A 3.9-Centimorgan-Resolution Human Single-Nucleotide Polymorphism Linkage Map and Screening Set (original) (raw)
Abstract
Recent advances in technologies for high-throughout single-nucleotide polymorphism (SNP)–based genotyping have improved efficiency and cost so that it is now becoming reasonable to consider the use of SNPs for genomewide linkage analysis. However, a suitable screening set of SNPs and a corresponding linkage map have yet to be described. The SNP maps described here fill this void and provide a resource for fast genome scanning for disease genes. We have evaluated 6,297 SNPs in a diversity panel composed of European Americans, African Americans, and Asians. The markers were assessed for assay robustness, suitable allele frequencies, and informativeness of multi-SNP clusters. Individuals from 56 Centre d'Etude du Polymorphisme Humain pedigrees, with >770 potentially informative meioses altogether, were genotyped with a subset of 2,988 SNPs, for map construction. Extensive genotyping-error analysis was performed, and the resulting SNP linkage map has an average map resolution of 3.9 cM, with map positions containing either a single SNP or several tightly linked SNPs. The order of markers on this map compares favorably with several other linkage and physical maps. We compared map distances between the SNP linkage map and the interpolated SNP linkage map constructed by the deCode Genetics group. We also evaluated cM/Mb distance ratios in females and males, along each chromosome, showing broadly defined regions of increased and decreased rates of recombination. Evaluations indicate that this SNP screening set is more informative than the Marshfield Clinic’s commonly used microsatellite-based screening set.
Introduction
The approaches and technologies used for linkage analysis of human diseases have undergone several major advances since RFLPs were first used as genetic markers. Currently, the primary resources for linkage analysis are robust and highly informative microsatellite markers (Weber and May 1989; Weissenbach et al. 1992; Sheffield et al. 1995). However, the ability to scale up the typing of microsatellite markers to very high throughput is limited, since electrophoretic separation must be done to accurately determine fragment sizes. SNPs are an abundant resource in the human genome and in other genomes. What they lack in informativeness (maximum heterozygosity is 0.5), they make up for in abundance, uniformity, global genomic distribution, and adaptability to massively parallel genotyping technology. Therefore, the use of SNPs as genetic markers offers the promise of lower-cost very-high-throughput genotyping.
The high density of SNPs in the human genome, together with the relatively narrow width of regions in linkage disequilibrium (LD), have led to the proposal that SNPs be employed for genomewide association screening (Lander 1996; Risch and Merikangas 1996; Collins et al. 1997; Kruglyak 1997). However, considerably less attention has been paid to the potential utility of SNPs for linkage-based genome scans, an approach that can be put into practice immediately. Several studies have established the theoretical rationale that predicts that a high-density genomewide SNP-marker set could be superior to the current microsatellite sets for genome scanning by linkage (Kruglyak 1997; Wilson and Sorant 2000; Weber and Broman 2001; Goddard and Wijsman 2002). Kruglyak (1997) showed that 700–900 SNPs spaced equally throughout the genome would have the same statistical power as the standard 350–400 mapped microsatellite markers and that 3,000 SNPs at 1-cM spacing throughout the genome would be far superior in information content (IC). Other theoretical studies and computer simulations have demonstrated that two- and three-locus composite diallelic markers can provide linkage information, similar to highly informative multiallelic markers (Wilson and Sorant 2000; Goddard and Wijsman 2002).
Numerous academic groups and biotechnology companies have pushed forward with the development of new technologies and methods to greatly accelerate SNP genotyping (Dearlove 2002; Tsuchihashi and Dracopoli 2002), and work is under way on new analytical methods that can take advantage of genomewide SNP-genotype data (Gudbjartsson et al. 2000; Markianos et al. 2001; Abecasis et al. 2002). The success that the SNP Consortium (TSC) and other groups have had in the discovery of millions of SNPs throughout the human genome and the precise physical mapping of these SNPs onto the genome have provided the starting material for a high-density genomewide SNP-marker set (Wang et al. 1998; Sachidanandam et al. 2001; Venter et al. 2001). However, a map of such diallelic markers has, to our knowledge, not yet been constructed. By use of existing STR markers as anchors, we have evaluated the polymorphism and robustness of >20,000 candidate SNPs for construction of a SNP linkage marker set. A subset of ∼3,000 SNPs were selected for map construction and were genotyped in 56 families from the CEPH (Dausset et al. 1990) pedigree panel. The final SNP linkage maps contain 716 SNP clusters (i.e., groups of physically close markers) and 332 singleton SNPs.
Methods
We sought to build a SNP map composed of clusters of two to four physically close SNPs located every ∼5 cM throughout the genome, interspersed with singleton SNPs between the clusters. STRs from the Marshfield linkage map were used to help identify the SNPs to be used for map construction. These STR anchors were not themselves included in the map construction. A large set of candidate SNPs was initially evaluated for polymorphism and genotyping success in a diversity panel (phase I). A subset of SNPs best suited for mapping were then selected and genotyped in a panel of CEPH pedigrees for map construction (phase II). Celera Genomics and Motorola Life Sciences (now Amersham Biosciences) provided genotyping services for the present project.
Phase I SNP Selection
All STR markers present both in the UniSTS database and on the Marshfield linkage map (see the Center for Medical Genetics Web site) (Broman et al. 1998) were identified and localized on the August 2001 Golden Path genome assembly (see the UCSC Genome Bioinformatics Web site) (Lander et al. 2001). A set of 1,345 anchor STRs were selected at ∼2.5-cM intervals on the Marshfield map. In the TSC database (Single Nucleotide Polymorphisms for Biomedical Research), all SNPs located within 100 kb of an anchor STR were identified. SNPs located in regions with high repeat content (defined as regions with >30% repeat content in the 500-bp flanking regions) were excluded from further use, owing to difficulties in the design of unique PCR primers. For evaluation of each SNP cluster, 10–20 SNPs were selected; for evaluation of each singleton map position, 2 SNPs were selected. In this manner, >20,000 SNPs were initially identified and assigned to either Celera Genomics or Motorola Life Sciences for evaluation and genotyping in the TSC allele-frequency panel. These SNPs were chosen to yield as complete and even coverage of all 100-kb clusters as possible. For a few STR sites, there were not enough TSC SNPs with successful assay designs to meet these goals. For these STR sites, Celera performed a search of its database and supplemented the TSC collection with 104 candidate SNPs from dbSNP and 39 SNPs discovered by the Celera human genome sequencing program (Venter et al. 2001). Each SNP evaluated by Motorola was confirmed by sequencing for accuracy in three diverse DNA samples, as well as by analyzing the cluster patterns for all samples tested in scatter plots, and only accurately performing SNPs progressed through the project.
SNP Evaluation in the Allele-Frequency Panel
Ninety individuals from the TSC allele-frequency panel were genotyped using 6,297 SNPs. The panel is composed of 30 European Americans, 30 African Americans, and 30 Asians (10 Chinese and 20 Japanese). (For details on the TSC panel, see the Allele Frequency Panels Web site.) Celera Genomics genotyped 5,446 SNPs, and Motorola Life Sciences genotyped 851 SNPs. The allele frequency of each SNP was determined in each of the three populations. Only SNPs that were polymorphic in two of the three populations were candidates for phase II. An expectation-maximization algorithm, as implemented in the program Hapfreqs (Goddard et al. 1996), was used to obtain maximum-likelihood estimates of haplotype frequencies, which were used to determine the composite (haplotype) heterozygosity of each SNP cluster for each population. Using the program LD (software by M. Eberle and L. Kruglyak; for download, see the Kruglyak Lab Web site), we estimated the _r_2 measure of LD for all pairs of markers within each cluster (Carlson et al. 2003). Initially, we anticipated that cluster LD would be used in the SNP-selection procedure, but, since nearly all sets of SNPs that were evaluated for each cluster demonstrated quite low LD, it was not ultimately part of the SNP-selection criteria. For each singleton STR anchor, the SNP with the highest heterozygosity (averaged across all populations) was selected for submission to phase II of the project. For each cluster, the three SNPs that provided both the highest total composite heterozygosity (averaged across all populations) and the smallest population-specific deviations from the highest total composite heterozygosity were selected for submission to phase II. In some cases, the number of SNPs per cluster is either fewer or more than three.
Celera Genotyping
SNPs assigned to Celera Genomics were mapped to the Celera reference human genome sequence, and 300 bp of flanking sequence on both sides of each SNP was imported into the _Taq_Man assay-design program. A pipelined version of Primer Express (Applied Biosystems), modified for this purpose, was used to design both the PCR primers and the _Taq_Man minor groove binder (MGB) probes. One allelic probe was labeled with the fluorescent FAM dye, and the other was labeled with the fluorescent VIC dye. PCRs were set up with pipetting robots (BioCube [Protedyne] and Biomek 2000 [Beckman-Coulter]). The PCRs were run in _Taq_Man Universal Master Mix without uracil-DNA-glycosylase (Applied Biosystems), with PCR primer concentrations of 900 nM and _Taq_Man MGB-probe concentrations of 200 nM. Reactions were performed in a 384-well format in a total reaction volume of 5 μl with 1.0 ng of genomic DNA. The plates were then placed in a thermal cycler (PE 9700; Applied Biosystems) and were heated at 95° C for 10 min, followed by 50 cycles of 95°C for 15 s and 60°C for 1 min, with a final soak at 25°C. The _Taq_Man assay plates were transferred from the thermal cyclers to the Prism 7900HT instruments (Applied Biosystems), in which the fluorescence intensity in each well of the plate was read. Fluorescence data files from each plate were analyzed by automated allele-calling software (Heil et al. 2002) and were reviewed by a skilled operator, and genotyping results were exported directly into an Oracle database from which genotype results were delivered electronically to TSC. Individual genotypes that were ambiguous (i.e., did not fall clearly into a genotype cluster) were designated as missing data, and data for SNPs that did not clearly form separate genotype clusters were excluded from further analysis. In addition, SNPs for which all genotypes appeared as heterozygotes were excluded, and SNPs with strong deviation from Hardy-Weinberg proportions were flagged. Genomic DNA samples for phase I were purchased from the TSC allele-frequency DNA panels.
Motorola CodeLink Genotyping
PCR primer pairs were designed to amplify SNP target sequences in a 32-plex format, using proprietary in silico processes. Automated PCR-primer design included source sequence filtering against vector and repetitive sequences. Amplicon length was set for 90–122 bp. Primer mixtures were made using Qiagen BioRobot 9604 and Biomek FX robots. All primer pairs were tested in uniplex PCRs, and poor performers were rejected prior to inclusion in 32-plex formatting. Automated PCR setup was performed with a Tecan Genesis robot. Plates containing primer sets for each of the 32-plex PCRs in individual wells were manufactured to ensure uniformity and repeatability of target generation. Assay PCR included a nucleotide mixture containing a 1:8 ratio of dUTP to dTTP and was performed on Tetrad thermal cyclers (MJ Research). PCR amplicons (targets) were pooled and purified using QIAquick 96 PCR Purification Kit and BioRobot 3000 (Qiagen). Target fragmentation was performed with uracil-DNA-glycosylase (New England Biolabs). Each array type was supplied with an amplicon target from a complementary set of eight pooled 32-plex PCRs.
Four different CodeLink bioarrays were specifically designed and manufactured for the SNP Consortium Linkage Map Project. Each of the four bioarray designs contained 256 different pairs of SNP probes, for a total of 1,024 SNPs. Each CodeLink SNP bioarray had four identical and separately assayable arrays on a single slide. A reaction chamber (FlexChamber; Motorola Life Sciences) maintained each array as an independent set of SNP-genotype assays, allowing for minimal assay volumes and permitting the testing of a different samples in each chamber. The CodeLink SNP assay utilizes a DNA polymerase–mediated, allele-specific extension of anchored oligonucleotide probes attached to a three-dimensional activated matrix. At their 3′ ends, each pair of allele-specific oligonucleotide probes are complementary to the polymorphic position of a SNP. Pooled and fragmented multiplex target DNA was added to the CodeLink SNP-genotyping reagents, including a biotin-labeled acyclo terminator nucleotide (PerkinElmer) mixture and Thermo Sequenase DNA polymerase (Amersham). The complete target-reaction mixture was loaded on the bioarrays, and chambers were sealed. Loaded bioarrays were then placed on a Hybaid Omnislide, and the CodeLink SNP thermal-cycling program was engaged. Bioarrays were then washed and labeled with streptavidin–Alexa Fluor 532 conjugate (Molecular Probes). Stained bioarrays were scanned on an Axon Scanner A. Genotype data analysis was performed on the scanned chip images by using a proprietary CodeLink SNP-calling algorithm. The phase I samples were genotyped in duplicate, and consensus genotypes were generated.
DNA Samples
Genomic DNA samples for phase I (the diversity sample) were obtained from the allele-frequency DNA panels (Coriell Cell Repository), and genomic DNA for phase II (the CEPH pedigrees) was obtained from the Fondation Jean Dausset–CEPH and Coriell.
Genotype Cleaning
We used a modified version of the Ilink program (Lathrop et al. 1984; Cottingham et al. 1993; Schäffer et al. 1994; Terwilliger and Ott 1994; Gordon et al. 2001) to obtain an average estimate of the rate of genotyping error on the data that we received from Celera Genomics and Motorola Life Sciences. This program uses the Elston-Stewart algorithm to maximize the likelihood as a function of the SNP allele frequencies and three error parameters—_v_1, _v_2, and _v_3 (the probability that a true homozygote is incorrectly coded as a heterozygote, the probability that a true homozygote is incorrectly coded as another homozygote, and the probability that a true heterozygote is incorrectly coded as a homozygote, respectively). This approach uses the same error model as that developed by Sobel et al. (2002), which assumes that error rates are independent of the particular alleles at each locus. For diallelic loci, this is the most general model possible under this assumption. The PedCheck program (O’Connell and Weeks 1998) was used to identify non-Mendelian inheritance and to search for pedigrees with an unusual number of Mendelian inconsistencies, which can be a clue to errors in pedigree structure or sample switching.
The error-detection approach in Merlin (Abecasis et al. 2002) was used to search for Mendelian-consistent genotype errors within each of the SNP clusters. Merlin applies a maximum-likelihood approach for identification of specific genotypes that cause larger-than-expected changes in the multipoint likelihood of the data (Abecasis et al. 2002). Such genotypes are likely to be erroneous. Our SNP clusters span ⩽100 kb, and recombination events within a region of 100 kb are relatively rare. Therefore, recombination events could not be used to determine the order of markers within clusters; instead, we relied on observed physical order, as identified in assembled DNA sequence. Because this step was performed early in our project, within-cluster marker order was determined from the National Center for Biotechnology Information (NCBI) build 28 (February 2002) assembly.
Once our maps were completed, we used the SimWalk2 (Sobel and Lange 1996; Sobel et al. 2002) program to search for Mendelian-consistent erroneous genotypes across all autosomal mapped SNPs. For the relatively large CEPH pedigrees, SimWalk2 applies a Monte Carlo Markov-chain approach with general error models, to compute posterior mistyping probabilities for each genotype. It identifies both genotypes that imply improbable recombination patterns and genotypes likely to be erroneous on the basis of the observed allele frequencies. Because the SNP clusters were previously cleaned and because SimWalk2 analysis is extremely time-consuming, we used only one SNP (the most informative) to represent each SNP cluster for the present analysis. Although SimWalk2 is slow, this method could be used to analyze all 56 of the CEPH pedigrees that we studied, whereas other programs, such as Mendel and Merlin, could not analyze all pedigrees. Because SimWalk2 cannot analyze X-linked markers, we used Merlin to search for erroneous genotypes for the X chromosome. Because of reduced memory needs for the analysis of the X chromosome, Merlin was able to evaluate all 56 pedigrees.
Linkage-Map Construction
For construction of the SNP linkage map, a common set of 42 CEPH pedigrees were genotyped by both Celera Genomics and Motorola Life Sciences. In addition to these 42, Celera genotyped 6 pedigrees, and Motorola genotyped 8 pedigrees, for a total of 56 CEPH pedigrees utilized for genotyping. All of the pedigrees were selected from the complete CEPH panel, to maximize sibship size and presence of four grandparents. Celera genotyped 661 individuals, and Motorola genotyped 609–639 individuals. These panels include 770–842 potentially informative meioses.
The SNPs assigned to each chromosome were tested for linkage to other SNPs on the same chromosome. A LOD threshold of 3.0 was used to determine linkage groups. The MultiMap (Matise et al. 1994) and Cri-Map (Lander and Green 1987) programs were used for construction of the linkage maps, and the Kosambi map function was applied. Markers within clusters were assigned to haplotype systems as defined and implemented in Cri-Map. SNP clusters were placed on the map before singleton SNPs, to maximize unique placement of clusters. A LOD threshold of 3.0 (2.0 for the X chromosome) was used for initial map construction; this was followed by a final round of mapping, to include a few additional markers, using a LOD threshold of 2.0. The statistical support for the order of the resulting maps was evaluated by the Flips function (as defined in Cri-Map). The maps were further confirmed by removal and remapping of each marker on every map. In the present analysis, any markers that do not remap to their postulated position are removed from the map.
The rates of recombination per unit of physical distance (recombination intensities) were initially determined as the map size of each interval divided by the length (in bp) of each map interval. Recombination intensities are expressed as cM/Mb. Owing both to errors present in the data and to the well-known tendency of derivative estimators to amplify noise, plots of the raw breakage intensity showed a highly fluctuating pattern (data not shown). To more precisely determine the rate of change of genetic distance relative to sequence distance, we used the software Locfit (Loader 1999) to fit a local quadratic regression to the map distance as a function of the sequence distance. The local quadratic functions were based on Gaussian-weighted sliding windows containing a constant proportion of markers. The linear component, or the first derivative, was extracted from the local quadratic fit at each marker position. This local slope can be thought of as a good approximation to the first derivative of the underlying function that relates map distance to sequence distance. We used generalized cross-validation to select the best values of the tuning parameter (Craven and Wahba 1979).
Comparison with Other Linkage and Physical Maps
The physical position of each SNP was determined by searching, with BLAST, for sequence homology to the NCBI build 28 (February 2002), build 29 (May 2002), and build 30 (August 2002) genome assemblies and the Celera component 4 (C4) (May 2001) human genome assembly (Venter et al. 2001) for each chromosome. The order of SNPs on our linkage maps was compared with that of the Marshfield and deCode linkage maps (Broman et al. 1998; Kong et al. 2002). Since no STR markers were actually included in our linkage maps, for these comparisons, we used the closest microsatellite markers from the Marshfield map that were identified near each mapped SNP as proxies. The interpolated physical positions of SNPs on the deCode map were obtained from the deCode map supplemental Web data (A High-Resolution Recombination Map of the Human Genome). The order of markers on our map was compared with other maps by using the method of identification of the longest common subsequence (LCS) (Agarwala et al. 2000), a method commonly used in computer science. This approach identifies the largest subset of markers that have the same relative order on both maps. The level of concordance between two maps is determined as the length of the LCS divided by the number of markers compared (i.e., the number of markers in common between two maps). The LCS provides a measure that is easily compared across multiple map comparisons.
Evaluation of IC
Multilocus polymorphic information content (MPIC) (Goddard and Wijsman 2002) was computed for each cluster of SNPs under the assumption of zero phase information (MPICmin) and under the assumption of complete phase information (MPICmax). IC was measured using the Genehunter program, version 2.1 (Markianos et al. 2001), for several maps and data sets: the Marshfield version 10 STR screening set, using genotypes from eight CEPH pedigrees (1331, 1332, 1347, 1362, 1413, 1416, 884, and 102); complete SNP maps from the present study, using genotypes from the same set of eight CEPH pedigrees; and a modified SNP map of chromosome 12 (from the present study) with a subset of only 47 markers, also using the same set of eight CEPH pedigrees. Genotypes for the Marshfield STR screening set map were obtained from the Center for Medical Genetics Web site. Genotypes were simulated for STR markers for the nuclear-pedigree and sib-pair analyses of chromosome 22, using the Simulate program (Terwilliger et al. 1993).
Results
The project was divided into two phases: Phase I consisted of the initial genotyping and evaluation of a large sample of SNPs in the TSC allele-frequency panel, to identify a subset of informative and robust SNPs for the construction of linkage maps in phase II; the phase I data alone are of interest, because they provide information to analyze distributions of LD in 538 clusters of SNPs spanning the genome (Clark et al. 2003 [in this issue]). Phase II consisted of the genotyping of this subset of SNPs in a large sample of CEPH pedigrees and the subsequent construction of the SNP linkage map.
Phase I SNP Selection
TSC selected 19,602 SNPs to be evaluated by Celera Genomics for phase I of the present project. These were distributed among 1,345 SNP clusters (groups of SNPs spanning <100 kb) spaced at 2.5-cM intervals throughout the genome. Of these, 15,469 SNPs could be mapped to the Celera genome sequence, and 11,447 passed the _Taq_Man assay-design pipeline. A total of 6,000 of these SNPs (2–8 per cluster) were selected for evaluation in 90 individuals from the TSC allele-frequency panel. Of these, 554 failed _Taq_Man assay synthesis, and 453 were either homozygous or heterozygous in all samples, resulting in 4,993 successful _Taq_Man SNP assays. Celera generated a total of 490,140 genotypes for phase I.
Motorola Life Sciences CodeLink evaluated 2,917 SNPs from 335 SNP clusters with an average cluster spacing of ∼10 cM, for phase I of the present project. The incoming SNP sequences were filtered for highly repetitive regions and motifs, resulting in a culling of the original allotment to 2,222 SNPs (24% reduction). Of these, 1,517 passed the CodeLink primer- and probe-design processes, and SNP content was selected for evaluation from 331 of the 335 SNP clusters. One thousand twenty-four SNPs (1–4 per cluster) were selected for evaluation in the 90 individuals from the TSC allele-frequency panel. Of these, 61 SNPs could not be amplified by PCR, and 166 failed owing to poor performance on the bioarray. One hundred six SNPs were monomorphic in all three populations tested, 85 were monomorphic in two populations, and 606 SNPs were polymorphic in at least two of the three populations tested. A total of 76,590 genotypes for phase I of the project were generated using CodeLink.
SNP Evaluation in the Allele-Frequency Panel
The average observed minor-allele frequencies were 0.253, 0.236, and 0.247 in the European American, African American, and Asian population samples, respectively. The average composite heterozygosity in the SNP clusters among the SNPs chosen for phase II (see the “Methods” section) was 0.64 in European Americans, 0.66 in African Americans, and 0.61 in Asians. The average pairwise LD within clusters among the SNPs chosen for mapping in phase II was _r_2=0.1. Details on the distribution of allele frequencies and LD in these populations can be found in the companion article (Clark et al. 2003 [in this issue]).
Premapping Genotype Cleaning
Considerable effort was made to remove erroneous and problematic genotypes. As described in the “Methods” section, the Ilink, PedCheck, Merlin, and SimWalk2 computer programs were used to assess and identify genotypes that are likely to be erroneous or changed from parental form by either mutation or gene conversion. The application of multiple methods allowed us to detect both Mendelian-inconsistent and Mendelian-consistent errors. Problematic genotypes (due to errors, mutations, and gene conversions) were excluded from analysis and from the Cri-Map–format data files that we have made publicly available.
Genotype errors and error rates were identified with three approaches. A modified version of the Ilink program was used to estimate the average probability of genotype error in the initial uncleaned data to be 0.003 ± 0.008 (averaged over 52 SNPs). Comprehensive evaluation of genotypes by the PedCheck program identified a minimum of 1,940 Mendelian-inconsistent genotypes, resulting in an estimated error rate of 0.001. Although non-Mendelian inheritance can clearly be identified within a pedigree, it is often not possible to determine which specific individual or individuals in the pedigree have an erroneous genotype. Therefore, for each marker, the genotypes were deleted for all members of any pedigree that were determined to have non-Mendelian inheritance (i.e., the data is “overcleaned”). This approach resulted in the deletion of 1.6% of the genotypes. It is impossible to estimate the actual error rate from these data. Evaluation of the PedCheck results did not identify any errors in pedigree structure. Merlin was used next, to search for Mendelian-consistent erroneous genotypes within our SNP clusters. This step identified and deleted 4,581 likely erroneous genotypes.
Postmapping Genotype Cleaning
Once our maps were completed, we used the SimWalk2 program to search for Mendelian-consistent erroneous genotypes across all autosomal mapped SNPs, and we used the Merlin program to search for Mendelian-consistent erroneous genotypes across the X-linked mapped SNPs. A total of 2,579 likely erroneous genotypes were identified and removed by this step (error rate 0.004). This cleaning step reduced the total length of our maps by 6.5%. Both Broman et al. (1998) and Kong et al. (2002) applied similar postmapping cleaning steps for the Marshfield and deCode linkage maps, which resulted in map reductions of 25% and 11%, respectively. The final sex-averaged length of the SNP linkage map is 3,707 cM, with 4,415 cM observed in females and 2,642 cM observed in males. By design, the SNP linkage map spans the same physical length as do the Marshfield linkage maps. Our total map length is 1% longer than the Marshfield map, which is 3,672 cM in sex-averaged length. If the CIs surrounding these estimates of map length are considered, then the length of the SNP linkage map is consistent with the length of the Marshfield map.
SNP Linkage Map
Using the criteria described in the “Methods” section, we selected a subset of 2,988 SNPs from phase I for genotyping in the 56 CEPH pedigrees, with 1.48 million genotypes generated by Celera Genomics and 550,000 genotypes generated by Motorola Life Sciences. A total of 2,825 of these SNPs were informative in CEPH pedigrees. The average minor-allele frequency of these SNPs in the CEPH pedigrees was 0.28. Of these, a total of 2,771 SNP markers were confirmed, by linkage analysis, as belonging to 1 of the 23 chromosome-specific linkage groups (table 1) and were used for map construction.
Table 1.
Description of the SNP Linkage Maps
| Map Length(cM)c | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Chromosome | No. ofSNPs inLinkageGroup | No. ofMapPositionsa | No. ofSNPson Mapb | No. ofClusters | SexAveraged | Female | Male | MapResolution(cM)d | PhysicalLength(Mb) | cM/MbRatio |
| 1 | 242 | 93 | 204 | 69 | 277.7 | 353.8 | 191.4 | 3.0 | 239.74 | 1.16 |
| 2 | 210 | 81 | 176 | 58 | 259.9 | 337.9 | 175.2 | 3.2 | 237.39 | 1.09 |
| 3 | 185 | 77 | 156 | 50 | 220.7 | 203.2 | 160.9 | 2.9 | 197.40 | 1.12 |
| 4 | 159 | 57 | 124 | 42 | 216.0 | 275.1 | 159.7 | 3.9 | 190.04 | 1.14 |
| 5 | 162 | 64 | 130 | 43 | 211.3 | 255.0 | 164.3 | 3.4 | 179.99 | 1.17 |
| 6 | 155 | 57 | 126 | 43 | 193.1 | 235.5 | 142.5 | 3.4 | 169.69 | 1.14 |
| 7 | 138 | 56 | 118 | 38 | 182.6 | 226.1 | 140.0 | 3.3 | 153.76 | 1.19 |
| 8 | 141 | 57 | 113 | 31 | 160.1 | 210.8 | 109.8 | 2.9 | 138.82 | 1.15 |
| 9 | 130 | 47 | 96 | 32 | 158.4 | 190.8 | 130.9 | 3.4 | 131.04 | 1.21 |
| 10 | 136 | 54 | 116 | 37 | 176.3 | 211.1 | 131.9 | 3.3 | 133.36 | 1.32 |
| 11 | 107 | 43 | 87 | 31 | 152.6 | 187.3 | 116.5 | 3.6 | 130.66 | 1.17 |
| 12 | 127 | 47 | 97 | 29 | 170.6 | 210.5 | 126.3 | 3.7 | 131.48 | 1.30 |
| 13 | 85 | 36 | 69 | 20 | 121.5 | 149.3 | 97.2 | 3.5 | 94.08 | 1.29 |
| 14 | 100 | 33 | 82 | 28 | 115.3 | 126.1 | 86.3 | 3.6 | 81.48 | 1.42 |
| 15 | 84 | 29 | 59 | 17 | 120.5 | 143.0 | 93.5 | 4.3 | 75.95 | 1.59 |
| 16 | 99 | 34 | 78 | 25 | 137.6 | 168.8 | 105.8 | 4.2 | 90.08 | 1.53 |
| 17 | 90 | 28 | 65 | 19 | 126.8 | 157.2 | 94.8 | 4.7 | 79.99 | 1.59 |
| 18 | 99 | 37 | 81 | 27 | 124.8 | 149.4 | 91.0 | 3.5 | 75.56 | 1.65 |
| 19 | 67 | 25 | 43 | 14 | 120.5 | 128.1 | 114.6 | 5.0 | 57.89 | 2.08 |
| 20 | 81 | 31 | 74 | 24 | 106.4 | 128.8 | 85.0 | 3.5 | 58.43 | 1.82 |
| 21 | 51 | 18 | 44 | 11 | 66.6 | 79.9 | 51.2 | 3.9 | 31.86 | 2.09 |
| 22 | 55 | 22 | 50 | 18 | 78.9 | 78.2 | 72.8 | 3.8 | 32.01 | 2.46 |
| X | 68 | 22 | 35 | 10 | 208.6 | 208.6 | … | 9.9 | 144.61 | 1.44 |
| Overall | 2,771 | 1,048 | 2,223 | 716 | 3,706.8 | 4,414.5 | 2,641.6 | 3.9 | 2,855.31 | 1.4 |
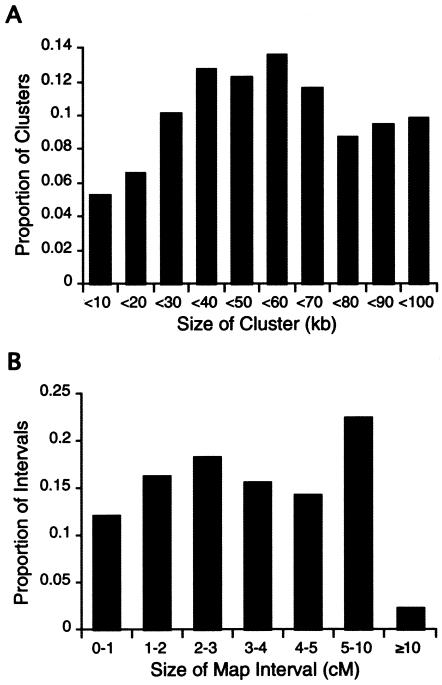
The SNP linkage maps contain a total of 2,223 SNP markers mapped, as singletons or clusters (i.e., groups of SNPs that lie within 100 kb on the NCBI build 31 genome assembly), to 1,048 unique map positions (table 1). Of these, 1,891 markers comprise 716 SNP clusters, and the remainder (332) are singleton SNPs. The average map resolution is 3.9 cM between map positions, with 76% of the intervals being <5 cM (fig. 1_A_). The statistical support for relative local marker order ranges in odds from 100:1 to >1040:1. An additional 558 SNPs could not be assigned unique map positions, owing to lack of significant linkage support and/or lack of consistent evidence confirming membership in a 100-kb SNP cluster. These markers are therefore localized to LOD 3 map intervals or bins, instead of to specific map positions. Of the SNP clusters, 54% contain three SNPs, 42% contain two SNPs, 3% contain four SNPs, and the remaining 1% contain five to eight SNPs. The average distance spanned by markers within a cluster is 53 kb, and the distribution of cluster sizes is shown in figure 1_B_. In >770 meioses, the majority of the clusters (66%) have zero recombination events between SNPs, 20% have one recombination event between SNPs, and the remaining 14% have two or more recombination events between SNPs.
Figure 1.
Map features. A, Distribution of sizes of autosomal map intervals. B, Distribution of sizes of SNP clusters.
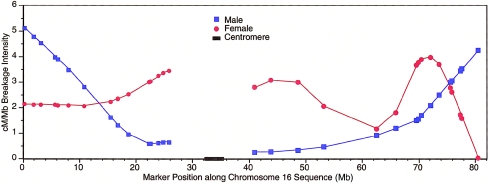
Although these SNP maps are not dense enough to study how recombination rates vary along each chromosome at a fine scale, we plotted the ratio of cM/Mb in females and males for each chromosome (see the “Methods” section), to observe broad trends. One example plot, for chromosome 16, is shown as figure 2; plots for the remaining chromosomes are available at the corresponding author’s Web site (Additional Data for the TSC SNP Linkage Map). We observed 1.4 cM/Mb when averaged over the entire genome, although this varied greatly both by chromosome and by specific chromosome region (table 1 and fig. 2). As has been reported elsewhere, the ratio of cM/Mb increases with decreasing physical chromosome size (Lander et al. 2001; Kong et al. 2002). In addition, although every autosome showed increased recombination in females as compared with males, the average female-to-male length ratio, 1.7:1 cM, also varied considerably by chromosome and by specific chromosome region.
Figure 2.
Female and male smoothed cM/Mb ratios, plotted along chromosome 16. Data are plotted separately for the p and q arms.
Comparison with Other Linkage and Physical Maps
We compared the order of SNPs on our linkage maps with their observed order on the NCBI build 28 (February 2002), build 29 (May 2002), and build 30 (August 2002) genome assemblies and the Celera C4 (May 2001) human genome assembly (Venter et al. 2001) (table 2). The average concordances with each of these maps, as determined by the proportion of markers in the LCS (see the “Methods” section), were 93%, 92%, 99%, and 96%, respectively. The concordance between the SNP linkage maps and the build 30 genome sequence assembly ranges from 100%, on 14 chromosomes, to a low of 94%, on chromosome 16.
Table 2.
Percentage of Markers Whose Map Order Is Concordant between the SNP Linkage Map and Other Maps
| Concordant Markers(%) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Chromosome | No. of mappositions | NCBI Build 28Assembly | NCBI Build 29Assembly | NCBI Build 30Assembly | Celera C4Assembly | MarshfieldLinkage Map | deCodeLinkage Map | deCodeSNP Mapa |
| 1 | 93 | 86 | 87 | 99 | 94 | 97 | 98 | 98 |
| 2 | 81 | 95 | 85 | 97 | 97 | 97 | 98 | 98 |
| 3 | 77 | 97 | 91 | 99 | 97 | 98 | 100 | 99 |
| 4 | 57 | 96 | 91 | 96 | 96 | 98 | 98 | 100 |
| 5 | 64 | 76 | 84 | 100 | 98 | 98 | 96 | 100 |
| 6 | 57 | 89 | 96 | 96 | 94 | 100 | 100 | 99 |
| 7 | 56 | 92 | 100 | 100 | 98 | 96 | 100 | 100 |
| 8 | 57 | 93 | 94 | 100 | 96 | 98 | 97 | 100 |
| 9 | 47 | 93 | 98 | 100 | 97 | 93 | 100 | 100 |
| 10 | 54 | 96 | 87 | 100 | 93 | 94 | 100 | 100 |
| 11 | 43 | 85 | 93 | 98 | 95 | 100 | 97 | 97 |
| 12 | 47 | 93 | 90 | 100 | 93 | 100 | 100 | 100 |
| 13 | 36 | 97 | 100 | 100 | 94 | 97 | 100 | 100 |
| 14 | 33 | 97 | 97 | 97 | 97 | 96 | 100 | 97 |
| 15 | 29 | 96 | 88 | 100 | 100 | 100 | 100 | 100 |
| 16 | 34 | 85 | 62 | 94 | 97 | 91 | 96 | 98 |
| 17 | 28 | 85 | 83 | 100 | 100 | 100 | 100 | 100 |
| 18 | 37 | 94 | 96 | 97 | 100 | 100 | 96 | 97 |
| 19 | 25 | 91 | 92 | 100 | 95 | 91 | 100 | 97 |
| 20 | 31 | 100 | 100 | 100 | 93 | 96 | 96 | 100 |
| 21 | 18 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| 22 | 22 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| X | 22 | 100 | 100 | 100 | 89 | 95 | 100 | 100 |
| Overall | 1,048 | 93 | 92 | 99 | 96 | 97 | 99 | 99 |
We also compared the order of STR markers near SNPs on our SNP maps (see the “Methods” section) with the Marshfield and deCode STR linkage maps (Broman et al. 1998; Kong et al. 2002). The SNP linkage map has excellent concordance with both: on average, it is 97% concordant with the Marshfield maps and 99% concordant with the deCode maps. We identified a number of STR markers each of whose chromosomal assignment on one or both of the NCBI build 30 and Celera C4 assemblies is different from its chromosomal assignment as determined by linkage analysis (data available at the corresponding author’s Web site [Additional Data for the TSC SNP Linkage Map]).
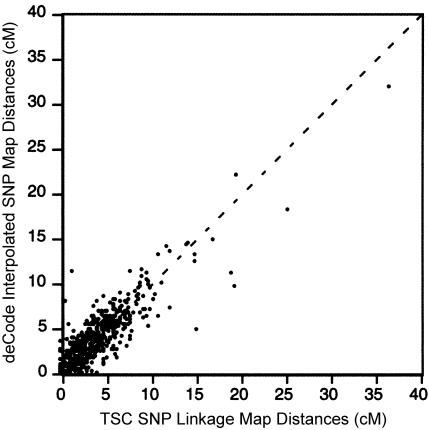
Finally, we compared the order and map distances of our SNP maps with the deCode interpolated SNP linkage maps. Approximately 2 million SNPs were placed onto the deCode linkage maps by using linear interpolation between their physical assembly and their genetic map (Kong et al. 2002). This approach arrived at estimated linkage-map positions for markers that were neither genotyped nor evaluated for recombination. On average, the order of markers is 99% concordant between our SNP map and the deCode interpolated SNP map (table 2). We compared map distances between markers in common over the 14 completely concordant chromosomes. The map distances are well correlated, with r equal to 0.92 (fig. 3). Of the 963 map intervals included in the present comparison, 43% differ by <0.1 cM, 32% differ by 0.1–1 cM, and 25% differ by >1 cM.
Figure 3.
Map distance for corresponding map intervals on the TSC SNP linkage map and the deCode interpolated SNP linkage map.
Evaluation of IC
The utility of our SNP map for linkage mapping was evaluated by two measures, MPIC (Goddard and Wijsman 2002) and IC (Kruglyak 1997). MPIC, an extension of the polymorphism information content (PIC) measure (Botstein et al. 1980), determines the probability of identifying which haplotype in a cluster of tightly linked loci is transmitted from a parent to an offspring. MPIC is partially a function of the amount of phase information obtainable within a given data set (see the “Methods” section). Because the amount of phase information is study specific, we calculated MPIC for each SNP cluster under both an assumption of availability of complete phase information (MPICmax) and an assumption of zero phase information (MPICmin). The median MPICmax was 0.65 (range 0.11–0.99), and the median MPICmin was 0.50 (range 0.11–0.59). For comparison, the median PIC from the Marshfield version 9 screening set is 0.68 (Goddard and Wijsman 2002).
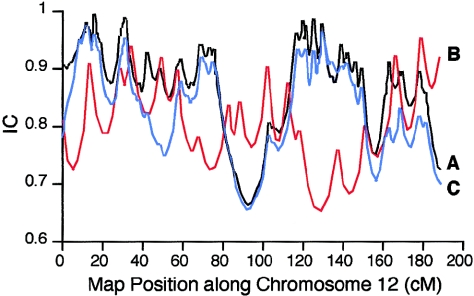
IC, as defined by Kruglyak (1997), is a measure of the fraction of inheritance information extracted by a linkage map, as compared with an infinitely dense map. We computed the IC of our SNP maps and of the Marshfield version 10 screening set. The Marshfield screening map was constructed using a subset of only eight of the CEPH pedigrees. For comparison, genotypes from only the same eight-pedigree subset were used when evaluating the IC of the SNP linkage map. The average, minimum, and maximum IC values for all chromosomes for both our SNP linkage maps and the Marshfield screening set are shown in table 3. In addition, detailed results for chromosome 12 are shown in figure 4. The Marshfield screening map of chromosome 12 has 17 markers, an average map resolution of 8.2 cM, and an average IC of 0.79 (fig. 4_B_). The SNP linkage map of chromosome 12 contains a total of 97 markers, distributed among 29 clusters and 18 singleton SNPs, with an average distance of 3.7 cM between the 47 map positions; the average IC of the SNP linkage map was 0.86 (fig. 4_A_).
Table 3.
IC of SNP Maps and Marshfield Version 10 Screening Set
| SNP Map | Marshfield Version 10 Screening Set | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Chromosome | No. of MapPositionsa | No. of SNPson Mapb | AverageIC | MinimumIC | MaximumIC | No. of Markerson Map | AverageIC | MinimumIC | MaximumIC |
| 1 | 93 | 204 | .91 | .71 | 1.00 | 30 | .79 | .67 | .95 |
| 2 | 81 | 176 | .91 | .80 | 1.00 | 25 | .79 | .63 | .96 |
| 3 | 77 | 156 | .91 | .74 | 1.00 | 25 | .83 | .68 | .98 |
| 4 | 57 | 124 | .88 | .75 | .99 | 22 | .78 | .67 | .92 |
| 5 | 64 | 130 | .89 | .74 | .98 | 20 | .81 | .66 | .97 |
| 6 | 57 | 126 | .88 | .75 | .98 | 21 | .75 | .45 | .95 |
| 7 | 56 | 118 | .90 | .72 | 1.00 | 21 | .83 | .72 | .95 |
| 8 | 57 | 113 | .91 | .75 | 1.00 | 19 | .82 | .61 | .95 |
| 9 | 47 | 96 | .88 | .80 | .98 | 18 | .80 | .63 | .93 |
| 10 | 54 | 116 | .90 | .72 | .99 | 21 | .82 | .66 | .95 |
| 11 | 43 | 87 | .89 | .72 | .99 | 20 | .79 | .58 | .96 |
| 12 | 47 | 97 | .86 | .66 | .99 | 17 | .79 | .65 | .95 |
| 13 | 36 | 69 | .89 | .77 | 1.00 | 12 | .81 | .68 | .97 |
| 14 | 33 | 82 | .88 | .68 | .98 | 13 | .77 | .62 | .96 |
| 15 | 29 | 59 | .85 | .71 | .99 | 13 | .82 | .67 | .98 |
| 16 | 34 | 78 | .86 | .75 | 1.00 | 15 | .81 | .73 | .94 |
| 17 | 28 | 65 | .88 | .59 | 1.00 | 13 | .79 | .65 | .95 |
| 18 | 37 | 81 | .88 | .75 | .99 | 14 | .82 | .66 | .97 |
| 19 | 25 | 43 | .84 | .68 | .97 | 10 | .77 | .65 | .93 |
| 20 | 31 | 74 | .90 | .73 | .99 | 11 | .77 | .43 | .94 |
| 21 | 18 | 44 | .92 | .82 | .98 | 7 | .77 | .67 | .93 |
| 22 | 22 | 50 | .88 | .73 | .98 | 8 | .82 | .73 | .97 |
| X | 22 | 35 | .65 | .46 | .97 | 17 | .79 | .64 | .96 |
| Overall | 1,048 | 2,223 | .88 | .73 | .99 | 392 | .80 | .64 | .95 |
Figure 4.
IC for chromosome 12, on SNP linkage maps and Marshfield screening set 10. A, Complete SNP map with 97 markers (black). B, Marshfield map with 17 markers (red). C, SNP map with 47 markers (blue).
It has been estimated that, as compared with STR markers, 1.7–2.5 times as many SNP markers are needed to obtain equivalent IC (Kruglyak 1997; Goddard and Wijsman 2002). Therefore, we also evaluated the IC of a SNP linkage map of chromosome 12, containing only 47 evenly spaced markers (2.8 times as many as the Marshfield screening map). Unlike the complete SNP map of chromosome 12, no markers in this subset are clustered, and the markers are more evenly distributed. The average IC of this 47-marker 3.7-cM SNP map was 0.82 (range 0.65–0.98) (fig. 4_C_). This result shows that our map of 47 SNP markers has an IC comparable to a map of 17 microsatellite markers. However, several factors affect estimates of IC, and analysis of other chromosomes indicates that the relationship between SNP and microsatellite informativeness is not simple.
We were curious how the IC of a SNP-marker set would compare to a microsatellite-marker set when used to analyze a collection of sib pairs, with and without parents available. To evaluate the SNP IC within nuclear pedigrees, we analyzed genotype data from chromosome 22, and we created sib pairs from the 42 commonly genotyped three-generation CEPH families by removing the grandparents and analyzing the first two offspring from each pedigree. For the microsatellite comparison, we simulated data for 42 nuclear pedigrees (two parents and two offspring each) for markers with the same allele frequencies and map features as those on the Marshfield version 10 screening set. When parents were included in the analyses, the average IC for the SNP set was 0.75, and the average IC for the microsatellite set was 0.74. When parents were not included in the analyses, the average IC for the SNP set was 0.41, and the average IC for the microsatellite set was 0.41. Although the IC for both SNPs and STRs drops considerably when only sib pairs (i.e., with parents unavailable) are analyzed, the IC of the 3.9-cM SNP set remains comparable to that of an 8.4-cM microsatellite screening set.
Discussion
Considerable effort is being put into the development of new methods for high-throughput SNP genotyping (Tsuchihashi and Dracopoli 2002). Some of these methods have achieved a level of affordability and ease of use such that genomewide linkage scans can now be performed using SNPs. However, these approaches cannot move forward without a SNP-based linkage map. It is anticipated that the successful identification of a set of SNPs tailored for linkage analysis, such as that presented here, will stimulate development of mass-produced (i.e., less expensive) means for large-scale genotyping with this same marker set.
The SNPs in the 3.9-cM linkage set presented here have been rigorously validated, including evaluation for assay robustness in at least one genotyping platform and estimation of allele frequencies in medium-sized samples from three different ethnic groups. The SNPs within each 100-kb cluster show low rates of recombination, as expected, and also show a low level of LD. These features maximize the SNP set's utility for linkage studies, although modifications to existing computer programs are required in order to easily accommodate both the presence of LD and nonzero recombination within marker clusters. For analyses using only a small number of SNPs, currently available programs such as Linkage (Lathrop et al. 1984) or Fastlink (Cottingham et al. 1993; Schäffer et al. 1994) can simultaneously accommodate both LD and recombination. For analyses involving larger sets of markers, until new program modifications are introduced, one resolution would be to eliminate genotypes that introduce recombination within clusters. Although many of these genotypes will accurately represent true recombination events and removal would introduce some small bias on the results, some apparent recombinants will be erroneous. This process is similar to the current practice, required by most linkage analysis programs, of removal of genotypes that introduce non-Mendelian segregation. In these cases, many such Mendelian inconsistencies are due to erroneous genotypes, but some are due to marker mutations; hence, the simple removal of a small subset of problematic genotypes is a commonly accepted practice in linkage studies. Developers of genetic-analysis software are aware of this issue, and we are confidant that they are pursuing changes that will allow for proper solutions to this issue.
Given a fixed limit on the total number of SNPs to be placed on this map, the relative utility of a dense map of uniform SNPs versus a less dense map of SNP clusters depends on the types of linkage analyses for which the map will be used (e.g., multipoint vs. two-point) and the types of pedigrees analyzed (e.g., small vs. large and/or complex). Since methods of linkage analysis continue to be improved and developed, we felt the best approach to be an intermediate one; therefore, we created a map that contains both SNP clusters and singleton SNPs. Each SNP cluster can be used as a composite marker for two-point linkage analysis, or multipoint analyses can be performed using SNPs from each cluster plus the singleton SNPs. This SNP map will make a useful tool for initial linkage screening. Follow-up finer mapping can proceed in the same manner as it does now after a linkage scan with STR markers, using a dense map that includes both STR and SNP markers. As the genome assemblies continue to improve, it will be possible to increase the resolution of this map by the addition of SNPs and/or SNP clusters that have insignificant support for linkage map locations to their most likely positions in cases in which these agree with locations identified on the physical map.
The median MPIC values across all clusters were 0.50–0.65, depending on the assumptions of availability of phase information. When complete phase information is available, the median SNP MPIC value is similar to the PIC value for a commonly used microsatellite screening set. Naturally, in the absence of phase information, SNP markers, even in clusters, show reduced MPIC values. The level of IC averaged across an entire SNP map, as assessed by the IC measure in Genehunter, was equal to or greater than that of the Marshfield screening set map for well-phased CEPH pedigrees, nuclear pedigrees (i.e., parents plus two offspring), and only sib pairs (i.e., two offspring with parents unavailable). Since expected LOD scores are closely correlated with IC (Kruglyak 1997), the observed increase in IC implies that the SNP linkage maps, as compared with current STR maps, should provide improved or equal average power to detect linkage.
A recent study by Rhodes et al. (2002) used SNPs to replicate previously reported microsatellite-based linkage for hereditary spastic paraplegia (HSP [MIM 182601]). HSP has previously been linked to the SPG4 gene, on chromosome 2, as well as to several other loci. Rhodes et al. (2002) evaluated one large family in which linkage to SPG4 has previously been shown (Reid et al. 1999) and in which the specific SPG4 mutation has been identified (Lindsey et al. 2000). Rhodes et al. (2002) genotyped 122 SNPs across chromosome 2 in 24 family members (11 of whom were affected). Of these SNPs, 101 overlap with SNPs in our screening set. Multipoint linkage analysis using these SNPs confirmed linkage to SPG4, with LOD scores that were equal to or greater than those reported in the previously published microsatellite-based analysis of the same family (results varied depending on the specific analysis performed) (Rhodes et al. 2002).
As compared with more-informative markers, individual SNPs suffer from one particular drawback: it is more difficult to detect genotyping errors by checking for Mendelian inheritance. To identify and remove problematic genotypes, we applied extensive procedures both before and after mapping. These procedures both reduced the number of apparent recombination events within our clusters and resulted in a significant reduction in overall map lengths. By design, the SNP linkage maps span the same physical distances as do the Marshfield linkage maps. The length of the SNP linkage map is within 1.1% of the length of the Marshfield map, supporting the conclusion that the map distances observed on these maps are reasonable estimates of the true underlying distance, at least as manifested in the CEPH individuals.
The order of STR markers near the SNPs on our maps is 97% concordant with the Marshfield maps and is 99% concordant with the recently published deCode linkage maps, providing additional support to the general accuracy, in marker order, of all three maps. The SNP map shows varying concordance with the sequence maps: 93% with NCBI build 28 (February 2002), 92% with NCBI build 29 (May 2002), 99% with NCBI build 30 (August 2002), and 96% with the Celera C4 (May 2001) assembly. In particular, note that the NCBI build 30 assembly shows a fit to the SNP linkage maps that is much improved over earlier builds. These data do not lead to any simple conclusions as to whether a linkage map or a sequence assembly is more accurate; it is likely that each has some errors for at least some chromosomes. However, the fact that our unique SNP data, large number of CEPH samples, and mapping approaches have independently produced a map that is highly concordant with the Marshfield and deCode linkage maps lends credibility to the overall accuracy of our SNP linkage map. Further credibility can be given to SNP-based mapping by the 99% average concordance between our linkage map and the deCode map; both were constructed using large (and completely independent) pedigree data sets. The higher concordance between our linkage map and the deCode map, as opposed to between our map and the Marshfield map, is consistent with greater map accuracy due to larger sample sizes. Additionally, comparison with the deCode interpolated SNP linkage map shows that determination of genetic-map position by interpolation can result in fairly accurate estimates of position and map distances (fig. 3). However, because of substantial genomic variation in the ratio of recombination rate per physical distance, such interpolation will be only reasonably accurate when a dense linkage map is utilized. Since map distance misspecification has been shown to be a problem in terms of both false-positive (Daw et al. 2000) and false-negative (Halpern and Whittemore 1999) linkage-analysis results, caution should be exercised in the use of such interpolated distances.
Given the current state of the physical assembly of the human genome, any reasonably dense linkage map can be used to examine how the rate of recombination varies across the genome and how it varies between females and males. Although a denser map is required in order to draw conclusions on a fine scale, the SNP maps presented here are useful in order to make broad observations about variation in recombination intensity. Chromosome 16 is shown here as an example of such plots (fig. 2), which we have made for each autosome. From our graphs, we can make some broad observations. First, we note that the average ratio of cM/Mb differs for each chromosome, ranging from 1.2, on chromosome 2, to 2.46, on chromosome 22 (table 2) and generally increasing as chromosome size decreases. This ratio also varies, sometimes by severalfold, along each chromosome (for a plot of chromosome 16, see fig. 2; plots for the other chromosomes are available at the corresponding author’s Web site [Additional Data for the TSC SNP Linkage Map]). In addition, although the genomewide average ratio of female-to-male recombination intensity is 1.7, this also varies considerably along each chromosome. We note that, at every metacentric p-telomeric end and at most metacentric and acrocentric q-telomeric ends, there is a higher rate of recombination in males than in females. In the most extreme case (chromosome 18), the telomeric rate of male recombination is eight times greater than the female rate. On approximately one-half of the chromosomes, there appears to be a notable increase in female recombination intensity, followed by a sharp decrease near the q telomeres. Any further study of these phenomena would require detailed knowledge of the physical location of the telomeres and would be better suited to a denser linkage map.
Clusters of SNPs provide the greatest information for linkage mapping if they exhibit low levels of LD. In this way, selecting a group of clustered SNPs provides the greatest number of haplotypes with the most even frequency distribution. However, by selecting SNPs with low LD, we may enrich the set for those that exhibit recombination within clusters. In fact, the CEPH pedigree data revealed numerous likely recombination events within clusters. The relationships between intracluster LD, inferred population recombination rates, and observed intracluster recombinants is explored in further detail in the companion article (Clark et al. 2003 [in this issue]). Additional work is under way by other groups to improve the resolution of the SNP map in regions with large gaps, especially on the X chromosome, and, as the sequencing and assembly of the human genome progresses, it will be possible to further refine the positions of those SNPs for which unique map positions could not be identified.
This SNP linkage map provides an IC equal to or better than a commonly used map for genome screening (Marshfield version 10) and provides a critical initial resource needed in the pursuit of SNP-based linkage screening in humans. Given a customized high-throughput genotyping platform—such as those currently under production by Applied Biosystems, Motorola Life Sciences, and Illumina—a whole-genome scan in a study with 1,000 individuals can be performed in as little as several days to a few weeks. This improvement in genotyping time is especially useful in the study of genes for complex traits, for which larger sample sizes are required in order to find significant signals. Further data from the present study, including the linkage maps and the genotypes used to evaluate the SNPs and prepare these maps, can be found at the SNP Consortium Linkage Map Project Web site. The genotype data have also been submitted to the CEPH Genotype Database.
With several currently available techniques for SNP genotyping, a whole-genome scan for a disease locus could be completed with greater efficiency than most laboratories currently achieve using STR markers. The SNP linkage map described here, together with advancing technologies for high-throughput SNP-based genotyping, provides the necessary tools for highly efficient, robust, and rapid linkage-based genome screening.
Acknowledgments
We thank the following individuals for valuable technical assistance: Victoria Appleton, Roxane Bonner, Amish Gandhi, Michael Gaskin, Penny Gwynne, Ken Harris, Miguel Hernandez, Xiangyang Kong, Annalee Ledesma, William Lee, Sandra Lew, Kerri McWeeny, Tom Perez, Gudmundur Thorisson, and Jeff Zdunek. We also thank Karl Broman, Linda Brzustowicz, Norman Doggett, Julie Douglas, Derek Gordon, Leigh Pascoe, Greg Schuler, Steve Sherry, Eric Sobel, and Daniel Weeks, as well as the anonymous reviewers, for helpful scientific input. The Laboratory for Computer Science Research of the Division of Computer and Information Sciences at Rutgers provided valuable time on their system. This project would not have been possible without the public resources provided by the American Diabetes Association (for the Japanese samples in the diversity panel), the Center for Medical Genetics at the Marshfield Clinic, NCBI, TSC, and deCode Genetics. This project was funded by TSC and Motorola Life Sciences.
Electronic-Database Information
URLs for data presented herein are as follows:
- Additional Data for the TSC SNP Linkage Map, http://compgen.rutgers.edu/SNPmap/ (for additional data from the construction and analysis of the TSC SNP linkage map)
- Allele Frequency Panels, http://snp.cshl.org/allele_frequency_project/panels.shtml
- BLAST, http://www.ncbi.nlm.nih.gov/BLAST/
- Center for Medical Genetics, http://research.marshfieldclinic.org/genetics/ (for Marshfield linkage maps)
- CEPH Genotype Database, http://www.cephb.fr/cephdb/
- dbSNP Home Page, http://www.ncbi.nlm.nih.gov/SNP/
- High-Resolution Recombination Map of the Human Genome, A, http://www.nature.com/ng/journal/v31/n3/suppinfo/ng917_S1.html [DOI] [PubMed]
- Kruglyak Lab, http://www.fhcrc.org/labs/kruglyak/Downloads/ (for the program LD)
- Locfit, http://cm.bell-labs.com/cm/ms/departments/sia/project/locfit/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for HSP) [PubMed]
- Single Nucleotide Polymorphisms for Biomedical Research, http://snp.cshl.org/ (for the SNP database)
- SNP Consortium Linkage Map Project, The, http://snp.cshl.org/linkage_maps/
- UCSC Genome Bioinformatics (“Golden Path”), http://genome.ucsc.edu
- UniSTS, http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=unists
References
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR (2002) Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 30:97–101 [DOI] [PubMed] [Google Scholar]
- Agarwala R, Applegate DL, Maglott D, Schuler GD, Schäffer AA (2000) A fast and scalable radiation hybrid map construction and integration strategy. Genome Res 10:350–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botstein D, White RL, Skolnick M, Davies RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet 32:314–331 [PMC free article] [PubMed] [Google Scholar]
- Broman KW, Murray JC, Sheffield VC, White RL, Weber JL (1998) Comprehensive human genetic maps: individual and sex-specific variation in recombination. Am J Hum Genet 63:861–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson CS, Eberle MA, Rieder MJ, Smith JD, Kruglyak L, Nickerson DA (2003) Additional SNPs and linkage-disequilibrium analyses are necessary for whole-genome association studies in humans. Nat Genet 33:518–521 [DOI] [PubMed] [Google Scholar]
- Clark AG, Nielsen R, Signorovitch J, Matise TC, Glanowski S, Heil J, Winn-Deen ES, Holden AL, Lai E (2003) Linkage disequilibrium and inference of ancestral recombination in 538 single-nucleotide polymorphism clusters across the human genome. Am J Hum Genet 72:285–300 (in this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins FS, Guyer MS, Charkravarti A (1997) Variations on a theme: cataloging human DNA sequence variation. Science 278:1580–1581 [DOI] [PubMed] [Google Scholar]
- Cottingham RW Jr, Idury RM, Schäffer AA (1993) Faster sequential genetic linkage computations. Am J Hum Genet 53:252–263 [PMC free article] [PubMed] [Google Scholar]
- Craven P, Wahba G (1979) Smoothing noisy data with spline functions. Numerische Mathematik 31:377–403 [Google Scholar]
- Dausset J, Cann H, Cohen D, Lathrop M, Lalouel J-M, White R (1990) Centre d’Etude du Polymorphisme Humain (CEPH): collaborative genetic mapping of the human genome. Genomics 6:575–577 [DOI] [PubMed] [Google Scholar]
- Daw EW, Thompson EA, Wijsman EM (2000) Bias in multipoint linkage analysis arising from map misspecification. Genet Epidemiol 19:366–380 [DOI] [PubMed] [Google Scholar]
- Dearlove A (2002) High throughput genotyping technologies. Brief Funct Genomics Proteomics 1:139–150 [DOI] [PubMed] [Google Scholar]
- Goddard KA, Wijsman EM (2002) Characteristics of genetic markers and maps for cost-effective genome screens using diallelic markers. Genet Epidemiol 22:205–220 [DOI] [PubMed] [Google Scholar]
- Goddard KA, Yu CE, Oshima J, Miki T, Nakura J, Piussan C, Martin GM, Schellenberg GD, Wijsman EM (1996) Toward localization of the Werner syndrome gene by linkage disequilibrium and ancestral haplotyping: lessons learned from analysis of 35 chromosome 8p11.1–21.1 markers. Am J Hum Genet 58:1286–1302 [PMC free article] [PubMed] [Google Scholar]
- Gordon D, Heath SC, Liu X, Ott J (2001) A transmission/disequilibrium test that allows for genotyping errors in the analysis of single-nucleotide polymorphism data. Am J Hum Genet 69:371–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudbjartsson DF, Jonasson K, Frigge ML, Kong A (2000) Allegro, a new computer program for multipoint linkage analysis. Nat Genet 25:12–13 [DOI] [PubMed] [Google Scholar]
- Halpern J, Whittemore AS (1999) Multipoint linkage analysis: a cautionary note. Hum Hered 49:194–196 [DOI] [PubMed] [Google Scholar]
- Heil J, Glanowski S, Scott J, Winn-Deen E, McMullen I, Wu L, Gire C, Sprague A (2002) An automated computer system to support ultra high throughput SNP genotyping. Pac Symp Biocomput 30–40 [DOI] [PubMed] [Google Scholar]
- Kong A, Gudbjartsson DF, Sainz J, Jonsdottir GM, Gudjonsson SA, Richardsson B, Sigurdardottir S, Barnard J, Hallbeck B, Masson G, Shlien A, Palsson ST, Frigge ML, Thorgeirsson TE, Gulcher JR, Stefansson K (2002) A high-resolution recombination map of the human genome. Nat Genet 31:241–247 [DOI] [PubMed] [Google Scholar]
- Kruglyak L (1997) The use of a genetic map of biallelic markers in linkage studies. Nat Genet 17:21–24 [DOI] [PubMed] [Google Scholar]
- Lander ES (1996) The new genomics: global views of biology. Science 274:536–539 [DOI] [PubMed] [Google Scholar]
- Lander ES, Green P (1987) Construction of multi-locus genetic linkage maps in humans. Proc Natl Acad Sci USA 84:2363–2367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, et al (2001) Initial sequencing and analysis of the human genome. Nature 409:860–921 [DOI] [PubMed] [Google Scholar]
- Lathrop GM, Lalouel JM, Julier C, Ott J (1984) Strategies for multilocus linkage analysis in humans. Proc Natl Acad Sci USA 81:3443–3446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsey JC, Lusher ME, McDermott CJ, White KD, Reid E, Rubinsztein DC, Bashir R, Hazan J, Shaw PJ, Bushby KM (2000) Mutation analysis of the spastin gene (SPG4) in patients with hereditary spastic paraparesis. J Med Genet 37:759–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loader C (1999) Local regression and likelihood. Springer Verlag, New York [Google Scholar]
- Markianos K, Daly MJ, Kruglyak L (2001) Efficient multipoint linkage analysis through reduction of inheritance space. Am J Hum Genet 68:963–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matise TC, Perlin M, Chakravarti A (1994) Automated construction of genetic linkage maps using an expert system (MultiMap): a human genome linkage map. Nat Genet 6:384–390 [DOI] [PubMed] [Google Scholar]
- O’Connell JR, Weeks DE (1998) PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 63:259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid E, Grayson C, Rogers MT, Rubinsztein DC (1999) Locus-phenotype correlations in autosomal dominant pure hereditary spastic paraplegia: a clinical and molecular genetic study of 28 United Kingdom families. Brain 122:1741–1755 [DOI] [PubMed] [Google Scholar]
- Rhodes M, Day J, Dong P, Scafe C, Dailey D, Gilbert G, Wang Y, Laig-Webster N, Su X, Koehler R, Avi-Itshak H, Ziegle J, Wogan L, McMullen I, Spier E, De La Vega F, Reid E, Brooking J, Dudbridge F, Dearlove A, Weaver T, Yuille M (2002) A validated set of SNPs for linkage mapping studies. Am J Hum Genet Suppl 71:442 [Google Scholar]
- Risch N, Merikangas K (1996) The future of genetic studies of complex human diseases. Science 273:1516–1517 [DOI] [PubMed] [Google Scholar]
- Sachidanandam R, Weissman D, Schmidt SC, Kakol JM, Stein LD, Marth G, Sherry S, et al (2001) A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature 409:928–933 [DOI] [PubMed] [Google Scholar]
- Schäffer AA, Gupta SK, Shriram K, Cottingham RW Jr (1994) Avoiding recomputation in linkage analysis. Hum Hered 44:225–237 [DOI] [PubMed] [Google Scholar]
- Sheffield VC, Weber JL, Buetow KH, Murray JC, Even DA, Wiles K, Gastier JM, et al (1995) A collection of tri- and tetranucleotide repeat markers used to generate high quality, high resolution human genome-wide linkage maps. Hum Mol Genet 4:1837–1844 [DOI] [PubMed] [Google Scholar]
- Sobel E, Lange K (1996) Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker-sharing statistics. Am J Hum Genet 58:1323–1337 [PMC free article] [PubMed] [Google Scholar]
- Sobel E, Papp JC, Lange K (2002) Detection and integration of genotyping errors in statistical genetics. Am J Hum Genet 70:496–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger JD, Ott J (1994) Handbook of human genetic linkage. Johns Hopkins University Press, Baltimore [Google Scholar]
- Terwilliger JD, Speer M, Ott J (1993) Chromosome-based method for rapid computer simulation in human genetic linkage analysis. Genet Epidemiol 10:217–224 [DOI] [PubMed] [Google Scholar]
- Tsuchihashi Z, Dracopoli NC (2002) Progress in high throughput SNP genotyping methods. Pharmacogenomics J 2:103–110 [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, et al (2001) The sequence of the human genome. Science 291:1304–1351 [DOI] [PubMed] [Google Scholar]
- Wang DG, Fan JB, Siao CJ, Berno A, Young P, Sapolsky R, Ghandour G, et al (1998) Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science 280:1077–1082 [DOI] [PubMed] [Google Scholar]
- Weber JL, Broman KW (2001) Genotyping for human whole-genome scans: past, present, and future. Adv Genet 42:77–96 [DOI] [PubMed] [Google Scholar]
- Weber JL, May PE (1989) Abundant class of human DNA polymorphisms which can be typed using the polymerase chain reaction. Am J Hum Genet 44:388–396 [PMC free article] [PubMed] [Google Scholar]
- Weissenbach J, Gyapay G, Dib C, Vignal A, Morissette J, Millasseau P, Vaysseix G, Lathrop M (1992) A second-generation linkage map of the human genome. Nature 359:794–801 [DOI] [PubMed] [Google Scholar]
- Wilson AF, Sorant AJ (2000) Equivalence of single- and multilocus markers: power to detect linkage with composite markers derived from biallelic loci. Am J Hum Genet 66:1610–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]