PGRMC1 and PAQR4 are promising molecular targets for a rare subtype of ovarian cancer - PubMed (original) (raw)
PGRMC1 and PAQR4 are promising molecular targets for a rare subtype of ovarian cancer
Kamila Kozłowska-Tomczyk et al. Open Life Sci. 2024.
Abstract
The heterogeneity of ovarian cancer (OC) has made developing effective treatments difficult. Nowadays, hormone therapy plays a growing role in the treatment of OC; however, hormone modulators have had only limited success so far. To provide a more rigorous foundation for hormonal therapy for different OC subtypes, the current study used a series of bioinformatics approaches to analyse the expression profiles of genes encoding membrane progesterone (PGRMC1, progestins and the adipoQ receptor [PAQR] family), and androgen (zinc transporter member 9 [ZIP9], OXER1) receptors. Our work investigated also their prognostic value in the context of OC. We found differences in expression of ZIP9 and OXER1 between different OC subtypes, as well as between patient tumour and normal tissues. Expression of mRNA encoding PAQR7 and PAQR8 in a panel of OC cell lines was below the qPCR detection limit and was downregulated in tumour tissue samples, whereas high expression of PGRMC1 and PAQR4 mRNA was observed in rare subtypes of OC cell lines. In addition, chemical inhibition of PGRMC1 reduced the viability of rare OCs represented by COV434 cells. In conclusion, PGRMC1 and PAQR4 are promising targets for anticancer therapy, particularly for rare subtypes of OC. These findings may reflect differences in the observed responses of various OC subtypes to hormone therapy.
Keywords: OXER1; PGRMC1; ZIP9; bioinformatics; cell lines; mPR; ovarian cancer; prognosis.
© 2024 the author(s), published by De Gruyter.
Conflict of interest statement
Conflict of interest: Authors state no conflict of interest.
Figures
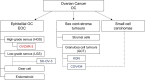
Figure 1
Schematic presentation of the ovarian cancer subtypes classification including tested cancer cell lines. The cell line representing a common type is marked in red, and those representing rare types are in blue.
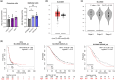
Figure 2
Comparison of ZIP9 mRNA levels in (a) epithelial (HOSEpiC, SK-OV-3, OVCAR-3) and granulosa (HGrC1, COV434, KGN) ovarian normal and cancer cell lines, and (b) in ovarian cancer and normal ovarian tissues (based on analysis of open-source data from the TCGA and GTEx databases using the GEPIA online tool). (c) Correlation between ZIP9 expression and tumour stage in patients with ovarian cancer patients. (d) KM survival plots based on ZIP9 expression, showing PFS, OS, and PPS of ovarian cancer patients with different tumour expression of ZIP9. The HR is indicated, along with the 95% confidence interval in brackets. *p ≤ 0.05 and **p ≤ 0.01.
Figure 3
Comparison of OXER1 mRNA levels in (a) epithelial (HOSEpiC, SK-OV-3, OVCAR-3) and granulosa (HGrC1, COV434, KGN) ovarian normal and cancer cells and (b) ovarian cancer and normal ovarian tissue (based on analysis of open source from the TCGA and GTEx mRNA databases using the GEPIA online tool). (c) Correlation between expression of OXER1 and tumour stage of OC patients and (d) KM survival curves related to OXER1 expression, showing PFS, OS, and PPS of OC patients with different tumour expression of OXER1. The HR is indicated, along with the 95% confidence interval in brackets. *p ≤ 0.05 and **p ≤ 0.01.
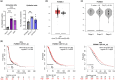
Figure 4
Comparison of PGRMC1 mRNA levels in (a) epithelial (HOSEpiC, SK-OV-3, OVCAR-3) and granulosa (HGrC1, COV434, KGN) ovarian normal and cancer cell lines and (b) ovarian cancer and normal ovarian tissues (based on analysis of open-source data from the TCGA and GTEx mRNA data using the GEPIA online tool). (c) The correlation between PGRMC1 expression and OC stage. (d) KM survival curves related to PGRMC1 expression, showing PFS, OS, and PPS of OC patients with different tumour expression of PGRMC1. The HR is indicated, along with the 95% confidence interval in brackets. *p ≤ 0.05, **p ≤ 0.01, and ***p ≤ 0.001.
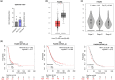
Figure 5
Comparison of PAQR4 mRNA levels in (a) epithelial (HOSEpiC, SK-OV-3, OVCAR-3) and granulosa (HGrC1, COV434, KGN) ovarian normal and cancer cell lines and (b) ovarian cancer and normal ovarian tissues (based on analysis of open-source data from the TCGA and GTEx mRNA databases using the GEPIA online tool). (c) The correlation between PAQR4 expression and stage of OC. (d) KM survival curves related to PAQR4 expression, showing PFS, OS, and PPS of OC patients with different tumour expression of PAQR4. The HR is indicated, along with the 95% confidence interval in brackets. *p ≤ 0.05 and ****p ≤ 0.0001.
Figure 6
Comparison of PAQR7 mRNA levels in (a) epithelial (HOSEpiC, SK-OV-3, OVCAR-3) and granulosa (HGrC1, COV434, KGN) ovarian normal and cancer cell lines and (b) in ovarian cancer and normal ovarian tissues (based on analysis of open-source data from the TCGA and GTEx mRNA database using the GEPIA online tool). (c) The correlation between PAQR7 expression and tumour stage in OC patients. (d) KM survival curves related to PAQR7 expression, showing PFS, OS, and PPS of OC patients with different tumour expression of PAQR7. The HR is indicated, with the 95% confidence interval in brackets. *p ≤ 0.05 and **p ≤ 0.01.
Figure 7
Comparison of PAQR8 mRNA levels in (a) epithelial (HOSEpiC, SK-OV-3, OVCAR-3) ovarian normal and cancer cell lines and (b) ovarian cancer and normal ovarian tissues (based on analysis of open-source TCGA and GTEx mRNA data using the GEPIA online tool). (c) The correlation between expression of PAQR8 and stages of OC. (d) KM survival curves related to PAQR8 expression, showing PFS, OS, and PPS of ovarian cancer patients with different tumour expression of PAQR8. The HR is indicated, with the 95% confidence interval in brackets. *p ≤ 0.05.
Figure 8
AG-205 reduces cell viability in PGRMC1-positive cells. (a) Comparison of PGRMC1 protein level in rare OC (SK-OV-3, COV434) and non-cancer (HGrC1) cells. Dose-dependent effect on cell viability in COV434 and HGrC1 cells following 0, 0.1, 1, 10, 50, and 100 μM AG-205 treatment for (b) 24 h and (c) 48 h. (d) A bright-field microscope analysis following 0, 10 and 50 μM AG-205 treatment for 48 h in COV434 cells (20× magnification). (e) PGRMC1 mRNA expression following 50 μM AG-205 treatment in COV434 cells. **p ≤ 0.01 and ****p ≤ 0.0001.
Similar articles
- Expression of membrane progesterone receptors (mPR/PAQR) in ovarian cancer cells: implications for progesterone-induced signaling events.
Charles NJ, Thomas P, Lange CA. Charles NJ, et al. Horm Cancer. 2010 Aug;1(4):167-76. doi: 10.1007/s12672-010-0023-9. Horm Cancer. 2010. PMID: 21761364 Free PMC article. - PAQR6 Upregulation Is Associated with AR Signaling and Unfavorite Prognosis in Prostate Cancers.
Yang M, Li JC, Tao C, Wu S, Liu B, Shu Q, Li B, Zhu R. Yang M, et al. Biomolecules. 2021 Sep 18;11(9):1383. doi: 10.3390/biom11091383. Biomolecules. 2021. PMID: 34572596 Free PMC article. - Nonclassical progesterone signalling molecules in the nervous system.
Petersen SL, Intlekofer KA, Moura-Conlon PJ, Brewer DN, Del Pino Sans J, Lopez JA. Petersen SL, et al. J Neuroendocrinol. 2013 Nov;25(11):991-1001. doi: 10.1111/jne.12060. J Neuroendocrinol. 2013. PMID: 23763432 Review. - Progesterone signaling mediated through progesterone receptor membrane component-1 in ovarian cells with special emphasis on ovarian cancer.
Peluso JJ. Peluso JJ. Steroids. 2011 Aug;76(9):903-9. doi: 10.1016/j.steroids.2011.02.011. Epub 2011 Mar 1. Steroids. 2011. PMID: 21371489 Free PMC article. Review.
References
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018 Nov;68(6):394–424. 10.3322/caac.21492. Epub 2018 Sep 12. Erratum in: CA Cancer J Clin. 2020 Jul;70(4):313. PMID: 30207593. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous