Mutations of the RNA-Specific Adenosine Deaminase Gene (DSRAD) Are Involved in Dyschromatosis Symmetrica Hereditaria (original) (raw)
Abstract
Dyschromatosis symmetrica hereditaria (DSH) (also called “reticulate acropigmentation of Dohi”) is a pigmentary genodermatosis of autosomal dominant inheritance characterized by a mixture of hyperpigmented and hypopigmented macules distributed on the dorsal aspects of the hands and feet. To determine the gene responsible for this disease, we performed a genomewide search in three families with DSH and mapped the DSH locus to chromosome 1q21.3. The mutations involved in causing DSH have been identified in the gene that encodes double-stranded RNA-specific adenosine deaminase (DSRAD) as the disease gene.
Dyschromatosis symmetrica hereditaria (DSH [MIM 127400]) (also called “reticulate acropigmentation of Dohi”) was first described by Toyama (1910, 1929) and has been reported mainly in Japan, although it occurs in families of every ethnic origin all over the world (Oyama et al. 1999). Patients with DSH have pinpoint, pea-sized hyperpigmented and hypopigmented macules on the back of their hands and the tops of their feet (fig. 1). The face is spared, apart from a few scattered small discrete, pigmented macules. These abnormalities are otherwise asymptomatic and do not affect the general health of the patient. The prevalence of DSH in the population of Japan is estimated to be ∼1.5 per 100,000. DSH generally shows an autosomal dominant pattern of inheritance, with high penetrance, but some patients with sporadic DSH have been reported. When similar skin lesions occur all over the body, the disease is discriminated from DSH and is named “dyschromatosis universalis hereditaria” (DUH [MIM 127500]). The clinical features of DSH and DUH are quite distinct and have not been found in the same pedigrees. Although clinical and morphological investigations of DSH and DUH have been reported, the cause and the pathogenesis of the diseases, as well as the pathological gene(s), have not yet been clarified. Until now, the DSH gene was hypothesized to be located on chromosome 9, because of another family with DSH and idiopathic torsion dystonia (the gene responsible for the latter disease is localized on chromosome 9) (Patrizi et al. 1994), but our early analyses of three Japanese families with DSH (as shown in fig. 2A–C) excluded chromosome 9 as a possibility (Kono et al. 2000).
Figure 1.
Small hyperpigmented and hypopigmented macules on the dorsal aspects of the hands and feet. Upper, Sixty-year-old female. Lower, Forty-year-old male.
Figure 2.
Pedigrees and haplotypes. A, Pedigree 1. B, Pedigree 2. C, Pedigree 3. D, Pedigree 4. Affected and unaffected individuals are represented by the blackened and unblackened symbols, respectively. An asterisk (*) indicates individuals who provided blood. Single asterisk (*) and double asterisk (**) indicate members in pedigrees 1, 2, and 3 who were utilized for linkage analysis. E, Haplotype analysis. The shared disease-associated haplotype is colored.
We then tried an entire genomewide scan, using 343 microsatellite markers for linkage analysis in the three families with DSH (41 affected and 47 unaffected individuals, as shown in fig. 2A–C), using the method detailed elsewhere (Kono et al. 2000). In brief, genotyping errors or misspecified relationships within the pedigrees were checked by the PedCheck software package. DNA fragment–length analysis on PCR products of microsatellite markers from the Linkage Mapping Set (Perkin Elmer) was performed, using a Macintosh Centris 650 (Apple Japan) personal computer, with 672 Genescan software and Genotype, version 1.1 (both Perkin Elmer products), as described in the manufacturer’s manuals. The allele size of each marker was rounded, using the GAS package, version 2.0 (as found on the Oxford University Web site). Calculations for linkage analysis were performed with the FASTLINK software package, version 4.0. (Lathrop et al. 1984; Cottingham et al. 1993; Schaffer et al. 1994), under an assumed genetic model as follows: autosomal dominant, gene frequency of 0.0001, marker-allele frequency set to be equal, complete penetrance, zero phenocopy rate, and no sex difference. Two-point LOD scores were calculated with the MLINK module in the FASTLINK software package. Kosambi map function was used to convert the recombination fractions to map distances. Haplotype analyses were performed using the SimWalk2 software. The study was approved by the ethics committee of Nagoya University School of Medicine.
In a genomewide scan for linkage analysis of three pedigrees, we obtained cumulative LOD scores >3.0 with satellite markers D1S502, D1S252, D1S498, and D1S484 in chromosome 1q (table 1; fig. 3). To determine a smaller interval containing the DSH locus, haplotype analysis was performed, and the results suggested that the gene responsible for DSH lies between D1S2715 and D1S2777 (figs. 2E and 3). To further refine the localization, we identified novel SNPs and integrated the genetic and physical maps of the region. The final DSH genetic interval, of ∼500 kb, was bound proximally by the IL6R gene and distally by the KCNN3 gene, at chromosome 1q21.3. Between those two genes, seven genes are mapped on the Entrez Genome Map View, NCBI Web site: namely, NICE-5, cholinergic receptor, double-strand RNA-specific adenosine deaminase (DSRAD), and four hypothetical proteins (LOC126669, LOC126668, LOC343053, and LOC343054).
Table 1.
Cumulative Two-Point LOD Scores
| LOD at θ = | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Marker | 0 | .05 | .1 | .15 | .2 | .25 | .3 | .35 | .4 | .45 | .5 | _Z_max | θmax |
| D1S424 | −∞ | .17 | 1.15 | 1.51 | 1.59 | 1.51 | 1.30 | 1.01 | .66 | .29 | 0 | 1.59 | .20 |
| D1S206 | −∞ | −1.21 | .56 | 1.29 | 1.59 | 1.64 | 1.51 | 1.25 | .90 | .48 | 0 | 1.64 | .25 |
| D1S502 | −∞ | 3.81 | 4.88 | 5.02 | 4.75 | 4.23 | 3.51 | 2.65 | 1.68 | .71 | 0 | 5.02 | .15 |
| D1S252 | −∞ | 5.46 | 5.96 | 5.78 | 5.29 | 4.61 | 3.77 | 2.82 | 1.79 | .78 | 0 | 5.96 | .10 |
| D1S498 | −∞ | 4.49 | 4.43 | 4.09 | 3.62 | 3.05 | 2.42 | 1.75 | 1.06 | .45 | 0 | 4.49 | .05 |
| D1S484 | −∞ | 1.80 | 2.99 | 3.34 | 3.31 | 3.05 | 2.62 | 2.07 | 1.42 | .71 | 0 | 3.34 | .15 |
| D1S196 | −∞ | −.76 | .46 | .99 | 1.21 | 1.25 | 1.17 | .98 | .72 | .40 | 0 | 1.25 | .25 |
| D1S218 | −∞ | −7.17 | −4.03 | −2.40 | −1.41 | −.77 | −.36 | −.11 | .02 | .05 | 0 | .05 | .45 |
Figure 3.
Genetic map position for microsatellite markers. A, Marker used in the linkage analyses, as shown in table 1. B, Markers used in the haplotype analyses, as shown in figure 2E. cM = Genetic distance. D1S468 and D1S423 are the nearest markers to the telomeres of the p and the q arms, respectively.
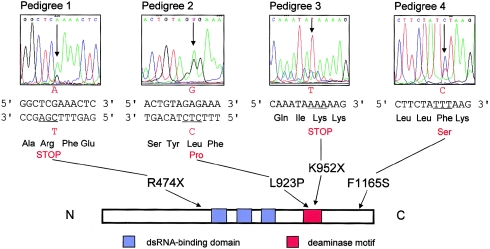
To detect pathological mutations in the seven candidate genes, four affected individuals from each of pedigrees 1–3 and pedigree 4 (a fourth family with DSH added to the study, with 10 affected and 14 unaffected individuals) were screened by single-strand conformational polymorphism analysis (SSCP), which revealed mutant heteroduplexes made by hybridizing PCR fragments of the DSRAD gene. Every patient in each pedigree showed the mutant SSCP pattern of exons 2, 10, 10, and 15 in pedigrees 1, 2, 3, and 4, respectively. Direct sequence analysis of the PCR products showed that they were heterozygous for mutations of Arg474STOP (CGA→TGA), Leu923Pro (CTC→CCC), Lys952STOP (AAA→TAA), and Phe1165Ser (TTT→TCT) in pedigrees 1, 2, 3, and 4, respectively (fig. 4). None of those four mutations were found in the blood samples of any of the 55 unaffected individuals in any of the four pedigrees or in the 116 unrelated, normally pigmented Japanese adults who were surveyed. Thus, we concluded that those four mutations are not polymorphic but are the pathologic ones that cause the disease.
Figure 4.
Mutations of the DSRAD gene found in patients with DSH. Antisense sequences are shown for pedigrees 1 and 2. L923P and K952X are located in the deaminase domain (red box). N = N-terminal; C = C-terminal.
DSRAD is composed of 1,226 amino acid residues, with a calculated molecular mass of 139 kDa (O’Connell et al. 1995). It catalyzes the deamination of adenosine to inosine in double-stranded RNA substrates (Bass and Weintraub 1988; Wagner et al. 1989), which results in the creation of alternative splicing sites (Rueter et al. 1999) or alterations of codons and thus leads to functional changes in proteins. DSRAD is expressed ubiquitously (O’Connell et al. 1995), but only three target genes for DSRAD—including ionotropic glutamate receptor (Higuchi et al. 1993; Lomeli et al. 1994), the serotonin receptor 2C subtype in the brain (Burns et al. 1997), and hepatitis delta virus antigen in the liver (Polson et al. 1996)—are known. Recently, it was reported that DSRAD expression was increased in the spleen, thymus, and peripheral lymphocytes of endotoxin-treated mice (Yang et al. 2003). The target genes and the induction of the enzyme in lymphocytes seem not to be involved in the pathogenesis of DSH.
Since the DSRAD gene has 15 exons, and since three copies of the dsRNA-binding domains and the putative deaminase domain are reported to be located in exons 2–7 and 9–14, respectively (Wang et al. 1995), it is understandable that truncated proteins with no functional activity would be synthesized from genes with a nonsense mutation in exon 2 or in exon 10, as found in pedigrees 1 and 3, respectively.
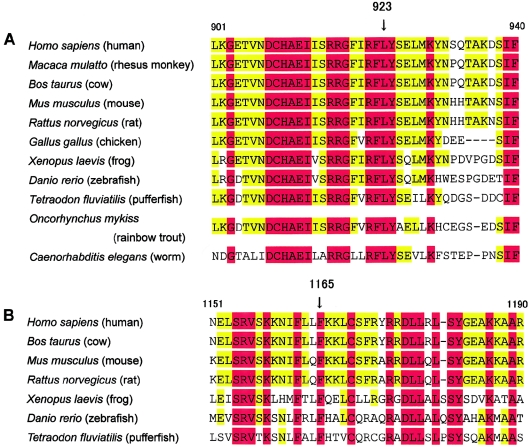
Since the amino acid residue at 923 in exon 10 is also located in the putative deaminase domain, and since the 4–amino acid sequence containing it is absolutely conserved among all 11 species, as shown in figure 5A, the leucine at 923 is suspected to play an important role in the conformation of the catalytic site of the enzyme, and the mutation of Leu923Pro probably compromises enzyme activity. On the other hand, the missense mutation at 1165 in exon 15 is a little bit difficult to explain, because exon 15 is not involved in the catalytic domain. Phe1165 is supposed to contribute to the structure of the enzyme, an expectation supported by evidence that Phe1165 is conserved among all available sequences (fig. 5B); therefore, its mutation may induce an unstable tertiary structure of the protein that results in defective activity.
Figure 5.
Amino acid alignment of various species around the two missense mutations. Human DSRAD amino acid residues 901–940 (A) and 1151–1190 (B) are shown aligned to known DSRAD orthologues. Residues L, at 923, and F, at 1165, are conserved in all species compared in the figure. Residues conserved among all available sequences and among all mammalian orthologues are shown with red or yellow background, respectively. All data were cited from GenBank.
Heterozygosity for the Dsrad knockout causes embryonic lethality in mice (Wang et al. 2000). In contrast, patients with DSH have a good prognosis, with only the dyschromatosis. The prognostic difference between the two species may depend on differences between them. The reason the skin lesions are localized specifically on the backs of the hands and on the tops of the feet is difficult to explain pathogenetically, since DSRAD is ubiquitously expressed all over the skin. In the case of piebaldism—an autosomal dominant disease resulting from mutations in the c-KIT proto-oncogene (Fleischman et al. 1991; Giebel and Spritz 1991)—white patches on the forehead, chest, abdomen, and extremities are induced by the reduced activity of the c-kit receptor, which is required for the migration of melanocyte precursors throughout the skin during the early fetal period (Nishikawa et al. 1991). Similarly, we speculate that, when melanoblasts migrate from the neural crest to the skin during development, a greater reduction in DSRAD activity might occur at anatomic sites most distant from the neural crest. The failure of correct RNA editing may induce the differentiation of melanoblasts to hyperactive or hypoactive melanocytes, then colonizing in an irregular distribution in the skin lesions.
During the preparation of this manuscript, the locus for DSH was reported, by Zhang et al. (2003), to be at chromosome 1q11-1q21 and, by Xing et al. (2003), to be at chromosome 6q24.2-q25.2. The patients of Xing et al. (2003) showed dyschromatosis over almost their entire bodies, which suggests that they have DUH. If so, there is no contradiction between the two studies; that is, the DSH gene is located on chromosome 1, whereas the DUH gene is located on chromosome 6.
Acknowledgments
We thank the members of the families for their collaboration. This work was supported, in part, by Grants-in-Aids for Scientific Research from The Ministry of Education, Culture, Sports, Science and Technology of Japan.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- Entrez Genome Map Viewer, NCBI, http://www.ncbi.nlm.nih.gov/mapview/maps.cgi (for physical map of chromosome 1q)
- GenBank, http://www.ncbi.nlm.nih.gov/Entrez/ (for the cDNA of human DSRAD [accession number NM_001111], the genome of human DSRAD [accession number NT_004668], and the DSRAD-related amino acid sequences or EST of rhesus monkey [accession number CB553335], cow [accession number CB449783], mouse [accession number BAC40888], rat [accession number NP_112268], chicken [accession number BU437798], frog [accession number T30340], zebrafish [accession number NP_571671], pufferfish [accession number AAF69674], rainbow trout [accession number CA359632], and worm [accession number T16913])
- Genetic Analysis System (GAS), http://users.ox.ac.uk/~ayoung/gas.html (for allele size of microsatellite markers [package 2.0, Oxford University, 1993–1995])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for DSRAD and DSH)
- PedCheck, http://watson.hgen.pitt.edu/register/docs/pedcheck.html (for checking genotyping errors and misspecified relationships within pedigrees)
- SimWalk2, http://watson.hgen.pitt.edu/docs/SW2_Overview.html (for haplotype analyses)
References
- Bass BL, Weintraub H (1988) An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell 55:1089–1098 [DOI] [PubMed] [Google Scholar]
- Burns CM, Chu H, Rueter SM, Hutchinson LK, Canton H, Sanders-Bush E, Emeson RB (1997) Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature 387:303–308 [DOI] [PubMed] [Google Scholar]
- Cottingham RW Jr, Idury RM, Schaffer AA, (1993) Faster sequential genetic linkage computations. Am J Hum Genet 53:252–263 [PMC free article] [PubMed] [Google Scholar]
- Fleischman RA, Saltman DL, Stastny V, Zneimer S (1991) Deletion of the c-kit protooncogene in the human developmental defect piebald trait. Proc Natl Acad Sci USA 88:10885–10889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giebel LB, Spritz RA (1991) Mutation of the KIT (mast/stem cell growth factor receptor) protooncogene in human piebaldism. Proc Natl Acad Sci USA 88:8696–8699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi M, Single FN, Kohler M, Sommer B, Sprengel R, Seeburg PH (1993) RNA editing of AMPA receptor subunit GluR-B: a base-paired intron exon structure determines position and efficiency. Cell 75:1361–1370 [DOI] [PubMed] [Google Scholar]
- Kono M, Miyamura Y, Matsunaga J, Tomita Y (2000) Exclusion of linkage between dyschromatosis symmetrica hereditaria and chromosome 9. J Dermatol Sci 22:88–95 [DOI] [PubMed] [Google Scholar]
- Lathrop GM, Lalouel JM, Julier C, Ott J (1984) Strategies for multilocus linkage analysis in humans. Proc Natl Acad Sci USA 81:3443–3446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomeli H, Mosbacher J, Melcher T, Hoger T, Geiger JRP, Kuner T, Monyer H, Higuchi M, Bach A, Seeburg PH (1994) Control of kinetic properties of AMPA channels by nuclear RNA editing. Science 266:1709–1713 [DOI] [PubMed] [Google Scholar]
- Nishikawa S, Kusakabe M, Yoshinaga K, Ogawa M, Hayashi S, Kunisada T, Era T, Sakakura T, Nishikawa S (1991) In utero manipulation of coat color formation by a monoclonal anti-c-kit antibody: two distinct waves of c-kit-dependency during melanocyte development. EMBO J 10:2111–2118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell MA, Krause S, Higuchi M, Hsuan JJ, Totty NF, Jenny A, Keller W (1995) Cloning of cDNAs encoding mammalian double-stranded RNA-specific adenosine deaminase. Mol Cell Biol 15:1389–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyama M, Shimizu H, Ohata Y, Tajima S, Nishikawa T (1999) Dyschromatosis symmetrica hereditaria (reticulate acropigmentation of Dohi): report of a Japanese family with the condition and a literature review of 185 cases. Br J Dermatol 140:491–496 [DOI] [PubMed] [Google Scholar]
- Patrizi A Manneschi V, Pini A, Baioni E, Ghetti P (1994) Dyschromatosis symmetrica hereditaria associated with idiopathic torsion dystonia: a case report. Acta Derm Venereol 74:135–137 [DOI] [PubMed] [Google Scholar]
- Polson AG, Bass BL, Casey JL (1996) RNA editing of hepatitis delta virus antigenome by dsRNA-adenine deaminase. Nature 380:454–455 [DOI] [PubMed] [Google Scholar]
- Rueter SM, Dawson TR, Emeson RB (1999) Regulation of alternative splicing by RNA editing. Nature 399:75–80 [DOI] [PubMed] [Google Scholar]
- Schaffer AA, Gupta SK, Shriram K, Cottingham RW Jr (1994) Avoiding recomputation in linkage analysis. Hum Hered 44:225–237 [DOI] [PubMed] [Google Scholar]
- Toyama I (1910) An unknown disorder of hyperpigmentation (in Japanese). Jap J Dermatol Urol 10:644 [Google Scholar]
- ——— (1929) Dyschromatosis symmetrica hereditaria (in Japanese). Jap J Dermatol Urol 29:95–96 [Google Scholar]
- Wang Q, Khillan J, Gadue P, Nishikura K (2000) Requirement of the RNA editing deaminase ADAR1 gene for embryonic erythropoiesis. Science 290:1765–1768 [DOI] [PubMed] [Google Scholar]
- Wang Y, Zeng Y, Murray JM, Nishikura K (1995) Genomic organization and chromosomal location of the human dsRNA adenosine deaminase gene: the enzyme for glutamate-activated ion channel RNA editing. J Mol Biol 254:184–195 [DOI] [PubMed] [Google Scholar]
- Wagner RW, Smith JE, Cooperman BS, Nishikura K (1989) A double-stranded RNA unwinding activity introduces structural alterations by means of adenosine to inosine conversions in mammalian cells and Xenopus eggs. Proc Natl Acad Sci USA 86:2647–2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Q, Wang M, Chen X, Feng G, Ji H, Yang J, Gao J, Qin W, Qian X, Wu S, He L (2003) A gene locus responsible for dyschromatosis symmetrica hereditaria (DSH) maps to chromosome 6q24.2-q25.2. Am J Hum Genet 73:377–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J-H, Luo X, Nie Y, Su Y, Zhao Q, Kabir K, Zhang D, Rabinovici R (2003) Widespread inosine-containing mRNA in lymphocytes regulated by ADAR1 in response to inflammation. Immunology 109:15–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XJ, Gao M, Li M, Li M, Li C-R, Cui Y, He P-P, Xu S-J, Xiong X-Y, Wang Z-X, Yuan W-T, Yang S, Huang W (2003) Identification of a locus for dyschromatosis symmetrica hereditaria at chromosome 1q11-1q21. J Invest Dermatol 120:776–780 [DOI] [PubMed] [Google Scholar]