Residues in the alternative reading frame tumor suppressor that influence its stability and p53-independent activities - PubMed (original) (raw)
Residues in the alternative reading frame tumor suppressor that influence its stability and p53-independent activities
Anne di Tommaso et al. Exp Cell Res. 2009.
Abstract
The Alternative Reading Frame (ARF) protein suppresses tumorigenesis through p53-dependent and p53-independent pathways. Most of ARF's anti-proliferative activity is conferred by sequences in its first exon. Previous work showed specific amino acid changes occurred in that region during primate evolution, so we programmed those changes into human p14ARF to assay their functional impact. Two human p14ARF residues (Ala(14) and Thr(31)) were found to destabilize the protein while two others (Val(24) and Ala(41)) promoted more efficient p53 stabilization and activation. Despite those effects, all modified p14ARF forms displayed robust p53-dependent anti-proliferative activity demonstrating there are no significant biological differences in p53-mediated growth suppression associated with simian versus human p14ARF residues. In contrast, p53-independent p14ARF function was considerably altered by several residue changes. Val(24) was required for p53-independent growth suppression whereas multiple residues (Val(24), Thr(31), Ala(41) and His(60)) enabled p14ARF to block or reverse the inherent chromosomal instability of p53-null MEFs. Together, these data pinpoint specific residues outside of established p14ARF functional domains that influence its expression and signaling activities. Most intriguingly, this work reveals a novel and direct role for p14ARF in the p53-independent maintenance of genomic stability.
Figures
Figure 1
Evolutionary changes in the N-terminus of primate p14ARF proteins. A, alignment of amino acid sequences of exon 1β of different primate species. Seven different sequences are aligned from human (HU, Homo sapiens), great apes (GA, both Pan paniscus [pygmy chimpanzee] and Gorilla gorilla [gorilla]), old world monkeys (OM, both Papio ursinus [baboon] and Macaca arctoides [stump-tail macaque]), new world monkey (NM, Ateles belzebuth [long-haired spider monkey]) and prosimian (Pro, Varecia variegate [lemur]). p14ARF residues that are identical in mouse p19ARF are indicated by bolded lines above the human sequence. B, Schematic of the primate evolutionary tree depicting the timeline of divergence between various primates. M, millions of years.
Figure 2
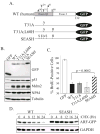
Expression and activity of GFP-tagged p14ARF mutants in human U20S cells. A, schematic of the C-terminal GFP-tagged forms of p14ARF that were assayed. B, immunoblot analysis of p53, Mdm2, NPM, tubulin (loading control), and GFP or ARF-GFP expression levels in whole cell lysates. C, BrdU incorporation in ARF-positive cells showing SEASH ARF-GFP is statistically impaired in blocking DNA synthesis compared to other p14ARF forms. Data were quantified from 3 or more experiments (error bars representing the standard deviation from the mean). D, half-life analyses showing SEASH residues from new world monkeys stabilize p14ARF. Cells expressing WT or SEASH p14ARF-GFP were treated with cycloheximide (CHX) for the indicated times, and total cell lysates were analyzed by immunoblotting for ARF and GAPDH (loading control).
Figure 3
Simian-specific residues Ala31 and Ser14 stabilize the p14ARF protein. A, schematic of the untagged forms of p14ARF assayed throughout the remainder of the study. B, Western blot of ARF and GAPDH (loading control) showing differential expression levels of the indicated ARF forms following efficient and equivalent transduction (> 95%) of MSCV-ARF-IRES-GFP retroviruses into _ARF_-null MEFs. C, half-life analyses of the WT p14ARF and modified forms in human U20S cells. Cells were treated with cycloheximide (CHX) for the indicated times two days after transfection with the indicated ARF construct. Representative Western blots of ARF and GAPDH (loading control) expression in whole cell lysates are shown, with the calculated half-lives denoted in parentheses to the left of each set of blots. Half-lives were determined by quantification of blots using ImageJ software.
Figure 4
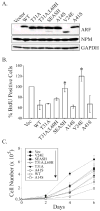
Limited impact of simian ARF residues on p53-dependent cell cycle arrest. (A, B and D), _ARF_−/− MEFs were infected with MSCV-IRES-GFP vector control or the indicated MSCV-ARF-IRES-GFP retroviruses and analyzed two days later. A, representative Western blots examining expression of ARF, GAPDH (loading control), GFP (indicator of equivalent infection), endogenous NPM and regulators of p53 signaling (p53, Mdm2 and p21). B, representative histograms of the DNA content in successfully infected cells showing similar cell cycle inhibitory activities for the different ARF forms. G1 and G2-M peaks are shaded in gray while the percentage of cells in S phase cells is denoted and highlighted graphically in black. Notably, when results from 3 or more experiments were pooled together, no statistical difference in S phase reduction were seen between any of the modified ARF forms (Fig. S3). C, bar graph depicting the relative ability of each ARF protein to stimulate p53 transcriptional activity in U20S cells co-expressing a p53-luciferase reporter construct. D, bar graph comparing the percent of BrdU incorporation in ARF-positive cells (vector controls normalized to 100%). In both C and D, data were averaged from at least three independent experiments and subjected to student’s t-test analyses. Error bars represent standard deviations from the mean, and asterisks (*) indicate statistically significant differences (p<0.05) for the sample compared to vector control and human ARF.
Figure 5
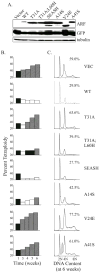
Differential p53-independent anti-proliferative activities conferred by human versus simian-specific ARF residues. MEFs lacking p53, Mdm2 and ARF were infected with the indicated retroviruses and assays begun 72 to 96 hours post-infection. A, representative Western blots showing relative expression levels of the different ARF forms, NPM and GAPDH (loading control) in the samples. B, bar graph comparing the percent of BrdU incorporation in ARF-positive cells (vector [vec] controls normalized to 100%). Error bars represent the standard deviation from the mean for three or more experiments. Asterisks (*) denote statistically significant differences (p<0.05) exist between the indicated samples and human ARF. _C_, representative growth curves of successfully infected populations (>95% GFP-positive) expressing vector (Vec) or the indicated p14ARF proteins. Note that the key is aligned to correspond directly to the position of each curve in the plot, with Vec cells at the top (most rapidly proliferating) and A14S expressors (most slowly proliferating) at the bottom.
Figure 6
Differential p53-independent effects of human versus simian-specific ARF residues on chromosomal instability. MEFs lacking p53, Mdm2 and ARF were infected with the indicated retroviruses and maintained on a 3T3 protocol for 6 weeks. Representative data from several independent experiments are shown in A–C. A, Western blots confirming sustained expression of the different ARF forms and GFP in the populations at 3 weeks post-infection. Tubulin served as a loading control. B, comparative bar graphs of the percent tetraploidy in each population over time, as measured by flow cytometric analyses of DNA content. Increases (diagonally striped bars) or decreases (open bars) relative to the percent tetraploidy at week 1 after infection (filled bars) are depicted. C, histograms of DNA content for each population at week 6 post-infection are shown, with the percent of tetraploid cells designated. Although not shown, flow cytometric analyses of GFP fluorescence revealed all populations remained 88 to 99 percent GFP positive by week 6.
Similar articles
- ARF function does not require p53 stabilization or Mdm2 relocalization.
Korgaonkar C, Zhao L, Modestou M, Quelle DE. Korgaonkar C, et al. Mol Cell Biol. 2002 Jan;22(1):196-206. doi: 10.1128/MCB.22.1.196-206.2002. Mol Cell Biol. 2002. PMID: 11739734 Free PMC article. - p53-Dependent and -independent functions of the Arf tumor suppressor.
Sherr CJ, Bertwistle D, DEN Besten W, Kuo ML, Sugimoto M, Tago K, Williams RT, Zindy F, Roussel MF. Sherr CJ, et al. Cold Spring Harb Symp Quant Biol. 2005;70:129-37. doi: 10.1101/sqb.2005.70.004. Cold Spring Harb Symp Quant Biol. 2005. PMID: 16869746 Review. - CARF is a novel protein that cooperates with mouse p19ARF (human p14ARF) in activating p53.
Hasan MK, Yaguchi T, Sugihara T, Kumar PK, Taira K, Reddel RR, Kaul SC, Wadhwa R. Hasan MK, et al. J Biol Chem. 2002 Oct 4;277(40):37765-70. doi: 10.1074/jbc.M204177200. Epub 2002 Aug 1. J Biol Chem. 2002. PMID: 12154087 - ARF-BP1/Mule is a critical mediator of the ARF tumor suppressor.
Chen D, Kon N, Li M, Zhang W, Qin J, Gu W. Chen D, et al. Cell. 2005 Jul 1;121(7):1071-83. doi: 10.1016/j.cell.2005.03.037. Cell. 2005. PMID: 15989956 - DNA damage, p14ARF, nucleophosmin (NPM/B23), and cancer.
Gjerset RA. Gjerset RA. J Mol Histol. 2006 Sep;37(5-7):239-51. doi: 10.1007/s10735-006-9040-y. Epub 2006 Jul 20. J Mol Histol. 2006. PMID: 16855788 Review.
Cited by
- Novel PDGFRB rearrangement in multifocal infantile myofibromatosis is tumorigenic and sensitive to imatinib.
Hassan M, Butler E, Wilson R, Roy A, Zheng Y, Liem P, Rakheja D, Pavlick D, Young LL, Rosenzweig M, Erlich R, Ali SM, Leavey PJ, Parsons DW, Skapek SX, Laetsch TW. Hassan M, et al. Cold Spring Harb Mol Case Stud. 2019 Oct 23;5(5):a004440. doi: 10.1101/mcs.a004440. Print 2019 Oct. Cold Spring Harb Mol Case Stud. 2019. PMID: 31645346 Free PMC article. - Oncogenic RABL6A promotes NF1-associated MPNST progression in vivo.
Kohlmeyer JL, Kaemmer CA, Lingo JJ, Voigt E, Leidinger MR, McGivney GR, Scherer A, Koppenhafer SL, Gordon DJ, Breheny P, Meyerholz DK, Tanas MR, Dodd RD, Quelle DE. Kohlmeyer JL, et al. Neurooncol Adv. 2022 Apr 9;4(1):vdac047. doi: 10.1093/noajnl/vdac047. eCollection 2022 Jan-Dec. Neurooncol Adv. 2022. PMID: 35571990 Free PMC article. - The ARF tumor suppressor prevents chromosomal instability and ensures mitotic checkpoint fidelity through regulation of Aurora B.
Britigan EM, Wan J, Zasadil LM, Ryan SD, Weaver BA. Britigan EM, et al. Mol Biol Cell. 2014 Sep 15;25(18):2761-73. doi: 10.1091/mbc.E14-05-0966. Epub 2014 Jul 23. Mol Biol Cell. 2014. PMID: 25057018 Free PMC article. - NMI mediates transcription-independent ARF regulation in response to cellular stresses.
Li Z, Hou J, Sun L, Wen T, Wang L, Zhao X, Xie Q, Zhang SQ. Li Z, et al. Mol Biol Cell. 2012 Dec;23(23):4635-46. doi: 10.1091/mbc.E12-04-0304. Epub 2012 Oct 3. Mol Biol Cell. 2012. PMID: 23034180 Free PMC article. - Dual Role of the Alternative Reading Frame ARF Protein in Cancer.
Fontana R, Ranieri M, La Mantia G, Vivo M. Fontana R, et al. Biomolecules. 2019 Mar 4;9(3):87. doi: 10.3390/biom9030087. Biomolecules. 2019. PMID: 30836703 Free PMC article. Review.
References
- Lowe SW, Sherr CJ. Tumor suppression by Ink4a-Arf: progress and puzzles. Curr Op Genet Dev. 2003;13:77–83. - PubMed
- Quelle DE, Zindy F, Ashmun RA, Sherr CJ. Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell. 1995;83:993–1000. - PubMed
- Ruas M, Peters G. The p16INK4a/CDKN2A tumor suppressor and its relatives. Biochim Biophys Acta. 1998;1378:F115–77. - PubMed
- Carnero A, Hudson JD, Price CM, Beach DH. p16(INK4A) and p19(ARF) act in overlapping pathways in cellular immortalization. Nat Cell Biol. 2000;2:148–155. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous