What we could do now: molecular pathology of bladder cancer (original) (raw)
Abstract
There is much information on the genetic alterations that contribute to the development of bladder cancer. Because it is hypothesised that the genotype of the cancer cell plays a major role in determining phenotype, this genetic information should impact on clinical practice. To date however, this has not happened. Some of the alterations identified in bladder cancer have clear associations with outcome—for example, mutational inactivation of the cell cycle regulator proteins p53 and the retinoblastoma protein (Rb). However, as single markers, these events have insufficient predictive power to be applied in the management of individual patients. The use of panels of markers is a potential solution to this problem. Examples of suitable panels include those genes/proteins with known impact on specific cell cycle checkpoints or with impact on cellular phenotypes, such as immortalisation, invasion, or metastasis. To evaluate such marker panels, large tumour series will be needed—for example, archival samples from completed clinical trials. The use of these valuable resources will require coordination of sample provision. This might involve central collection and distribution of tissue blocks, sections, or tissue arrays and the provision of patient follow up information to laboratories participating in a study. With the availability of microarray technologies, including cDNA and comparative genomic hybridisation arrays, the transcriptome and genome of transitional cell carcinomas of different phenotypes can be compared and will undoubtedly provide a wealth of information with potential diagnostic and prognostic uses. Although these studies can be initiated using small local tissue collections, high quality collection of fresh tissues from new clinical trials will be crucial for proper evaluation of associations with clinical outcome. Funding for molecular pathological studies to date has been poor. To begin to translate molecular information from the laboratory to the clinic and to make maximum use of valuable urological patient resources in the UK, adequate funding and scientific energy are required. Whereas the latter is not in doubt, present funding for this type of translational research is inadequate.
Keywords: bladder cancer, molecular pathology
A great deal of information has accumulated on the genetics of transitional cell carcinoma (TCC) in recent years. Numerous genes and genetic changes involved in tumour development have been identified and the literature abounds with papers that show clear associations of specific genetic changes or gene mutations with clinical parameters. Therefore, one might predict that this must have generated novel laboratory tests (for screening, diagnosis, prediction of prognosis, etc) or some prospective studies or clinical trials. However, not only is little of this information currently being applied in the clinical setting but also, on closer examination of the data, it is clear that additional information will be needed before relevant applications emerge. This review examines the crucial clinical issues involved in the management of TCC, presents a summary of the current state of knowledge of molecular genetic alterations in bladder cancer, describes attempts made to apply genetic markers in the clinic, and then considers the way forward both in terms of what could be applied now and what further information is needed.
Problems in clinical management of TCC
What are the questions relating to the clinical management of this disease that might be dealt with by the application of novel molecular genetic markers? There are several situations during the diagnosis and selection of appropriate treatment for TCC where objective markers could potentially transform clinical practice. Because it is postulated that the genetic changes that contribute to the development of these tumours collectively determine tumour phenotype, markers based on genetic changes should provide robust diagnostic and predictive power.
Urothelial tumours can be classified into two groups based on histopathology and clinical behaviour. At presentation, more than 80% of bladder tumours are non-muscle invasive papillary tumours (pTa or pT1). Patients with such superficial tumours frequently develop recurrences (approximately 70%), often over the course of many years, but progression to muscle invasion occurs in only 10–20% of cases. The tumours that do progress are usually those that show superficial invasion (pT1) at diagnosis. In contrast, the 20% of tumours that show muscle invasion at the time of diagnosis have a much less favourable prognosis and often progress rapidly.
Superficial papillary and invasive bladder tumours are widely believed to have different natural histories, based both on histopathological and genetic information. Most invasive bladder tumours have no known papillary precursor, are solid invasive lesions, and are commonly associated with carcinoma in situ (CIS) elsewhere in the bladder. The proposed progression pathway for these lesions is therefore urothelial atypia → dysplasia → CIS → invasive TCC. Genetic analyses have shown that CIS contains a spectrum of genetic alterations (such as TP53 mutation and loss of heterozygosity (LOH) of 3p, 8p, 13q, and 17p), similar to that seen in invasive TCC and very distinct from that seen in low grade papillary TCC, where only LOH of chromosome 9 is common. The proposed pathway for development of this last category of tumours is urothelial hyperplasia → urothelial atypia (G1–3) → low grade papillary TCC.
For superficial bladder cancer, identification of tumours that will progress has long been perceived as a potential application of genetic studies. If such patients could be identified at presentation, more aggressive treatment might result in a cure. The debate about choice of cystectomy, particularly for pT1 tumours, has raged for many years and, although it is clear that this can achieve complete cure, the associated morbidity cautions against this approach except where a very high probability of progression is predicted. Therefore, the identification of such patients is a priority. Markers that show very strong association with progression will also be useful at diagnosis, with the potential to identify those tumours that are invasive but for which an inadequate tissue sample failed to show invasion. For many pTa tumours, most of which are classified as carcinoma rather than papilloma, current management involves repeated hospital attendance for cystoscopy. For those patients with truly benign disease that will not recur, this practice creates unnecessary anxiety and is costly for the health service. Markers to identify tumours with different potentials to recur will have great impact for this large group of patients. Because bladder cancer is often multifocal and some tumours arise within a urothelium that shows widespread urothelial atypia, objective methods are needed to assess the status of the urothelium remaining after resection of all overt tumours, possibly by the assessment of irrigation specimens. For example, selection of patients for BCG treatment might be made objectively on the basis of the extent of genetic changes, rather than subjectively, based on visible histopathological changes.
For the 20% of tumours that show muscle invasion (> T2) with or without metastatic disease (N+M+), the issues at diagnosis are different. If radiotherapy or chemotherapy is given in addition to surgery, response rates are not high and the prediction of this and the possible identification of the optimum treatment would avoid ineffective, expensive, and unpleasant treatment. The major cause of treatment failure and death is the presence of unrecognised distant metastatic disease, and markers that could identify either the presence of distant metastases or predict those primary tumours most likely to metastasise early would be beneficial.
Finally, of potential benefit for all patients whose bladders are preserved will be markers for disease monitoring, based on urine analysis. At present, grade and stage are the best prognostic indicators in TCC. The crucial roles for molecular markers are in those circumstances where morphologically similar tumours behave in dramatically different ways in the patient.
What we know: genetics of TCC
Genetic studies to date have attempted to identify the spectrum of genetic changes that occurs during urothelial transformation and to elucidate in more detail the natural history of bladder tumours with different clinical outcome.1 Several known oncogenes and tumour suppressor genes are mutated in TCC (table 1 ▶).2–33 These include the genes encoding several key G1 checkpoint proteins (p16, p14ARF, retinoblastoma protein (Rb), p53, and cyclin D1) which are altered in many other tumour types.
Table 1.
Genes genetically altered in transitional cell carcinoma
| Gene (cytogenetic location) | Alteration | Frequency/clinical association |
|---|---|---|
| Oncogenes | ||
| HRAS (11p15) | Activating mutation | 10–15% overall (high grade)2–4 |
| FGFR3 (4p16) | Activating mutation | 30–40%5 6 |
| ERBB2 (17q ) | Amplification/overexpression | Amplified 10–14% high grade/stage7–11 |
| CCND1(11q13) | Amplification/overexpression | 10–20% all grades and stages12–14 |
| MDM2 (12q13) | Amplification/overexpression | Amplified 1–4%15 16 |
| Tumour suppressor genes | ||
| INK4A-ARF (9p21) | Homozygous deletion/methylation/mutation | 20–30% high grade/stage17–19 |
| LOH 60% all grades/stages | ||
| Immortalisation in vitro20 | ||
| RB1 (13q14) | Deletion/mutation | 10–15% overall21–23 |
| 37% muscle invasive | ||
| TP53 (17p13) | Deletion/mutation | 70% muscle invasive24–26 |
| High grade and stage | ||
| PTEN (10q23) | Homozygous deletion/mutation | 10q LOH in 35% muscle invasive27 28 |
| 6.6% superficial | ||
| PTCH (9q22) | Deletion/mutation | LOH 60% all grades/stages29 |
| Mutation frequency low | ||
| TSC1 (9q34) | Deletion/mutation | LOH 60% all grades/stages30 |
| Mutation frequency low | ||
| *DBCCR1 (9q32–33) | Deletion/methylation | LOH 60% all grades/stages31 32 |
| *DCC/SMAD (18q) | Deletion | LOH 30% high grade/stage33 |
| No mutation analysis to date |
In addition to alterations to known genes, many non-random genetic alterations have been identified in TCC. Regions commonly showing deletion and predicted to contain tumour suppressor genes have been identified by LOH analyses. These are listed in table 2 ▶.34–46 Whole genome analysis has been carried out by comparative genomic hybridisation (CGH) (table 3 ▶).47–53 Although this technique is not able to detect very small deletions and thus cannot be used to map critical deleted regions, it has nevertheless confirmed the deletion of the same chromosome arms as identified by LOH.53 The major strength of CGH is the detection of small regions of high level amplification, which are predicted to identify the location of relevant oncogenes. Some of the amplifications identified by CGH have confirmed the amplification of known oncogenes such as CCND1 (cyclin D) at 11q13 and ERBB2 at 17q21, whereas others potentially identify the locations of novel oncogenes (table 3 ▶). It should be noted that most changes commonly identified in TCC are found predominantly in muscle invasive tumours and hence represent potential prognostic markers for superficial tumours.
Table 2.
Common regions of deletion detected by loss of heterozygosity analysis in transitional cell carcinoma
| Cytogenetic location | Frequency | Association with clinical parameters |
|---|---|---|
| 3p | 48% | Stage34 |
| 4p | 22% | None35 36 |
| 4q | 24% | High grade/stage36 |
| 8p | 23% | High grade/stage37–40 |
| 9q | 60% | None41–43 |
| 11p | 40% | Grade44 45 |
| 11q | 15% | None45 |
| 14q | 10–40% | Stage46 |
Table 3.
Comparative genomic hybridisation findings in transitional cell carcinoma47–52
| Tumour stage | Losses | Gains | Amplification |
|---|---|---|---|
| Ta | 9p, 9q, Y | 1q, 17 | 11q |
| T1 | 2q, 4p, 4q, 5q, 6q, 8p, 9p, 9q, 10q, 11p, 11q, 13q, 17p, 18q, Y | 1q, 3p, 3q, 5p, 6p, 8q, 10p, 17q, 20q | 1q22–24, 3p24–25, 6p22, 8p12, 8q22, 10p12–14, 10q22–23, 11q13, 12q12–21, 17q21, 20q13 |
| T2–4 | As for T1 + 15q | As for pT1 + 7p, Xq | As for pT1 |
Progress in the Human Genome Project, together with the development of new technologies such as microarray techniques to analyse global changes in gene expression54, 55 or gene copy number,56 will undoubtedly lead to the identification of the genes targeted by these deletions and amplifications within a relatively short time. Meanwhile, all of the common genetic alterations can be identified with relative ease, often using polymerase chain reaction (PCR) based techniques, and can therefore be assessed and explored as potential molecular markers with clinical usefulness. In addition, the expression status of many known genes can be assessed by immunohistochemistry. Therefore, there is no technological barrier to the detailed analysis of multiple markers in either retrospectively or prospectively collected tumour panels.
Molecular genetic markers as prognostic indicators in TCC
Many studies have assessed the relation between individual genetic changes and clinical parameters, particularly tumour grade and stage. The mutation of some genes has been assessed in relation to clinical outcome. Many studies have been retrospective and have examined only a single gene or marker. This is illustrated well by studies in which the predictive value of TP53 or RB gene mutation has been assessed. Most studies of TP53 have used immunohistochemical detection of the p53 protein as a surrogate for gene inactivation by mutation. Mutant p53 has an increased half life and can be detected with ease, whereas normal physiological concentrations of the wild-type protein are undetectable. Mutation frequencies of TP53 are higher in high grade and high stage tumours than in low grade superficial tumours,24–26, 57 and this is reflected in a clear association with outcome.58–62 Several studies have documented a significant reduction in progression free interval for p53 positive tumours. Similarly, absence of the Rb protein, indicative of gene mutation, is found more frequently in tumours of high grade and stage and is clearly associated with poor outcome.2–23 These results for bladder cancer are reiterated in many studies of these two genes in a range of tumour types.
Given such clear association with outcome, can these genes be used as predictive markers upon which to make clinical decisions? Unfortunately, the answer is no. The examination of data for single markers reveals insufficient predictive power upon which to base decisions for individual patients. This is disappointing and applies not only to bladder tumours but to single marker studies in all types of tumour. A vast number of single gene studies with highly significant p values have been published during the past 10 years but this has contributed little to clinical practice.
Improvement of predictive power
This lack of predictive power is not surprising in light of our current understanding of neoplastic development and progression. Multiple genetic and epigenetic events contribute to tumour development, and analyses to date indicate that few tumours are genetically identical. It follows therefore that the same tumour phenotype may be generated in various ways.
To reach a point where our genetic knowledge can be applied efficiently, the tumour genotype–phenotype connection must be examined in different ways. One possible approach is to consider those phenotypes which, based on our knowledge of cell biology, biochemistry, and tumour behaviour in vivo, may be considered likely to confer a selective advantage. Examples of these include cellular immortalisation, escape from G1 checkpoint control, loss of DNA damage checkpoints (G2/M), loss of normal DNA repair mechanisms and/or apoptotic response, and ability to invade normal tissues. For all of these phenotypes, a set of genes with potential impact can be listed, any one of which when altered in a tumour may confer the same phenotype.
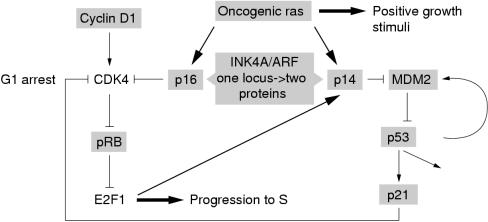
An example is the potential escape from G1 checkpoint control by mutational activation or inactivation of one of a group of genes that impact on this control mechanism. Two pathways, the pRb and p53 pathways, involving several common molecules, provide G1 checkpoint control and regulate the response to DNA damage or inappropriate mitogenic signalling.63, 64 The INK4A/ARF locus and the two proteins it encodes, p16 and p14ARF,65 play a pivotal role in these control mechanisms (fig 1 ▶). In one pathway, p16 inhibits the activity of cyclin dependent kinase (CDK)–cyclin complexes that act to phosphorylate the Rb protein. Phosphorylation of Rb causes the release of the transcription factor E2F1, which initiates the transcription of genes required for progression into S phase. In the second pathway, p14ARF upregulates p53.66, 67 p53 can induce G1 arrest or apoptosis in a cell type dependent manner via the upregulation of another CDK inhibitor, p21/WAF1.64 p53 stabilisation via p14ARF appears to be via binding to MDM2,67 a protein that interacts with p53 and targets it for degradation via the ubiquitin pathway.68 The recent findings that the expression of p14ARF is in turn regulated by E2F1,69 and that the expression of a ras oncogene results in the upregulation of p14ARF70 and p16,71 provide evidence for a fail safe mechanism to protect the cell from aberrant stimuli (fig 1 ▶). Escape from G1 checkpoint control can theoretically be achieved via inactivation of RB or INK4A or by overexpression of CCND1 but, in fact, inactivation of TP53 or p14/ARF is also required to inactivate this fail safe mechanism. Of the nine proteins shown in fig 1 ▶, six commonly show genetic alteration in TCC. In addition, alteration of p21 expression has been found, with loss of expression reported as an independent predictor of progression and maintenance of expression reported to abrogate the effect of p53 alteration.72 MDM2 amplification has also been reported in a small proportion of TCCs,15, 16 and alternatively spliced transcripts, lacking the p53 binding domain have been found in tumours of high grade and stage.73 In this context, it is of interest that a recent report showed that 19 of 19 bladder tumour cell lines studied had lesions in both the Rb and p53 pathways.74 Eleven had mutations in exons 5–11 of TP53 and concomitant loss of expression of the Rb protein. Seven had wild-type TP53 or a mutation in exons 1–4 (predicted to affect the transactivation function only) and concomitant alterations in INK4A and ARF.
Figure 1.
pRb and p53 pathways and control of the G1 checkpoint. The two proteins encoded by the INK4A/ARF locus, p16 and p14ARF, are negative regulators of the Rb and p53 pathways, respectively. p14ARF is regulated by the transcription factor E2F1, which is an RB effector. Genes commonly altered in transitional cell carcinoma are shown boxed in grey. CDK, cyclin dependent kinase.
Predictions from this model for G1 checkpoint control are that complete dysregulation requires inactivation of RB or INK4A and p14/ARF or TP53 and that the phenotype generated by alteration to either TP53 or RB alone will be less aggressive than when both pathways are altered. Already this prediction has been confirmed by three studies of TCC, which have shown that tumours with inactivation of TP53 and RB have a worse prognosis than tumours with alteration of either gene alone.75–77 The impressive results from these retrospective studies have led to the initiation of some new clinical trials based on stratification by TP53 and RB status. As yet, no studies have attempted a comprehensive assessment of all the key proteins involved in the G1 checkpoint, but clearly this should now be initiated.
Only through detailed study by numerous laboratories of the role of the various genes involved in cell cycle progression from G1 to S has current knowledge reached the point where we can assemble a relevant panel of genes to use as markers of the dysregulated phenotype in this way. For some phenotypes, understanding of the key proteins involved is more rudimentary. The ideal situation would be to use a single marker, possibly a final effector in a pathway, to identify a specific phenotype. This might be possible, even in situations where the underlying mechanisms have not been elucidated. One potential example of this type of marker has been described in the bladder. During the differentiation of the normal urothelium, a highly regulated pattern of uroplakin and cytokeratin expression has been documented. One of the proteins expressed only in the superficial cells and occasional intermediate cells of the normal differentiated urothelium is cytokeratin 20. In a study of 51 non-invasive papillary tumours,78 10 tumours with normal cytokeratin 20 staining pattern were identified and none of these recurred. In contrast, all tumours that did recur showed an abnormal staining pattern (staining in all layers of the epithelium). A much larger study will be needed to confirm the apparent predictive power of this single marker. However, this study exemplifies a situation in which it seems possible to measure a cellular phenotype (normal urothelial differentiation) altered in tumour cells with propensity to recur, by means of a single protein marker. In this instance, we have no understanding of the molecular basis of the differentiation process involved. Although understanding the underlying processes involved in tumour related cell phenotypes, such as immortalisation and escape from cell cycle checkpoints, yields potentially powerful panels of markers, it will be simpler in the long term if single protein surrogates, easily measurable by immunohistochemistry, can be identified for these phenotypes also.
The future
What types of study can be started now? The possibilities include both retrospective studies to validate and test the predictive power of marker panels and prospective studies where high quality tissue samples could be collected from patients in clinical trials. The latter would enable novel approaches to marker identification to be used and would allow measurements only applicable to fresh tissue (such as RNA expression analysis) to be associated with clinical outcome.
Sufficient information is already available for studies of small to moderate numbers of markers (phenotype associated panels) to be initiated, and this is feasible in research laboratories with a repertoire of basic molecular techniques. Such studies could readily include PCR based genetic analyses and immunohistochemistry on archival, paraffin wax embedded tissues. The use of material from patients entered into completed clinical trials, where available, will provide robust follow up data. Because several laboratories are likely to be interested in carrying out such studies, and the stored tissue samples from patients entered into trials are inevitably scattered throughout the country, a necessary prerequisite will be the establishment of a central repository of tissue blocks from where marker studies can be coordinated. The best location for such a central resource is debatable, but an appropriate research laboratory with access to microdissection and tissue arraying facilities79 and close links with a specialist pathologist might be appropriate. It is predicted that the proper use of archival material will provide a wealth of clinically useful information. Several large trials have been carried out in TCC and relevant technology is in place in several laboratories in the UK; thus, there are no barriers to such studies apart from the provision of funding.
To date, small local collections of tissue samples have provided the material for most of the marker studies carried out. These have been very valuable and, although the coordination of countrywide resources as outlined above will be important, the value of properly collected local samples must not be overlooked. For bladder cancer, appropriate samples include snap frozen tumour tissue (to provide frozen sections for dissection and extraction of macromolecules and for certain immunohistochemical analyses), a venous blood sample (for genetic comparisons with the tumour—for example, LOH analyses), and urine samples at each visit (for protein analyses and genetic and other analyses on cells in the sediment). Such collections will continue to fuel local research and, with future coordination of these smaller collections, may provide large sample sets for collaborative studies. Funding for state of the art tissue collection and storage is generally inadequate, and this will require improvement in the future if the best use is to be made of our valuable urological patient resources in the UK.
The ideal funding mechanism for studies of this type is not clear. To date, funding for multiple marker analyses in specific tumour types has not been easy to obtain. Many published studies represent minor research projects carried out either in laboratories whose major interest and funding centres around a particular gene or pathway, or in histopathology laboratories with an interest in testing new antibodies with potential prognostic application. Some studies have been carried out in laboratories with an interest in the biology of a specific tumour type, who inevitably assess known oncogenes and tumour suppressor genes to measure the frequency of involvement in the tumour of interest. These comments are not intended to condemn such studies, but to illustrate the point that studies funded with the stated major objective of finding markers or panels of markers with clinical usefulness have to date been few and far between. The key question is how can that be changed. Where will such funding come from? Is this likely to be funded by the major cancer charities, the research councils or the National Health Service? Should funding decisions be driven by academic excellence rather than potential application or vice versa? Certainly, translational research is currently perceived as less likely to generate scientific output in journals with the highest impact factors. Hopefully, this situation may be short lived because exciting developments in the field of molecular pathology should increase the impact of some pathology journals.
Within larger organisations and specialist laboratories, there is the potential to apply large scale screening strategies to identify novel markers and, with appropriate bioinformatics support, to maximise the potential of the information obtained. For example, both cDNA expression arrays and array CGH are now available and represent powerful methods to compare tumours at the mRNA and genetic levels. To obtain the optimum information from these technologies, highly pure tumour cell samples are needed, and these can be obtained using one of the various microdissection systems available, such as laser capture microdissection.80 Given the vast amount of information obtained from each array analysis, it will be important to select tumour panels carefully, with guidance from an expert histopathologist. Once we have identified patterns of expression or levels of DNA over-representation or under-representation that allow tumours to be subclassified,81 larger panels of tumours can be studied and high throughput analysis for specific diagnostic purposes can be envisaged. Alternatively, the information obtained may identify simple tests—for example, small numbers of proteins for immunocytochemical analysis, which can be carried out in the histopathology laboratory.
Assuming that relevant markers for diagnostic, prognostic, and disease monitoring are identified, it will be important to establish methods that can be applied routinely in hospital laboratories. At this stage, direct links between histopathology and hospital genetics laboratories may be the means to implement the relevant tests.
Conclusions
My conclusion is that to date the available genetic information on TCC has had little impact in the clinic. The reasons for this are not entirely clear. The inadequate power of single markers must partly explain this but it seems likely that many factors, mostly circumstantial, including lack of funding, conspire to reduce the chances of translation into the clinic. Existing tissue collections and archival resources should be used to their full potential and this will require appropriate funding for coordinating mechanisms. Ongoing tissue collection, preferably in conjunction with clinical trials, is a priority. As far as choice of appropriate analyses and markers is concerned, small to moderate sized panels of markers should be assembled, and these should be logical—that is, based on current knowledge of a particular biochemical pathway or cell phenotype. Finally, carefully selected panels of tumours should be used for whole genome analysis and expression profiling at both the RNA and protein level. The potential of such studies to change the clinical management of bladder cancer is clear. Resources and scientific energy are all that is now required.
References
- 1.Knowles MA. The genetics of transitional cell carcinoma: progress and potential clinical application. BJU Int 1999;84:412–27. [DOI] [PubMed] [Google Scholar]
- 2.Knowles MA, Williamson M. Mutation of H-ras is infrequent in bladder cancer: confirmation by single-strand conformation polymorphism analysis, designed restriction fragment length polymorphisms, and direct sequencing. Cancer Res 1993;53:133–9. [PubMed] [Google Scholar]
- 3.Fitzgerald JM, Ramchurren N, Rieger K, et al. Identification of H-ras mutations in urine sediments complements cytology in the detection of bladder tumors. J Natl Cancer Inst 1995;87:129–33. [DOI] [PubMed] [Google Scholar]
- 4.Ooi A, Herz F, Ii S, et al. Ha-ras codon 12 mutation in papillary tumors of the urinary bladder: a retrospective study. Int J Oncol 1994;4:85–90. [DOI] [PubMed] [Google Scholar]
- 5.Cappellen D, De Oliveira C, Ricol D, et al. Frequent activating mutations of FGFR3 in human bladder and cervix carcinomas. Nat Genet 1999;23:18–20. [DOI] [PubMed] [Google Scholar]
- 6.Sibley K, Cuthbert-Heavens D, Knowles MA. Loss of heterozygosity at 4p16.3 and mutation of FGFR3 in transitional cell carcinoma. Oncogene 2001;20:686–91. [DOI] [PubMed] [Google Scholar]
- 7.Coombs LM, Pigott DA, Sweeney E, et al. Amplification and over-expression of c-erbB-2 in transitional cell carcinoma of the urinary bladder. Br J Cancer 1991;63:601–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sato K, Moriyama M, Mori S, et al. An immunohistologic evaluation of c-erbB-2 gene product in patients with urinary bladder carcinoma. Cancer 1992;70:2493–8. [DOI] [PubMed] [Google Scholar]
- 9.Sauter G, Moch H, Moore D, et al. Heterogeneity of erbB-2 gene amplification in bladder cancer. Cancer Res 1993;53:2199–203. [PubMed] [Google Scholar]
- 10.Underwood M, Bartlett J, Reeves J, et al. C-erbB-2 gene amplification: a molecular marker in recurrent bladder tumors? Cancer Res 1995;55:2422–30. [PubMed] [Google Scholar]
- 11.Mellon JK, Lunec J, Wright C, et al. c-erbB-2 in bladder cancer: molecular biology, correlation with epidermal growth factor receptors and prognostic value. J Urol 1996;155:321–6. [DOI] [PubMed] [Google Scholar]
- 12.Proctor AJ, Coombs LM, Cairns JP, et al. Amplification at chromosome 11q13 in transitional cell tumours of the bladder. Oncogene 1991;6:789–95. [PubMed] [Google Scholar]
- 13.Bringuier PP, Tamimi Y, Schuuring E, et al. Expression of cyclin D1 and EMS1 in bladder tumours: relationship with chromosome 11q13 amplification. Oncogene 1996;12:1747–53. [PubMed] [Google Scholar]
- 14.Shin KY, Kong G, Kim WS, et al. Overexpression of cyclin D1 correlates with early recurrence in superficial bladder cancers. Br J Cancer 1997;75:1788–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lianes P, Orlow I, Zhang Z-F, et al. Altered patterns of MDM2 and TP53 expression in human bladder cancer. J Natl Cancer Inst 1994;86:1325–30. [DOI] [PubMed] [Google Scholar]
- 16.Habuchi T, Kinoshita H, Yamada H, et al. Oncogene amplification in urothelial cancers with p53 gene mutation or MDM2 amplification. J Natl Cancer Inst 1994;86:1331–5. [DOI] [PubMed] [Google Scholar]
- 17.Williamson MP, Elder PA, Shaw ME, et al. p16 (CDKN2) is a major deletion target at 9p21 in bladder cancer. Hum Mol Genet 1995;4:1569–77. [DOI] [PubMed] [Google Scholar]
- 18.Orlow I, Lacombe L, Hannon GJ, et al. Deletion of the p16 and p15 genes in human bladder tumors. J Natl Cancer Inst 1995;87:1524–9. [DOI] [PubMed] [Google Scholar]
- 19.Cairns P, Polascik TJ, Eby Y, et al. Frequency of homozygous deletion at p16/CDKN2 in primary human tumours. Nat Genet 1995;11:210–12. [DOI] [PubMed] [Google Scholar]
- 20.Yeager TR, DeVries S, Jarrard DF, et al. Overcoming cellular senescence in human cancer pathogenesis. Genes Dev 1998;12:163–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cairns P, Proctor AJ, Knowles MA. Loss of heterozygosity at the RB locus is frequent and correlates with muscle invasion in bladder carcinoma. Oncogene 1991;6:2305–9. [PubMed] [Google Scholar]
- 22.Cordon-Cardo C, Wartinger D, Petrylak D, et al. Altered expression of the retinoblastoma gene product: prognostic indicator in bladder cancer. J Natl Cancer Inst 1992;84:1251–6. [DOI] [PubMed] [Google Scholar]
- 23.Logothetis CJ, Xu H-J, Ro JY, et al. Altered expression of retinoblastoma protein and known prognostic variables in locally advanced bladder cancer. J Natl Cancer Inst 1992;84:1256–61. [DOI] [PubMed] [Google Scholar]
- 24.Habuchi T, Takahashi R, Yamada H, et al. Influence of cigarette smoking and schistosomiasis on p53 gene mutation in urothelial cancer. Cancer Res 1993;53:3795–9. [PubMed] [Google Scholar]
- 25.Sidransky D, von Eschenbach A, Tsai YC, et al. Identification of p53 gene mutations in bladder cancers and urine samples. Science 1991;252:706–9. [DOI] [PubMed] [Google Scholar]
- 26.Spruck CH, III, Rideout WM, III, Olumi AF, et al. Distinct pattern of p53 mutations in bladder cancer: relationship to tobacco usage. Cancer Res 1993;53:1162–6. [PubMed] [Google Scholar]
- 27.Aveyard JS, Skilleter A, Habuchi T, et al. Somatic mutation of PTEN in bladder carcinoma. Br J Cancer 1999;80:904–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cairns P, Evron E, Okami K, et al. Point mutation and homozygous deletion of PTEN/MMAC1 in primary bladder cancers. Oncogene 1998;16:3215–18. [DOI] [PubMed] [Google Scholar]
- 29.McGarvey TW, Maruta Y, Tomaszewski JE, et al. PTCH gene mutations in invasive transitional cell carcinoma of the bladder. Oncogene 1998;17:1167–72. [DOI] [PubMed] [Google Scholar]
- 30.Hornigold N, Devlin J, Davies AM, et al. Mutation of the 9q34 gene TSC1 in bladder cancer. Oncogene 1999;18:2657–61. [DOI] [PubMed] [Google Scholar]
- 31.Habuchi T, Yoshida O, Knowles MA. A novel candidate tumour suppressor locus at 9q32–33 in bladder cancer: localisation of the candidate region within a single 840kb YAC. Hum Mol Genet 1997;6:913–19. [DOI] [PubMed] [Google Scholar]
- 32.Habuchi T, Luscombe M, Elder PA, et al. Structure and methylation-based silencing of a gene (DBCCR1) within a candidate bladder cancer tumor suppressor region at 9q32–q33. Genomics 1998;48:277–88. [DOI] [PubMed] [Google Scholar]
- 33.Brewster SF, Gingell JC, Browne S, et al. Loss of heterozygosity on chromosome 18q is associated with muscle-invasive transitional cell carcinoma of the bladder. Br J Cancer 1994;70:697–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Presti JC, Jr, Reuter VE, Galan T, et al. Molecular genetic alterations in superficial and locally advanced human bladder cancer. Cancer Res 1991;51:5405–9. [PubMed] [Google Scholar]
- 35.Elder PA, Bell SM, Knowles MA. Deletion of two regions on chromosome 4 in bladder carcinoma: definition of a critical 750kB region at 4p16.3. Oncogene 1994;9:3433–6. [PubMed] [Google Scholar]
- 36.Polascik TJ, Cairns P, Chang WYH, et al. Distinct regions of allelic loss on chromosome 4 in human primary bladder carcinoma. Cancer Res 1995;55:5396–9. [PubMed] [Google Scholar]
- 37.Knowles MA, Shaw ME, Proctor AJ. Deletion mapping of chromosome 8 in cancers of the urinary bladder using restriction fragment length polymorphisms and microsatellite polymorphisms. Oncogene 1993;8:1357–64. [PubMed] [Google Scholar]
- 38.Takle LA, Knowles MA. Deletion mapping implicates two tumor suppressor genes on chromosome 8p in the development of bladder cancer. Oncogene 1996;12:1083–7. [PubMed] [Google Scholar]
- 39.Ohgaki K, Iida A, Ogawa O, et al. Localization of tumor suppressor gene associated with distant metastasis of urinary bladder cancer to a 1-Mb interval on 8p22. Genes Chromosomes Cancer 1999;25:1–5. [DOI] [PubMed] [Google Scholar]
- 40.Choi C, Kim MH, Juhng SW, et al. Loss of heterozygosity at chromosome segments 8p22 and 8p11.2–21.1 in transitional-cell carcinoma of the urinary bladder. Int J Cancer 2000;86:501–5. [DOI] [PubMed] [Google Scholar]
- 41.Habuchi T, Devlin J, Elder PA, et al. Detailed deletion mapping of chromosome 9q in bladder cancer: evidence for two tumour suppressor loci. Oncogene 1995;11:1671–4. [PubMed] [Google Scholar]
- 42.Simoneau M, Aboulkassim TO, LaRue H, et al. Four tumor suppressor loci on chromosome 9q in bladder cancer: evidence for two novel candidate regions at 9q22.3 and 9q31. Oncogene 1999;18:157–63. [DOI] [PubMed] [Google Scholar]
- 43.Czerniak B, Chaturvedi V, Li L, et al. Superimposed histologic and genetic mapping of chromosome 9 in progression of human urinary bladder neoplasia: implications for a genetic model of multistep urothelial carcinogenesis and early detection of urinary bladder cancer. Oncogene 1999;18:1185–96. [DOI] [PubMed] [Google Scholar]
- 44.Fearon ER, Feinberg AP, Hamilton SH, et al. Loss of genes on the short arm of chromosome 11 in bladder cancer. Nature 1985;318:377–80. [DOI] [PubMed] [Google Scholar]
- 45.Shaw ME, Knowles MA. Deletion mapping of chromosome 11 in carcinoma of the bladder. Genes Chromosomes Cancer 1995;13:1–8. [DOI] [PubMed] [Google Scholar]
- 46.Chang WY-H, Cairns P, Schoenberg MP, et al. Novel suppressor loci on chromosome 14q in primary bladder cancer. Cancer Res 1995;55:3246–9. [PubMed] [Google Scholar]
- 47.Simon R, Burger H, Brinkschmidt C, et al. Chromosomal aberrations associated with invasion in papillary superficial bladder cancer. J Pathol 1998;185:345–51. [DOI] [PubMed] [Google Scholar]
- 48.Richter J, Jiang F, Gorog JP, et al. Marked genetic differences between stage pTa and stage pT1 papillary bladder cancer detected by comparative genomic hybridization. Cancer Res 1997;57:2860–4. [PubMed] [Google Scholar]
- 49.Richter J, Beffa L, Wagner U, et al. Patterns of chromosomal imbalances in advanced urinary bladder cancer detected by comparative genomic hybridization. Am J Pathol 1998;153:1615–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Richter J, Wagner U, Schraml P, et al. Chromosomal imbalances are associated with a high risk of progression in early invasive (pT1) urinary bladder cancer. Cancer Res 1999;59:5687–91. [PubMed] [Google Scholar]
- 51.Hovey RM, Chu L, Balazs M, et al. Genetic alterations in primary bladder cancers and their metastases. Cancer Res 1998;58:3555–60. [PubMed] [Google Scholar]
- 52.Zhao J, Richter J, Wagner U, et al. Chromosomal imbalances in noninvasive papillary bladder neoplasms (pTa). Cancer Res 1999;59:4658–61. [PubMed] [Google Scholar]
- 53.Kallioniemi A, Kallioniemi O-P, Citro G, et al. Identification of gains and losses of DNA sequences in primary bladder cancer by comparative genomic hybridisation. Genes Chromosomes Cancer 1995;12:213–19. [DOI] [PubMed] [Google Scholar]
- 54.Schena M, Shalon D, Heller R, et al. Parallel human genome analysis: microarray-based expression monitoring of 1000 genes. Proc Natl Acad Sci U S A 1996;93:10614–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.DeRisi J, Penland L, Brown PO, et al. Use of a cDNA microarray to analyse gene expression patterns in human cancer. Nat Genet 1996;14:457–60. [DOI] [PubMed] [Google Scholar]
- 56.Pollack JR, Perou CM, Alizadeh AA, et al. Genome-wide analysis of DNA copy-number changes using cDNA microarrays. Nat Genet 1999;23:41–6. [DOI] [PubMed] [Google Scholar]
- 57.Williamson MP, Elder PA, Knowles MA. The spectrum of TP53 mutations in bladder carcinoma. Genes Chromosomes Cancer 1994;9:108–18. [DOI] [PubMed] [Google Scholar]
- 58.Lipponen PK. Over-expression of p53 nuclear oncoprotein in transitional-cell bladder cancer and its prognostic value. Int J Cancer 1993;53:365–70. [DOI] [PubMed] [Google Scholar]
- 59.Esrig D, Elmajian D, Groshen S, et al. Accumulation of nuclear p53 and tumor progression in bladder cancer. N Engl J Med 1994;331:1259–64. [DOI] [PubMed] [Google Scholar]
- 60.Soini Y, Turpeenniemi-Hujanen T, Kamel D, et al. p53 immunohistochemistry in transitional cell carcinoma and dysplasia of the urinary bladder correlates with disease progression. Br J Cancer 1993;68:1029–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sarkis AS, Zhang Z-F, Cordon-Cardo C, et al. p53 nuclear overexpression and disease progression in Ta bladder carcinoma. Int J Oncol 1993;3:355–60. [DOI] [PubMed] [Google Scholar]
- 62.Sarkis AS, Dalbagni G, Cordon-Cardo C, et al. Nuclear overexpression of p53 protein in transitional cell bladder carcinoma: a marker for disease progression. J Natl Cancer Inst 1993;85:53–9. [DOI] [PubMed] [Google Scholar]
- 63.Sherr CJ. Cancer cell cycles. Science 1996;274:1672–7. [DOI] [PubMed] [Google Scholar]
- 64.Levine AJ. p53, the cellular gatekeeper for growth and division. Cell 1997;88:323–31. [DOI] [PubMed] [Google Scholar]
- 65.Chin L, Pomerantz J, DePinho RA. The INK4a/ARF tumor suppressor: one gene—two products—two pathways. Trends Biochem Sci 1998;23:291–6. [DOI] [PubMed] [Google Scholar]
- 66.Pomerantz J, Schreiber-Agus N, Liegeois NJ, et al. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2's inhibition of p53. Cell 1998;92:713–23. [DOI] [PubMed] [Google Scholar]
- 67.Zhang Y, Xiong Y, Yarbrough WG. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell 1998;92:725–34. [DOI] [PubMed] [Google Scholar]
- 68.Haupt Y, Maya R, Kazaz A, et al. Mdm2 promotes the rapid degradation of p53. Nature 1997;387:296–99. [DOI] [PubMed] [Google Scholar]
- 69.Bates S, Phillips AC, Clark PA, et al. p14ARF links the tumour suppressors RB and p53. Nature 1998;395:124–5. [DOI] [PubMed] [Google Scholar]
- 70.Palmero I, Pantoja C, Serrano M. p19ARF links the tumour suppressor p53 to Ras. Nature 1998;395:125–6. [DOI] [PubMed] [Google Scholar]
- 71.Serrano M, Lin AW, McCurrach ME, et al. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 1997;88:593–602. [DOI] [PubMed] [Google Scholar]
- 72.Stein JP, Ginsberg DA, Grossfeld GD, et al. Effect of p21WAF1/CIP1 expression on tumor progression in bladder cancer. J Natl Cancer Inst 1998;90:1072–9. [DOI] [PubMed] [Google Scholar]
- 73.Sigalas I, Calvert AH, Anderson JJ, et al. Alternatively spliced mdm2 transcripts with loss of p53 binding domain sequences: transforming ability and frequent detection in human cancer. Nat Med 1996;2:912–17. [DOI] [PubMed] [Google Scholar]
- 74.Markl ID, Jones PA. Presence and location of TP53 mutation determines pattern of CDKN2A/ARF pathway inactivation in bladder cancer. Cancer Res 1998;58:5348–53. [PubMed] [Google Scholar]
- 75.Grossman HB, Liebert M, Antelo M, et al. p53 and RB expression predict progression in T1 bladder cancer. Clin Cancer Res 1998;4:829–34. [PubMed] [Google Scholar]
- 76.Cote RJ, Dunn MD, Chatterjee SJ, et al. Elevated and absent pRb expression is associated with bladder cancer progression and has cooperative effects with p53. Cancer Res 1998;58:1090–4. [PubMed] [Google Scholar]
- 77.Cordon-Cardo C, Zhang Z-F, Dalbagni G, et al. Cooperative effects of p53 and pRB alterations in primary superficial bladder tumors. Cancer Res 1997;57:1217–21. [PubMed] [Google Scholar]
- 78.Harnden P, Mahmood J, Southgate J. Cytokeratin 20 expression redefines uroepithelial papillomas of the bladder. Lancet 1999;353:974–7. [DOI] [PubMed] [Google Scholar]
- 79.Kononen J, Bubendorf L, Kallioniemi A, et al. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med 1998;4:844–7. [DOI] [PubMed] [Google Scholar]
- 80.Emmert-Buck MR, Bonner RF, Smith PD, et al. Laser capture microdissection. Science 1996;274:998–1001. [DOI] [PubMed] [Google Scholar]
- 81.Alizadeh A, Elsen M, Davis R, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 2000;403:503–11. [DOI] [PubMed] [Google Scholar]