Saccharomyces cerevisiae Rab-GDI Displacement Factor Ortholog Yip3p Forms Distinct Complexes with the Ypt1 Rab GTPase and the Reticulon Rtn1p (original) (raw)
Abstract
Rab GTPases are crucial regulators of organelle biogenesis, maintenance, and transport. Multiple Rabs are expressed in all cells, and each is localized to a distinct set of organelles, but little is known regarding the mechanisms by which Rabs are targeted to their resident organelles. Integral membrane proteins have been postulated to serve as receptors that recruit Rabs from the cytosol in a complex with the Rab chaperone, GDI, to facilitate the dissociation of Rab and GDI, hence facilitating loading of Rabs on membranes. We show here that the yeast (Saccharomyces cerevisiae) Golgi Rab GTPase Ypt1p can be copurified with the integral membrane protein Yip3p from detergent cell extracts. In addition, a member of the highly conserved reticulon protein family, Rtn1p, is also associated with Yip3p in vivo. However, Ypt1p did not copurify with Rtn1p, indicating that Yip3p is a component of at least two different protein complexes. Yip3p and Rtn1p are only partially colocalized in cells, with Yip3p localized predominantly to the Golgi and secondarily to the endoplasmic reticulum, whereas Rtn1p is localized predominantly to the endoplasmic reticulum and secondarily to the Golgi. Surprisingly, the intracellular localization of Rabs was not perturbed in _yip3_Δ or _rtn1_Δ mutants, suggesting that these proteins do not play a role in targeting Rabs to intracellular membranes. These data indicate that Yip3p may have multiple functions and that its interaction with Rabs is not critical for their recruitment to organelle membranes.
The biogenesis and maintenance of membrane-bound organelles depend on many different cellular pathways that are regulated by Rab GTPases. Every eukaryotic cell expresses a family of Rab GTPases that are each distinctly localized to different organelles. The general function of Rabs is to regulate interorganelle trafficking and intracellular organelle transport by facilitating the formation of protein complexes at particular sites on organelle membranes (38, 56). Thus, localization of Rabs is critical for their function, but very little is known about how they are specifically targeted within the cell. Pioneering studies have identified _cis_-acting protein sequence features in a small number of Rabs that are necessary and sufficient for targeting these Rabs in vivo to their resident membranes (5, 10, 15, 49). General membrane association is conferred by two C20 geranyl-geranyl lipids that are attached to two COOH-terminal cysteine residues within the prenylation motif (53). Double prenylation of Rab is required for proper organelle targeting, as monoprenylated Rabs (in which one of the prenylated Cys residues is eliminated by mutagenesis) are mistargeted to the endoplasmic reticulum (ER) (6, 19). Because prenylation is a general modification of all Rabs, it alone cannot confer organelle-specific targeting. For several human endocytic Rabs, the C-terminal ∼30 amino acids, which include the prenylated cysteine residues, can target these Rabs to the appropriate endosomal organelles (10, 49). Of the protein sequences of all Rabs, this region is the most diverse and has been termed the “hypervariable” region (35). It is thought that organelle-specific factors may recognize the hypervariable regions of different Rabs and facilitate localization of Rabs via unknown mechanisms.
After prenylation, newly synthesized Rab is delivered to cellular membranes in a dimeric complex with a Rab-specific chaperone, Rab escort protein (2). However, Rab is only transiently associated with its resident membrane, as it is continuously exchanged with the cytosol via the action of a second Rab-specific chaperone, Rab GDP dissociation inhibitor (GDI) (17, 18). Because many Rab GTPases are localized to transport carriers that fuse with target organelles, ongoing membrane trafficking distributes Rabs throughout the cell, so they must be continually retrieved and delivered back to their resident organelles. The role of Rab-GDI is thought to be in Rab recycling, where GDI extracts inactive Rab-GDP from membranes to the cytosol, the GDI-Rab complex is targeted to the appropriate organelle, and, upon the dissociation of the GDI-Rab complex, Rab is loaded onto the organelle membrane (11, 17, 48, 52, 54). Where known, GDI binds Rabs with relatively high affinities, with equilibrium dissociation constants on the order of ∼50 nM (39, 44), leading to the hypothesis that the dissociation of Rab from GDI is facilitated by factors that displace GDI from Rab, referred to as GDI displacement factors (GDFs) (13). A mammalian protein, prenylated Rab acceptor (PRA1; also called Yip3) (31), exhibits GDF activity for endosomal Rab-GDI complexes in vitro, and the depletion of PRA1/Yip3 by small interfering RNA results in the accumulation of Rab9 in the cytosol (47). No other Rab GDFs are known, nor are any proteins that regulate PRA1/Yip3.
The genome of the budding yeast Saccharomyces cerevisiae encodes 11 Rab GTPases that are localized to organelles of the secretory and endocytic pathways (29). Several yeast integral membrane proteins that appear to bind Rabs in a prenylation signal-dependent manner have been identified: Yip1p, Yop1p/Yip2p, Yip3p, Yip4p, Yip5p, and Yif1p (7-9, 55). The protein sequences and predicted topologies of Yip1p, Yip4p, and Yip5p are related, and each of these proteins has been reported to bind multiple Rabs in a prenylation signal-dependent manner (9). Yip1p appears to be required for the budding of COPII vesicles from the endoplasmic reticulum and for the fusion of these vesicles with the Golgi in vitro (3, 22). Furthermore, Yip1p associates with the Rab GTPase Ypt1p (55), and inactivation of a mutant Yip1 protein in vivo leads to the depletion of Ypt1p from the Golgi and its accumulation in the cytosol, suggesting that Yip1p may be a GDF for Ypt1p (6). Yip3p has no primary sequence relationship to the YIP1 protein family; however, like the other YIP proteins, it has a large COOH-terminal hydrophobic region, and it binds multiple Rabs (7).
Because the number of Yip proteins in yeast and other organisms is smaller than the number of Rab proteins, it has been suggested that combinatorial interactions among Yip proteins could generate sufficient diversity to accommodate Rab-specific GDF functions (37, 47). In support of this, Yip1p is associated with Yif1p, Yop1p, and Yip3p, and two-hybrid interactions have been reported between other Yip proteins (8, 25, 51). As a first step in exploring a potential role for yeast Yip3p in the localization of Rab proteins, we have identified the organelles where Yip3p is localized, identified proteins that are associated with Yip3p in vivo, and tested several predictions of the model that yeast Yip3p is a GDF for Rabs in vivo.
MATERIALS AND METHODS
Microbiology methods.
All yeast strains used in this study were derived from SEY6210 (_MAT_α _ura3_-_52 his3-Δ_200 _trp1-_Δ_901 lys2_-_801 suc2-_Δ_9 leu2_-3,11). The strain used for the large-scale Yip3p-myc immunopurification was TVY614 (_MAT_α YIP3::13xMyc::_KanMX ura3_-_52 his3-Δ_200 _trp1-_Δ_901 lys2_-_801 suc2-_Δ_9 leu2_-3,11 pep4Δ::LEU2 prb1Δ::HISG prc1Δ::HIS3). DNA sequences encoding epitope tags were amplified by PCR and then integrated into the genome by transformation using standard methods (36). Primary transformants were screened by immunoblotting, and then the presence of the epitope tag at the correct locus was confirmed by PCR of genomic DNA using primers to amplify the appropriate locus. All strains were grown on synthetic complete medium lacking particular nutrients as required to select and maintain plasmids.
Fluorescence microscopy.
Strains were grown in selective medium until the culture reached an optical density at 600 nm (OD600) of between 0.3 and 0.5. The GFP-Rtn1p images (see Fig. 3 and 7) were captured using a Nikon Eclipse E800 microscope fitted with a cooled, high-resolution charge-coupled-device camera. Cells were stained with Hoechst 33258 (Molecular Probes, Eugene, OR) by adding Hoechst at 500 μg/ml to the growth medium and incubating for 10 min at room temperature. Cells were then immediately visualized. Some images (see Fig. 5) were captured using an Olympus IX70 fluorescence microscope fitted with a 100× lens objective controlled through a Deltavision deconvolution workstation. Images were deconvoluted and then manipulated in Adobe Photoshop (v. 7.0) as described previously (43). The plasmid encoding Sec7p-RFP has been described previously (8).
FIG. 3.
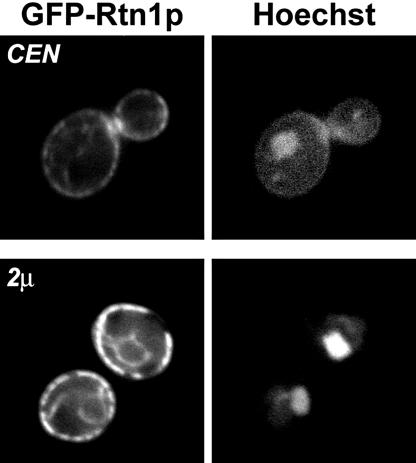
GFP-Rtn1p is localized to the endoplasmic reticulum and to the Golgi. An _rtn1_Δ strain was transformed with a single-copy CEN plasmid or a multicopy 2μ plasmid containing a GFP-RTN1 gene, and GFP-Rtn1p was visualized by fluorescence microscopy. To identify nuclei, cells were stained with Hoechst.
FIG. 7.
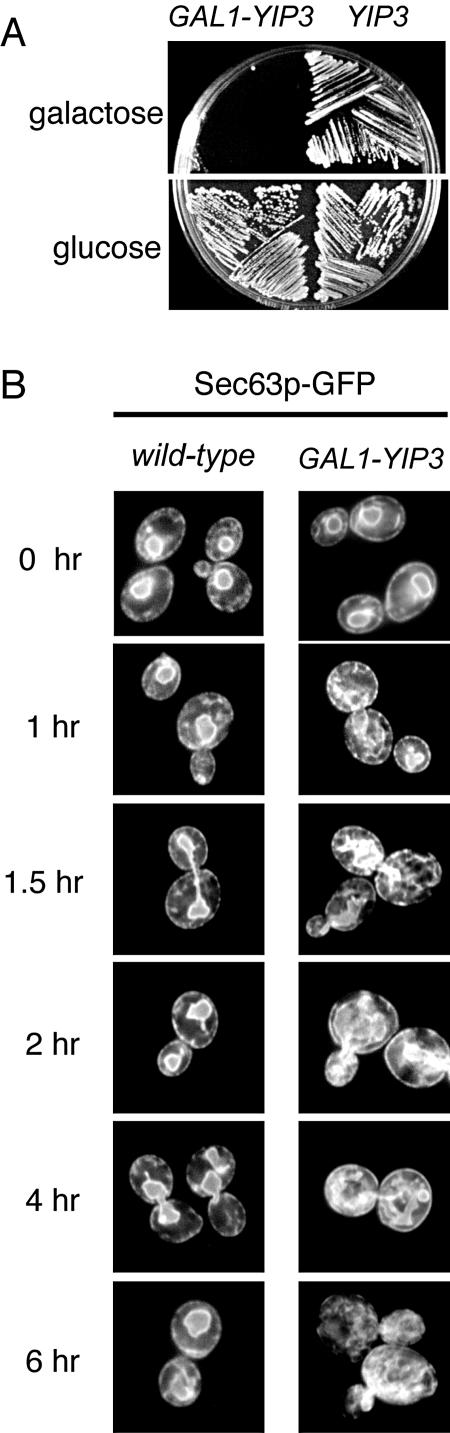
Overproduction of Yip3p is lethal and leads to an expansion of the endoplasmic reticulum. (A) The native YIP3 promoter was replaced with the inducible GAL1 promoter, and this strain was grown on plates containing galactose or glucose as the carbon sources for 3 days. The growth of a wild-type strain that expresses YIP3 from its native promoter on galactose and glucose is shown for comparison. (B) A plasmid containing a SEC63-GFP fusion gene, which allows membranes of the endoplasmic reticulum to be visualized by fluorescence microscopy, was transformed into a wild-type strain and the GAL1-YIP3 strain. Each strain was grown in a glucose-containing medium and then shifted to a galactose-containing medium. Cells were then visualized by fluorescence microscopy at the indicated time points. The morphology of the endoplasmic reticulum in wild-type cells did not change over time, whereas in cells that overproduce Yip3p, ER membranes appear to proliferate with increasing time of Yip3p overproduction.
FIG. 5.
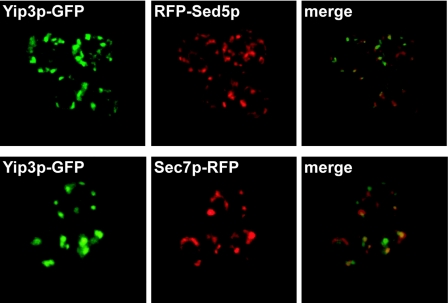
Yip3p is localized throughout the Golgi and to the endoplasmic reticulum. A strain expressing Yip3p-GFP was transformed with a single-copy CEN plasmid that expresses an RFP-Sed5p fusion protein, and each protein was visualized by fluorescence microscopy. To compare the localizations of Yip3p-GFP and Sec7-RFP, each gene was tagged at the COOH terminus with GFP or red fluorescent protein so that each gene was expressed from the native promoter, and each protein was visualized by fluorescence microscopy. The images of each fluorescent protein are shown, and an overlay of the two images is shown in the images labeled “merge.” Note that the signal of Yip3p-GFP in the endoplasmic reticulum is very faint because the Golgi puncta fluoresce too intensely to obtain a properly exposed micrograph.
The red fluorescent protein NH2-terminal fusion to SED5 was constructed using PCR to amplify the cloned T4.DsRed gene (4) using the following oligonucleotides: ACCACCTGGACCACCCAGAAATAAATGATGTCTACCTTC AG and GGAAACAGCTATGACCATG. The SED5 gene was amplified from genomic DNA using the following oligonucleotides: TTCTGGGTGGTCCAGGTGGTATGAACATAAAGGATAGAACTTCAG and TACCGGGCCCCCCCTCGAGGTCGACTGTTAATGCGGCGCCTATCT. Expression of RFP-SED5 was driven by 452 bp of the YOP1 promoter and contains the endogenous gene terminator (441 bp) in vector pRS316 (46).
Protein purification and identification.
For the large-scale Yip3p-myc immunopurification, 4 liters of a strain expressing Yip3p-myc under the control of the native YIP3 promoter was grown in yeast extract-peptone-dextrose (YPD) medium to an OD600 of 4. A control wild-type strain that did not express epitope-tagged Yip3p-myc was grown and processed in parallel. The cells were harvested and washed once with distilled water, weighed, and then frozen at −80°C until use. This produced approximately 25.2 g of Yip3p-myc cells and 23 g of wild-type cells. The cell pellets were thawed in 30 ml lysis buffer (1 mM KH2PO4, 10 mM Na2HPO4, 137 mM NaCl, 2.7 mM KCl, 1 mM EDTA, 3 mM MgCl2, 1 mM dithiothreitol, pH 7.4) containing protease inhibitors (Complete protease inhibitor tablet; Roche), and then cells were lysed by two passages through an EmulsiFlex-C5 high-pressure homogenizer (Avestin, Inc., Ottawa, Canada) at a pressure limit of 25,000 lb/in2. The cell extracts were centrifuged at 30,000 × g for 60 min (4°C), and the supernatants (S30 fractions) and pellets (P30 fractions) were harvested. Yip3p-myc was distributed equally between the P30 and S30 fractions. The P30 fractions were resuspended in 30 ml fresh lysis buffer containing 1% octylglucoside by mechanically disrupting the pellet with a pipette and then incubated with tumbling for 1 h at 22°C. Material that was not solubilized was removed by centrifugation, and the supernatant was recovered. No detergent was added to the S30 fraction. To clear the extracts of proteins that bind Sepharose, 1 ml of Sephadex G-25 (packed volume) was added to each supernatant fraction, and the tubes were incubated for 2 h at 4°C with tumbling. The beads were removed by centrifugation (2,000 × g for 5 min), and the supernatants were recovered. To purify Yip3p-myc, 250 μl (packed volume) of 9E10 affinity matrix (Covance, Berkeley, CA) was added to each fraction and tumbled overnight at 4°C. The 9E10 beads were recovered by centrifugation (2 min at 120 × g) and washed once with 5 ml lysis buffer. All buffer was removed with a needle and syringe, and 200 μl protein sample buffer was added to each pellet. After boiling for 3 min, the sample buffer was collected by piercing the bottom of the microcentrifuge tube with a needle, placing the tube in a new microcentrifuge tube, and then centrifuging for 30 seconds. Approximately 90% of each sample was run in a 12% polyacrylamide-sodium dodecyl sulfate gel and stained with Coomassie blue, and protein bands of interest were excised with a razor blade. Matrix-assisted laser desorption/ionization-time of flight analysis after in-gel trypsin proteolysis was done by the University of Massachusetts Medical School Laboratory for Proteomic Mass Spectrometry. Protein Prospector software (12) was used to compare the mass spectrometry results with the yeast proteome database.
Small-scale immunopurifications were carried out with approximately 25 OD600 units of cells harvested at OD600s of 0.5 to 0.7. Cells were washed once with water and resuspended in 200 μl phosphate-buffered saline lysis buffer containing 0.5% Tween 20 and protease inhibitors, and then 200 μl of glass beads was added. The cell suspension was vortexed for 2 min to disrupt the cells, and then 1 ml of lysis buffer-Tween was added and the mixture was tumbled for 8 min at room temperature. Cellular debris was collected by centrifugation (21,000 × g for 5 min at room temperature), and 1.1 ml of supernatant was harvested. The concentration of protein in each sample was determined (to assure that all strains were lysed to the same degree), and 10% of each sample was removed and precipitated with trichloracetic acid to serve as a loading control. For immunopurification experiments from mixed extracts, 550 μl of each strain was combined. To each sample, 40 μl of anti-myc or anti-hemagglutinin (HA) affinity matrix (Covance, Berkeley, CA) was added, and the samples were tumbled overnight at 4°C. The beads were washed twice with lysis buffer containing 0.1% Tween 20, and bound proteins were eluted with sample buffer by heating at 70°C for 30 min. Analysis of purified proteins was done by immunoblotting. For mixing experiments, only half the amount of sample from control purifications (that is, one strain that expressed both tagged proteins) was loaded for comparison to the mixed sample.
For visualizing proteins by immunoblotting, an antiserum to Ypt1p was kindly supplied by Gerry Waters (Princeton University) and was used at a dilution of 1:5,000, the antiserum to Sec4p was kindly supplied by Pat Brennwald (University of North Carolina School of Medicine) and was used at a dilution of 1:1,000, and the antiserum to Ypt7p was kindly supplied by Scott Emr (University of California, San Diego School of Medicine). The antiserum to Vps21p was raised in a rabbit by Cocalico Biologicals, Inc. (Reamstown, PA) using full-length, recombinant Vps21p as the antigen. The crude antisera to Vps21p and Ypt7p were affinity purified on recombinant Vps21p and Ypt7p using previously described methods (45) and used at dilutions of 1:100 (anti-Vps21p) or 1:500 (anti-Ypt7p). The antisera to Dpm1p and Vps10p were purchased from Molecular Probes (Eugene, OR) and were each used at a concentration of 4 μg/ml.
RESULTS AND DISCUSSION
Yip3p forms distinct complexes with the reticulon Rtn1p and the Rab GTPase Ypt1p.
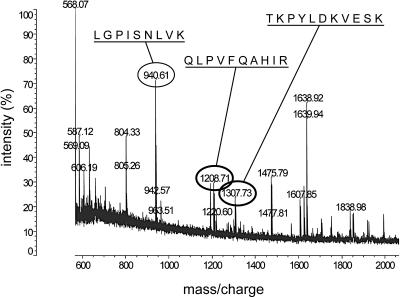
All characterized yeast Yip proteins have been reported to interact with other proteins, and in some cases these interactions are essential for their function (7-9, 32). Thus, we sought to identify proteins that may be associated with Yip3p in vivo. To do so, we purified Yip3p from cell extracts and identified proteins that copurified with it. To facilitate purification, the native YIP3 locus was tagged at the COOH terminus with myc epitope tags in a strain with deletions of the genes encoding several abundant vacuolar proteases. Yip3p-myc was immunopurified from an extract of these cells using anti-myc antibodies. An equivalent purification from a strain in which Yip3p was not tagged was used for comparison to identify proteins that specifically copurified Yip3p-myc, and these were tentatively identified by mass spectrometry. Six proteins were observed to copurify with Yip3p-myc but not with untagged Yip3p. Matrix-assisted laser desorption/ionization-time of flight analysis of one ∼45-kDa copurifying protein that migrated on gels identified three masses that matched the masses of peptides derived from the uncharacterized YDR233c open reading frame (Fig. 1). Although this protein has not yet been characterized, it has recently been named Rtn1p because its primary sequence contains a reticulon homology domain (RHD) (34). Two other proteins that copurified with Yip3p-myc were tentatively identified as Fba1p (fructose 2,6-bisphosphate aldolase) and Rpl12b (a ribosomal protein), and they were not pursued further because they are common contaminants of proteins purified in this manner (23). Another protein that copurified with Yip3p-myc was identified as Ape1p, a vacuolar protease. In subsequent experiments, we have not found any role for Yip3p in Ape1p biogenesis, so the copurification of Ape1p and Yip3p may not reflect a physiologically significant association. So far we have not investigated the other two proteins that copurified with Yip3p, and the analysis presented here focuses on Rtn1p.
FIG. 1.
Identification of Rtn1p as a Yip3p-associated protein. Yip3p-myc was immunopurified from wild-type cells, and copurifying proteins were tentatively identified by mass spectrometry. The portion of the mass/charge spectrum containing three peaks (circled) derived from the YDR233c open reading frame encoding Rtn1p is shown. The predicted amino acid sequences of the three peaks are indicated. These peptides cover approximately 10% of the Rtn1p protein sequence.
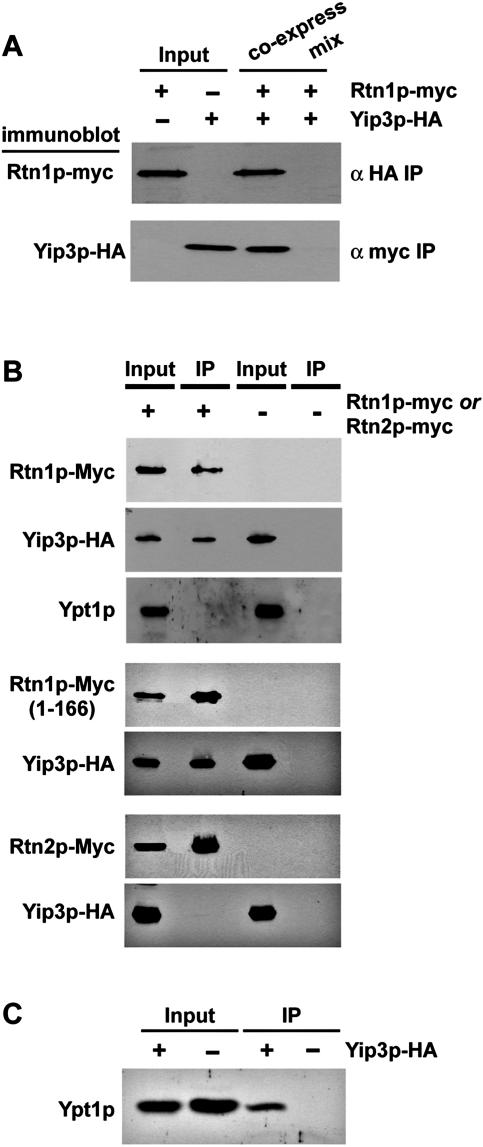
To confirm that the 45-kDa protein that copurified with Yip3p was Rtn1p, we carried out several additional experiments. Because in the original purification Rtn1p and Yip3p copurified in the absence of detergent (see Materials and Methods), as a more stringent criterion to confirm copurification, detergent (0.5% Tween 20) was included in the lysis buffer for this and all follow-up experiments. To make sure that the copurification of Yip3p and Rtn1p was not dependent on the myc epitope tag on Yip3p, the chromosomal YIP3 gene was tagged with the HA epitope at its COOH terminus, and the chromosomal RTN1 gene was similarly tagged with the myc epitope. We then confirmed that Rtn1p-myc could be copurified with antibodies to Yip3p-HA (Fig. 2A). In addition, the purification of Rtn1p-myc with anti-myc antibodies resulted in the copurification of Yip3p-HA (Fig. 2A and B). In each of these experiments, we also determined whether the copurification of Yip3p and Rtn1p required that both tagged proteins were coexpressed in the same cell. For this, cell extracts were made from strains that expressed Yip3-HA or Rtn1p-myc, and then the extracts were mixed and each protein was purified with anti-myc or anti-HA antibodies. Rtn1p and Yip3p copurified only when both tagged proteins were expressed in the same cell (Fig. 2A). This observation suggests that the Yip3p-Rtn1p complex forms in vivo, that it does not dissociate during purification, and that it cannot be reconstituted at a low concentration in a cell lysate.
FIG. 2.
Yip3p can be purified in distinct complexes with the reticulon Rtn1p and the Rab GTPase Ypt1p in vivo. (A) Copurification of Yip3p-HA and Rtn1p-myc was confirmed by immunoprecipitation of Yip3-HA or Rtn1-myc, followed by immunoblotting with the reciprocal antibody (“co-express” lane). To determine whether the complex containing Rtn1p and Yip3p was formed in vivo or after cell lysis during the purification, detergent extracts prepared from strains expressing only one of the tagged proteins were mixed, and then Rtn1p-myc was purified. No Yip3p-HA was observed to copurify with Rtn1p-myc under these conditions. (B) Yeast strains expressing epitope-tagged Yip3p-HA and Rtn1p-myc, Yip3-HA and a truncated version of Rtn1p lacking the region of the C-terminal cytoplasmic domain (amino acids 166 to 295), or Yip3p-HA and Rtn2p-myc were lysed in buffer containing 0.5% Tween 20, and anti-myc antibodies were used to purify Rtn1p-myc or Rtn2p-myc. The purified material was probed with anti-HA antibodies or an anti-Ypt1p antiserum, revealing that Yip3p-HA, but not Ypt1p, copurified with Rtn1p-myc and truncated Rtn1p (amino acids 1 to 208)-myc but not with Rtn2p-myc. In control experiments, no copurification of Yip3p-HA was observed when Rtn1p or Rtn2p was not epitope tagged. (C) Cells expressing Yip3-HA were lysed in buffer containing 1% Tween 20, and Yip3p-HA was immunopurified. The purified material was probed with antibodies to Ypt1p. +, present; −, absent.
Reticulon proteins are highly conserved in eukaryotic evolution, but their functions are largely obscure (33, 34). The RHD is defined as a region of approximately 200 amino acids that contains two membrane-spanning hydrophobic segments that are separated by a loop of 60 to 70 amino acids that is probably oriented to the lumen (34). The most divergent regions of reticulon proteins are the NH2-terminal and C-terminal extensions surrounding the RHD (33). Besides the RHD, Rtn1p contains a region near the carboxy terminus (amino acids 266 to 295) containing heptad repeats that is predicted to form an amphipathic alpha helix (30). This region may bind other proteins via coiled-coil interactions. Based on topology analyses of other reticulons, this region of Rtn1p is predicted to be oriented to the cytoplasm (20, 21). We generated a truncated form of Rtn1p that lacks the entire C-terminal cytoplasmic domain, including the coiled-coil motif (amino acids 166 to 295), and tested whether this protein could associate with Yip3p-HA (Fig. 2B). This truncated Rtn1p copurified with Yip3p-myc as well as with the full-length Rtn1p, indicating that the cytoplasmic domain of Rtn1p is dispensable for binding to Yip3p. Since the NH2-terminal region of Rtn1p is comprised essentially of just the RHD, these results suggest that the Yip3p-binding region of Rtn1p is located in the RHD.
The yeast genome encodes a second reticulon family protein, Rtn2p, which was not identified as a Yip3p-associated protein in the large-scale Yip3p purification. To directly test for the association of Yip3p and Rtn2p, we tagged Rtn2p (with myc epitope tags at its COOH terminus) in the Yip3-HA strain, immunopurified Yip3p-HA, and then assayed the purified material for copurifying Rtn2p-myc by immunoblotting. In this experiment, Rtn2p was not detected as a Yip3p-binding protein (Fig. 2B), although Rtn2p-myc was clearly expressed. Taken together, the results of all the purification experiments indicate that Rtn1p and Yip3p are specifically associated in the cell. Moreover, these results indicate that the reticulon domains of Rtn1p and Rtn2p are not functionally equivalent.
No Rab GTPases were identified as abundant proteins that copurified with Yip3p. Because the copurifying proteins in the large-scale experiments were detected by staining with silver or Coomassie blue, we tested whether any Rab proteins could be detected in the Yip3p purified fractions by more-sensitive immunoblotting analysis with antibodies to native or green fluorescent protein (GFP)-tagged Rabs. We purified Yip3p-HA from detergent extracts and probed the purified material with antibodies to Ypt1, Ypt7, Vps21, Sec4p, GFP-Ypt31p, and GFP-Ypt32p. Approximately 5% of the total Ypt1p in the cell could be copurified with Yip3p-HA (Fig. 2C), but only trace amounts (less than 0.1% of input) of the other Rabs could be detected in the Yip3-HA immunoprecipitates (not shown). These results suggest that a small proportion of membrane-associated Ypt1p is associated with Yip3p-HA at the steady state. Human PRA1/Yip3 has been reported to stably associate with Rab1A (a human ortholog of Ypt1p) but not to stimulate dissociation from GDI (47).
The finding that Ypt1p and Rtn1p can be copurified with Yip3p led us to test whether all three proteins are components of a single complex or whether these interactions represent distinct complexes. To test this, Rtn1p-myc was purified from detergent extracts of cells that also expressed Yip3p-HA, and the purified material was probed with antibodies to Ypt1p and to Yip3p-HA (Fig. 2B and C). No Ypt1p copurified with Rtn1p-myc. These results indicate that Yip3p is a component of at least two distinct protein complexes, one containing the Ypt1 Rab GTPase and another containing Rtn1p.
Yip3p and Rtn1p are localized to the ER and the Golgi apparatus.
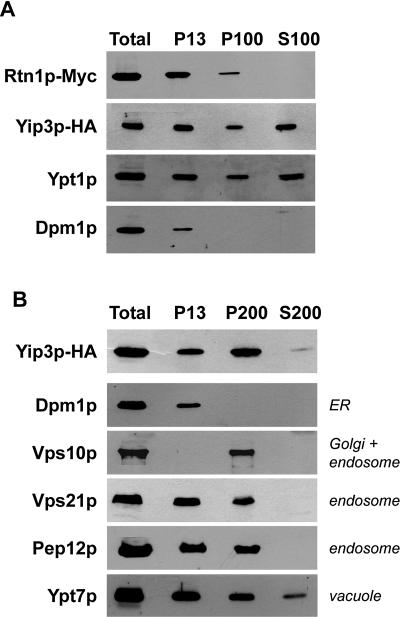
Based on the finding that Yip3p and Rtn1p can be copurified, we predicted that at least a portion of these proteins should be localized to the same organelles. Most characterized reticulon proteins are localized to the endoplasmic reticulum (34). To determine where Rtn1p is localized within the cell, the RTN1 gene was cloned into single-copy CEN and multicopy 2μ NH2-terminal GFP fusion vectors which were then transformed into an _rtn1_Δ strain, and GFP-Rtn1p was visualized by fluorescence microscopy. In cells expressing GFP-Rtn1p from a CEN vector, Rtn1p-GFP fluorescence was observed predominantly in the cortical endoplasmic reticulum (Fig. 3). In some cells, labeling of several punctate organelles was also observed. When GFP-Rtn1p was expressed from the 2μ vector, cortical and nuclear ERs were more intensely labeled, as were several cytoplasmic puncta which probably represent Golgi compartments. These results were corroborated by subcellular fractionation experiments in which the majority of Rtn1p-myc was observed in the low-speed particulate fraction, which is enriched with the ER protein Dpm1p, and a small amount was also observed in the P100 fraction, which contains Golgi markers (Fig. 4A).
FIG. 4.
Subcellular fractionation of Yip3p and Rtn1p. (A) A strain expressing Yip3p-HA and Rtn1p-myc was converted to spheroplasts and then lysed by Dounce homogenization. The extract was cleared of intact cells, and the extract was centrifuged consecutively at 13,000 × g for 10 min to generate the P13 particulate fraction and at 100,000 × g for 1 hour to generate the P100 and S100 fractions. Equal cell equivalents of each fraction were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then probed with antibodies to the indicated proteins. (B) A strain expressing Yip3-HA was fractionated as described for panel A, but the supernatant from the 13,000 × g centrifugation was then subjected to a 200,000 × g centrifugation for 2 h. The identity of the protein in each set of lanes is indicated to the left, and the steady-state localization of that protein is indicated to the right.
To determine where Yip3p is localized within the cell, we inserted GFP immediately upstream of the YIP3 termination codon in the chromosomal locus and examined these cells by fluorescence microscopy (Fig. 5). In most cells, Yip3p-GFP localized to numerous puncta, but many cells also exhibited faint GFP signals on ER membranes. Note that the ER Yip3p-GFP signal is difficult to observe in Fig. 5 because the ER signal is faint compared to the bright puncta. The cytoplasmic puncta correspond mainly to Golgi compartments because many of the Yip3-GFP puncta colocalized with RFP-Sed5p, a marker of the early Golgi, or with Sec7p-RFP, a marker of the late Golgi (Fig. 5). It appears that all of the punctate Yip3p-GFP signal colocalizes with Sed5p- or Sec7p-containing Golgi compartments, although the possibility that some Yip3p may be localized to other punctate organelles, such as endosomes, cannot be ruled out. Localization to the Golgi is consistent with the observation that Yip3p is associated with Ypt1p, which is also localized to the Golgi (41).
As an independent test of Yip3p localization, we examined the subcellular fractionation profile of Yip3p-HA and compared it to marker proteins of several different organelles (Fig. 4B). Consistent with the localization of Yip3p to the ER and the Golgi, Yip3-HA was found to be distributed between the 13,000 × g and the 100,000 × g fractions. Surprisingly, a significant amount of Yip3p-HA was present in the 100,000 × g supernatant (S100) fraction. The amounts of Yip3p in the S100 fraction varied slightly between experiments, and nearly all of the Yip3p in the S100 could be pelleted by centrifugation at 200,000 × g for 2 h (Fig. 4B), suggesting that this pool of Yip3p is either in a large complex or associated with small membranes. Yip3 proteins from other organisms have also been observed to fractionate in a similar manner, and this has been interpreted to indicate that the protein aggregates upon cell lysis (1, 24, 47). No Ypt1p copurified with Yip3p-HA purified from the S100 fraction, suggesting that this pool of Yip3p does not associate with Rab in the cytosol, and overproduction of Rtn1p did not affect the fractionation behavior of Yip3p (data not shown).
The fluorescence microscopy and fractionation experiments indicate that Yip3p is localized throughout the Golgi apparatus, with a small amount localized to the ER. On the other hand, Rtn1p appears to localize predominantly to the ER, with only a small amount localized to the Golgi at steady state. Based on the coimmunopurification experiments, we estimate that approximately 10% of Rtn1p can be copurified with Yip3p, which is consistent with the amounts of Yip3p and Rtn1p that appear to overlap in localization. The localizations of Yip3p-GFP and GFP-Rtn1p were not significantly affected by the deletion or the overproduction of the other protein, so it seems unlikely that the Yip3p-Rtn1p complex is involved in the targeting of either protein within the secretory pathway. At present we do not know if the Yip3p-Rtn1p complex is dynamic. For example, Yip3p might interact with Rtn1p and Ypt1p, as well as other proteins, such as Yip1p (7), in a sequential manner. Alternatively, the Yip3p-Rtn1p and Yip3p-Ypt1p complexes could form independently of each other and have distinct functions. Because the Yip3p-Ypt1p complex does not contain Rtn1p, one possible role for Rtn1p could be in inhibiting the association of Yip3p and Ypt1p, thereby restricting their association to the Golgi, where Yip3p and Ypt1p are localized at steady state.
Localization of Rab GTPases is not affected in a yip3 deletion mutant.
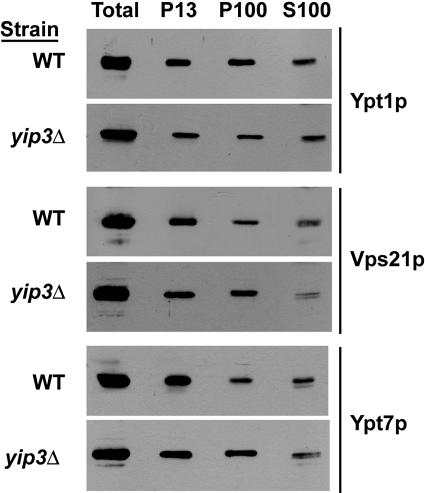
Human PRA1/Yip3 has been shown to act as a Rab GDF in vitro, and the depletion of PRA1/Yip3 results in a reduction in the amount of membrane-associated Rab9, with a concomitant increase in the cytosolic pool of Rab9 (47). As yeast and human Yip3p are clearly orthologous, these results led us to predict that the membrane-associated pools of one or more yeast Rabs will be depleted in a _yip3_Δ mutant. We tested this with subcellular fractionation of wild-type and _yip3_Δ mutant cells, followed by immunoblotting with antibodies to various native Rabs (Ypt1p, Vps21p, Ypt7p, and Sec4p) or GFP-tagged Rabs (GFP-Ypt31p, GFP-Ypt32p, and GFP-Ypt6p) that were expressed from single-copy CEN plasmids. The results are shown only for Ypt1p, Vps21p, and Ypt7p because Yip3p is associated with Ypt1p in vivo (Fig. 2C) and human PRA1/Yip3 is active only on endosomal Rabs (47). Surprisingly, no changes in the fractionation pattern of any Rab could be observed in _yip3_Δ cells compared to wild-type cells (Fig. 6). In addition, no changes in the localizations of GFP-tagged Ypt1p, Ypt31p, Ypt32p, Ypt6p, Vps21p, or Ypt7p were observed in a _yip3_Δ mutant, as determined by fluorescence microscopy. Similarly, no changes in Rab localization were observed in an _rtn1_Δ mutant.
FIG. 6.
Localization of Rab GTPases is not affected in a _yip3_Δ mutant. Wild-type and _yip3_Δ strains were fractionated as described in the legend to Fig. 4A, and the relative amounts of the indicated Rab GTPases in each fraction were determined by immunoblotting.
Given the observed mislocalization of Rab9 in cells depleted of PRA1/Yip3 in human tissue culture cells (47) and the degree to which yeast and human Yip3p is conserved, it is surprising that all Rabs that we have examined are properly localized to membrane fractions in a _yip3_Δ mutant. One possibility is that both Yip3p and Rtn1p have GDF activity, so that Rtn1p can compensate for the loss of Yip3p. To test this possibility, we constructed a _yip3_Δ _rtn1_Δ double mutant and found that this mutant was viable, indicating that Rtn1p and Yip3p do not form an essential pair of genes, as would be expected if they had overlapping functions in intracellular targeting of essential Rabs such as Ypt1p. At least five other proteins (Yip1p, Yip4p, Yip5p, Yif1p, and Yop1) that interact with Rab GTPases in a manner similar to that of Yip3p have been described, so if any of these proteins function as Rab GDFs there could be considerable redundancy in the Rab membrane-loading machinery (7-9, 32, 55). Consistent with this idea, using a previously established Rab membrane-loading assay that monitors GDI-mediated membrane targeting of Rabs onto semipurified cellular membranes (18), we have observed that membranes prepared from wild-type, _yip3_Δ, _yip4_Δ, _yip5_Δ, and _yop1_Δ cells (all nonessential YIP genes) are indistinguishable in their acceptor activities for Ypt1p, Vps21p, Ypt7p, GFP-Ypt31p, and GFP-Ypt32p (data not shown). In addition, in fractionation experiments with _yip4_Δ, _yip5_Δ, and _yop1_Δ mutant strains, the membrane and cytosol pools of all Rabs tested were unchanged compared to those of wild-type cells (data not shown). We have also tested for synthetic lethality of the _yip3_Δ _yip4_Δ, _yip3_Δ _yip5_Δ, and _yip3_Δ _yop1_Δ double mutants and found that these strains are viable with no significant growth defects (not shown). Although our results so far do not support a model in which the physiologic function of yeast Yip3p is as a Rab GDF, this activity will need to be tested directly using purified yeast Yip3p and purified GDI-Rab complexes, as has been done for human PRA1/Yip3p (47).
Overproduction of YIP3 leads to expansion of the ER.
To examine the consequences of YIP3 and RTN1 overproduction, we replaced the native promoters of these genes with the strong, inducible GAL1 promoter and examined the growth of these strains under inducing conditions (Fig. 7A). Overproduction of Yip3p, but not of Rtn1p (data not shown), severely inhibited cell growth. The simultaneous overproduction of Rtn1p and Yip3p did not rescue the toxic effects of Yip3p overproduction (data not shown), suggesting that Rtn1p does not negatively regulate the function of Yip3p under these conditions. The distribution of Ypt1p between the cytosol and intracellular membranes in Yip3p-overproducing cells, as determined by subcellular fractionation, was unchanged compared to that of control cells (data not shown), suggesting that the toxic effects of Yip3p overproduction were not due to a loss of Ypt1p function.
The effects of Yip3p overproduction on the structures of secretory organelles were examined by observing GFP-tagged markers of ER and Golgi membranes in a time course of Yip3p overproduction. Membranes of the ER were observed by expressing Sec63p-GFP, an ER resident integral membrane protein (16), and Golgi membranes were visualized with Sec7-GFP, a marker of late Golgi membranes (42). Under noninducing conditions, the appearance of each of these GFP-tagged proteins was typical for ER and Golgi membranes of wild-type cells (Fig. 7B; Sec7-GFP not shown). In contrast, beginning 1.5 h after the induction of Yip3p expression, striking effects on the structure of the ER were observed, and the membranes of the ER appeared to proliferate (Fig. 7B). Starting at 2 hours postinduction, the Sec7-GFP Golgi pattern changed from a punctate distribution to a haze that was distributed throughout the cytoplasm (not shown), indicating either that Sec7p-GFP had been released from Golgi membranes or that these membranes had vesiculated. In either case, this likely arises as a consequence of the effects of YIP3 overproduction on the early secretory pathway. Another Yip3p-binding protein, Yip1p, is required for export from the ER, so we tested whether simultaneous overproduction of Yip1p rescued the growth defect of Yip3p-overproducing cells and found that it did not (data not shown). These results demonstrate that the overproduction of Yip3p impacts the structure of the ER, and it is likely that this is due to inhibition of ER-to-Golgi transport, as it is well documented that ER membranes proliferate in mutants that are deficient in the budding of COPII vesicles from the ER (28).
One important aspect of the work presented here is the discovery that the reticulon Rtn1p is associated with Yip3p in vivo. Although the reticulon protein family was described several years ago and is highly conserved in eukaryotic evolution (33, 34), very little is known about the functions of these proteins. The best-characterized reticulon is Nogo, a protein that is expressed in oligodendrocytes and is a component of myelin sheaths, where it functions as a neurite growth inhibitor (40). However, the function of Nogo in the regulation of neurite growth is due to a unique portion of the protein that differs from all other reticulons, so Nogo may not serve as a paradigm for investigating the functions of typical reticulons (33, 34). Nearly all proteins identified so far that bind typical reticulons have been identified by yeast two-hybrid screening; these include transport vesicle coat proteins (26), SNARE Tlg1p (25), the endocytosis protein RME-1 (27), Bcl proteins that regulate apoptosis (50), and a lipid biosynthetic enzyme (14). The experiments described here demonstrate by affinity purification that the yeast reticulon Rtn1p is associated with Yip3p, a Golgi protein that binds Rab GTPases, and this finding expands the repertoire of trafficking machinery components that interact with reticulons. To test for the roles of Yip3p and Rtn1p in interorganelle transport, we directly analyzed the trafficking and sorting of a variety of biosynthetic and endocytic cargo molecules (CPY, CPS, ALP, Ste2p, Ste3p, and Ape1p) in _rtn1_Δ and _yip3_Δ mutants. We did not observe any defects in the sorting, transport, or steady-state localization of these proteins. Thus, despite the large amount of evidence that suggests that Yip3p and reticulons interact with components of the membrane-trafficking machinery, it is still not clear if, or how, these interactions regulate membrane trafficking. Nonetheless, the work presented here establishes for the first time that Yip3p is associated with the reticulon Rtn1p and the Rab GTPase Ypt1p, and future work will need to explore the interplay between these proteins and the mechanism by which they participate in ER and Golgi membrane dynamics.
Acknowledgments
This work was supported by grants to C.G.B. from the National Institutes of Health (GM61221), the University of Pennsylvania Cancer Center Pilot Projects Program, and the University of Pennsylvania Research Foundation and by an equipment grant from the National Institutes of Health to J. Sanger (1-S10-RR17879-01).
The technical assistance of Gerard Joe and Megan Speare is appreciated. We thank Erfei Bi for the use of his microscope, Mickey Marks for critical reading of the manuscript, and Mickey Marks, Mark Lemmon, Margaret Chou, and Erfei Bi for helpful discussions.
REFERENCES
- 1.Abdul-Ghani, M., P. Y. Gougeon, D. C. Prosser, L. F. Da-Silva, and J. K. Ngsee. 2001. PRA isoforms are targeted to distinct membrane compartments. J. Biol. Chem. 276**:**6225-6233. [DOI] [PubMed] [Google Scholar]
- 2.Alexandrov, K., H. Horiuchi, O. Steele-Mortimer, M. C. Seabra, and M. Zerial. 1994. Rab escort protein-1 is a multifunctional protein that accompanies newly prenylated rab proteins to their target membranes. EMBO J. 13**:**5262-5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrowman, J., W. Wang, Y. Zhang, and S. Ferro-Novick. 2003. The Yip1p.Yif1p complex is required for the fusion competence of endoplasmic reticulum-derived vesicles. J. Biol. Chem. 278**:**19878-19884. [DOI] [PubMed] [Google Scholar]
- 4.Bevis, B. J., and B. S. Glick. 2002. Rapidly maturing variants of the Discosoma red fluorescent protein (DsRed). Nat. Biotechnol. 20**:**83-87. [DOI] [PubMed] [Google Scholar]
- 5.Brennwald, P., and P. Novick. 1993. Interactions of three domains distinguishing the ras-related GTP-binding proteins Ypt1 and Sec4. Nature 362**:**560-563. [DOI] [PubMed] [Google Scholar]
- 6.Calero, M., C. Z. Chen, W. Zhu, N. Winand, K. A. Havas, P. M. Gilbert, C. G. Burd, and R. N. Collins. 2003. Dual prenylation is required for Rab protein localization and function. Mol. Biol. Cell 14**:**1852-1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calero, M., and R. N. Collins. 2002. Saccharomyces cerevisiae Pra1p/Yip3p interacts with Yip1p and Rab proteins. Biochem. Biophys. Res. Commun. 290**:**676-681. [DOI] [PubMed] [Google Scholar]
- 8.Calero, M., G. R. Whittaker, and R. N. Collins. 2001. Yop1p, the yeast homolog of the polyposis locus protein 1, interacts with Yip1p and negatively regulates cell growth. J. Biol. Chem. 276**:**12100-12112. [DOI] [PubMed] [Google Scholar]
- 9.Calero, M., N. J. Winand, and R. N. Collins. 2002. Identification of the novel proteins Yip4p and Yip5p as Rab GTPase interacting factors. FEBS Lett. 515**:**89-98. [DOI] [PubMed] [Google Scholar]
- 10.Chavrier, P., J. P. Gorvel, E. Stelzer, K. Simons, J. Gruenberg, and M. Zerial. 1991. Hypervariable C-terminal domain of rab proteins acts as a targeting signal. Nature 353**:**769-772. [DOI] [PubMed] [Google Scholar]
- 11.Chou, J. H., and R. Jahn. 2000. Binding of Rab3A to synaptic vesicles. J. Biol. Chem. 275**:**9433-9440. [DOI] [PubMed] [Google Scholar]
- 12.Clauser, K. R., P. Baker, and A. L. Burlingame. 1999. Role of accurate mass measurement (+/− 10 ppm) in protein identification strategies employing MS or MS/MS and database searching. Anal. Chem. 71**:**2871-2882. [DOI] [PubMed] [Google Scholar]
- 13.Dirac-Svejstrup, A. B., T. Sumizawa, and S. R. Pfeffer. 1997. Identification of a GDI displacement factor that releases endosomal rab GTPases from rab-GDI. EMBO J. 16**:**465-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Di Sano, F., B. Fazi, G. Citro, P. E. Lovat, G. Cesareni, and M. Piacentini. 2003. Glucosylceramide synthase and its functional interaction with RTN-1C regulate chemotherapeutic-induced apoptosis in neuroepithelioma cells. Cancer Res. 63**:**3860-3865. [PubMed] [Google Scholar]
- 15.Dunn, B., T. Stearns, and D. Botstein. 1993. Specificity domains distinguish the ras-related GTPases Ypt1 and Sec4. Nature 362**:**563-565. [DOI] [PubMed] [Google Scholar]
- 16.Fehrenbacher, K. L., D. Davis, M. Wu, I. Boldogh, and L. A. Pon. 2002. Endoplasmic reticulum dynamics, inheritance, and cytoskeletal interactions in budding yeast. Mol. Biol. Cell 13**:**854-865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garrett, M. D., J. E. Zahner, C. M. Cheney, and P. J. Novick. 1994. GDI1 encodes a GDP dissociation inhibitor that plays an essential role in the yeast secretory pathway. EMBO J. 13**:**1718-1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilbert, P. M., and C. G. Burd. 2001. GDP dissociation inhibitor domain II required for Rab GTPase recycling. J. Biol. Chem. 276**:**8014-8020. [DOI] [PubMed] [Google Scholar]
- 19.Gomes, A. Q., B. R. Ali, J. S. Ramalho, R. F. Godfrey, D. C. Barral, A. N. Hume, and M. C. Seabra. 2003. Membrane targeting of Rab GTPases is influenced by the prenylation motif. Mol. Biol. Cell 14**:**1882-1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.GrandPre, T., F. Nakamura, T. Vartanian, and S. M. Strittmatter. 2000. Identification of the Nogo inhibitor of axon regeneration as a Reticulon protein. Nature 403**:**439-444. [DOI] [PubMed] [Google Scholar]
- 21.Hamada, N., J. Iwahashi, K. Suzuki, H. Ogi, T. Kashiwagi, K. Hara, M. Toyoda, T. Yamada, and T. Toyoda. 2002. Molecular cloning and characterization of the mouse reticulon 3 cDNA. Cell. Mol. Biol. (Noisy-le-Grand) 48**:**163-172. [PubMed] [Google Scholar]
- 22.Heidtman, M., C. Z. Chen, R. N. Collins, and C. Barlowe. 2003. A role for Yip1p in COPII vesicle biogenesis. J. Cell Biol. 163**:**57-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ho, Y., A. Gruhler, A. Heilbut, G. D. Bader, L. Moore, S. L. Adams, A. Millar, P. Taylor, K. Bennett, K. Boutilier, L. Yang, C. Wolting, I. Donaldson, S. Schandorff, J. Shewnarane, M. Vo, J. Taggart, M. Goudreault, B. Muskat, C. Alfarano, D. Dewar, Z. Lin, K. Michalickova, A. R. Willems, H. Sassi, P. A. Nielsen, K. J. Rasmussen, J. R. Andersen, L. E. Johansen, L. H. Hansen, H. Jespersen, A. Podtelejnikov, E. Nielsen, J. Crawford, V. Poulsen, B. D. Sorensen, J. Matthiesen, R. C. Hendrickson, F. Gleeson, T. Pawson, M. F. Moran, D. Durocher, M. Mann, C. W. Hogue, D. Figeys, and M. Tyers. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415**:**180-183. [DOI] [PubMed] [Google Scholar]
- 24.Hutt, D. M., L. F. Da-Silva, L. H. Chang, D. C. Prosser, and J. K. Ngsee. 2000. PRA1 inhibits the extraction of membrane-bound rab GTPase by GDI1. J. Biol. Chem. 275**:**18511-18519. [DOI] [PubMed] [Google Scholar]
- 25.Ito, T., K. Tashiro, S. Muta, R. Ozawa, T. Chiba, M. Nishizawa, K. Yamamoto, S. Kuhara, and Y. Sakaki. 2000. Toward a protein-protein interaction map of the budding yeast: a comprehensive system to examine two-hybrid interactions in all possible combinations between the yeast proteins. Proc. Natl. Acad. Sci. USA 97**:**1143-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iwahashi, J., and N. Hamada. 2003. Human reticulon 1-A and 1-B interact with a medium chain of the AP-2 adaptor complex. Cell. Mol. Biol. (Noisy-le-grand) 49**:**OL467-OL471. [Online.] [PubMed] [Google Scholar]
- 27.Iwahashi, J., I. Kawasaki, Y. Kohara, K. Gengyo-Ando, S. Mitani, Y. Ohshima, N. Hamada, K. Hara, T. Kashiwagi, and T. Toyoda. 2002. Caenorhabditis elegans reticulon interacts with RME-1 during embryogenesis. Biochem. Biophys. Res. Commun. 293**:**698-704. [DOI] [PubMed] [Google Scholar]
- 28.Kaiser, C. A., and R. Schekman. 1990. Distinct sets of SEC genes govern transport vesicle formation and fusion early in the secretory pathway. Cell 61**:**723-733. [DOI] [PubMed] [Google Scholar]
- 29.Lazar, T., M. Gotte, and D. Gallwitz. 1997. Vesicular transport: how many Ypt/Rab-GTPases make a eukaryotic cell? Trends Biochem. Sci. 22**:**468-472. [DOI] [PubMed] [Google Scholar]
- 30.Lupas, A., M. Van Dyke, and J. Stock. 1991. Predicting coiled coils from protein sequences. Science 252**:**1162-1164. [DOI] [PubMed] [Google Scholar]
- 31.Martincic, I., M. E. Peralta, and J. K. Ngsee. 1997. Isolation and characterization of a dual prenylated Rab and VAMP2 receptor. J. Biol. Chem. 272**:**26991-26998. [DOI] [PubMed] [Google Scholar]
- 32.Matern, H., X. Yang, E. Andrulis, R. Sternglanz, H. H. Trepte, and D. Gallwitz. 2000. A novel Golgi membrane protein is part of a GTPase-binding protein complex involved in vesicle targeting. EMBO J. 19**:**4485-4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oertle, T., M. Klinger, C. A. Stuermer, and M. E. Schwab. 2003. A reticular rhapsody: phylogenic evolution and nomenclature of the RTN/Nogo gene family. FASEB J. 17**:**1238-1247. [DOI] [PubMed] [Google Scholar]
- 34.Oertle, T., and M. E. Schwab. 2003. Nogo and its paRTNers. Trends Cell Biol. 13**:**187-194. [DOI] [PubMed] [Google Scholar]
- 35.Pereira-Leal, J. B., and M. C. Seabra. 2001. Evolution of the Rab family of small GTP-binding proteins. J. Mol. Biol. 313**:**889-901. [DOI] [PubMed] [Google Scholar]
- 36.Petracek, M. E., and M. S. Longtine. 2002. PCR-based engineering of yeast genome. Methods Enzymol. 350**:**445-469. [DOI] [PubMed] [Google Scholar]
- 37.Pfeffer, S., and D. Aivazian. 2004. Targeting Rab GTPases to distinct membrane compartments. Nat. Rev. Mol. Cell Biol. 5**:**886-896. [DOI] [PubMed] [Google Scholar]
- 38.Pfeffer, S. R. 2001. Rab GTPases: specifying and deciphering organelle identity and function. Trends Cell Biol. 11**:**487-491. [DOI] [PubMed] [Google Scholar]
- 39.Schalk, I., K. Zeng, S.-K. Wu, E. A. Stura, J. Matteson, M. Huang, A. Tandon, I. A. Wilson, and W. E. Balch. 1996. Structure and mutational analysis of Rab GDP dissociation inhibitor. Nature 381**:**42-48. [DOI] [PubMed] [Google Scholar]
- 40.Schwab, M. E. 2004. Nogo and axon regeneration. Curr. Opin. Neurobiol. 14**:**118-124. [DOI] [PubMed] [Google Scholar]
- 41.Segev, N., J. Mulholland, and D. Botstein. 1988. The yeast GTP-binding YPT1 protein and a mammalian counterpart are associated with the secretion machinery. Cell 52**:**915-924. [DOI] [PubMed] [Google Scholar]
- 42.Seron, K., V. Tieaho, C. Prescianotto-Baschong, T. Aust, M. O. Blondel, P. Guillaud, G. Devilliers, O. W. Rossanese, B. S. Glick, H. Riezman, S. Keranen, and R. Haguenauer-Tsapis. 1998. A yeast t-SNARE involved in endocytosis. Mol. Biol. Cell 9**:**2873-2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Setty, S. R., M. E. Shin, A. Yoshino, M. S. Marks, and C. G. Burd. 2003. Golgi recruitment of GRIP domain proteins by Arf-like GTPase 1 is regulated by Arf-like GTPase 3. Curr. Biol. 13**:**401-404. [DOI] [PubMed] [Google Scholar]
- 44.Shapiro, A. D., and S. R. Pfeffer. 1995. Quantitative analysis of the interactions between prenyl rab9, GDP dissociation inhibitor-α, and guanine nucleotides. J. Biol. Chem. 270**:**11085-11090. [DOI] [PubMed] [Google Scholar]
- 45.Shin, M. E., K. D. Ogburn, O. A. Varban, P. M. Gilbert, and C. G. Burd. 2001. FYVE domain targets Pib1p ubiquitin ligase to endosome and vacuolar membranes. J. Biol. Chem. 276**:**41388-41393. [DOI] [PubMed] [Google Scholar]
- 46.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122**:**19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sivars, U., D. Aivazian, and S. R. Pfeffer. 2003. Yip3 catalyses the dissociation of endosomal Rab-GDI complexes. Nature 425**:**856-859. [DOI] [PubMed] [Google Scholar]
- 48.Soldati, T., A. D. Shapiro, A. B. D. Svejstrup, and S. R. Pfeffer. 1994. Membrane targeting of the small GTPase rab9 is accompanied by nucleotide exchange. Nature 369**:**76-78. [DOI] [PubMed] [Google Scholar]
- 49.Stenmark, H., A. Valencia, O. Martinez, O. Ullrich, B. Goud, and M. Zerial. 1994. Distinct structural elements of rab5 define its functional specificity. EMBO J. 13**:**575-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tagami, S., Y. Eguchi, M. Kinoshita, M. Takeda, and Y. Tsujimoto. 2000. A novel protein, RTN-XS, interacts with both Bcl-XL and Bcl-2 on endoplasmic reticulum and reduces their anti-apoptotic activity. Oncogene 19**:**5736-5746. [DOI] [PubMed] [Google Scholar]
- 51.Uetz, P., L. Giot, G. Cagney, T. A. Mansfield, R. S. Judson, J. R. Knight, D. Lockshon, V. Narayan, M. Srinivasan, P. Pochart, A. Qureshi-Emili, Y. Li, B. Godwin, D. Conover, T. Kalbfleisch, G. Vijayadamodar, M. Yang, M. Johnston, S. Fields, and J. M. Rothberg. 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403**:**623-627. [DOI] [PubMed] [Google Scholar]
- 52.Ullrich, O., H. Horiuchi, C. Bucci, and M. Zerial. 1994. Membrane association of Rab5 mediated by GDP-dissociation inhibitor and accompanied by GDP/GTP exchange. Nature 368**:**157-160. [DOI] [PubMed] [Google Scholar]
- 53.Walworth, N. C., B. Goud, A. K. Kabcenell, and P. J. Novick. 1989. Mutational analysis of SEC4 suggests a cyclical mechanism for the regulation of vesicular traffic. EMBO J. 8**:**1685-1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wilson, A. L., R. A. Erdman, and W. A. Maltese. 1996. Association of rab1B with GDP-dissociation inhibitor (GDI) is required for recycling but not initial membrane targeting of the rab protein. J. Biol. Chem. 271**:**10932-10940. [DOI] [PubMed] [Google Scholar]
- 55.Yang, X., H. T. Matern, and D. Gallwitz. 1998. Specific binding to a novel and essential Golgi membrane protein (Yip1p) functionally links the transport GTPases Ypt1p and Ypt31p. EMBO J. 17**:**4954-4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zerial, M., and H. McBride. 2001. Rab proteins as membrane organizers. Nat. Rev. Mol. Cell Biol. 2**:**107-117. [DOI] [PubMed] [Google Scholar]