BCL6b mediates the enhanced magnitude of the secondary response of memory CD8+ T lymphocytes (original) (raw)
Abstract
A characteristic of the secondary response of CD8+ T cells that distinguishes it from the primary response is the generation of greater numbers of effector cells. Because effector CD8+ T cells are derived from a pool of less differentiated, replicating cells in secondary lymphoid organs, and because IL-2 mediates effector differentiation, the enhanced secondary response may reflect the enlargement of this generative pool by the transient repression of IL-2-mediated differentiation. We have examined for this function the transcriptional repressor BCL6b, a homologue of BCL6 that represses IL-2-induced B cell differentiation. BCL6b is expressed in a small subset of antigen-experienced CD8+ T cells. Ectopic expression of BCL6b in CD8+ T cells diminishes their growth in response to IL-2 in vitro. Female mice in which the BCL6b gene has been interrupted have normal primary responses of CD8+ T cells to infection with vaccinia expressing the H-Y epitope, Uty, but Uty-specific, BCL6b–/–, memory CD8+ T cells have diminished recall proliferative responses to this epitope in vitro. BCL6b–/– mice also have normal primary CD8+ T cell responses to influenza infection, but nucleoprotein peptide-specific, BCL6b–/–, memory CD8+ T cells have a cell autonomous defect in the number of effector cells generated in response to reinfection. Therefore, BCL6b is required for the enhanced magnitude of the secondary response of memory CD8+ T cells.
Keywords: BCL6-associated zinc finger protein, influenza, vaccinia, interleukin 2
The secondary response of CD8+ T cells has three distinguishing characteristics that enable it to control viral infections more efficiently than does the primary response: immediate antiviral defense, more rapid production of new effector CD8+ T cells, and the generation of greater numbers of effector CD8+ T cells. The mechanisms involved in the first and second characteristics have been defined: the immediate antiviral response is mediated by “effector” memory CD8+ T cells that are maintained by the cytokines IL-7 and IL-15 (1–3) and by CD4+ T cells (4), and the accelerated production of new effector CD8+ T cells reflects the relatively high frequency of “central” memory CD8+ T cells that replicate in response to viral antigens (5). The mechanism underlying the capacity of the secondary response for generating larger numbers of effector CD8+ T cells, however, remains unknown but presumably must be related to the factors that govern antigen-driven proliferation of CD8+ T cells.
The proliferation of CD8+ T cells during a primary response has two, probably sequential, phases. The first phase occurs in secondary lymphoid tissue, is independent of the cytokines IL-2 (6), IL-4 (7), IL-7 (8), or IL-15 (9), and is not associated with differentiation to an effector cell, as indicated by the inability of cells generated in the absence of IL-2 or IL-15 signaling to make IFN-γ or exhibit immediate cytotoxic T cell activity (9). The second phase correlates with emigration of the activated CD8+ T cells to peripheral, nonlymphoid tissues, may be mediated by IL-2, and is associated with the acquisition of effector functions by the responding CD8+ T cells (10). This phase may correspond to the IL-2-dependent proliferation and differentiation of CD8+ T cells that has been shown to be programmed by the prior ligation of the T cell receptor (TCR) (11). Because the number of cellular divisions mediated by IL-2 signaling is finite (11), and religation of the TCR after cellular stimulation by IL-2 causes activation-induced cell death (12), it is likely that variability in the number of effector CD8+ T cells that are generated during a primary response is determined mainly by the first phase of proliferation in the secondary lymphoid tissues draining sites of infection.
Although a similar, two-phase pattern of proliferation for memory CD8+ T cells has not been shown, some findings indicate its occurrence here also. The first, lymphoid tissue phase may be inferred from the finding that memory CD8+ T cells that have not passed through an effector stage of development, and therefore have maintained the property of naive CD8+ T cells for localizing to lymphoid tissues, mediate secondary proliferative responses (9). Furthermore, even memory CD8+ T cells that may have been effector cells but have reverted to a high level of CD62L expression, which promotes homing to lymph nodes, have more robust secondary proliferative responses than do memory cells with low CD62L (13). The occurrence of the second, peripheral tissue phase is indicated simply by the finding of differentiated effector cells during a secondary response. Therefore, as with the primary response, the magnitude of the secondary CD8+ T cell response may be determined by how many cells comprise the antigen-dependent, cytokine-independent phase of proliferation in the secondary lymphoid organs that drain sites of infection.
One means by which the size of the proliferating pool of CD8+ T cells in secondary lymphoid organs might be increased would be to prevent their emigration by suppressing IL-2-dependent differentiation to effector cells. An example of this process has been found with germinal center B lymphocytes that express the transcriptional repressor BCL6, which inhibits IL-2 mediated, signal transducers and activators of transcription (STAT)-dependent differentiation of activated B cells to plasma cells (14). Without BCL6, the antigen-stimulated clones of B cells do not accumulate to form rapidly replicating clusters of cells, termed germinal centers (15–17). Because this process is analogous to that being proposed for increasing the cytokine-independent pool of replicating memory CD8+ T cells, we have examined a homologue of BCL6, BCL6b, also termed BAZF (18), for its possible role in the ability of memory CD8+ T cells to generate large numbers of effector CD8+ T cells in secondary immune responses.
Materials and Methods
Real-Time and Single-Cell RT-PCR. Total RNA was extracted from sorted human CD8 T cells with Absolutely RNA RT-PCR (Stratagene), and reverse transcription was carried out by using the ProSTAR First-Strand RT-PCR Kit (Stratagene). Total RNA was purified from sorted murine CD8 T cells. The amount of amplicon generated during the PCR was measured by using an Applied Biosystems 7700 (Applied Biosystems). The sequences of the primers and TaqMan probes used were as follows: murineBCL6b forward, 5′-ATCTTCCTAAATGGAGGTGAGTTGTC-3′; murine BCL6b reverse, 5′-TTCGAACCCAGGCAAACC-3′; murine BCL6b probe, 5′-FAM-CCACCAAGGAGGCCCAGCATCA-TAMRA-3′; hypoxanthine guanine phosphoribosyltransferase (HPRT) forward, 5′-TTAAGCAGTACAGCCCCAAAATG-3′; HPRT reverse, 5′-CAAACTTGTCTGGAATTTCAAATCC-3′; HPRT probe, 5′-FAMCCTTTTCACCAGCAAGCTTGCAACCTT-TAMRA-3′; human BCL6b forward, 5′-ACCCCTCAGAGCACACAAGG-3′; human BCL6b reverse, 5′-GGCCCCGGAAAATTGAATAG-3′; human BCL6b probe, 5′-FAM-AGTTCTCATCGCCTGCAGTGGCTTCT-TAMRA-3′; β-actin forward, 5′-TCACCCACACTGTGCCCATCTACGA-3′; β-actin reverse, 5′-CAGCGGAACCGCTCATTGCCAATGG-3′; and β-actin probe, 5′-FAM-ATGCCCXCCCCCATGCCATCCTGCGT-TAMRA-3′.
Single cells were sorted into 15 μl of buffer A, One-Step RT-PCR buffer (Qiagen, Crawley, U.K.), containing 0.75 μl of RNase OUT (Invitrogen). Plates were immediately frozen and stored at –70°C until use. Each RT-PCR consisted of 15 μl of cell lysates in buffer A and 15 μl of Mastermix [3 μl of 5× Qiagen OneStep RT-PCR buffer/1.2 μl of Qiagen OneStep RT-PCR Enzyme Mix/0.45 μlof 25 mM dNTPs (Amersham Pharmacia)/1.5 μl of RNase OUT (Invitrogen)/0.6 μl of HotStarTaq DNA polymerase (Qiagen)/4.65 μl of Nuclease-Free Water (Ambion, Huntingdon, U.K.)/0.6 μl of of each 15-μM primer]. Primers for CD3ε mRNA were as follows: forward, 5′-TGA AGT AAT GAG CTG GCT GCG-3′; reverse, 5′-AGC AGA CGT AGT AGC CAC TGT CCT-3′. Primers for BCL6b mRNA were as follows: forward, 5′-GCA GCA GTG AAG AAG GAA CCA-3′; reverse, 5′-TGC GGC TGT GTG TTT TTA GGT-3′. RT-PCR was performed by using a GeneAmp 9700 thermal cycler (Applied Biosystems): 60 min at 50°C; 15 min at 95°C; and 45–50 cycles of 30 s at 94°C, 30 s at 55°C, 45 s at 72°C; and 5 min at 72°C. Second PCR was run with 1/25 dilution of an RT-PCR in 50 μl of HotstarTaq Mastermix. The semi-nested forward primer (0.6 μM) for CD3ε was 5′-AAA GTC TCC ATC TCA GGA ACC AGT-3′; and reverse primer for BCL6b was 5′-AGG TCG GTT AAA ACG TCG TCC-3′. The PCR program was as follows: 15 min at 95°C; 40 cycles of 30 s at 94°C, 30 s at 55°C, 45 s at 72°C; and 5 min at 72°C. The identity of all BCL6b PCR products was confirmed by sequencing.
To determine the sensitivity of the single-cell RT-PCR, purified BCL6b mRNA was prepared by using Ambion RT-PCR Competitor Construction kit and measured by using Rediplate 96 Ribogreen RNA quantitation kit (Invitrogen). Two-fold dilutions of BCL6b mRNA, alone or spiked into lysates of single CD8+CD44lowCD62L+ cells, were assayed by using the single-cell RT-PCR method.
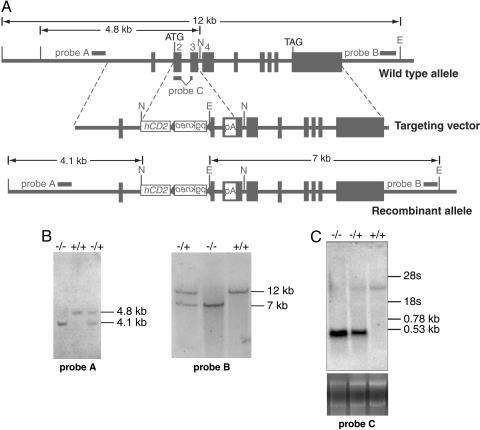
Gene Targeting. A 12-kb _Eco_RI fragment containing the BCL6b gene was isolated from a 129-mouse genomic DNA library. A targeting vector was constructed in which nucleotides 14–114 of the second exon of BCL6b were replaced with an hCD2 reporter (gift of D. Littman, Skirball Institute, New York University, New York) followed by, in reverse orientation, a loxP-flanked neomycin resistance cassette (gift of A. MacKenzie, Medical Research Council Laboratory of Molecular Biology, Cambridge, U.K.). In addition, three copies of the SV40 polyA signal were inserted into the _Acc_I site of exon 3. The linearized construct was injected into E14 129 mouse ES cells. G418-resistant clones were screened by PCR for homologous recombination by using a 5′ flanking sense primer 5′-GCCACATCGTCATCCAGGGTC-3′ and an antisense primer from the hCD2 gene 5′-TGGAGACTGCACCTTTGGAAG-3′ followed by confirmation using Southern blot and the 3′ flanking probe (Fig. 1). Targeted clones were injected into C57BL/6 (B6) blastocysts. Chimeric 129 males were crossed with 129 females, and germ-line transmission of the targeted allele was detected by Southern blot and by PCR using the primers 5′-ATGCTGAGTCCGCTATCTGATG-3′ (sense), 5′-GAACTGCCTTGTGTGCTCTTA-3′ (antisense, wild-type), and 5′-TGGAGACTGCACCTTTGGAAG-3′ (antisense, mutant).
Fig. 1.
Wild-type and targeted BCL6b alleles. (A) BCL6b genomic locus before gene targeting, the targeting vector, and after targeting. Black boxes indicate exons, and pA represents the inserted SV40 polyadenylation signal. E, _Eco_RI; N, _Nco_I. (B) Southern blot of _Eco_RI- and _Nco_I-digested genomic DNA from –/–, +/–, and +/+ mice using probes A and B that flank the 5′ and 3′ ends, respectively, of the BCL6b gene. (C) Northern blot of RNA prepared from the lungs of –/–, +/–, and +/+ mice using a cDNA probe of sequences of exons 2–3 up to the inserted polyadenylation signal.
Viral Infection of Mice. For analysis of CD8+ T cell responses to the H-Y Uty epitope, mice were infected intravenously with 3 × 106 plaque-forming units of recombinant vaccinia/Uty (19). Purified CD4+ and CD8+ T cells were prepared by magnetic-activated cell sorting (MACS) beads (Miltenyi Biotec, Cologne, Germany). In some in vitro cultures, 20 units of murine IL-2 (mIL-2) (R & D Systems) was added on days 1, 3, and 5. Purified memory CD4+ T cells were stimulated with splenocytes taken 2 days after infection of mice with vaccinia. To detect anti-vaccinia antibody responses, ELISA plates that had been coated with lysates of vaccinia-infected TK cells were incubated with dilutions of murine serum, followed by washing and addition of biotin anti-mouse IgG1 and biotin anti-mouse IgG2a (BD Biosciences). Intracellular IFN-γ was detected in CD8+ T cells stimulated with the 10–6 M Uty peptide (WMHHNMDLI) as described (BD Biosciences). In vitro cytolytic activity of restimulated splenocytes of vaccinia-Uty immunized mice was evaluated by a standard 5-h 51Cr release assay by using EL-4 target cells pulsed with 1 μM Uty-peptide or control NP peptide (ASNENMDAM). Db/Uty-specific CD8+ T cells were detected by the binding of Db/NP tetramers (ProImmune, Oxford).
Mice were infected intranasally with 40 hemagglutinating units (HAUs) of the A/WSN strain of influenza, or infected i.p. with 10,000 HAU of the A/x31 strain. Db/NP-specific CD8+ T cells were enumerated by the binding of Db/NP tetramers. The capacity of CD8+ T cells from mice previously infected with influenza to produce IFN-γ was assessed by intracellular cytokine staining after in vitro stimulation with the H-2 Db-restricted NP peptide, ASNENMETM.
Retroviral Expression of BCL6b in Murine CD8+ T Cells. The retroviral vector pMIG was a generous gift from J. C. Zuniga-Pflücker (Sunnybrook and Women's College Health Sciences Centre, Toronto). The cDNA encoding murine BCL6b (18) was mutated by substituting alanine for Ser-160 and Ser-186 to alter a potential PEST (proline, glutamate, serine, and threonine) domain, and inserted into pMIG by means of _Xho_I and _Eco_RI sites. The pCL-ECO packaging construct was a gift from F. Randow (Medical Research Council Laboratory of Molecular Biology, Cambridge, U.K.).
Moloney murine leukemia virus (MMLV) expressing either EGFP alone (pMIG-IRES-EGFP) or with BCL6b (pMIG-BCL6b-IRES-EGFP) was produced by transiently transfecting the ecotropic Phoenix packaging cell line (G. Nolan, Stanford University School of Medicine, Stanford, CA) at 70% confluency with an equimolar ratio of pMIG and the packaging construct, pCL-ECO, by using 20 μg of total DNA using a modified calcium-phosphate transfection protocol (20). All retroviral stocks used had titers >3 × 106 infectious units/ml, as assessed by infection of NIH 3T3 cells.
Replicate samples of 1 × 105 CD8+ T cells from the spleens of 12- to 16-week-old male OT-I Rag–/– transgenic mice (21) were cultured with 2 × 105 A20 cells that had been treated with 500 μg/ml mitomycin C (Sigma–Aldrich), in 0.2 ml of complete medium containing 200 ng/ml anti-CD3ε (clone 145-2C11, BD Biosciences) and 5 μg/ml anti-IL2 (clone JES6-1A12, eBioscience, San Diego) in 96-well flat bottom tissue culture plates. Two days postactivation, CD8+ T cells were restimulated by using the original conditions in 50 μl of complete medium and infected with the pMIG-IRES-EGFP and pMIG-BCL6b-IRES-EGFP retroviruses, respectively, by addition of 150 μl of viral supernatant per 1 × 105 cells and 6 μg/ml protamine sulfate. After 16 h, the cells were washed and replated in the presence of anti-CD3ε and anti-IL-2. Viable CD8+ T cells were measured by flow cytometry using counting beads (Caltag, South San Francisco, CA).
Results
Expression of BCL6b by Memory CD8+ T Cells. BCL6b is expressed in the lung and heart of mice, but its expression in specific populations of CD8+ T cells has not been examined quantitatively (18). To determine whether BCL6b is expressed by CD8+ T cells, populations of naive and memory CD8+ T cells were purified from human peripheral blood, and their content of BCL6b mRNA was measured by real-time quantitative PCR. Memory CD8+ T cells, defined by their expression of the CD45RA– isoform and CD28, had 20-fold more BCL6b mRNA than did naive CD8+ T cells (Table 1). When naive and central and effector memory CD8+ T cells were purified based on their pattern of expression of CD45RA and CCR7, both central and effector subpopulations were found to have increased expression of the transcription factor. These findings with human lymphocytes were extended by measuring mRNA levels in murine memory CD8+ T cells taken from the spleens of mice that had been infected 60 days previously with lymphocytic choriomeningitis virus (22). Both central and effector memory CD8+ T cells, as distinguished by high and low expression of CD62L, respectively, had ≈10-fold higher levels of BCL6b mRNA than did naive CD8+ T cells (Table 1).
Table 1. Expression of BCL6b by human and murine CD8+ T cells as assayed by real-time RT-PCR.
| mRNA ratio | ||
|---|---|---|
| CD8+ T cells | BCL6b/actin | BCL6b/HPRT |
| Human | ||
| Naive (CD45RA+, CD28+) | 0.6 × 10-4 | |
| Memory (CD45RA-, CD28+) | 12 × 10-4 | |
| Naive (CD45RA+, CCR7+) | 0.5 × 10-4 | |
| Central memory (CD45RA-, CCR7+) | 6.6 × 10-4 | |
| Effector memory (CD45RA-, CCR7-) | 5.5 × 10-4 | |
| Db/gp33-specific murine | ||
| Naive | 0.2 × 10-3 | |
| Central memory (CD62Lhigh) | 3 × 10-3 | |
| Effector memory (CD62Llow) | 2.7 × 10-3 |
The low level of BCL6b mRNA in human and murine memory CD8+ T cells, relative to actin or hypoxanthine phosphoribosyltransferase (HPRT) mRNA levels, suggested that only a subpopulation of these cells expressed the transcription factor. This possibility was evaluated by assaying BCL6b mRNA in single murine CD8+ T cells derived from populations of naive and antigen-stimulated cells. The detection limit of the RT-PCR assay was found to be eight copies of BCL6b mRNA, and successful sorting of a single T cell was positively controlled by finding CD3ε mRNA. BCL6b mRNA was not detected in 415 single, naive CD8+ T cells, taken either from C57BL6/J mice or from the F5 transgenic mouse strain expressing a TCR specific for a peptide from the influenza nucleoprotein in the context of H-2 Db and on a Rag–/– background (23) (Table 2). The identification of naive cells in the former population relied on the markers CD44low and CD62Lhigh, and F5 CD8+ T cells were highly unlikely to have responded to environmental antigens. In contrast, BCL6b mRNA was detected in 3/111 CD8+ T cells specific for the H-2 Db restricted H-Y epitope, Uty, that were purified from the spleens of female mice that had been infected 14 days earlier with vaccinia expressing the Uty peptide (Fisher's exact test, P = 0.009). Therefore, although different viral systems were used to assess BCL6b expression at the population and single-cell levels, these results are consistent with the possibility that the increase in BCL6b mRNA in memory CD8+ T cell populations (Table 1) reflects the presence of a small subpopulation of cells expressing higher levels of BCL6b.
Table 2. Expression of BCL6b mRNA by single CD8+ T cells.
| CD8+ T cell population | BCL6b+ cells/CD3ε+ cells |
|---|---|
| Naive | |
| Wild-type, CD44low, CD62Lhigh, splenic | 0/211 |
| F5 Rag1-/-, Db/NP+, splenic | 0/204 |
| Antigen-stimulated | |
| Wild-type, 14 days post-i.v. vaccinia/Uty, splenic, Db/Uty+ | 3/111* |
Suppression of IL-2-Dependent Growth of CD8+ T Cells by BCL6b. To determine whether BCL6b shares with BCL6 a capacity to regulate lymphocyte responses to IL-2, CD8+ T cells were infected with a bicistronic retroviral vector encoding BCL6b and EGFP. CD8+ T cells from OT-I, Rag–/– mice and expressing a transgenic TCR specific for the Db-restricted SIINFEKL peptide of ovalbumin were activated by culture with anti-CD3 and mitomycin C-treated A20 B lymphoma cells expressing FcγRII and CD70; the former served to cross-link T cell-bound anti-CD3, and the latter to ligate CD27 for initial proliferation and maintenance of viability. Neutralizing anti-IL-2 was included to suppress differentiation. On the second day, cells were infected with retroviral vectors directing the expression of either EGFP alone or with BCL6b. After two additional days of stimulation with anti-CD3 and A20 cells in the presence of anti-IL-2 the number of EGFP+, CD8+ T cells in the cultures was measured. IL-2, 200 units/ml, or anti-IL-2 was then added to replicate samples of cells that were restimulated again with anti-CD3 and A20 cells, and the cultures were continued for a further 72 h when the cell number measurements were repeated. Without addition of IL-2, the numbers of vector control CD8+ T cells decreased by approximately one-half, whereas addition of IL-2 caused an approximate doubling of their cell number (Table 3). The expression of BCL6b did not alter the loss of cells in the absence of IL-2 but significantly diminished the increase caused by addition of IL-2. Therefore, although we are unable to determine whether the amount of BCL6b expressed by these cells was physiological because there are no known primary or transformed T cell lines that express BCL6b to serve as reference standards, these findings indicate that BCL6b at least has the potential for suppressing IL-2-mediated signaling in CD8+ T cells.
Table 3. Inhibition by BCL6b of IL-2 induced CD8+ T cell growth.
| Change in EGFP+ cell number, % | ||
|---|---|---|
| Retrovirus | -IL-2 | +IL-2 |
| pMIG-IRES-EGFP | -53 ± 12 | 103 ± 19 |
| pMIG-BCL6b-IRES-EGFP | -47 ± 9* | -1 ± 10** |
Generation of BCL6b–**/**– Mice. The BCL6b gene was inactivated by replacing codons 1–33 in exon 2 with a human CD2-pgk/neo cassette, and inserting a polyadenylation signal at codon 95 in exon 3, leaving the first downstream, in-frame methionine at codon 119, which is distal to the poxvirus and zinc finger (POZ) domain (Fig. 1_A_). The locus was appropriately targeted, as assessed by Southern blot with probes distinguishing between the right and left arms of the wild-type and recombinant alleles (Fig. 1_B_). Northern blot analysis was performed on mRNA prepared from the lung of BCL6b–/–, BCL6b+/–, and BCL6b+/+ mice by using as the probe cDNA containing the 5′ coding sequence up to the site at which the polyA signal had been inserted. This probe identified the normal, 3.8-kb band in mRNA from a BCL6b+/+ mouse, and revealed a truncated transcript of 0.53 kb in mRNA from a BCL6b–/– mouse (Fig. 1_C_). The latter band did not hybridize with a human CD2 cDNA probe but did with a probe containing intronic sequences between exons 2 and 3 (data not shown), indicating that introduction of the hCD2 cDNA sequence and the pgk-neo cassette altered the normal splicing of the BCL6b gene. Removal of the loxP-flanked pgk-neo cassette by Cre recombinase did not correct this altered pattern of splicing (data not shown).
BCL6b–/– mice on the 129 background were viable and fertile, and homozygous progeny of heterozygous crosses occurred at the expected Mendelian frequency. BCL6b–/– mice had slightly lower percentages of single-positive CD4+ (8.8 ± 0.4% versus 11.9 ± 1.1%; P = 0.03) and CD8+ thymocytes (3.0 ± 0.3% versus 4.3 ± 0.3%; P = 0.01) than did BCL6b+/+ mice, but splenic CD4+ (7.3 ± 0.8 × 106 versus 8.9 ± 1.8 × 106, P = 0.44) and CD8+ T cells (2.2 ± 1.5 × 106 versus 3.4 ± 0.8 × 106, P = 0.19) did not significantly differ.
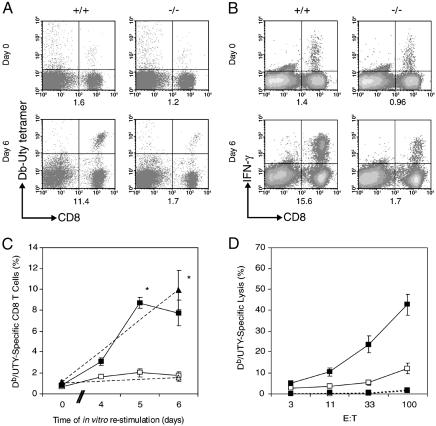
Analysis of the Response to in Vitro Restimulation of in Vivo Primed, BCL6b–**/**– CD8+ T Cells. BCL6b–/– and BCL6b+/+ female mice were intravenously infected with recombinant vaccinia virus expressing H-2Db-restricted H-Y peptide, Uty. Fourteen days later, the mice were assessed for the percentage of splenic CD8+ T cells that were capable of binding Db/Uty tetramers and of antigen-induced secretion of IFN-γ. Although naive mice have no detectable Uty-specific CD8+ T cells (data not shown), ≈1% of the CD8+ splenocytes from both groups of mice 14 days postinfection (p.i.) were Uty-specific (Fig. 2 A and B), indicating that BCL6b expression is not required for a normal primary CD8+ T cell response to this virus. The day 14 p.i. splenocytes were cultured at a 1:10 ratio with irradiated syngeneic male B cells for 6 days. During this period, the percentage of BCL6b+/+, Db/Uty-specific CD8+ T cells increased ≈8-fold, whereas the percentage of restimulated BCL6b–/–, Db/Uty-specific CD8+ T cells increased <2-fold. (Fig. 2 A and C). The capacity of the CD8+ T cells to synthesize IFN-γ in response to Uty-peptide stimulation was assessed at the beginning and end of the 6-day in vitro culture period and showed that the percentages of cells having this function were similar to percentages binding the Db/Uty tetramers (Fig. 2 B and C). The day 6, restimulated CD8+ T cells were also evaluated for their ability to lyse Uty-pulsed EL-4 cells. Restimulated CD8+ T cells from the BCL6b–/– mice had diminished cytotoxic T lymphocyte (CTL) function, which was proportional to their lower frequency of Db/Uty-specific CD8+ T cells (Fig. 2_D_). Similar results were observed when the experiment was repeated 40 days after in vivo priming with vaccinia/Uty (data not shown). Therefore, primed CD8+ T cells that were unable to express BCL6b in vivo are impaired in their subsequent ability to proliferate and generate CTL effectors during in vitro restimulation with antigen.
Fig. 2.
In vitro restimulation of Db/Uty-specific CD8+ T cells from mice that had been infected with vaccinia/Uty. (A and B) The frequency of Db/Uty-specific CD8+ T cells in splenocytes of BCL6b+/+ and BCL6b–/– mice 2 weeks post-vaccinia/Uty infection, as determined by binding of Db/Uty tetramers (A) or Uty peptide-induced synthesis of IFN-γ (B) before and after restimulation with irradiated male B cells for 6 days. The numbers under each panel are the percentages of total CD8+ T cells binding the Db/Uty tetramer or synthesizing IFN-γ in response to the Uty peptide. (C) Changes during in vitro restimulation in the percentages of CD8+ T cells from primed BCL6b+/+ mice (filled symbols) and BCL6b–/– mice (open symbols) that are Db/Uty tetramer+ (squares) or IFN-γ+ (triangles) (*, P < 0.05 for BCL6b+/+ vs. BCL6b–/–). (D) Cytotoxic activity of splenocytes from BCL6b+/+ (filled symbols) and BCL6b–/– mice (open symbols) after restimulation toward EL-4 target cells pulsed with Uty peptide (solid lines) or control peptide (dashed lines).
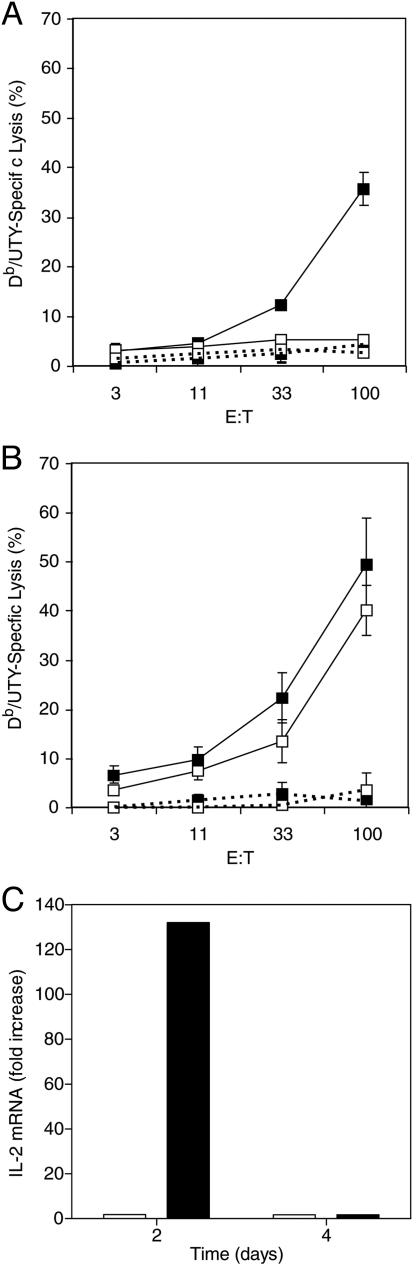
To evaluate further the possibility that the defect was intrinsic to BCL6b–/– CD8+ T cells, CD8+ T cells were purified from the spleens of BCL6b+/+ and BCL6–/– female mice that had been infected 2 weeks previously with vaccinia/Uty, and were restimulated for 6 days with syngeneic male B cells. Restimulated, BCL6+/+, CD8+ T cells, but not BCL6b–/–, CD8+ T cells, generated CTL effectors specific for Uty peptide-pulsed EL-4 cells (Fig. 3_A_). Generation of CTLs in vitro is usually dependent on IL-2, suggesting that the impaired response of the BCL6b–/– CD8+ T cells may have been caused by diminished production of this cytokine. In support of this possibility was the finding that, in a parallel set of restimulated cultures to which exogenous IL-2 had been added, the BCL6b–/–, CD8+ T cells generated CTL activity that was equivalent to that of the BCL6b+/+ cells (Fig. 3_B_). The relative capacity of primed BCL6b+/+ and BCL6b–/– CD8+ T cells to synthesize IL-2 was assessed in purified CD8+ T cells derived from female mice 40 days p.i. with vaccinia/Uty. Although the BCL6b+/+ and BCL6b–/– CD8+ T cells contained equivalent percentages of Uty-specific cells, 0.9% and 0.8%, respectively, restimulation in vitro with irradiated male B cells for 2 days caused CD8+ T cells from BCL6b+/+ mice to have a 131-fold increase in IL-2 mRNA relative to the cells at day 0, but CD8+ T cells from BCL6b–/– mice to have only a 1.7-fold increase (Fig. 3_C_). Thus, primed BCL6b–/– CD8+ T cells have an impaired secondary response in vitro because they have a diminished capacity for the production of IL-2.
Fig. 3.
In vitro restimulation of CD8+ T cells purified from mice that had been infected with vaccinia/Uty. (A and B) Cytotoxic activity toward EL-4 cells of purified CD8+ T cells from primed BCL6b+/+ (filled squares) and BCL6b–/–(open squares) mice after 6 days of restimulation with irradiated male B cells in the absence (A) and presence (B) of exogenous IL-2; solid and dashed lines, EL-4 cells pulsed with Uty and control peptide, respectively. Error bars indicate means ± SEM. (C) Levels of IL-2 mRNA in CD8+ T cells from primed BCL6+/+ (filled bars) and BCL6b–/– (open bars) mice induced by 2 and 4 days restimulation, as measured by real-time RT-PCR.
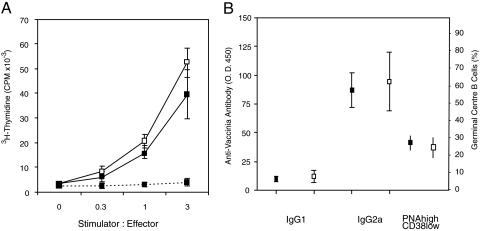
Priming of CD4+ T cells, which may be required for the development of replication-competent memory CD8+ T cells (24–27), was also assessed in BCL6b+/+ and BCL6b–/– mice 14 days p.i. with vaccinia/Uty. BCL6b+/+ and BCL6b–/– CD4+ T cells had similar in vitro, proliferative recall responses to irradiated splenocytes from vaccinia-infected mice (Fig. 4_A_). There were similar titers of serum IgG1 and IgG2a anti-vaccinia antibody and similar percentages of germinal center B cells in the spleens of BCL6b+/+ and BCL6b–/– mice (Fig. 4_B_); these two assays are relevant because they reflect B cell responses that are dependent on help from primed CD4+ T cells expressing CD40L, the mediator of CD4+ T cell help for CD8+ T cells (28–31). Thus, normal priming of CD4+ T cells occurred in the BCL6b–/– mice.
Fig. 4.
CD4+ T cell priming and T-dependent B cell responses induced by vaccinia/Uty infection. (A) Proliferation of CD4+ T cells purified from BCL6b+/+ (filled squares) and BCL6b–/– (open squares) mice, which had been infected with vaccinia/Uty 2 weeks previously, in response to increasing numbers of irradiated, vaccinia-infected (solid lines) or uninfected (dashed lines) splenocytes. (B) Titers of anti-vaccinia serum IgG1 and IgG2a antibodies, and percentages of splenic germinal center B cells in BCL6b+/+ mice (filled symbols) and BCL6b–/– mice (open symbols) 2 weeks p.i. Error bars indicate means ± SEM.
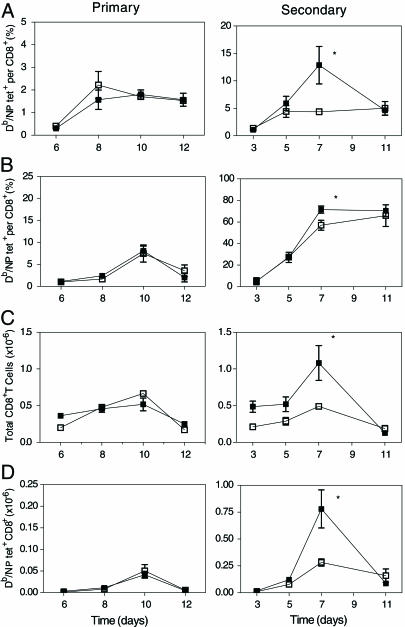
Analysis of the Secondary in Vivo Response of BCL6b–**/**– CD8+ T Cells to Influenza Virus Infection. BCL6b+/+ and BCL6b–/– mice were infected intranasally with the A/WSN strain primarily or secondarily 5 weeks after i.p. infection with the A/x31 strain. CD8+ T cells specific for the NP366 epitope of influenza nucleoprotein in the context of H-2 Db were identified by their capacity to bind Db/NP-tetramers. During the first 12 days p.i. of the primary response, the BCL6b+/+ and BCL6b–/– mice had similar increments in the percentage of CD8+ T cells specific for the Db/NP epitope in the mediastinal lymph nodes (Fig. 5_A_) and lungs (Fig. 5_B_), and in the total number of Db/NP-specific CD8+ T cells in the lungs (Fig. 5_D_). During a secondary infection with influenza, both BCL6b+/+ and BCL6b–/– mice exhibited the accelerated kinetics and immunodominance of CD8+ T cells specific for the Db/NP epitope that are typical of a normal secondary response. However, the BCL6b–/– mice were impaired in the magnitude of their secondary response. The peak percentage of Db/NP-specific CD8+ T cells in the mediastinal lymph nodes of BCL6b–/– mice was only one-third that of BCL6b+/+ mice (Fig. 5_A_). This result was associated with reductions in the number of total CD8+ T cells in the lungs (Fig. 5_C_) and, therefore, in the number of Db/NP-specific CD8+ T cells in the lungs of the BCL6b–/– mice (Fig. 5_D_). Accordingly, although BCL6b is not required for the maintenance of memory CD8+ T cells that are capable of a rapid, proliferative secondary response, as indicated by the normal initial increase of the Db/NP-specific BCL6b–/–, CD8+ T cells in the secondary influenza infection, BCL6b is required for the increased magnitude of the secondary response of the memory Db/NP-specific CD8+ T cells.
Fig. 5.
Primary and secondary responses of Db/NP-specific CD8+ T cells in mice infected with influenza virus. BCL6b+/+ (filled symbols) and BCL6b–/– (open symbols) mice were infected intranasally with the A/WSN strain primarily or secondarily 5 weeks after i.p. infection with the A/x31 strain. At timed intervals, mice were killed to determine the percentage of CD8+ T cells from the mediastinal lymph nodes (A) and lungs (B) binding Db/NP tetramers, the number of all CD8+ T cells from the lungs (C), and the total number of CD8+ T cells from the lungs binding Db/NP tetramers (D). *, P < 0.05 for BCL6b+/+ vs. BCL6b–/–. Error bars indicate means ± SEM.
Because the proportion of all influenza-specific CD8+ T cells that are directed to the Db/NP epitope increases from 20–25% during a primary pulmonary infection to 70–80% during a secondary infection (32), the absence of BCL6b seems to reduce the magnitude of the secondary response of all influenza-specific CTLs to the level of a primary response. This consideration may account for the similar number of total CD8+ T cells in the lungs of BCL6b–/– mice during primary and secondary infections (Fig. 5_C_).
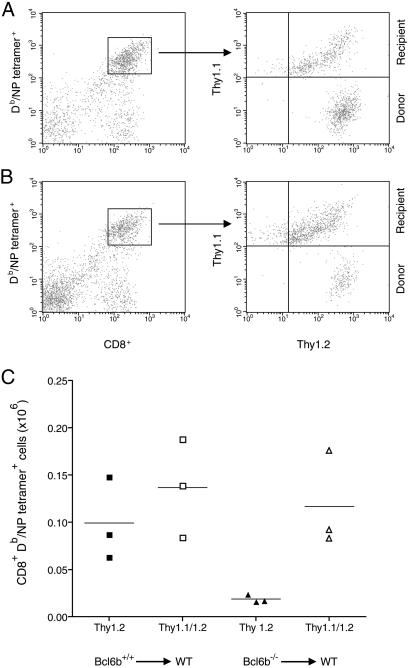
To determine whether the diminished secondary expansion of BCL6b–/– memory CD8+ T cells was caused by an abnormality intrinsic to these cells, an adoptive transfer experiment was performed. CD8+ T cells were purified from spleens of BCL6b+/+ and BCL6b–/– mice that had been primarily and secondarily infected with the A/WSN and A/x31 strains of influenza, respectively, 10 and 8 weeks earlier. The BCL6b+/+ and BCL6b–/– pools of Thy1.2+, CD8+ splenocytes were adjusted to contain 10% Db/NP-specific memory CD8+ T cells, and 2 × 106 cells from each pool were adoptively transferred to BCL6b+/+, Thy1.1+/1.2+, naive mice. One day later, the mice were challenged intranasally with the A/x31 strain of influenza, and 6 days p.i., the lungs of the mice were assessed for the number of Thy1.2+ and Thy1.1/1.2+ CD8+ T cells binding Db/NP tetramers. The adoptively transferred BCL6b+/+, Db/NP-specific memory CD8+ T cells generated 5-fold more effector CD8+ T cells than did the BCL6b–/– memory cells, whereas the recipient, naive CD8+ T cells in the two groups were equally capable of producing effector cells (Fig. 6). Therefore, BCL6b–/– memory CD8+ T cells mediate an impaired recall response, even when in a BCL6b+/+ cellular environment.
Fig. 6.
Response of adoptively transferred BCL6b+/+ and BCL6b–/– memory CD8+ T cells to influenza infection of naive BCL6b+/+ mice. Thy1.2+ CD8+ T cells purified from BCL6b+/+ and BCL6b–/– mice previously infected with influenza, and containing equivalent numbers of Db/NP-specific memory cells, were adoptively transferred to naive, Thy1.1/1.2+, BCL6b+/+ mice. One day later, the mice were intranasally infected with influenza, and 6 days p.i., the Db/NP-specific, CD8+T cells were evaluated for the expression of Thy1.1 and Thy1.2. Representative FACS plots are shown for recipient mice that had received memory CD8+ T cells from BCL6b+/+ (A) and BCL6b–/– (B) mice. The total number of donor and recipient Db/NP-specific CD8+ T cells recovered from the lungs of individual recipient mice is shown in C. P < 0.025 for BCL6b+/+ vs. BCL6b–/–.
Discussion
The lifetime of memory CD8+ T cells can be divided into at least three distinct phases: their development during or after primary infections, their maintenance after primary infections, and their response to secondary infections. The last phase can be further subdivided into acute effector responses by memory cells having an immediate capacity for IFN-γ secretion or CTL function after TCR ligation, and proliferative responses by memory cells for the purpose of generating new effector cells. In the present study, we have found that the transcriptional repressor BCL6b has an essential role in the ability of memory CD8+ T cells to produce large numbers of effector cells during secondary responses. The diminished secondary response of BCL6b–/– memory CD8+ T cells was manifest both in the mediastinal lymph nodes, which may be the major site of memory cell replication, and in the lungs to which progeny of the replicating cells in the lymph nodes migrate during their differentiation to effector cells. This finding indicates that BCL6b augments the number of effector cells by promoting the replicative function of memory CD8+ T cells in secondary lymphoid tissue.
The absence of BCL6b expression did not alter the primary response of CD4+ or CD8+ T cells, diminish the number of memory CD8+ T cells that were maintained after primary responses, or impair the ability of memory CD8+ T cells to initiate the proliferative phase of the secondary response. Thus, the role of BCL6b is distinct from that of the cytokines IL-7 and IL-15, which promote the development of memory cells during the transition from a primary response to the memory phase and their maintenance until reinfection (reviewed in ref. 1), and from that of signaling by CD8αα, which may be required for the expression of the IL-7 receptor-α on activated CD8+ T cells and their development into memory CD8+ T cells (33). The role of BCL6b in enhancing the magnitude of the secondary CD8+ T cell response overlaps in part with the function of CD4+ T cells (28–30), although a recent study has emphasized that CD4+ T cells serve to maintain the number of memory cells rather than to program them for optimal secondary proliferative responses (4). The absence of CD4+ CD25+ regulatory T cells, which also can influence primary and secondary CD8+ T cell antiviral responses (34), in CD4+ T cell-deficient mice may have contributed to these variable outcomes in secondary CD8+ T cell responses in CD4+ T cell-deficient mice. Nevertheless, in the present study, CD4+ T cells were normally primed by vaccinia infection in the BCL6b–/– mice, indicating that a dysfunction of BCL6b–/– CD4+ T cells is not responsible for the defect in secondary proliferative responses of BCL6b–/– CD8+ T cells. We have also found that BCL6b–/– CD4+ T cells respond normally to Salmonella infection, which causes a marked expansion of these cells (data not shown). The expression of BCL6b in memory CD8+ T cells, both human (Table 1) and murine (Tables 1 and 2), and the finding that purified BCL6b–/– memory CD8+ T cells exhibited impaired recall responses, whether assessed in vitro (Fig. 3) or in vivo after adoptive transfer to BCL6b+/+ mice (Fig. 6), also support the likelihood that defect is CD8+ T cell autonomous.
The mechanism by which BCL6b promotes the secondary proliferative response of memory CD8+ T cells may be related to its ability to suppress IL-2 signaling in primary CD8+ T cells that had been transduced with a retroviral vector expressing the transcription factor (Table 3). IL-2 can serve as a growth factor for antigen-stimulated CD8+ T cells, but studies in mice unable either to express or to respond to IL-2 have shown that, whereas its role in the expansion of CD8+ T cells is nonessential (6, 9, 10), it may have a nonredundant function in their differentiation to effector cells. Therefore, the major effect of an inhibitor of cellular responses induced by IL-2 may be to retard the differentiation of activated CD8+ T cells while not diminishing proliferation that is mediated by other factors in secondary lymphoid organs. The capacity of BCL6b+/+ memory CD8+ T cells to produce more IL-2 than BCL6b–/– memory cells also is consistent with the transcription factor maintaining a subset of relatively less differentiated cells, because the potential for IL-2 production is characteristic of central memory as compared with effector memory and effector CD8+ T cells (13, 35, 36). The expression of BCL6b in a small subset of memory CD8+ T cells was demonstrated by single-cell PCR (Table 3), raising the possibility that the BCL6b-expressing and IL-2-competent subsets may be overlapping. Even if this subset had been prevented from responding to IL-2 in vitro, their enhanced production of the cytokine would have mediated the proliferation of the much greater number of memory cells not expressing BCL6b. This effect of BCL6b would be analogous to that of its homologue, BCL6, which represses the differentiation of germinal center B cells without impairing their proliferation and, in so doing, enlarging the population of replicating B cells. The purpose of self-renewal in BCL6-expressing germinal center B cells is to permit the process of somatic hypermutation of Ig genes to lead to affinity maturation of the antibody response whereas, in memory CD8+ T cells, self-renewal mediated by BCL6b may increase the pool of cells that are replicating in the secondary lymphoid organs, which would ultimately increase the production of effector cells.
Previous work suggesting that BCL6 itself may have a role in memory CD8+ T cells did not examine the response of BCL6–/– mice to secondary infections (37, 38). Furthermore, although the present study does not exclude a function for BCL6 in memory CD8+ T cells, the diminished recall responses of BCL6b–/– memory CD8+ T cells, both in vitro and in vivo, indicates that BCL6b has a nonredundant function. We have not been able to reproduce the reported abnormality of naive BCL6b–/– CD4 T cells to stimulation by anti-CD3 in vitro (39) (data not shown). Furthermore, the normal in vitro recall response to vaccinia antigens of CD4+ T cells from vaccinia/Uty-primed BCL6b–/– mice (Fig. 4) and the normal primary response of CD4+ T cells to infection of BCL6b–/– mice with Salmonella indicate that BCL6b does not have an essential role in vivo in the antigenic stimulation of naive CD4+ T cells.
In summary, BCL6b has been found to be expressed in memory CD8+ T cells, to have the capacity to suppress an IL-2-induced response in primary CD8+ T cells, and to be required for the increased number of effector CD8+ T cells that is characteristic of a secondary response. Thus, as had been proposed (40), interposing a phase of self-renewal in memory cells by expression of BCL6b promotes a secondary CD8+ T cell response.
Acknowledgments
We thank Philip Stevenson and Paul Digard for help with influenza infections, Jenefer Blackwell for help with Salmonella infections, and Elizabeth Simpson and Diane Scott for help with in vitro studies. This work was supported by grants from the Wellcome Trust (to D.T.F.), the National Institutes of Health (to D.T.F. and R.A.), and the Cancer Research Institute (to V.C.).
Author contributions: P.M.M., P.J.H., A.I.T., J.M.C., J.L.M., M.C., and Y.M. performed research; P.M.M., P.J.H., A.I.T., J.M.C., M.C., Y.M., and D.T.F. analyzed data; M.J.P., V.C., R.A., and S.M.K. contributed new reagents/analytic tools; D.T.F. designed research; and D.T.F. wrote the paper.
This contribution is part of the special series of Inaugural Articles by members of the National Academy of Sciences elected on May 1, 2001.
Abbreviations: TCR, T cell receptor; p.i., postinfection; CTL, cytotoxic T lymphocyte. See accompanying Biography on page 7415.
References
- 1.Schluns, K. S. & Lefrancois L. (2003) Nat. Rev. Immunol. 3**,** 269–279. [DOI] [PubMed] [Google Scholar]
- 2.Lodolce, J. P., Boone, D. L., Chai, S., Swain, R. E., Dassopoulos, T., Trettin, S. & Ma, A. (1998) Immunity 9**,** 669–676. [DOI] [PubMed] [Google Scholar]
- 3.Kennedy, M. K., Glaccum, M., Brown, S. N., Butz, E. A., Viney, J. L., Embers, M., Matsuki, N., Charrier, K., Sedger, L., Willis, C. R., et al. (2000) J. Exp. Med. 191**,** 771–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun, J. C., Williams, M. A. & Bevan, M. J. (2004) Nat. Immunol. 5**,** 927–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wherry, E. J. & Ahmed, R. (2004) J. Virol. 78**,** 5535–5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kundig, T. M., Schorle, H., Bachmann, M. F., Hengartner, H., Zinkernagel, R. M. & Horak, I. (1993) Science 262**,** 1059–1061. [DOI] [PubMed] [Google Scholar]
- 7.Bachmann, M. F., Schorle, H., Kuhn, R., Muller, W., Hengartner, H., Zinkernagel, R. M. & Horak, I. (1995) J. Virol. 69**,** 4842–4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schluns, K. S., Kieper, W. C., Jameson, S. C. & Lefrancois, L. (2000) Nat. Immunol. 1**,** 426–432. [DOI] [PubMed] [Google Scholar]
- 9.Yu, A., Zhou, J., Marten, N., Bergmann, C.C., Mammolenti, M., Levy, R. B. & Malek, T. R. (2003) J. Immunol. 170**,** 236–242. [DOI] [PubMed] [Google Scholar]
- 10.D'Souza, W. N., Schluns, K. S., Masopust, D. & Lefrancois, L. (2002) J. Immunol. 168**,** 5566–5572. [DOI] [PubMed] [Google Scholar]
- 11.Kaech, S. M. & Ahmed, R. (2001) Nat. Immunol. 2**,** 415–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lenardo M. J. (1991) Nature 353**,** 858–861. [DOI] [PubMed] [Google Scholar]
- 13.Wherry, E. J., Teichgraber, V., Becker, T. C., Masopust, D., Kaech, S. M., Antia, R., von Andrian, U. H. & Ahmed, R. (2003) Nat. Immunol. 4**,** 225–234. [DOI] [PubMed] [Google Scholar]
- 14.Reljic, R., Wagner, S. D., Peakman, L. J. & Fearon, D. T. (2000) J. Exp. Med. 192**,** 1841–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dent, A. L., Shaffer, A. L., Yu, X., Allman, D. & Staudt L. M. (1997) Science 276**,** 589–592. [DOI] [PubMed] [Google Scholar]
- 16.Ye, B. H., Cattoretti, G., Shen, Q., Zhang, J., Hawe, N., de Waard, R., Leung, C., Nouri-Shirazi, M., Orazi, A., Chaganti, R. S., et al. (1997) Nat. Genet. 16**,** 161–170. [DOI] [PubMed] [Google Scholar]
- 17.Fukuda, T., Yoshida, T., Okada, S., Hatano, M., Miki, T., Ishibashi, K., Okabe, S., Koseki, H., Hirosawa, S., Taniguchi, M., et al. (1997) J. Exp. Med. 186**,** 439–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okabe, S., Fukuda, T., Ishibashi, K., Kojima, S., Okada, S., Hatano, M., Ebara, M., Saisho, H. & Tokuhisa, T. (1998) Mol. Cell. Biol. 18**,** 4235–4244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salio, M., Palmowski, M. J., Atzberger, A., Hermans, I. F. & Cerundolo, V. (2004) J. Exp. Med. 199**,** 567–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Costa, G. L., Benson, J. M., Seroogy, C. M., Achacoso, P., Garrison Fathman, C. & Nolan, G. P. (2000) J. Immunol. 164**,** 3581–3590. [DOI] [PubMed] [Google Scholar]
- 21.Hogquist, K. A., Jameson, S. C., Heath, W. R., Howard, J. L., Bevan, M. J. & Carbone, F. R. (1994) Cell 76**,** 17–27. [DOI] [PubMed] [Google Scholar]
- 22.Kaech, S. M., Hemby, S., Kersh, E. & Ahmed, R. (2002) Cell 111**,** 837–851. [DOI] [PubMed] [Google Scholar]
- 23.Mamalaki, C., Murdjeva, M., Tolaini, M., Norton, T., Chandler, P., Townsend, A., Simpson, E. & Kioussis, D. (1996) Dev. Immunol. 4**,** 299–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Belz, G. T., Wodarz, D., Diaz, G., Nowak, M. A. & Doherty, P. C. (2002) J. Virol. 76**,** 12388–12393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Janssen, E. M., Lemmens, E. E., Wolfe, T., Christen, U., von Herrath, M. G. &Schoenberger, S. P. (2003) Nature 421**,** 852–856. [DOI] [PubMed] [Google Scholar]
- 26.Sun, J. C. & Bevan, M. J. (2003) Science 300**,** 339–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shedlock, D. J. & Shen, H. (2003) Science 300**,** 337–339. [DOI] [PubMed] [Google Scholar]
- 28.Schoenberger, S. P., Toes, R. E., van der Voort, E. I., Offringa, R. & Melief, C. J. (1998) Nature 393**,** 480–483. [DOI] [PubMed] [Google Scholar]
- 29.Ridge, J. P., Di Rosa, F. & Matzinger, P. (1998) Nature 393**,** 474–478. [DOI] [PubMed] [Google Scholar]
- 30.Bennett, S. R., Carbone, F. R., Karamalis, F., Flavell, R. A., Miller, J. F. & Heath, W. R. (1998) Nature 393**,** 478–480. [DOI] [PubMed] [Google Scholar]
- 31.Bourgeois, C., Rocha, B. & Tanchot, C. (2002) Science 297**,** 2060–2063. [DOI] [PubMed] [Google Scholar]
- 32.Belz, G. T., Xie, W. & Doherty, P. C. (2001) J. Immunol. 166**,** 4627–4633. [DOI] [PubMed] [Google Scholar]
- 33.Madakamutil, L. T., Christen, U., Lena, C. J., Wang-Zhu, Y., Attinger, A., Sundarrajan, M., Ellmeier, W., von Herrath, M. G., Jensen, P., Littman, D. R. & Cheroutre, H. (2004) Science 304**,** 590–593. [DOI] [PubMed] [Google Scholar]
- 34.Suvas, S., Kumaraguru, U., Pack, C. D., Lee, S. & Rouse, B. T. (2003) J. Exp. Med. 198**,** 889–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sallusto, F., Lenig, D., Forster, R., Lipp, M. & Lanzavecchia, A. (1999) Nature 401**,** 708–712. [DOI] [PubMed] [Google Scholar]
- 36.Huster, K. M., Busch, V., Schiemann, M., Linkemann, K., Kerksiek, K. M., Wagner, H. & Busch, D. H. (2004) Proc. Natl. Acad. Sci. USA 101**,** 5610–5615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ichii, H., Sakamoto, A., Hatano, M., Okada, S., Toyama, H., Taki, S., Arima, M., Kuroda, Y. & Tokuhisa, T. (2002) Nat. Immunol. 3**,** 558–563. [DOI] [PubMed] [Google Scholar]
- 38.Ichii, H., Sakamoto, A., Kuroda, Y. & Tokuhisa, T. (2004) J. Immunol. 173**,** 883–891. [DOI] [PubMed] [Google Scholar]
- 39.Takamori, M., Hatano, M., Arima, M., Sakamoto, A., Fujimura, L., Hartatik, T., Kuriyama, T. & Tokuhisa, T. (2004) Int. Immunol. 16**,** 1439–1449. [DOI] [PubMed] [Google Scholar]
- 40.Fearon, D. T., Manders, P. & Wagner, S. D. (2001) Science 293**,** 248–250. [DOI] [PubMed] [Google Scholar]