Dissecting the cell to nucleus, perinucleus and cytosol - PubMed (original) (raw)
Dissecting the cell to nucleus, perinucleus and cytosol
Tattym E Shaiken et al. Sci Rep. 2014.
Erratum in
- Author Correction: Dissecting the cell to nucleus, perinucleus and cytosol.
Shaiken TE, Opekun AR. Shaiken TE, et al. Sci Rep. 2021 Apr 22;11(1):9178. doi: 10.1038/s41598-021-88968-0. Sci Rep. 2021. PMID: 33888848 Free PMC article. No abstract available.
Abstract
Cells have been described under the microscope as organelles containing cytoplasm and the nucleus. However, an unnoted structure exists between the cytoplasm and the nucleoplasm of eukaryotic cells. In addition to the nuclear envelope, there exists a perinuclear region (PNR or perinucleus) with unknown composition and function. Until now, an investigation of the role of the perinucleus has been restricted by the absence of a PNR isolation method. This manuscript describes a perinucleus isolation technique on the basis of its unique compact organization. The perinucleus was found to contain approximately 15 to 18% of the total proteins of the mammalian cell, almost half of the proteins of nuclei. Using four different normal and cancer cell lines, it was shown that the composition of PNR is highly dynamic. Application of the method showed that translocation of the p53 tumor-suppressor protein to the perinucleus in immortalized MEF cells is correlated with the translocation of p53-stabilizing protein, nucleophosmin (B23), to the PNR. Herein, the concept of the perinuclear region is advanced as a formal, identifiable structure. The roles of the perinucleus in maintaining genome integrity, regulation of gene expression and understanding of malignant transformation are discussed.
Figures
Figure 1. Phase contrast images of MDA-MB-435 cells and isolated nuclei.
(A) MDA-MB-435 cells. (B) Nuclei isolated in isotonic buffer A. Cytoplasm of the cell at this stage was removed by detergent-containing buffer; approximately 65% of proteins were extracted to the cytosol; nuclei contain perinuclear region proteins; nucleoli are visible. (C) Nuclei after extraction of the perinuclear region with buffer B. The core nucleus does not collapse after removal of the perinuclear region proteins; approximately 20% of total cellular proteins were extracted with the perinuclear fractionation; nucleoli are visible. (D) Nuclei isolated with the classical method of using hypotonic buffer. The shape of nuclei varies; some “fibrous” structures around nuclei are visible; nuclei contain nucleoli.
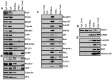
Figure 2. Patterns of protein distribution in MDA-MB-435 cells.
Cyt. Cont is a control for cytosolic proteins obtained with the 0.3% Chaps buffer cell lysis (far left panel of bands) for CDS method; Nuc. Cont is a control for nuclear proteins obtained with classical method of nuclei isolation in hypotonic buffer (far right panel of bands) for CDS method. (A) Proteins of cytosol: proteins extracted by regular lysis buffer and Buffer A from cytoplasm. They are not detected in perinuclear and nuclear fractions. Nuclear fractions were obtained with new and classical nuclei extraction techniques (B) Proteins detected in the cytosol and the perinuclear fraction: proteins were detected as cytosolic proteins with both cellular lysis technique; in addition, these proteins also appeared in the perinuclear fraction by extraction with Buffer B. (C) Proteins of perinuclear fraction: proteins are detected only in the perinuclear fraction by buffer B extraction. (D) Proteins detected in nuclear and perinuclear fractions: transcription factors (that supposedly belong to the nuclear fraction) also appeared in the perinuclear fraction. (E) Nuclear proteins: proteins were detected in the nuclear fraction. Nuclear proteins were obtained with both new and classical nuclei isolation techniques. The PVDF membranes were cropped into two halves and the high and low molecular weight proteins were shown correspondingly.
Figure 3. Patterns of protein distribution in HeLa cells.
Cyt. Cont is a control for cytosolic proteins obtained with the 0.3% Chaps buffer cell lysis (far left panel of bands) for the CDS method; Nuc. Cont is a control for nuclear proteins obtained with the classical method of nuclei isolation in hypotonic buffer (far right panel of bands) for the CDS method. (A) Proteins of cytosol: proteins extracted by regular lysis buffer and Buffer A from the cytoplasm. They are not detected in the perinuclear and the nuclear fractions. Nuclear fractions were obtained with new and classical nuclei extraction techniques (B) Proteins detected in the cytosol and the perinuclear fraction: proteins were detected as cytosolic proteins with both cellular lysis technique; in addition, these proteins also appeared in the perinuclear fraction by extraction with Buffer B. (C) Proteins of perinuclear fraction: proteins are detected only in perinuclear fraction by buffer B extraction. p53 protein was detected with long exposure. (D) Proteins detected in the nuclear and the perinuclear fractions: transcription factor CREB appeared in both fractions. (E) Nuclear proteins: proteins were detected in nuclear fraction. Nuclear proteins were obtained with new and classical nuclei isolation techniques. The PVDF membranes were cropped into two halves and the high and low molecular weight proteins were shown correspondingly.
Figure 4. Patterns of protein distribution in MEF cells.
Cyt. Cont is a control for cytosolic proteins obtained with the 0.3% Chaps buffer cell lysis (far left panel of bands) for the CDS method; Nuc. Cont is a control for nuclear proteins obtained with classical method of nuclei isolation in hypotonic buffer (far right panel of bands) for the CDS method. (A) Proteins of cytosol: proteins extracted by regular lysis buffer and Buffer A from the cytoplasm. They are not detected in perinuclear and nuclear fractions. Nuclear fractions were obtained with new and classical nuclei extraction techniques. (B) Proteins detected in the cytosol and the perinuclear fraction: proteins were detected as cytosolic proteins with both cellular lysis technique; in addition, these proteins also appeared in the perinuclear fraction by extraction with Buffer B. (C) Proteins of the perinuclear fraction: proteins are detected only in the perinuclear fraction by buffer B extraction. CBP protein was detected with long exposure. (D) Proteins detected in nuclear and perinuclear fractions: transcription factor CREB appeared in both fractions, in addition Ras and p53 proteins were detected. (E) Nuclear proteins: Nuclear proteins were obtained with new and classical nuclei isolation techniques. The PVDF membranes were cropped into two halves and the high and low molecular weight proteins were shown correspondingly.
Figure 5. Patterns of protein distribution in Primary MEF cells.
Cyt. Cont is a control for cytosolic proteins obtained with the 0.3% Chaps buffer cell lysis (far left panel of bands) for the CDS method; Nuc. Cont is a control for nuclear proteins obtained with the classical method of nuclei isolation in hypotonic buffer (far right panel of bands) for the CDS method. (A) Proteins of cytosol: proteins extracted by regular lysis buffer and Buffer A from the cytoplasm. They are not detected in the perinuclear and the nuclear fractions. Nuclear fractions were obtained with new and classical nuclei extraction techniques. (B) Proteins detected in the cytosol and the perinuclear fractions: proteins were detected as cytosolic proteins with both cellular lysis technique; in addition, these proteins also appeared in the perinuclear fraction by extraction with Buffer B. (C) Proteins of the perinuclear fraction: proteins are detected only in the perinuclear fraction by buffer B extraction. Ras, p53 and CBP proteins were detected with long exposure. (D) Proteins detected in the nuclear and the perinuclear fractions: transcription factor CREB appeared in both the nuclear and the the perinuclear fractions. (E) Nuclear proteins: proteins were detected in nuclear fraction. Nuclear proteins were obtained with new and classical nuclei isolation techniques. The PVDF membranes were cropped into two halves and the high and low molecular weight proteins were shown correspondingly.
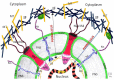
Figure 6. Model view of the perinucleus of cell.
The nuclear boundary of the perinucleus is limited with the nuclear lamina where the N termini of SUN1 and SUN2 form a network with the nuclear lamina. The C termini of SUN proteins interact with the KASH domains of nesprins, that are located at the outer nuclear membrane, where it conntects to plectins (LINC complex). Plectin interacts with intermediate filaments, microtubules and the actin cytoskeleton. This filamentous region on the cytoplasmic side represents the outer boundary of the perinucleus. Note that the Sun 2 proteins form the nuclear boundaries and Plectin 1, from the cytosolic borders, were partially distributed in PNF, representing delimiting boundaries of the perinucleus. The core nucleus embedded into nucleoplasm, and supported by rigid lamin structure, does not collapse after extracting the perinucleus by CDS technique. KEY: CHRM – chromatin, IF – intermediate filaments, INM – inner nuclear membrane, LN – lamin, MT – microtubules, NPC – nuclear pore complex, Nn – Nesprin proteins, ONM – outer nuclear membrane, PL – plectin proteins, PNS – perinuclear space, Sn – Sun proteins.
Similar articles
- REAP+: A single preparation for rapid isolation of nuclei, cytoplasm, and mitochondria.
Dos Santos B, Bion MC, Goujon-Svrzic M, Maher P, Dafre AL. Dos Santos B, et al. Anal Biochem. 2024 Apr;687:115445. doi: 10.1016/j.ab.2023.115445. Epub 2023 Dec 20. Anal Biochem. 2024. PMID: 38135241 - Translocational inefficiency of intracellular proteins in senescence of human diploid fibroblasts.
Lim IK, Hong KW, Kwak IH, Yoon G, Park SC. Lim IK, et al. Ann N Y Acad Sci. 2001 Apr;928:176-81. doi: 10.1111/j.1749-6632.2001.tb05647.x. Ann N Y Acad Sci. 2001. PMID: 11795508 Review. - Role of the nucleophosmin (NPM) portion of the non-Hodgkin's lymphoma-associated NPM-anaplastic lymphoma kinase fusion protein in oncogenesis.
Bischof D, Pulford K, Mason DY, Morris SW. Bischof D, et al. Mol Cell Biol. 1997 Apr;17(4):2312-25. doi: 10.1128/MCB.17.4.2312. Mol Cell Biol. 1997. PMID: 9121481 Free PMC article. - Physical and functional interaction of the TPL2 kinase with nucleophosmin.
Kanellis DC, Bursac S, Tsichlis PN, Volarevic S, Eliopoulos AG. Kanellis DC, et al. Oncogene. 2015 May 7;34(19):2516-26. doi: 10.1038/onc.2014.183. Epub 2014 Jul 7. Oncogene. 2015. PMID: 24998852 - Regulation of nuclear transport: central role in development and transformation?
Poon IK, Jans DA. Poon IK, et al. Traffic. 2005 Mar;6(3):173-86. doi: 10.1111/j.1600-0854.2005.00268.x. Traffic. 2005. PMID: 15702986 Review.
Cited by
- Nanomechanical Insight of Pancreatic Cancer Cell Membrane during Receptor Mediated Endocytosis of Targeted Gold Nanoparticles.
Kulkarni T, Mukhopadhyay D, Bhattacharya S. Kulkarni T, et al. ACS Appl Bio Mater. 2021 Jan 18;4(1):984-994. doi: 10.1021/acsabm.0c01443. Epub 2020 Dec 30. ACS Appl Bio Mater. 2021. PMID: 34913031 Free PMC article. - Nuclear Localization of Integrin Cytoplasmic Domain-associated Protein-1 (ICAP1) Influences β1 Integrin Activation and Recruits Krev/Interaction Trapped-1 (KRIT1) to the Nucleus.
Draheim KM, Huet-Calderwood C, Simon B, Calderwood DA. Draheim KM, et al. J Biol Chem. 2017 Feb 3;292(5):1884-1898. doi: 10.1074/jbc.M116.762393. Epub 2016 Dec 21. J Biol Chem. 2017. PMID: 28003363 Free PMC article. - The SUMO protease SENP3 regulates mitochondrial autophagy mediated by Fis1.
Waters E, Wilkinson KA, Harding AL, Carmichael RE, Robinson D, Colley HE, Guo C. Waters E, et al. EMBO Rep. 2022 Feb 3;23(2):e48754. doi: 10.15252/embr.201948754. Epub 2022 Jan 7. EMBO Rep. 2022. PMID: 34994490 Free PMC article. - EDA ligand triggers plasma membrane trafficking of its receptor EDAR via PKA activation and SNAP23-containing complexes.
Yao Y, Yang R, Zhu J, Schlessinger D, Sima J. Yao Y, et al. Cell Biosci. 2023 Jul 10;13(1):128. doi: 10.1186/s13578-023-01082-8. Cell Biosci. 2023. PMID: 37430358 Free PMC article. - Non-metabolic functions of phosphofructokinase-1 orchestrate tumor cellular invasion and genome maintenance under bevacizumab therapy.
Lim YC, Jensen KE, Aguilar-Morante D, Vardouli L, Vitting-Seerup K, Gimple RC, Wu Q, Pedersen H, Elbaek KJ, Gromova I, Ihnatko R, Kristensen BW, Petersen JK, Skjoth-Rasmussen J, Flavahan W, Rich JN, Hamerlik P. Lim YC, et al. Neuro Oncol. 2023 Feb 14;25(2):248-260. doi: 10.1093/neuonc/noac135. Neuro Oncol. 2023. PMID: 35608632 Free PMC article.
References
- Toy-Miou-Leong M., Cortes C. L., Beaudet A., Rostene W. & Forgez P. Receptor trafficking via the perinuclear recycling compartment accompanied by cell division is necessary for permanent neurotensin cell sensitization and leads to chronic mitogen-activated protein kinase activation. J. Biol. Chem. 279, 12636–12646 (2004). - PubMed
- Nakamura K., Senda T., Sato K., Mori S. & Moriyama M. Accumulation of BCL10 at the perinuclear region is required for the BCL10-mediated nuclear factor-kappa B activation. Pathobiology. 72, 191–202 (2005). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous