Direct regulation of LAMP1 by tumor-suppressive microRNA-320a in prostate cancer - PubMed (original) (raw)
Direct regulation of LAMP1 by tumor-suppressive microRNA-320a in prostate cancer
Atsushi Okato et al. Int J Oncol. 2016 Jul.
Abstract
Advanced prostate cancer (PCa) metastasizes to bone and lymph nodes, and currently available treatments cannot prevent the progression and metastasis of the disease. Therefore, an improved understanding of the molecular mechanisms of the progression and metastasis of advanced PCa using current genomic approaches is needed. Our miRNA expression signature in castration-resistant prostate cancer (CRPC) revealed that microRNA-320a (miR‑320a) was significantly reduced in cancer tissues, suggesting that miR‑320a may be a promising anticancer miRNA. The aim of this study was to investigate the functional roles of miR‑320a in naïve PCa and CRPC cells and to identify miR‑320a-regulated genes involved in PCa metastasis. The expression levels of miR‑320a were significantly reduced in naïve PCa, CRPC specimens, and PCa cell lines. Restoration of mature miR‑320a in PCa cell lines showed that miR‑320a significantly inhibited cancer cell migration and invasion. Moreover, we found that lysosomal-associated membrane protein 1 (LAMP1) was a direct target of miR‑320a in PCa cells. Silencing of LAMP1 using siRNA significantly inhibited cell proliferation, migration, and invasion in PCa cells. Overexpression of LAMP1 was observed in PCa and CRPC clinical specimens. Moreover, downstream pathways were identified using si-LAMP1-transfected cells. The discovery of tumor-suppressive miR‑320a-mediated pathways may provide important insights into the potential mechanisms of PCa metastasis.
Figures
Figure 1
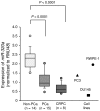
Expression levels of miR-320a in clinical prostate cancer specimens and cell lines. Expression levels of miR-320a were determined by RT-PCR. RNU48 was used for normalization.
Figure 2
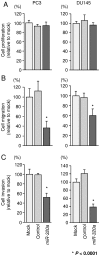
Effects of miR-320a restoration on the proliferation, migration, and invasion of PCa cells. (A) Cell proliferation was determined 72 h after transfection using XTT assays. (B) Cell migration activity was determined 48 h after transfection using Boyden chamber migration assays. (C) Cell invasion activity was determined 48 h after transfection using Matrigel invasion assays. *P<0.0001. Experiments were performed triplicate. Bars indicate SDs.
Figure 3
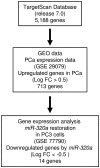
Strategy for identification of candidate target genes of miR-320a. TargetScan Release 7.0 was used to identify genes containing the miR-320a seed sequence in the 3′-UTR. Next, we analyzed the gene expression dataset using the GEO database (GEO accession no. GSE29079). Finally, we attempted to identify miR-320a target genes using _miR-320a_-transfected PC3 cells (GEO accession no. GSE77790).
Figure 4
Expression of LAMP1 in clinical PCa specimens using a tissue microarray. (A) Representative immunohistochemical staining for LAMP1 in a prostate cancer tissue microarray (Table III; nos. 8, 32, 61 and 70). LAMP1 was strongly expressed in PCa. (B) Comparison of immunohisto-chemical staining of LAMP1 in prostate cancer tissues using IHC scores. LAMP1 expression was significantly higher in PCa tissues than in normal prostate tissues.
Figure 5
Expression of LAMP1 in clinical CRPC specimens. Representative immunostaining for LAMP1 in CRPC clinical specimens. PSA and AR were used to confirm prostate cancer metastasis.
Figure 6
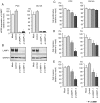
Direct regulation of LAMP1 by miR-320a. (A) LAMP1 mRNA expression was determined at 96 h after transfection of miR-320a. GAPDH was used as an internal control. (B) LAMP1 protein expression 72 h after transfection with miR-320a. GAPDH was used as a loading control. (C) Luciferase reporter assays using a vector encoding the putative miR-320a target site in the LAMP1 3′-UTR (wild-type and deletion constructs). *P=0.0038, **P=0.0002, and ***P<0.0001. Experiments were performed in triplicate. Bars indicate SDs.
Figure 7
Effects of silencing LAMP1 on cell proliferation, migration, and invasion in PCa cell lines. (A) LAMP1 mRNA expression was determined at 72 h after transfection with si-LAMP1. GAPDH was used as an internal control. (B) LAMP1 protein expression was evaluated by western blotting at 72 h after transfection with si-LAMP1. GAPDH was used as a loading control. (C) Cell proliferation was determined by XTT assays. (D) Cell migration activity was determined by Boyden chamber migration assays. (E) Cell invasion activity was determined by Matrigel invasion assays. *P<0.0001. Experiments were performed in triplicate. Bars indicate SDs.
Similar articles
- MicroRNA expression signature of castration-resistant prostate cancer: the microRNA-221/222 cluster functions as a tumour suppressor and disease progression marker.
Goto Y, Kojima S, Nishikawa R, Kurozumi A, Kato M, Enokida H, Matsushita R, Yamazaki K, Ishida Y, Nakagawa M, Naya Y, Ichikawa T, Seki N. Goto Y, et al. Br J Cancer. 2015 Sep 29;113(7):1055-65. doi: 10.1038/bjc.2015.300. Epub 2015 Sep 1. Br J Cancer. 2015. PMID: 26325107 Free PMC article. - Dual strands of pre-miR‑150 (miR‑150‑5p and miR‑150‑3p) act as antitumor miRNAs targeting SPOCK1 in naïve and castration-resistant prostate cancer.
Okato A, Arai T, Kojima S, Koshizuka K, Osako Y, Idichi T, Kurozumi A, Goto Y, Kato M, Naya Y, Ichikawa T, Seki N. Okato A, et al. Int J Oncol. 2017 Jul;51(1):245-256. doi: 10.3892/ijo.2017.4008. Epub 2017 May 17. Int J Oncol. 2017. PMID: 28534948 - MicroRNA-205 inhibits cancer cell migration and invasion via modulation of centromere protein F regulating pathways in prostate cancer.
Nishikawa R, Goto Y, Kurozumi A, Matsushita R, Enokida H, Kojima S, Naya Y, Nakagawa M, Ichikawa T, Seki N. Nishikawa R, et al. Int J Urol. 2015 Sep;22(9):867-77. doi: 10.1111/iju.12829. Epub 2015 Jun 7. Int J Urol. 2015. PMID: 26059417 - Microdissecting the role of microRNAs in the pathogenesis of prostate cancer.
Ayub SG, Kaul D, Ayub T. Ayub SG, et al. Cancer Genet. 2015 Jun;208(6):289-302. doi: 10.1016/j.cancergen.2015.02.010. Epub 2015 Mar 2. Cancer Genet. 2015. PMID: 26004033 Review. - The roles of microRNAs in the progression of castration-resistant prostate cancer.
Kojima S, Goto Y, Naya Y. Kojima S, et al. J Hum Genet. 2017 Jan;62(1):25-31. doi: 10.1038/jhg.2016.69. Epub 2016 Jun 9. J Hum Genet. 2017. PMID: 27278789 Review.
Cited by
- Integrative proteomics and n-glycoproteomics reveal the synergistic anti-tumor effects of aspirin- and gemcitabine-based chemotherapy on pancreatic cancer cells.
Li X, Kong R, Hou W, Cao J, Zhang L, Qian X, Zhao L, Ying W. Li X, et al. Cell Oncol (Dordr). 2024 Feb;47(1):141-156. doi: 10.1007/s13402-023-00856-z. Epub 2023 Aug 28. Cell Oncol (Dordr). 2024. PMID: 37639207 - miR‑539 suppresses proliferation and induces apoptosis in renal cell carcinoma by targeting high mobility group A2.
Ye ZH, Gui DW. Ye ZH, et al. Mol Med Rep. 2018 Apr;17(4):5611-5618. doi: 10.3892/mmr.2018.8578. Epub 2018 Feb 8. Mol Med Rep. 2018. PMID: 29436648 Free PMC article. - Exosomal MicroRNA-320a Derived From Mesenchymal Stem Cells Regulates Rheumatoid Arthritis Fibroblast-Like Synoviocyte Activation by Suppressing CXCL9 Expression.
Meng Q, Qiu B. Meng Q, et al. Front Physiol. 2020 May 26;11:441. doi: 10.3389/fphys.2020.00441. eCollection 2020. Front Physiol. 2020. PMID: 32528301 Free PMC article. - Investigating differential miRNA expression profiling using serum and urine specimens for detecting potential biomarkers for early prostate cancer diagnosis.
Hasanoğlu S, Göncü B, Yücesan E, Atasoy S, Kayalı Y, Özten Kandaş N. Hasanoğlu S, et al. Turk J Med Sci. 2021 Aug 30;51(4):1764-1774. doi: 10.3906/sag-2010-183. Turk J Med Sci. 2021. PMID: 33550766 Free PMC article. - Molecular pathogenesis of interstitial cystitis based on microRNA expression signature: miR-320 family-regulated molecular pathways and targets.
Arai T, Fuse M, Goto Y, Kaga K, Kurozumi A, Yamada Y, Sugawara S, Okato A, Ichikawa T, Yamanishi T, Seki N. Arai T, et al. J Hum Genet. 2018 May;63(5):543-554. doi: 10.1038/s10038-018-0419-x. Epub 2018 Mar 12. J Hum Genet. 2018. PMID: 29531336
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous