Intracellular CD24 disrupts the ARF-NPM interaction and enables mutational and viral oncogene-mediated p53 inactivation - PubMed (original) (raw)
Runhua Liu 1, Peiying Ye 2, Chunshu Wong 3, Guo-Yun Chen 2, Penghui Zhou 4, Kaoru Sakabe 2, Xincheng Zheng 5, Wei Wu 5, Peng Zhang 6, Taijiao Jiang 6, Michael F Bassetti 7, Sandro Jube 2, Yi Sun 7, Yanping Zhang 8, Pan Zheng 2, Yang Liu 2
Affiliations
- PMID: 25600590
- PMCID: PMC4300525
- DOI: 10.1038/ncomms6909
Intracellular CD24 disrupts the ARF-NPM interaction and enables mutational and viral oncogene-mediated p53 inactivation
Lizhong Wang et al. Nat Commun. 2015.
Abstract
CD24 is overexpressed in nearly 70% human cancers, whereas TP53 is the most frequently mutated tumour-suppressor gene that functions in a context-dependent manner. Here we show that both targeted mutation and short hairpin RNA (shRNA) silencing of CD24 retard the growth, progression and metastasis of prostate cancer. CD24 competitively inhibits ARF binding to NPM, resulting in decreased ARF, increase MDM2 and decrease levels of p53 and the p53 target p21/CDKN1A. CD24 silencing prevents functional inactivation of p53 by both somatic mutation and viral oncogenes, including the SV40 large T antigen and human papilloma virus 16 E6-antigen. In support of the functional interaction between CD24 and p53, in silico analyses reveal that TP53 mutates at a higher rate among glioma and prostate cancer samples with higher CD24 mRNA levels. These data provide a general mechanism for functional inactivation of ARF and reveal an important cellular context for genetic and viral inactivation of TP53.
Figures
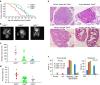
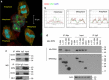
Figure 1. Cd24 promotes onset and progression of prostate cancer in TRAMP mice.
(a) Onset of palpable prostate tumour development in Cd24 +/+ , Cd24 +/− and Cd24 −/− TRAMP mice was observed over 35 weeks. _P_-value of log-rank tests are Cd24 WT versus Cd24 +/−, _P_=0.0001; Cd24 WT versus Cd24 −/−, _P_=0.0017; Cd24 +/− versus Cd24 −/−, P_=0.51. (b) Representative images of prostates from 30-week-old Cd24 +/+ (n_=7), Cd24 +/− (_n_=11) and Cd24 −/− (_n_=9) TRAMP mice. (c) Prostate sizes in 30-week-old TRAMP mice in Cd24 +/+ , Cd24 +/− and Cd24 −/− TRAMP mice. The lower abdominal area was divided into 0.4-mm-thick slices and MRI images were acquired to provide continuous images of the whole prostate. The surface area of the prostate slices was traced by segmenting and summed to estimate the total volume. Analysis of variance (ANOVA) tests revealed an extremely significant Cd24 gene dose effect on the prostate size (_P_=0.00026). (d) The weights of 35-week-old Cd24 +/+ (_n_=9), Cd24 +/− (_n_=22) and Cd24 −/− (_n_=10) TRAMP prostates. ANOVA tests revealed an extremely significant Cd24 gene-dose effect on the prostate weight (_P_=0.00011). (e) Representative histology of prostate carcinomas (PCa) that developed in Cd24 +/+ Cd24 +/−, and Cd24 −/− TRAMP mice. The data show a difference in the degree of differentiation in tumour morphology. (f) Cd24 promotes progression (left panel) and metastasis (right panel) of prostate cancer in the TRAMP model. The prostate tissue was examined double blind and classified into the following categories: normal prostate, hyperplasia prostate (HP), intraepithelial neoplasia in the prostate (PIN), moderate (mod) to well-differentiated (well diff) PCa and poorly differentiated PCa. Cancer was scored metastatic if one or more distal metastases to either the lungs, kidneys or liver were detected histologically. Examples of tumours at each grade are shown in e, whereas that of normal prostate and hyperplasia are shown in Supplementary Fig. 1. Pearson’s _χ_2 test was used to determine whether the Cd24 geneotypes affected the progression to poorly differentiated PCa (left panel) or distal metastasis (right panel).
Figure 2. Cd24 in non-haematopoietic cells contributes to tumour growth.
(a) Overexpression of CD24 mRNA in human prostate cancer tissue based on analysis of two data cohort deposited in Oncomine.com. Two reported cohorts with a sufficient number of matched cancer and normal tissue were analysed. _P_-values were calculated by Student’s _t_-tests. (b) Expression of Cd24 in Cd24 +/+ PCa but not Cd24 −/− PCa or normal Cd24 +/+ prostate epithelia. Data shown are representative images and have been reproduced three times. (c) Cd24 levels in TRAMP prostate are gene-dose dependent. Age-matched Cd24 +/+ and Cd24 +/− prostate were analysed for Cd24 levels by western blot. (d) Diagram of experimental design. Eight-week-old Cd24 +/+ TRAMP mice were lethally irradiated and transplanted with 5 × 106 bone marrow cells at 1 day after irradiation. The prostate size was measured by MRI at 30 weeks of age. (e) Successful replacement of haematopoietic cells from the donor bone marrow based on Cd24 expression. (f) Means and s.d. of the prostate volume in the bone marrow chimera mice.
Figure 3. CD24 promotes growth of DU145 cells.
(a) CD24 mRNA levels in three human prostate cancer cell lines are displaced as % of glyceraldehyde 3-phosphate dehydrogenase (GAPDH). (b) Efficient gene silencing of the CD24 gene based on the protein (insert, representative images that have been reproduced three times) and RNA levels. (c) ShRNA silencing eliminates cell surface CD24 expression as determined by flow cytometry. Data shown are representative profiles and have been reproduced three times. (d) ShRNA silencing of CD24 reduces the growth rate as measured by the 3-(4,5-dimethylthiazol-2-yl) 2,5-diphenyltetrazolium bromide assay. Means and s.d. of the relative cell proliferation are shown on the given days. The study has been reproduced three times. (e) Reduced growth rate of tumour cells based on colony size. Data shown are representative images and have been reproduced three times. (f) ShRNA silencing of CD24 reduced the tumour growth in the Il2rg −/− NOD.SCID (NSG) mice. The mice received a subcutaneous injection of 5 × 106 tumour cells. Data shown are means and s.d. of the tumour volumes. (g) Weights of tumours at day 39. Error bars, s.d. Scr _n_=14, Sh1, _n_=9, Sh2 _n_=9 for f and g. Substantially similar data were obtained when the cells were injected into nu/nu mice.
Figure 4. CD24 reduces levels of p53 and p21 and promotes tumour cell proliferation.
(a) Heat-map depiction of alterations in gene expression in DU145 cells following CD24 gene silencing. RNA isolated from five independent cultures of Scr, three cultures of Sh1 and three cultures of Sh2 were compared using Affymeatrix Human U133 Plus 2.0 microarrays. Only those genes that met the statistical threshold of P<0.001 and mean alterations that reached the cutoff of >150% or<66% of Scr levels are displayed. The genes are listed in Supplementary Data set and the pathway analysis summary of affected genes is provided in Supplementary Table 1. (b) CD24 silencing increases p21/CDKN1A mRNA. Error bars, s.d. _n_=3. The data have been reproduced three times. (c) CD24 silencing increased levels of p53 and p21 proteins, but not p27 protein. Representative images of western blots are shown and the data have been reproduced three times. (d) CD24 silencing reduces cell cycle re-entry after starvation. Scr, Sh1 and Sh2 clones were starved for 48 h in serum-free medium and then shifted into serum containing medium. The cells were harvested and analysed for cell cycle progression based on DNA content. These data have been reproduced three times. (e) Ectopic expression of CD24 in LNCaP cell line decreases levels of p53 and p21, but not p27. Representative images of western blots are shown and the data have been reproduced two times. (f) CD24 transduction increases proliferation of LNCaP cells. Data shown are triplicate means and s.d. of triplicates and have been reproduced twice. LSD, least significant difference.
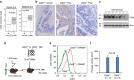
Figure 5. Intracellular CD24 and proliferation of prostate cancer cells.
(a) Measurement of cell surface and total cellular CD24 by flow cytometry. The prostate cancer cell lines PC3, DU145 and LNCaP were stained with anti-CD24 with or without permeabilization to measure either cell surface (top) or total cellular CD24 (bottom). (b) Analysis of CD24 in the cytoplasmic and plasma membrane fraction (Cp+Cm), nucleoplasm (Np) and the insoluble fraction that includes chromatin by western blot. The efficiency of the fractionation was monitored by the presence of β-actin (Cp+Cm+Np), Lamin (Np) and H1.5 (chromatin). (c) Confocal microscopy reveals dynamic upregulation and localization of CD24 during mitosis. Note alterations of CD24 distribution and signal intensity at different phases during mitosis. See Supplementary Fig. 6 for single-colour images. (d) Upregulation of CD24 during mitosis as revealed by western blot of cellular lysates prepared from nocodazale-synchronized DU145 cells. (e) Diagram of CD24-GFP and GFP-CD24 constructs. The N-terminus-tagged CD24 was produced by inserting the GFP-coding sequence after the signal peptide, whereas the C-terminus-tagged CD24 was generated by inserting the GFP-coding sequence before the stop codon of CD24. (f) Tagging the C-terminus with GFP prevents cell surface localization of CD24. The images show the distinct distribution of green fluorescence in cells transfected with either CD24-GFP (C) or GFP-CD24 (N). (g) Verification of cell surface versus intracellular CD24 expression by cell surface staining. CD24-GFP (C) was not present on the surface of DU145 cells as it was not detected by an anti-CD24 mAb added to non-permeabilized cells. CD24-GFP (N) was used as positive control for cell surface CD24. (h) Verification of fusion protein expression by western blot using anti-CD24 and anti-GFP antibodies. (i) Intracellular CD24 encoded by CD24-GFP (C) is at least as potent in promoting proliferation of DU145 cells ad GFP-CD24 (N). CD24-silenced DU145 cells were transfected with CD24-GFP (C) or GFP-CD24 (N) plasmids. After drug selection, an equal number of drug-resistant transfectants were plated onto 10-cm plates. The colonies were stained with crystal violet. All images have been reproduced three times.
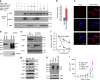
Figure 6. CD24 interacts with NPM during mitosis.
(a,b) Dynamic association between CD24 and NPM in DU145 cells. (a) Merged images displaying DNA (stained with DAPI, blue), NPM (green) and CD24 (red). Cells at prophase, metaphase and anaphase were identified by their chromatin distribution. The representative image was obtained from a Meta laser-scanning confocal microscope. (b) Overlay between CD24 (red) and NPM (green) signals of cells at the three indicated phases during mitosis. The intensity (y axis) of signals was simultaneously obtained from the laser-scanning cross-section with the major diameter and overlap of the red and the green signals was analysed using LSM image browser. (c) Reciprocal co-precipitation between CD24 and NPM in DU145 cells. The cells lysates were prepared after 2 h of release from nocodazale arrest. The cell lysates were precipitated with antibodies specific for either NPM or CD24 or IgG control. The precipitates were probed by western blot with indicated antibodies. (d) Localizing the CD24-binding domain in NPM to the regions of AA196-294 based on co-IP of Myc-tagged NPM fragments and HA-tagged CD24 in HA-CD24 and NPM-Myc co-transfected HEK293 cells. The association was confirmed by western blot with anti-HA and anti-Myc mAbs. All images have been reproduced three times. DAPI, 4',6-diamidino-2-phenylindole. Co-IP, co-immunoprecipitation; IgLC, immunoglobulin light chain; IP, immunoprecipitation; IB, immunoblot.
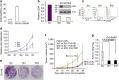
Figure 7. CD24 inhibits tumour-suppressor activity of ARF by antagonizing the NPM–ARF interaction.
(a) Recombinant CD24 associates with recombinant NPM and blocks the ARF–NPM interaction. Control GST and NPM–GST fusion proteins were incubated with indicated amounts of CD24-Fc and/or ARF. The GST or GST–NPM1-associated proteins were determined by western blot after pull-down with glutathione beads. (b,c) Proximity ligation assay reveals CD24 antagonism of NPM–ARF association. Scrambled or _CD24 shRNA_-transduced DU145 cells were compared for endogenous NPM–ARF association. Images were obtained using an Olympus BX61 microscope using a × 60 oil objective. Scale bar, 50 μm. Image J software was used to quantify fluorescent intensities. Mean fluorescent intensities were determined from more than 100 cells per slide. This experiment has been repeated three times. Data shown are means and s.d. of three independent repeats (b) and representative images (c). (d) Cd24 reduces Npm-associated p19Arf. Spleen lysates from WT and Cd24 −/− mice were precipitated with anti-Npm mAb. The amounts of Npm and p19Arf in the immunoprecipitates were determined by western blot. IgG heavy chain (IgHC) was used as loading controls. (e) Regulation of ARF levels and stability by endogenous CD24. Scrambled or shRNA-silenced DU145 cells were arrested at G2/M phase overnight and then treated with CHX for 0, 2 and 4 h. The levels of CD24 (upper panel) and p14ARF were analysed by western blot. (f) Decay kinetics of p14ARF are shown as % of ARF protein at different times after CHX treatment. (g) ShRNA silencing of CD24 does not affect the levels of the ARF E3 ligase ULF. (h) Ectopic expression of CD24 reduces ARF levels. U2OS cells that ectopically expressed CD24 alone, ARF alone or both were analysed for their CD24 and ARF levels by western blot. (i) CD24 expression alleviates ARF tumour-suppressor activity. Data shown are growth kinetics of U2OS cells transfected with the given plasmids. Data shown are means and s.d. of cell numbers during the 6-day period. All data in this figure have been reproduced two or three times.
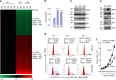
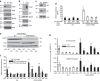
Figure 8. CD24 silencing restores transcriptional activity of p53.
(a) Effect of CD24 silencing on expression of NPM, ARF, MDM2 and p53. Scrambled (Scr) or CD24_-shRNA-silenced DU145 cells were analysed for CD24, NPM, ARF, MDM2 and p53 by western blot. (b) In ARF-deficient U2OS cells, ectopic expression of CD24 had no effect on the levels of MDM2 and p53. Actin levels were used as loading control. (c) Endogenous mutant p53 is responsible for the upregulation of p21 in CD24-silenced DU145 cells. (d) In DU145 cells, silencing CD24 restores transcriptional induction of the p21 promoter. DU145 cells transduced with Scr or shRNA for either_TP53 or CD24 were transfected with a luciferase reporter containing the p21 promoter. The WT reporter has an intact p53-binding site, whereas the mutant promoter has the site deleted. (e,f) CD24 silencing restores transcriptional activity of multiple TP53 mutants. DU145 cells in which the endogenous TP53 was silenced by shRNA were transduced with Scr or CD24 shRNA lentivirus. The stable cell lines were then transfected with p53 mutants in conjunction with WT or mutant p21 promoter reporter constructs. p53 levels (e) were determined by western blot, while luciferase activity (f) was measured at 72 h after transfection. The mutants used were p53R273H, p53V143A, p53R280T and p53R175H. Numbers on the x axis indicate the amino-acid positions of the p53 mutations. (g) As in e and f, except that the endogenous p21 and MDM2 mRNA were measured by quantitative PCR. Data shown are means and s.d. of triplicate samples. All data have been reproduced two or three times.
Figure 9. CD24 is required for viral oncogene-mediated inactivation of p53 function.
(a) By increasing the overall levels of p53 protein, Cd24 silencing increases the proportion of SV40 T antigen-unbound p53. SV40 T antigen was tagged with Flag and introduced into the mouse prostate cancer cell line C3 with or without transduction of CD24 shRNA. The total cellular (left panel) or SV40 T antigen-associated or non-associated p53 (right panel) were determined by either western blot or anti-Flag immunoprecipitation followed by western blot. Note that CD24 was not co-precipitated. S: supernatant; P: pellet. (b) Cd24 silencing reveals p53 function in SV40 large T antigen (SV40 Tag)-transduced (top) and human papilloma viral protein E6 (HPV E6)-transduced C3 cells (bottom). SV40 Tag- or HPV E6-transduced C3 cells were transfected with WT or p53-binding site mutant p21 promoter linked with a luciferase reporter. The function of the endogenous Cd24 and Tp53 locus was confirmed by shRNA silencing of either genes. Data shown are means and s.d. of relative luciferase activity. (c) In the Tp53+/+ TRAMP model, targeted mutation of Cd24 increased the levels of p53 and p21. Data shown are photographs of immunohistochemical analysis of p21 and p53 levels from either Cd24 +/+ or Cd24 −/− prostate cancer samples. (d) Cd24 silencing increases the proportion of HPV E6-unbound p53, as in a, except that HPV E6 was used instead of SV40 TAg. All data in this figure have been reproduced two or three times. Co-IP, co-immunoprecipitation; Mut, mutant; WB, western blot.
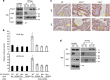
Figure 10. ShRNA silencing of CD24 restores tumour-suppressor activity of cancer-associated p53 mutants.
(a) DU145 cells whose endogenous TP53 was silenced were transduced with cDNAs of cancer-associated p53 mutants used in Fig. 8e,f and then cultured in six-well plates in the presence of neomycin for 2 weeks before the colonies were photographed and counted. Representative image of each transfectant is shown. (b) ShRNA silencing of CD24 reactivates the tumour-suppressor activity of p53R273H as assessed by the number of DU145 colonies visualized by crystal violet. Data shown are means and s.d. of triplicate samples and have been normalized against the vector control (artificially defined as 100%) from DU145 cells transduced with either Scr or CD24 ShRNA. The data have been reproduced twice. (c) In silico analysis revealed an association between CD24 levels and TP53 status. The expression and mutational status data were obtained from the TCGA database as detailed in the Methods section. Glioblastoma, low-grade brain glioma and prostate cancer samples were divided into CD24hi (above the mean) and CD24lo (below the mean) groups based on RNA-seq data. The frequencies of the samples with somatic Tp53 mutations were compared using either Pearson’s _χ_2 tests with correction for low counts of mutant samples in the prostate cancer cohort. CFU, colony-forming unit.
Similar articles
- Oncomir miR-125b suppresses p14(ARF) to modulate p53-dependent and p53-independent apoptosis in prostate cancer.
Amir S, Ma AH, Shi XB, Xue L, Kung HJ, Devere White RW. Amir S, et al. PLoS One. 2013 Apr 9;8(4):e61064. doi: 10.1371/journal.pone.0061064. Print 2013. PLoS One. 2013. PMID: 23585871 Free PMC article. - A CD24-p53 axis contributes to African American prostate cancer disparities.
Liu W, Zhang Y, Wei S, Bae S, Yang WH, Smith GJ, Mohler JL, Fontham ETH, Bensen JT, Sonpavde GP, Chen GY, Liu R, Wang L. Liu W, et al. Prostate. 2020 May;80(8):609-618. doi: 10.1002/pros.23973. Epub 2020 Mar 13. Prostate. 2020. PMID: 32168400 Free PMC article. - AKT regulates NPM dependent ARF localization and p53mut stability in tumors.
Hamilton G, Abraham AG, Morton J, Sampson O, Pefani DE, Khoronenkova S, Grawenda A, Papaspyropoulos A, Jamieson N, McKay C, Sansom O, Dianov GL, O'Neill E. Hamilton G, et al. Oncotarget. 2014 Aug 15;5(15):6142-67. doi: 10.18632/oncotarget.2178. Oncotarget. 2014. PMID: 25071014 Free PMC article. - Escape from p53-mediated tumor surveillance in neuroblastoma: switching off the p14(ARF)-MDM2-p53 axis.
Van Maerken T, Vandesompele J, Rihani A, De Paepe A, Speleman F. Van Maerken T, et al. Cell Death Differ. 2009 Dec;16(12):1563-72. doi: 10.1038/cdd.2009.138. Epub 2009 Sep 25. Cell Death Differ. 2009. PMID: 19779493 Review. - Regulation of the p14ARF-Mdm2-p53 pathway: an overview in breast cancer.
Agrawal A, Yang J, Murphy RF, Agrawal DK. Agrawal A, et al. Exp Mol Pathol. 2006 Oct;81(2):115-22. doi: 10.1016/j.yexmp.2006.07.001. Epub 2006 Aug 17. Exp Mol Pathol. 2006. PMID: 16919268 Review.
Cited by
- Targeting CD24 in Cancer Immunotherapy.
Chen W, Hu Z, Guo Z. Chen W, et al. Biomedicines. 2023 Nov 27;11(12):3159. doi: 10.3390/biomedicines11123159. Biomedicines. 2023. PMID: 38137380 Free PMC article. Review. - Regulation of NEIL1 protein abundance by RAD9 is important for efficient base excision repair.
Panigrahi SK, Hopkins KM, Lieberman HB. Panigrahi SK, et al. Nucleic Acids Res. 2015 May 19;43(9):4531-46. doi: 10.1093/nar/gkv327. Epub 2015 Apr 14. Nucleic Acids Res. 2015. PMID: 25873625 Free PMC article. - ZBTB28 inhibits breast cancer by activating IFNAR and dual blocking CD24 and CD47 to enhance macrophages phagocytosis.
Li L, Gong Y, Tang J, Yan C, Li L, Peng W, Cheng Z, Yu R, Xiang Q, Deng C, Mu J, Xia J, Luo X, Wu Y, Xiang T. Li L, et al. Cell Mol Life Sci. 2022 Jan 20;79(2):83. doi: 10.1007/s00018-021-04124-x. Cell Mol Life Sci. 2022. PMID: 35048182 Free PMC article. - Silencing of CD24 Enhances the PRIMA-1-Induced Restoration of Mutant p53 in Prostate Cancer Cells.
Zhang W, Yi B, Wang C, Chen D, Bae S, Wei S, Guo RJ, Lu C, Nguyen LL, Yang WH, Lillard JW, Zhang X, Wang L, Liu R. Zhang W, et al. Clin Cancer Res. 2016 May 15;22(10):2545-54. doi: 10.1158/1078-0432.CCR-15-1927. Epub 2015 Dec 28. Clin Cancer Res. 2016. PMID: 26712693 Free PMC article. - Reduced DAXX Expression Is Associated with Reduced CD24 Expression in Colorectal Cancer.
Chen YC, Lee TH, Tzeng SL. Chen YC, et al. Cells. 2019 Oct 12;8(10):1242. doi: 10.3390/cells8101242. Cells. 2019. PMID: 31614769 Free PMC article.
References
- Kristiansen G., Sammar M. & Altevogt P. Tumour biological aspects of CD24, a mucin-like adhesion molecule. J. Mol. Histol. 35, 255–262 (2004). - PubMed
- Sigurdsson S. et al.. Comprehensive evaluation of the genetic variants of interferon regulatory factor 5 (IRF5) reveals a novel 5 bp length polymorphism as strong risk factor for systemic lupus erythematosus. Hum. Mol. Genet 17, 872–881 (2008). - PubMed
- Sano A. et al.. CD24 expression is a novel prognostic factor in esophageal squamous cell carcinoma. Ann. Surg. Oncol. 16, 506–514 (2009). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA155235/CA/NCI NIH HHS/United States
- U54 CA118623/CA/NCI NIH HHS/United States
- R01 AI064350/AI/NIAID NIH HHS/United States
- R01 CA058033/CA/NCI NIH HHS/United States
- R01 CA171972/CA/NCI NIH HHS/United States
- R01 AG036690/AG/NIA NIH HHS/United States
- P50 CA093990/CA/NCI NIH HHS/United States
- CA171972/CA/NCI NIH HHS/United States
- R21 CA164688/CA/NCI NIH HHS/United States
- R21CA164688/CA/NCI NIH HHS/United States
- CA58033/CA/NCI NIH HHS/United States
- R01 CA167637/CA/NCI NIH HHS/United States
- AG036690/AG/NIA NIH HHS/United States
- U54 CA118948/CA/NCI NIH HHS/United States
- R21 AI105727/AI/NIAID NIH HHS/United States
- G12 MD007585/MD/NIMHD NIH HHS/United States
- AI64350/AI/NIAID NIH HHS/United States
- R01 CA171277/CA/NCI NIH HHS/United States
- R01 CA100302/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous