Activating mutations in the NT5C2 nucleotidase gene drive chemotherapy resistance in relapsed ALL - PubMed (original) (raw)
doi: 10.1038/nm.3078. Epub 2013 Feb 3.
Arianne Perez-Garcia, Zachary Carpenter, Hossein Khiabanian, Valeria Tosello, Maddalena Allegretta, Elisabeth Paietta, Janis Racevskis, Jacob M Rowe, Martin S Tallman, Maddalena Paganin, Giuseppe Basso, Jana Hof, Renate Kirschner-Schwabe, Teresa Palomero, Raul Rabadan, Adolfo Ferrando
Affiliations
- PMID: 23377281
- PMCID: PMC3594483
- DOI: 10.1038/nm.3078
Activating mutations in the NT5C2 nucleotidase gene drive chemotherapy resistance in relapsed ALL
Gannie Tzoneva et al. Nat Med. 2013 Mar.
Abstract
Acute lymphoblastic leukemia (ALL) is an aggressive hematological tumor resulting from the malignant transformation of lymphoid progenitors. Despite intensive chemotherapy, 20% of pediatric patients and over 50% of adult patients with ALL do not achieve a complete remission or relapse after intensified chemotherapy, making disease relapse and resistance to therapy the most substantial challenge in the treatment of this disease. Using whole-exome sequencing, we identify mutations in the cytosolic 5'-nucleotidase II gene (NT5C2), which encodes a 5'-nucleotidase enzyme that is responsible for the inactivation of nucleoside-analog chemotherapy drugs, in 20/103 (19%) relapse T cell ALLs and 1/35 (3%) relapse B-precursor ALLs. NT5C2 mutant proteins show increased nucleotidase activity in vitro and conferred resistance to chemotherapy with 6-mercaptopurine and 6-thioguanine when expressed in ALL lymphoblasts. These results support a prominent role for activating mutations in NT5C2 and increased nucleoside-analog metabolism in disease progression and chemotherapy resistance in ALL.
Figures
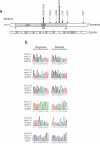
Figure 1. NT5C2 mutations in relapsed pediatric T-ALL
(a) Schematic representation of the structure of the NT5C2 protein. The haloacid dehalogenase (HAD) and the substrate binding domains (SB) are indicated. NT5C2 mutations identified in relapsed pediatric samples are shown. Filled circles represent heterozygous mutations. Multiple circles in the same amino acid position account for multiple patients with the same variant. (b) DNA sequencing chromatograms of paired diagnosis and relapse genomic T-ALL DNA samples showing representative examples of relapse specific heterozygous NT5C2 mutations, with the mutant allele sequence highlighted in red.
Figure 2. Structure-function analysis of the NT5C2 K359Q mutant protein
(a) Molecular surface representation of NT5C2 protein structure. The position of the NT5C2 K359Q mutation found is highlighted in red. The substrate inosine monophosphate (IMP) is depicted in purple; the ATP allosteric activator is shown in yellow. (b) Structure representation of the NT5C2 catalytic center and allosteric regulatory site devoid of substrate or ligands (PDB 2XCX). (c) Structure representation of the NT5C2 catalytic center and allosteric regulatory site bound to IMP and ATP, respectively (PDB 2XCW). (d) Structure representation of the NT5C2 K359Q mutant model corresponding to the catalytic center and allosteric regulatory sites. (e) Overlay of the structures shown in b–d. The white arrow indicates the repositioning of Phe354 from the inactive NT5C2 configuration to the active –ATP-bound NT5C2 and NT5C2 K359Q– structures. Mg2+ ions are depicted as green spheres.
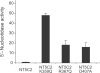
Figure 3. Increased 5'-IMP nucleotidase activity in NT5C2 mutant proteins
5'-Nucleotidase activity levels of recombinant mutant proteins relative to wild type NT5C2 control are shown. Data are means ± s.d.
Figure 4. Expression of NT5C2 mutations in ALL cells induces resistance to chemotherapy with 6-MP and 6-TG
(a) Viability assays in CCRF-CEM and CUTLL1 T-ALL cells expressing wild type NT5C2, relapse-associated mutant NT5C2 alleles or a red fluorescent protein control (RFP), treated with increased concentrations of 6-mercaptopurine (6-MP). (b) 6-Thioguanine (6-TG) dose response cell viability curves. Data is shown as means ± s.d.
Comment in
- Haematological cancer: Driver of relapse in ALL identified.
Razzak M. Razzak M. Nat Rev Clin Oncol. 2013 Apr;10(4):184. doi: 10.1038/nrclinonc.2013.28. Epub 2013 Feb 19. Nat Rev Clin Oncol. 2013. PMID: 23419958 No abstract available. - Therapeutic resistance: ALL-important mutations.
Seton-Rogers S. Seton-Rogers S. Nat Rev Cancer. 2013 Mar;13(3):151. doi: 10.1038/nrc3478. Nat Rev Cancer. 2013. PMID: 23429734 No abstract available. - Resistance revealed in acute lymphoblastic leukemia.
Aster JC, DeAngelo DJ. Aster JC, et al. Nat Med. 2013 Mar;19(3):264-5. doi: 10.1038/nm.3119. Nat Med. 2013. PMID: 23467232 No abstract available.
Similar articles
- Clonal evolution mechanisms in NT5C2 mutant-relapsed acute lymphoblastic leukaemia.
Tzoneva G, Dieck CL, Oshima K, Ambesi-Impiombato A, Sánchez-Martín M, Madubata CJ, Khiabanian H, Yu J, Waanders E, Iacobucci I, Sulis ML, Kato M, Koh K, Paganin M, Basso G, Gastier-Foster JM, Loh ML, Kirschner-Schwabe R, Mullighan CG, Rabadan R, Ferrando AA. Tzoneva G, et al. Nature. 2018 Jan 25;553(7689):511-514. doi: 10.1038/nature25186. Epub 2018 Jan 17. Nature. 2018. PMID: 29342136 Free PMC article. - Structure and Mechanisms of NT5C2 Mutations Driving Thiopurine Resistance in Relapsed Lymphoblastic Leukemia.
Dieck CL, Tzoneva G, Forouhar F, Carpenter Z, Ambesi-Impiombato A, Sánchez-Martín M, Kirschner-Schwabe R, Lew S, Seetharaman J, Tong L, Ferrando AA. Dieck CL, et al. Cancer Cell. 2018 Jul 9;34(1):136-147.e6. doi: 10.1016/j.ccell.2018.06.003. Cancer Cell. 2018. PMID: 29990496 Free PMC article. - Genetics and mechanisms of NT5C2-driven chemotherapy resistance in relapsed ALL.
Dieck CL, Ferrando A. Dieck CL, et al. Blood. 2019 May 23;133(21):2263-2268. doi: 10.1182/blood-2019-01-852392. Epub 2019 Mar 25. Blood. 2019. PMID: 30910786 Free PMC article. Review. - Pharmacologic Inhibition of NT5C2 Reverses Genetic and Nongenetic Drivers of 6-MP Resistance in Acute Lymphoblastic Leukemia.
Reglero C, Dieck CL, Zask A, Forouhar F, Laurent AP, Lin WW, Albero R, Miller HI, Ma C, Gastier-Foster JM, Loh ML, Tong L, Stockwell BR, Palomero T, Ferrando AA. Reglero C, et al. Cancer Discov. 2022 Nov 2;12(11):2646-2665. doi: 10.1158/2159-8290.CD-22-0010. Cancer Discov. 2022. PMID: 35984649 Free PMC article. - [Progress of Research on 6-Thioguanine versus 6-Mercaptopurine in childhood ALL].
Hou YJ, Zhao L, Liu XX, Ma YY. Hou YJ, et al. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2016 Apr;24(2):622-6. doi: 10.7534/j.issn.1009-2137.2016.02.059. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2016. PMID: 27151041 Review. Chinese.
Cited by
- The spleen as a sanctuary site for residual leukemic cells following ABT-199 monotherapy in ETP-ALL.
Di Grande A, Peirs S, Donovan PD, Van Trimpont M, Morscio J, Lintermans B, Reunes L, Vandamme N, Goossens S, Nguyen HA, Lavie A, Lock RB, Prehn JHM, Van Vlierberghe P, Ní Chonghaile T. Di Grande A, et al. Blood Adv. 2021 Apr 13;5(7):1963-1976. doi: 10.1182/bloodadvances.2021004177. Blood Adv. 2021. PMID: 33830207 Free PMC article. - Development of a chemical probe against NUDT15.
Zhang SM, Desroses M, Hagenkort A, Valerie NCK, Rehling D, Carter M, Wallner O, Koolmeister T, Throup A, Jemth AS, Almlöf I, Loseva O, Lundbäck T, Axelsson H, Regmi S, Sarno A, Krämer A, Pudelko L, Bräutigam L, Rasti A, Göttmann M, Wiita E, Kutzner J, Schaller T, Kalderén C, Cázares-Körner A, Page BDG, Krimpenfort R, Eshtad S, Altun M, Rudd SG, Knapp S, Scobie M, Homan EJ, Berglund UW, Stenmark P, Helleday T. Zhang SM, et al. Nat Chem Biol. 2020 Oct;16(10):1120-1128. doi: 10.1038/s41589-020-0592-z. Epub 2020 Jul 20. Nat Chem Biol. 2020. PMID: 32690945 Free PMC article. - Haematological cancer: Driver of relapse in ALL identified.
Razzak M. Razzak M. Nat Rev Clin Oncol. 2013 Apr;10(4):184. doi: 10.1038/nrclinonc.2013.28. Epub 2013 Feb 19. Nat Rev Clin Oncol. 2013. PMID: 23419958 No abstract available. - Mutational landscape, clonal evolution patterns, and role of RAS mutations in relapsed acute lymphoblastic leukemia.
Oshima K, Khiabanian H, da Silva-Almeida AC, Tzoneva G, Abate F, Ambesi-Impiombato A, Sanchez-Martin M, Carpenter Z, Penson A, Perez-Garcia A, Eckert C, Nicolas C, Balbin M, Sulis ML, Kato M, Koh K, Paganin M, Basso G, Gastier-Foster JM, Devidas M, Loh ML, Kirschner-Schwabe R, Palomero T, Rabadan R, Ferrando AA. Oshima K, et al. Proc Natl Acad Sci U S A. 2016 Oct 4;113(40):11306-11311. doi: 10.1073/pnas.1608420113. Epub 2016 Sep 21. Proc Natl Acad Sci U S A. 2016. PMID: 27655895 Free PMC article. - Deep sequencing and SNP array analyses of pediatric T-cell acute lymphoblastic leukemia reveal NOTCH1 mutations in minor subclones and a high incidence of uniparental isodisomies affecting CDKN2A.
Karrman K, Castor A, Behrendtz M, Forestier E, Olsson L, Ehinger M, Biloglav A, Fioretos T, Paulsson K, Johansson B. Karrman K, et al. J Hematol Oncol. 2015 Apr 24;8:42. doi: 10.1186/s13045-015-0138-0. J Hematol Oncol. 2015. PMID: 25903014 Free PMC article.
References
- Moricke A, et al. Long-term results of five consecutive trials in childhood acute lymphoblastic leukemia performed by the ALL-BFM study group from 1981 to 2000. Leukemia. 2010;24:265–284. - PubMed
- Koren G, et al. Systemic exposure to mercaptopurine as a prognostic factor in acute lymphocytic leukemia in children. N Engl J Med. 1990;323:17–21. - PubMed
- Relling MV, Hancock ML, Boyett JM, Pui CH, Evans WE. Prognostic importance of 6-mercaptopurine dose intensity in acute lymphoblastic leukemia. Blood. 1999;93:2817–2823. - PubMed
- Pui CH, Evans WE. Treatment of acute lymphoblastic leukemia. N Engl J Med. 2006;354:166–178. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials