19(1):311-6. doi: 10.14670/HH-19.311. Histol Histopathol. 2004. PMID: 14702199 Review. - [Role of the tumor suppressor ARF in oncogenesis].
Pimkina IuS, Dorosevich AE. Pimkina IuS, et al. Arkh Patol. 2009 Jan-Feb;71(1):60-3. Arkh Patol. 2009. PMID: 19514363 Review. Russian.
Cited by
- Chaperone-like protein p32 regulates ULK1 stability and autophagy.
Jiao H, Su GQ, Dong W, Zhang L, Xie W, Yao LM, Chen P, Wang ZX, Liou YC, You H. Jiao H, et al. Cell Death Differ. 2015 Nov;22(11):1812-23. doi: 10.1038/cdd.2015.34. Epub 2015 Apr 23. Cell Death Differ. 2015. PMID: 25909887 Free PMC article. - Epstein-Barr virus nuclear antigen 3C (EBNA3C) interacts with the metabolism sensing C-terminal binding protein (CtBP) repressor to upregulate host genes.
Ohashi M, Hayes M, McChesney K, Johannsen E. Ohashi M, et al. PLoS Pathog. 2021 Mar 15;17(3):e1009419. doi: 10.1371/journal.ppat.1009419. eCollection 2021 Mar. PLoS Pathog. 2021. PMID: 33720992 Free PMC article. - Elevated expression of HABP1 is correlated with metastasis and poor survival in breast cancer patients.
Niu M, Sun S, Zhang G, Zhao Y, Pang D, Chen Y. Niu M, et al. Am J Cancer Res. 2015 Feb 15;5(3):1190-8. eCollection 2015. Am J Cancer Res. 2015. PMID: 26045997 Free PMC article. - High molecular weight fibroblast growth factor 2 induces apoptosis by interacting with complement component 1 Q subcomponent-binding protein in vitro.
Hong X, Yu Z, Chen Z, Jiang H, Niu Y, Huang Z. Hong X, et al. J Cell Biochem. 2018 Nov;119(11):8807-8817. doi: 10.1002/jcb.27131. Epub 2018 Aug 29. J Cell Biochem. 2018. PMID: 30159917 Free PMC article. - p32/gC1qR is indispensable for fetal development and mitochondrial translation: importance of its RNA-binding ability.
Yagi M, Uchiumi T, Takazaki S, Okuno B, Nomura M, Yoshida S, Kanki T, Kang D. Yagi M, et al. Nucleic Acids Res. 2012 Oct;40(19):9717-37. doi: 10.1093/nar/gks774. Epub 2012 Aug 16. Nucleic Acids Res. 2012. PMID: 22904065 Free PMC article.
References
- Chappell JB, Greville GD. Effects of oligomycin on respiration and swelling of isolated liver mitochondria. Nature. 1961;190:502–504. - PubMed
- Dedio J, Jahnen-Dechent W, Bachmann M, Muller-Esterl W. The multiligand-binding protein gC1qR, putative C1q receptor, is a mitochondrial protein. J Immunol. 1998;160:3534–3542. - PubMed
- Enomoto T, Lindstrom MS, Jin A, Ke H, Zhang Y. Essential role of the B23/NPM core domain in regulating ARF binding and B23 stability. J Biol Chem. 2006;281:18463–18472. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA100302/CA/NCI NIH HHS/United States
- K01 CA087580-06/CA/NCI NIH HHS/United States
- K01 CA087580/CA/NCI NIH HHS/United States
- K01 CA087580-03/CA/NCI NIH HHS/United States
- K01 CA087580-07/CA/NCI NIH HHS/United States
- K01 CA087580-05/CA/NCI NIH HHS/United States
- K01 CA087580-02/CA/NCI NIH HHS/United States
- K01 CA087580-04/CA/NCI NIH HHS/United States
- K01 CA087580-01/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous