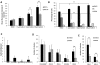Haploinsufficiency for ribosomal protein genes causes selective activation of p53 in human erythroid progenitor cells - PubMed (original) (raw)
. 2011 Mar 3;117(9):2567-76.
doi: 10.1182/blood-2010-07-295238. Epub 2010 Nov 10.
Anupama Narla, Katherine Lin, Ann Mullally, Nirmalee Abayasekara, Christine Megerdichian, Frederick H Wilson, Treeve Currie, Arati Khanna-Gupta, Nancy Berliner, Jeffery L Kutok, Benjamin L Ebert
Affiliations
- PMID: 21068437
- PMCID: PMC3062351
- DOI: 10.1182/blood-2010-07-295238
Haploinsufficiency for ribosomal protein genes causes selective activation of p53 in human erythroid progenitor cells
Shilpee Dutt et al. Blood. 2011.
Abstract
Haploinsufficiency for ribosomal protein genes has been implicated in the pathophysiology of Diamond-Blackfan anemia (DBA) and the 5q-syndrome, a subtype of myelodysplastic syndrome. The p53 pathway is activated by ribosome dysfunction, but the molecular basis for selective impairment of the erythroid lineage in disorders of ribosome function has not been determined. We found that p53 accumulates selectively in the erythroid lineage in primary human hematopoietic progenitor cells after expression of shRNAs targeting RPS14, the ribosomal protein gene deleted in the 5q-syndrome, or RPS19, the most commonly mutated gene in DBA. Induction of p53 led to lineage-specific accumulation of p21 and consequent cell cycle arrest in erythroid progenitor cells. Pharmacologic inhibition of p53 rescued the erythroid defect, whereas nutlin-3, a compound that activates p53 through inhibition of HDM2, selectively impaired erythropoiesis. In bone marrow biopsies from patients with DBA or del(5q) myelodysplastic syndrome, we found an accumulation of nuclear p53 staining in erythroid progenitor cells that was not present in control samples. Our findings indicate that the erythroid lineage has a low threshold for the induction of p53, providing a basis for the failure of erythropoiesis in the 5q-syndrome, DBA, and perhaps other bone marrow failure syndromes.
© 2011 by The American Society of Hematology
Figures
Figure 1
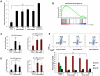
Decreased expression of RPS14 or RPS19 activates the p53 pathway. (A) Increased levels of total p53 protein in primary human bone marrow cells as analyzed by intracellular flow cytometry. (B) RPS14 shRNAs significantly increase the expression of downstream target genes of p53, as assessed by gene set enrichment analysis using a set of published p53 target genes. Genes are ranked according to their differential expression between cells expressing RPS14 shRNA and control shRNAs targeting the luciferase gene. Genes in the p53 gene set are marked with vertical bars, and the enrichment score is shown in green. (C-D) The mRNA expression of p21 and Bax in primary human bone marrow cells, relative to β-actin mRNA, was measured by quantitative real time PCR. (E) Cell cycle status was analyzed by flow cytometry using 7-AAD and BrdU. Numbers in the flow plot represent the percentage of cells in different phases of cell cycle. Results shown for each experiment are representative of 3 independent experiments performed in triplicate (mean ± SEM). *P < .05. **P < .01.
Figure 2
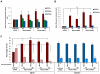
Lineage specificity of p53 accumulation, p21 levels, and cell cycle arrest. (A) Levels of p53 were determined by intracellular flow cytometry in cells expressing control (luciferase), RPS14, or RPS19 shRNAs. Lineage-specific activation of p53 protein was determined by staining for erythroid (CD71), megakaryocytic (CD41a), and myelomonocytic (CD11b) cell surface markers. (B) Levels of p21 were determined by intracellular flow cytometry in cells expressing control (luciferase), RPS14, or RPS19 shRNAs. Lineage-specific activation of p21 protein was determined by staining for erythroid (CD71) and myelomonocytic (CD11b) cell surface markers. (C) Primary human bone marrow cells were infected with control or ribosomal gene shRNAs and allowed to grow and differentiate over 5 days in the presence of cytokines supporting erythroid and myeloid differentiation. Cells were then labeled with BrdU, 7-AAD, and lineage markers (CD11b and CD71). Results in each bar graph are the composite data from 3 independent experiments performed in triplicate (mean ± SEM). *P < .05. **P < .01.
Figure 3
Partial knockdown of RPS14 leads to binding of RPL11 to HDM2. (A) Western blots showing the increased levels of p53 and RPL11 in A549 cells expressing the indicated shRNAs for 72 hours. Tubulin was used as a loading control. (B) Immunofluorescent images of discrete or disrupted nucleoli in A549 cells expressing the indicated shRNAs or cells treated with 10 ng/mL actinomycin D (Act. D) for 12 hours, respectively. Nucleoli were stained with an antibody against nucleophosmin (B23). Images were visualized with a fluorescent microscope (Olympus IX71) and with the 100× objective lens (Carl Zeiss) and analyzed using IPlab 3.6.5 software. Cells were mounted in Vectashield mounting medium containing 4,6-diamidino-2-phenylindole as counterstain for DNA. Images were visualized with Olympus PLanF1 using 100×/1.30 Oil objective (Carl Zeiss) and captured using SensiCam High Performance camera (The Cooke Corporation). (C) Immunoprecipitation of A549 cell lysates was performed using anti-HDM2 or normal rabbit IgG antibodies. Western blots show the levels of HDM2 and RPL11 in the immunoprecipitates.
Figure 4
Activation of p53 by nutlin-3 recapitulates the effects of RPS14 or RPS19 shRNAs. (A) Primary human CD34+ cells were treated with nutlin-3 for 72 hours and cultured in cytokines supporting erythroid and myeloid differentiation. Levels of p53 in erythroid and myeloid lineage cells were assessed by flow cytometry. (B) Cell cycle status of the nutlin-3-treated cells was analyzed by flow cytometry after labeling with BrdU and 7-AAD. (C) Primary human CD34+ cells were treated with nutlin-3 and cultured in cytokines supporting erythroid and myeloid differentiation for 10 days. The ratio of erythroid to myeloid cells was assessed by flow cytometric analysis GlyA (erythroid) and CD11b (myeloid) expression. (D) A total of 50 000 total bone marrow cells from mice injected with vehicle (dimethyl sulfoxide [DMSO]) or 93 mg/kg of nutlin-3 at alternate day for 5 days were plated on methylcellulose, and colonies were counted after 7 to 10 days in culture (mean ± SEM; n = 5 mice in each group). (E) Bone marrow cells from mice injected with vehicle (DMSO) or 93 mg/kg of nutlin-3 at alternate day for 5 days were plated on methylcellulose, and colonies were counted after 2 and 5 days in culture for CFU-E and BFU-E, respectively (mean ± SEM; n = 5 mice in each group). Results of all in vitro nutlin-3 treatment experiments are representative of 3 independent experiments performed in triplicate (mean ± SEM). *P < .05. **P < .01.
Figure 5
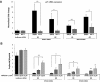
Treatment of cells with PFT-α rescues the erythroid phenotype of ribosomal gene shRNAs. (A) After infection of CD34+ cells with either a control (luciferase) gene or ribosomal gene shRNAs, cells were treated with different concentrations of PFT-α, an inhibitor of p53 function. After 72 hours, expression of p21 mRNA, relative to β-actin, was analyzed by quantitative RT-PCR. (B) The ratio of erythroid to myeloid lineage cells was also analyzed by flow cytometry using antibodies against surface markers GlyA (erythroid) and CD11b (myeloid) after 8 days in culture. Results of PFT-α experiments are representative of 2 independent experiments performed in triplicate (mean ± SEM). *P < .05. **P < .01.
Figure 6
p53 staining of bone marrow biopsies. Immunohistochemistry for p53 on histologic sections of bone marrow biopsies from patients with (A) aplastic anemia, (B) autoimmune hemolytic anemia, (C-D) DBA, and (E-F) MDS with del(5q). (A-F) Original magnification ×400. The samples were analyzed using an Olympus BX41 microscope with the objective lens of 40×/0.75 Olympus UPlanFL (Olympus). The pictures were taken using Olympus QColor5 and analyzed with acquisition software QCapture Pro v6.0 (QImaging) and Adobe Photoshop 6.0.
Figure 7
Colocalization of p53 and CD71 in 5q− and DBA patient samples. Histologic sections of bone marrow biopsies from patients were analyzed by double immunofluorescence for p53 and cell surface markers (CD71 or GlyA). The antibodies used are indicated in each panel. Bone marrow biopsies are shown from patients with hemolytic anemia (A-B), del(5q) MDS (C-D), and DBA (E-F). (A-F) Original magnification ×400. The stained slides were scanned using the TissueFAXS Plus automated microscopic workstation (Tissuegnostics) with a Zeiss Axiolmager Z1 using a 40× Zeiss objective using a high sensitivity digital monochrome camera for fluorescence microscopy (TissueGnostics), and captured using TissueFAXS 1.3 software (TissueGnostics). Final images were transferred into Adobe Photoshop 6.0 where TIF files were created. Scale bar represents 25 μm.
Comment in
- Drawing to a Diamond flush.
Ellis SR. Ellis SR. Blood. 2011 Mar 3;117(9):2558-9. doi: 10.1182/blood-2010-12-320036. Blood. 2011. PMID: 21372157
Similar articles
- Mice with a Mutation in the Mdm2 Gene That Interferes with MDM2/Ribosomal Protein Binding Develop a Defect in Erythropoiesis.
Kamio T, Gu BW, Olson TS, Zhang Y, Mason PJ, Bessler M. Kamio T, et al. PLoS One. 2016 Apr 4;11(4):e0152263. doi: 10.1371/journal.pone.0152263. eCollection 2016. PLoS One. 2016. PMID: 27042854 Free PMC article. - 5q- syndrome.
Boultwood J, Pellagatti A, Wainscoat JS. Boultwood J, et al. Curr Pharm Des. 2012;18(22):3180-3. doi: 10.2174/1381612811209023180. Curr Pharm Des. 2012. PMID: 22571696 Review. - Activation of the mTOR pathway by the amino acid (L)-leucine in the 5q- syndrome and other ribosomopathies.
Boultwood J, Yip BH, Vuppusetty C, Pellagatti A, Wainscoat JS. Boultwood J, et al. Adv Biol Regul. 2013 Jan;53(1):8-17. doi: 10.1016/j.jbior.2012.09.002. Epub 2012 Sep 13. Adv Biol Regul. 2013. PMID: 23031788 Review. - Rps14 haploinsufficiency causes a block in erythroid differentiation mediated by S100A8 and S100A9.
Schneider RK, Schenone M, Ferreira MV, Kramann R, Joyce CE, Hartigan C, Beier F, Brümmendorf TH, Germing U, Platzbecker U, Büsche G, Knüchel R, Chen MC, Waters CS, Chen E, Chu LP, Novina CD, Lindsley RC, Carr SA, Ebert BL. Schneider RK, et al. Nat Med. 2016 Mar;22(3):288-97. doi: 10.1038/nm.4047. Epub 2016 Feb 15. Nat Med. 2016. PMID: 26878232 Free PMC article. - L-Leucine improves the anaemia in models of Diamond Blackfan anaemia and the 5q- syndrome in a TP53-independent way.
Narla A, Payne EM, Abayasekara N, Hurst SN, Raiser DM, Look AT, Berliner N, Ebert BL, Khanna-Gupta A. Narla A, et al. Br J Haematol. 2014 Nov;167(4):524-528. doi: 10.1111/bjh.13069. Epub 2014 Aug 7. Br J Haematol. 2014. PMID: 25098371 Free PMC article.
Cited by
- Expansion of germline RPS20 mutation phenotype to include Diamond-Blackfan anemia.
Bhar S, Zhou F, Reineke LC, Morris DK, Khincha PP, Giri N, Mirabello L, Bergstrom K, Lemon LD, Williams CL, Toh Y, Elghetany MT, Lloyd RE, Alter BP, Savage SA, Bertuch AA. Bhar S, et al. Hum Mutat. 2020 Nov;41(11):1918-1930. doi: 10.1002/humu.24092. Epub 2020 Aug 30. Hum Mutat. 2020. PMID: 32790018 Free PMC article. - Translational control in cancer etiology.
Ruggero D. Ruggero D. Cold Spring Harb Perspect Biol. 2013 Feb 1;5(2):a012336. doi: 10.1101/cshperspect.a012336. Cold Spring Harb Perspect Biol. 2013. PMID: 22767671 Free PMC article. Review. - Inherited bone marrow failure syndromes in 2012.
Sakaguchi H, Nakanishi K, Kojima S. Sakaguchi H, et al. Int J Hematol. 2013 Jan;97(1):20-9. doi: 10.1007/s12185-012-1249-9. Epub 2012 Dec 28. Int J Hematol. 2013. PMID: 23271412 Review. - Frameshift mutation in p53 regulator RPL26 is associated with multiple physical abnormalities and a specific pre-ribosomal RNA processing defect in diamond-blackfan anemia.
Gazda HT, Preti M, Sheen MR, O'Donohue MF, Vlachos A, Davies SM, Kattamis A, Doherty L, Landowski M, Buros C, Ghazvinian R, Sieff CA, Newburger PE, Niewiadomska E, Matysiak M, Glader B, Atsidaftos E, Lipton JM, Gleizes PE, Beggs AH. Gazda HT, et al. Hum Mutat. 2012 Jul;33(7):1037-44. doi: 10.1002/humu.22081. Epub 2012 Apr 16. Hum Mutat. 2012. PMID: 22431104 Free PMC article. - Disruption of the 5S RNP-Mdm2 interaction significantly improves the erythroid defect in a mouse model for Diamond-Blackfan anemia.
Jaako P, Debnath S, Olsson K, Zhang Y, Flygare J, Lindström MS, Bryder D, Karlsson S. Jaako P, et al. Leukemia. 2015 Nov;29(11):2221-9. doi: 10.1038/leu.2015.128. Epub 2015 May 19. Leukemia. 2015. PMID: 25987256 Free PMC article.
References
- Draptchinskaia N, Gustavsson P, Andersson B, et al. The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nat Genet. 1999;21(2):169–175. - PubMed
- Ebert BL. Deletion 5q in myelodysplastic syndrome: a paradigm for the study of hemizygous deletions in cancer. Leukemia. 2009;23(7):1252–1256. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous