Catalysis of cis/trans isomerization in native HIV-1 capsid by human cyclophilin A (original) (raw)
Abstract
Packaging of cyclophilin A (CypA) into HIV-1 virions is essential for efficient replication; however, the reason for this is unknown. Incorporation is mediated through binding to the Gly-89–Pro-90 peptide bond of the N-terminal domain of HIV-1 capsid (CAN). Despite the fact that CypA is a peptidyl-prolyl cis/trans isomerase, catalytic activity on CAN has not been observed previously. We show here, using NMR exchange spectroscopy, that CypA does not only bind to CAN but also catalyzes efficiently the cis/trans isomerization of the Gly-89–Pro-90 peptide bond. In addition, conformational changes in CAN distal to the CypA binding loop are observed on CypA binding and catalysis. The results provide experimental evidence for efficient CypA catalysis on a natively folded and biologically relevant protein substrate.
An interaction between the HIV-1 Gag-encoded capsid (CA) protein and the prolyl isomerase cyclophilin A (CypA) is required for efficient HIV-1 replication (1). As a result of this interaction, CypA is packaged into HIV-1 virions during viral replication at a molar ratio of 1:10 CypA/CA (2–4). CypA is thought to be involved in an early step of HIV-1 replication (5, 6); however, its precise role has not been determined.
CypA belongs to the highly conserved cyclophilin family, members of which are abundantly expressed within many tissue types in both eukaryotic and prokaryotic organisms (7). Cyclophilins are peptidyl prolyl cis/trans isomerases (PPIases) (8, 9) that catalyze the cis/trans isomerization of prolyl peptide bonds (Xaa-Pro), where Xaa can be any amino acid. The cis/trans isomerization by CypA has been extensively characterized using test peptides in vitro (8,10–12). Despite the ubiquitous nature of PPIases, the biological significance of their isomerase activity in vivo remains elusive and controversial (13–15). In vitro evidence for isomerase activity exists mainly with respect to the acceleration of proline-limited steps in protein folding (7, 16). However, in vivo evidence for this activity is sparse (16). An example of a putative folding catalyst is the trigger factor, a ribosome-associated prolyl isomerase that binds nascent polypeptide chains during protein synthesis (17, 18). Human CypA, a cytosolic isomerase, initially gained recognition when it was found to be the target of the immunosuppressive drug cyclosporin A (CsA) (19, 20). The inhibition of PPIase activity by CsA is not related to its immunosuppressive role. Instead, the CypA/CsA complex inhibits the phosphatase calcineurin (21), which in turn suppresses T cell activation.
CypA binds to the Gly-89–Pro-90 (G89-P90) prolyl peptide bond located in a flexible loop within the N-terminal domain of CA (CAN) (22, 23). The CAN/CypA interface is formed exclusively through the CA loop sequence Pro-85–Pro-93 (22, 24). The significance of the CA/CypA interaction for HIV-1 virulence has eluded the field for almost a decade. Mutation of either G89 or P90, or addition of CsA, prevents CypA packaging into HIV-1 virions (3, 25), leading to the formation of noninfectious particles. Structural studies of CAN by NMR revealed that only the G89-P90 peptide bond exists in both a cis (14%) and trans (86%) conformation (23). Interestingly, only a few proteins are known to exhibit such conformational heterogeneity for a prolyl peptide bond in the folded state (ref. 26 and references therein) and, thus far, catalysis of those prolyl peptide bonds by PPIases could not be detected when assayed (27). An intriguing hypothesis for the role of CypA in HIV-1 virulence is that it catalyzes cis/trans isomerization about G89-P90 in CAN, resulting in a conformational change necessary for CA core disassembly (23). However, because only the trans conformation of G89-P90 is bound to CypA in the CAN/CypA cocrystal structure, a general view is that CypA most likely plays a chaperone role rather than a catalytic role in HIV-1 virulence (28–30). Here, we present direct experimental evidence for CypA catalysis of the isomerization about the G89-P90 peptide bond in CAN. To our knowledge, there is no previous direct evidence of CypA isomerase activity on a folded, biologically relevant protein substrate. In addition, we observe chemical shift changes for CAN residues distal from the CypA binding loop upon CypA wild-type (CypAWT) and CypAR55A binding that are indicative of conformational changes not predicted by the CAN/CypA cocrystal structure (22). The results discussed herein suggest that CypA may have a catalytic role in HIV-1 replication.
Materials and Methods
Protein Purification and Sample Preparation.
Vectors for expressing human CypA and CAN(1–146) have been described (24). CypAR55A was constructed using a quick-change protocol from Stratagene. Proteins were expressed in BL21 (DE3) Escherichia coli cells, induced with 1 mM isopropyl β-d-thiogalactoside (IPTG), and harvested after 3 h. Uniformly,15N-labeled CAN was grown in M9 minimal media supplemented with15NH4Cl. CypA was purified using an S-Sepharose column equilibrated with 25 mM Mes (pH 6.1)/5 mM β-mercaptoethanol, followed by a QP-Sepharose column equilibrated with 50 mM Tris (pH 6.8)/5 mM β-mercaptoethanol. CypA activity was measured by the coupled chymotrypsin assay (8). CAN was purified using an S-Sepharose column equilibrated with 100 mM citric acid (pH 4.5)/1 mM DTT, followed by a QP-Sepharose column equilibrated with 75 mM Tris (pH 8.0)/1 mM DTT. Peaks containing CAN were collected from the flow through. Finally, the buffer of each protein solution was exchanged to 50 mM Na2HPO4 (pH 6.5)/1 mM DTT containing 10% D2O.
NMR samples for 1H-15N heteronuclear exchange and three-dimensional (3D)15N edited nuclear Overhauser effect spectroscopy (NOESY)–heteronuclear single quantum coherence (HSQC) experiments were prepared with a molecular ratio of 12:1 (CAN/CypA). Samples inhibited by CsA (Sigma–Aldrich) were incubated overnight in the presence of excess CsA. For the titration of 15N CAN with unlabeled CypAWTand CypAR55A, a series of titration points were collected ranging from 1:12 CypA/CAN to 6:1.
NMR Experiments.
Two-dimensional (2D) 1H-15N heteronuclear exchange (31), 3D 15N edited NOESY-HSQC (32), and transverse relaxation-optimized spectroscopy (TROSY)-HSQC (33) spectra were collected on Varian INOVA 600 MHz and 500 MHz spectrometers at 25°C. All NMR spectra were processed with NMRPipe (34). The assignments for free CAN were provided by M. Summers (23). The CypA catalyzed exchange rate could be determined from the 3D 15N-edited NOESY-HSQC according to ref. 35, because the longitudinal relaxation times for the cis and trans isomer were measured to be equal. ANSIG software (36) was used to compare spectra of15N-labeled CAN in the absence and presence of unlabeled CypA. The chemical shift differences between the amide signals of CAN alone and in the presence of saturating CypA were calculated as follows:
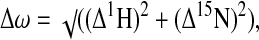
where Δ1H and Δ15N are the chemicals shift differences in the proton and nitrogen dimensions, respectively.
Results
Catalysis of Cis/Trans Isomerization of the G89-P90 Peptide Bond by CypA.
NMR exchange spectroscopy was used to detect catalysis of cis/trans isomerization in CAN by CypA. This technique identifies conformational exchange processes in a time regime of about 10−1 to 10 s between nuclei with different chemical shifts (37). In contrast to most spectroscopic methods, NMR can be used to measure dynamics under steady state or equilibrium conditions, making NMR exchange spectroscopy a powerful tool for analyzing the reversible cis/trans isomerization (11, 12). As previously shown, separate resonance signals can be observed for the cis (14%) and trans (86%) conformation of several amide resonances in close proximity to the G89-P90 peptide bond in CAN (23), because each conformation resides in distinct chemical environments, and because cis/trans isomerization is slow on the NMR time scale (rate constant of exchange,_k_ex < 0.1 s−1).
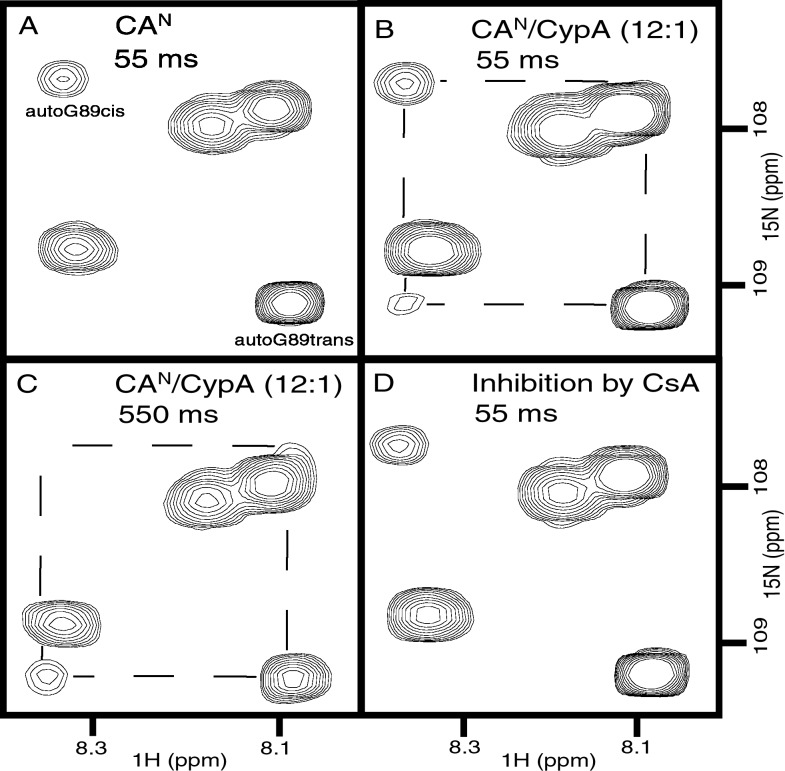
To detect catalysis of cis/trans interconversion by CypA, we performed 2D 1H-15N heteronuclear exchange experiments (31) on a uniformly15N-labeled CAN sample in the absence and presence of catalytic amounts of unlabeled CypA (Fig.1). Because the uncatalyzed isomerization rate is within the slow-time-scale regime, only the individual peaks (auto peaks) for the cis and trans conformer are observed in the absence of CypA. Addition of catalytic amounts of CypA results in the appearance of exchange peaks connecting G89 cis and trans auto peaks (Fig. 1 B and C). These results show that cis/trans isomerization about the G89-P90 peptide bond is efficiently catalyzed by CypA. Because of peak overlap, only one exchange peak is completely resolved in the exchange spectrum (Fig. 1 B and_C_). Exchange peaks are not observed after inhibition of the isomerase activity by CsA (Fig. 1D), or when the catalytically inactive CypAR55A mutant is used in place of CypAWT (data not shown), thus implicating CypA catalysis as the cause for these exchange peaks. These data provide direct evidence that CypA catalyzes the cis/trans isomerization about the G89-P90 peptide bond in native CAN.
Figure 1.
CypA catalysis of cis/trans isomerization at G89-P90 in folded CAN. Expansion of 1H-15N heteronuclear exchange spectra showing the amide signal of G89 for the trans and cis isomer, respectively. Chemical exchange between the cis and trans conformation of G89-P90 is slow in the absence of CypA (A) and is accelerated in the presence of catalytic amounts of CypA, as indicated by the appearance of exchange peaks between the cis and the trans auto peaks (B and_C_). The intensity of the exchange peak increases with longer mixing time and concurrently the less abundant cis auto peak decreases because of additional loss in magnetization from chemical exchange and longitudinal relaxation (C). Inhibition of isomerase activity by CsA results in a loss of the exchange peaks (D). The mixing times used in NMR experiments are indicated.
Quantification of Catalysis.
The 2D 1H-15N heteronuclear exchange experiment results (Fig. 1) are further substantiated by the exchange spectra obtained from a 3D 15N-edited NOESY-HSQC experiment (Fig. 2). A chemical exchange peak at 1H 8.0,1H 8.4, and 15N 109.3 ppm is clearly visible in the presence of catalytic amounts of CypA. From this experiment, the exchange rate is calculated as 10 ± 5 s−1 at 25°C from the ratio of the off-diagonal (exchange) peaks to the diagonal (auto) peaks (35). Only an upper limit of the uncatalyzed exchange rate can be estimated as 0.1 s−1, consistent with cis/trans isomerization rates of Gly-Pro peptide bonds measured in peptides (38). An apparent acceleration factor of at least 100-fold can be calculated for the CypA concentration used. The exchange peak is not observed after inhibition of the isomerase activity by CsA (Fig. 2B). In the absence of CypA, the exchange spectrum is identical to that obtained with CsA (data not shown).
Figure 2.
Quantification of CypA catalysis on CAN and inhibition by CsA. 1H/1H plane of a 3D15N-edited NOESY-HSQC shows the diagonal peak of amide G89trans at 8.0 (1H) and 109.3 ppm (15N). (A) An off-diagonal exchange peak between the amide proton of G89 in the trans and cis conformation is evidence for CypA catalysis and its intensity was used to calculate a cis/trans isomerization rate of 10 ± 5 s−1. (B) Addition of excess CsA to sample (A) results in a loss of the exchange peak because of inhibition of isomerase activity. Spectrum B is identical to a spectrum of CAN alone (data not shown).
Characterization of the Catalytically Active CAN/CypA Complex by NMR.
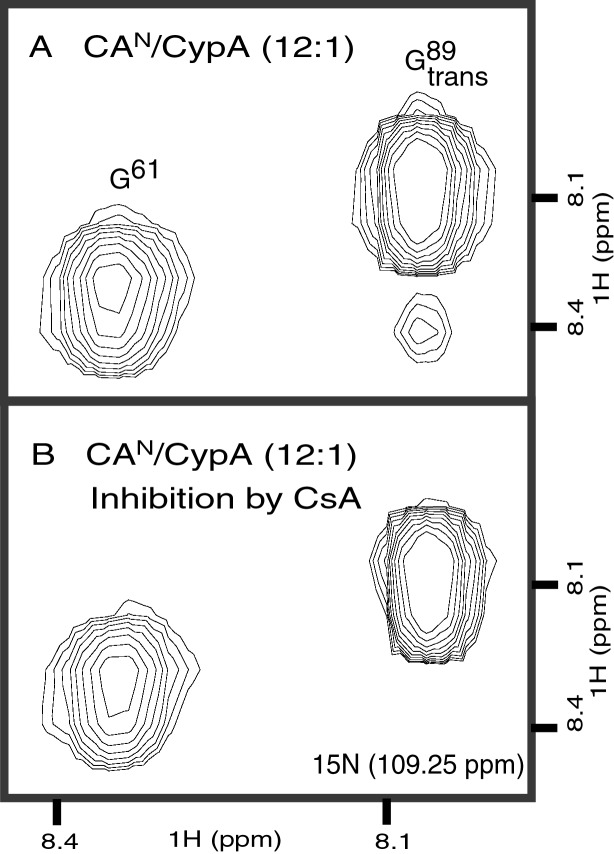
In the experiments showing the catalytic action of CypA (described above), only catalytic amounts of CypA were used. We now want to characterize the CAN/CypA complex and therefore performed NMR experiments at saturating concentrations of CypA. The catalysis of CAN at G89-P90 by CypA indicates that CypA must bind both the trans and cis conformations of that peptide bond. In contrast, only the trans conformation is seen in the cocrystal structure of the CAN/CypA complex. In an effort to gain structural information about the active CAN/CypA complex in solution, a titration of15N-labeled CAN with unlabeled CypA was performed. The expected molecular weight of the CAN/CypA complex is ≈34 kDa. Because of the relatively large molecular weight of the complex,1H-15N HSQC-TROSY (33) was used. The TROSY method effectively suppresses transverse (_T_2) relaxation, thereby resulting in improved spectral resolution for large macromolecules (39). On addition of increasing amounts of CypA, several amide resonances of CAN disappear and reappear at different chemical shift locations, indicating slow exchange between free and bound CAN. This finding is in agreement with a_K_D of 15 μM as measured by plasmon resonance biosensor and isothermal titration calorimetry experiments (24). The relatively high affinity between CypA and CAN enabled us to obtain a 95% saturation of CAN with CypA (Fig.3), but prevented us from unambiguously following the chemical shifts. A comparison of free CAN with the CAN/CypA complex identified all residues that change in chemical shift in the active complex (Fig. 3 A and B).
Figure 3.
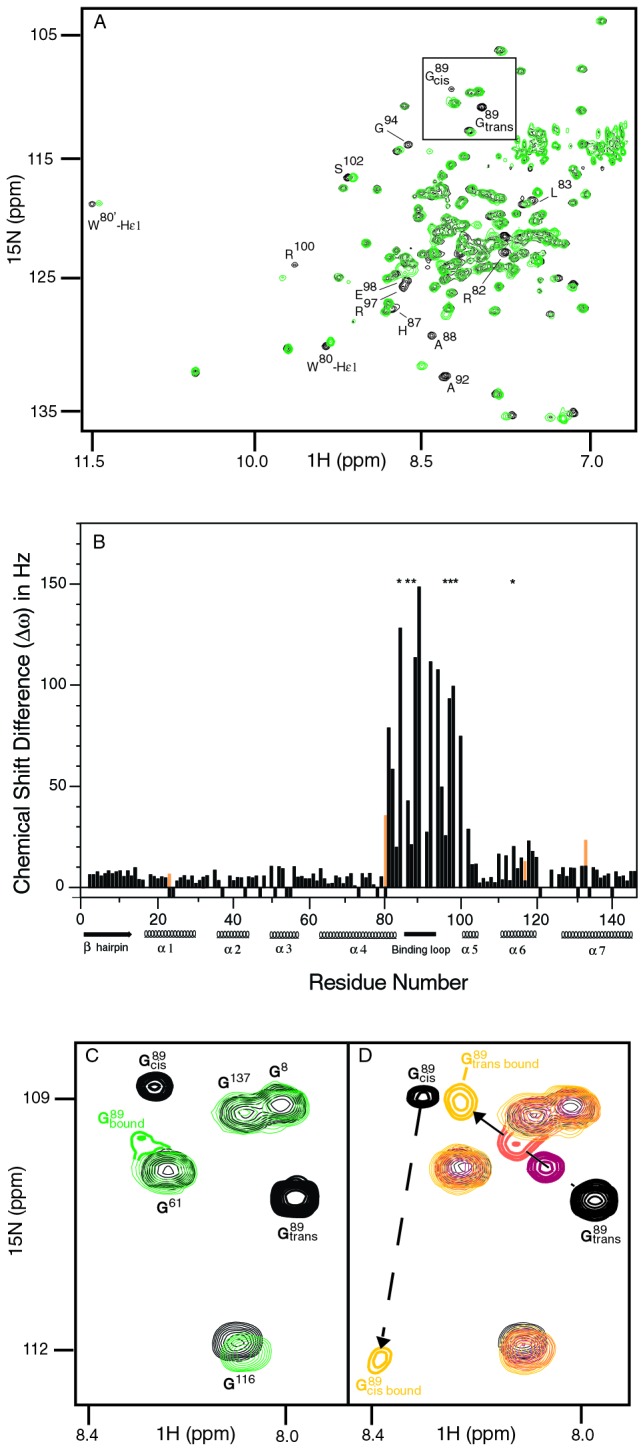
Characterization of conformational changes in CAN upon catalysis and/or binding to CypA. (A) Comparison of1H-15N HSQC-TROSY spectra for CANalone (black) and saturated with CypAWT (green) illustrates that residues remote from the CyPA binding loop are affected by CyPA binding and/or catalysis. (B) The magnitudes of chemical shift changes for the CAN amides on complex formation with CypAWT (Δω) are shown as a bar graph. Secondary structure elements and the CypA binding loop (85–93) are labeled. Residues for which Δω is at least 23 Hz, but which could not be unambiguously determined because of signal overlap, are labeled with an asterisk. Chemical shift changes for the side chain NH of Trp 23, 80, 117, and 133 are shown in orange. An expanded view of the G89 region, highlighted in the full spectrum A, is shown in_C_ and D. At saturating concentrations of CypAWT (CAN/CypA molar ratio of 1:2), the cis- and trans-bound forms resonate at an average chemical shift because of fast chemical exchange between the two isomers (C). Conversely, both the cis and trans conformers of G89 are observed in the enzyme-bound state when the titration is performed with catalytically inactive CyPAR55A(D). The chemical shift changes for G89transcan be followed on titration with CyPAR55A as shown for ratios of CAN/CypAR55A at 1:1 (red), 1:2 (orange), and 1:6 (yellow). However, for G89cis, the bound form is only observed at higher CyPAR55A/CANratios in agreement with a higher affinity of the CAN cis isomer for CypA.
Surprisingly, a number of amide signals in CANhave altered chemical shifts when bound to and catalyzed by CypA. Based on the CAN/CypA cocrystal structure, it has been proposed that only residues 85–93 within CAN are affected by CypA binding (22). However, in addition to the expected chemical shift changes within this flexible binding loop, chemical shift changes also occur within helices α 4, 5, and 6 (Fig. 4). Chemical shift changes are very sensitive markers for conformational changes; however, they do not provide a physical picture of the altered structure.
Figure 4.
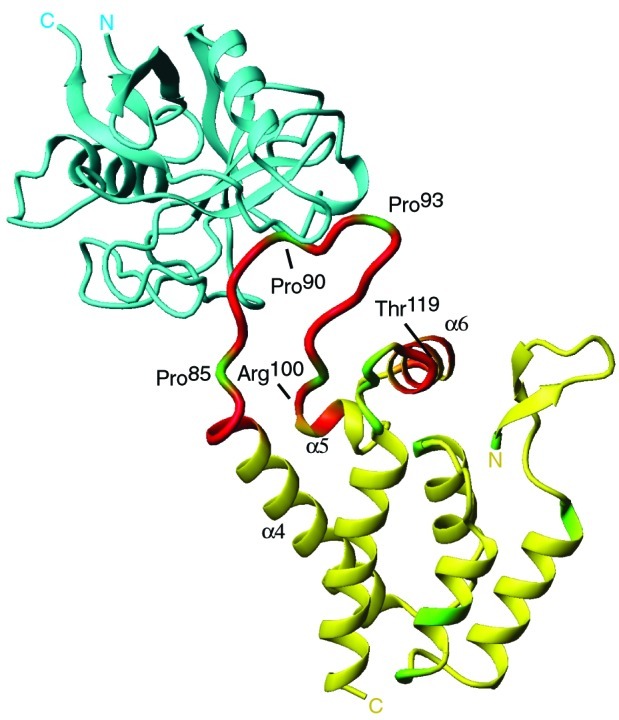
Identification of residues within CAN that change in chemical shift on binding and catalysis by CypA (22). CypA (blue) is shown bound to the flexible loop between Pro-85 and Pro-93 of CAN (yellow). Backbone amides of CAN that shift on CypA binding by more than 14 Hz are highlighted in red. These residues are not only located within the flexible loop (85–100), but also in α4, α5, and α6. Prolines within CAN (1–146) are shown in green.
Dissecting CypA Catalysis and Binding.
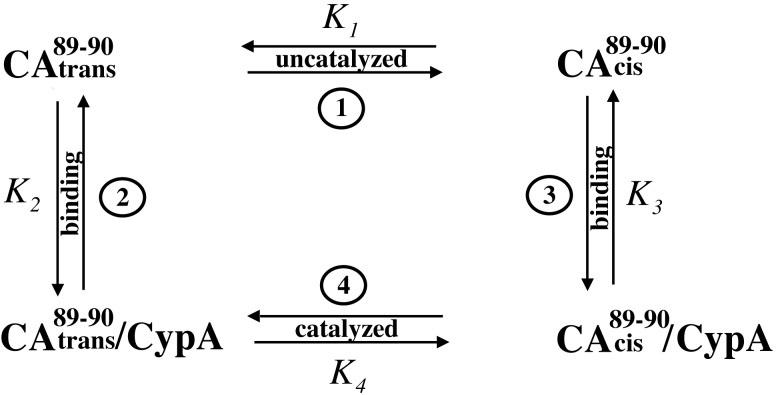
The minimal reaction scheme for CypA catalyzed cis/trans isomerization of CAN is shown in SchemeS1. In the presence of CypA, at least four different states of CAN exist in solution. However, the two CypA-bound forms of CAN cannot be separated because the cis/trans isomerization in the enzyme-bound states is fast relative to the chemical shift difference between the two states, Δω. Thus, when saturated with CypAWT, chemical shift averaging for the enzyme-bound cis and trans conformers of G89 is observed (Fig.3C, Scheme S1). To dissect binding from catalysis, CypAR55A was used with15N-labeled CAN in titration experiments similar to those described above for CypAWT. It was shown previously that the CypA active site mutation R55A exhibits <1% wild-type activity (40). As expected, the bound cis and trans conformations of CAN G89-P90 can now be separately detected in the complex with CypAR55A (Fig. 3D). Although the binding events (characterized by_K_2 and_K_3) do occur for CypAR55A and CAN, an approximate 100-fold reduction in binding affinity is observed for the mutant protein. An apparent _K_D of ≈1.3 mM was calculated based on the NMR titration experiments. The weak binding affinity of CypAR55A for CAN may explain why this mutant is not incorporated into HIV-1 virions (41, 42). At a molar ratio of 6:1 CypAR55A/CAN, approximately 60% of CAN is bound to CypAR55A, as compared with 95% CAN bound to CypA at a ratio of 2:1 CypA/CAN.
Scheme 1.
Four-state model of CypA catalysis on CAN. The capsid protein exists in two free forms with the G80-P90 peptide bond in the trans (CA ) and cis conformation (CA
) and cis conformation (CA ). Both forms bind to CypA and are interconverted on the enzyme. The rate of uncatalyzed cis/trans isomerization is slow, whereas the catalyzed rate is fast, resulting in an average chemical shift for the two enzyme-bound forms. The relative affinities of the two isomers of CAN for CypA determine the cis/trans equilibrium constant on the enzyme,_K_4.
). Both forms bind to CypA and are interconverted on the enzyme. The rate of uncatalyzed cis/trans isomerization is slow, whereas the catalyzed rate is fast, resulting in an average chemical shift for the two enzyme-bound forms. The relative affinities of the two isomers of CAN for CypA determine the cis/trans equilibrium constant on the enzyme,_K_4.
Overall, the same CAN amide signals shift when the titration is performed with CypAWT or with CypAR55A. In addition, these CAN residues shift toward similar chemical shift locations, taking into account that CAN is only ≈60% saturated by CypAR55A and that there may be minor effects due to the altered chemical environment as a result of the mutation. We therefore conclude that the mutant protein CypAR55A is a suitable model for a qualitative study of the binding events between CAN and CypAWT.
Different chemical shifts are detected for the backbone amides of residues 88, 89, 92, and 95 for the two enzyme-bound forms of CAN on titration with CypAR55A. This observation suggests that the chemical environment for residues in this region is different depending on whether they are bound to CypA in the cis or the trans conformation. For example, the chemical shifts for the cis and trans conformations of A92 are partially overlapped when CAN is free in solution, but a relatively large chemical shift difference between the cis and trans conformations is observed when bound to CypAR55A. For all other amides, no differences in chemical shift between the cis- and trans-bound states could be measured. In addition, the percentage of the cis conformer appears to increase from 14% free in solution to ≈45% when bound to CypAR55A based on the relative signal intensities of the trans- and cis-bound form of CAN (Fig. 3D). This population change corresponds to a 5-fold difference in binding affinity for the two conformers. A higher affinity of the cis conformer is further supported by the fact that the signal of G89 trans shifts with increasing concentrations of CypAR55A, whereas the signal of the cis conformer disappears and then reappears (Fig.3D). In other words, exchange between free and CypA-bound CAN is faster than the chemical shift difference for the trans isomer, but slower for the cis isomer. We note that the equilibrium constant between the CAN cis and trans isomer when bound to CypAWT(_K_4 in Scheme S1) cannot be directly measured based on the binding studies with CypAR55A. However, the titration experiments with CypAWT reveal that both isomers have comparable affinities: signals from the cis and trans isomer of CAN decrease about equally in intensity and later appear at an average chemical shift for the two enzyme-bound isomers during titration with CypAWT (Fig.3C). Furthermore, the average peak position for the two enzyme-bound isomers can be used to approximate the equilibrium constant _K_4 for the CA/CypAWT complex. For amides of residues 88, 89, 92, and 95, the average peak is located on a trajectory between the cis and trans-bound form of the CypAR55A/CA complex, but shifted toward the position of the trans-bound form (Figs.3 C and D). Because the average peak position is directly determined by the relative populations of the two states, a cis/trans equilibrium constant for the CA/CypAWT complex can be estimated as similar to that of CAN free in solution (14% cis/86% trans). The change in affinity of the two isomers of CAN for CypAR55A is likely a direct result of the mutation, because the side chain of R55 within CypA is in hydrogen bond distance to CAN (22).
Discussion
The detailed mechanism of how cyclophilin A promotes HIV-1 replication is not understood, although several mechanisms have been discussed in recent years (5, 6, 22, 23, 28, 30, 43–45). In light of the CAN/CypA cocrystal structure showing CA G89-P90 bound exclusively in the trans conformation (22), models depicting CypA as having a chaperone role in HIV-1 virulence have been suggested (28). In fact, a chaperone role has been ascribed to other PPIases (46), and there is speculation as to whether the isomerase activity of PPIases is relevant to their biological function in vivo (13, 15, 26). In contrast, our results show that CypA does not simply bind to CAN, but efficiently catalyzes the cis/trans isomerization about the G89-P90 bond. The finding that CypA can catalyze cis/trans isomerization in a folded protein may be of more general importance for elucidating the biological mode of action of PPIases on other protein substrates.
CypA Catalysis and HIV-1 Virulence.
What are the implications of CypA catalysis for HIV-1 virulence? The present data cannot answer the question of whether the catalytic activity or only pure binding by CypA is responsible for HIV-1 virulence. However, the identification of catalytic action by CypA on CAN justifies further consideration of a catalytic role for CypA in the enhancement of HIV-1 replication. Dissecting binding from catalysis in the CA/CypA interaction is difficult because mutations rendering CypA catalytically inactive also diminish the ability for Gag to incorporate CypA into HIV-1 virions (41, 42). The fact that the CypA/Gag ratio is substoichiometric in virions is consistent with CypA functioning as a catalyst. Increasing the CypA/Gag ratio to 1:1 ratio actually alters the efficacy of HIV-1 replication (29, 47). Furthermore, the kinetics of HIV-1 replication is largely influenced by the presence of CypA, where replication will still occur in the absence of CypA albeit at a slower rate (1). This finding suggests a kinetic control by CypA rather than an absolute requirement of CypA for replication, again consistent with a catalytic role.
An elegant experiment by Bukovsky et al. (48) provided_in vivo_ evidence that the CypA binding loop in CAN is not only responsible for CypA incorporation but is a functional target for CypA. Transfer of the segment between residues 86 and 93 from HIV CA into the corresponding position in simian immunodeficiency virus SIVmac resulted in efficient incorporation of CypA into SIVmac. These SIVmac viruses now showed HIV-1-like sensitivity toward a nonimmunosuppressive CsA analog. The authors further propose that the ability of residues C-terminal to P90 to form a type II turn determines whether CypA is required or will inhibit viral replication. These data support a model where this CAN type II turn is important for the functional role of CypA in HIV-1 replication, possibly for the destabilization of CA–CA interactions (48), which has also been proposed for the CsA-resistant CAA92E and CAG94D mutants (28, 49, 50). Our results lend support to this model. Interestingly, we observe chemical-shift differences between the cis and trans isomer when bound to CypAR55A for residues within 88–95, indicating that the chemical environment of the type II turn is sensitive to the conformation of the G89-P90 peptide bond. Thus, binding and catalysis by CypA could alter a conformation involving the type II tight turn formed by residues A92 through Q95. In fact, a crucial NOE indicative of a type II turn is absent in the CsA resistant mutant CAG94D (51). Alternatively, the binding event alone between CA and CypA may be sufficient for viral replication. Amide signals of residues distal from the CypA binding loop in CAN are shifted upon binding to CypA, suggesting that the interaction with CypA may result in conformational changes outside the flexible loop. Some of the observed changes in chemical shift could be a result of a hinge motion about CAN residues 86–98 (22). It is possible that the conformational changes due to binding could effectively disrupt CA–CA interactions in the virion core, and facilitate CA core disassembly. It remains to be determined whether the catalysis of CAN by CypA is relevant to HIV-1 virulence_in vivo_.
Catalysis of Cis/Trans Isomerization by Cyclophilin in a Folded Protein.
PPIases were originally discovered as enzymes that accelerate folding of certain proteins by catalyzing the rate limiting prolyl cis/trans isomerization (8). Their catalytic activity toward unfolded or partially folded proteins has been extensively studied in vitro (7, 15, 16) and has been suggested to be the biological function of the ribosome-associated PPIase trigger factor (17, 18). However, there are a growing number of examples where PPIases are involved in different cellular events (ref. 26 and references therein, refs. 52–55), in which they interact with protein substrates in their folded state. Consequently, the traditional view of PPIase-function needs to be extended to binding and catalysis of prolyl peptide bonds in native substrates.
There are two fundamentally different ways PPIases may control biological activity of a folded protein substrate: (i) proline-directed binding or (ii) catalysis of cis/trans isomerization (26). For the systems studied to date, the molecular mechanism of PPIase action is poorly understood, and the dissection between the two different modes of action has proven difficult. Moreover, there are a few reports on proteins containing prolyl peptide bonds exhibiting conformational heterogeneity in the folded state (ref.26 and references therein), a circumstance that most likely reflects the difficulty in detecting native state cis/trans isomerization. In Calbindin D9k, conformational heterogeneity for the G42-P43 peptide was detected (56); however, CypA catalysis was not observed (27).
PPIases have been implicated in the acceleration of cis/trans isomerization for several other proteins, where the conformation of the peptide bond appears to regulate the function (26). For the Ca2+-free form of a C-type mannose binding protein, addition of equimolar amounts of CypA slightly increased the isomerization rate (57). Using 8-anilino-1-naphthalenesulfonic acid (ANS) binding to probe induced-fit conformational changes associated with ATP binding in rabbit muscle adenylate kinase, Sheng et al. showed that CypA accelerates ANS binding (58). In both cases, the biological relevance of prolyl isomerization remains unclear. In vitro and in vivo evidence indicates a possible biological involvement of the FKBP12 PPIase in the inhibition of epidermal growth factor (EGF) (59). A more compelling example of a PPIase that accelerates cis/trans isomerization of prolyl peptide bonds in biologically relevant protein substrates is Pin1. Pin1 is essential for cell cycle regulation _in vivo_via its interaction with the mitosis specific protein cdc25 (60, 61). It is believed that Pin1 catalysis of cis/trans isomerization about phosphorylated Ser-Pro or Thr-Pro peptide bonds in cdc25 and tau, another protein substrate of Pin1, accelerates the dephosphorylation by the phosphatase PP2A (52, 62, 63). For all of the examples described above, the concentration of PPIase required to significantly accelerate cis/trans isomerization of the respective protein substrate was not in catalytic proportions, and rate enhancements were either very small (less than 2-fold; refs. 57 and 58) or the reaction involving the PPIase was completed on the time scale of minutes (52, 59). A recent report shows that substoichiometric amounts of Pin1 can catalyze a conformational change within cdc25 (62).
In contrast to these reports, the catalysis of HIV-1 CAN by CypA described here is directly visualized, and is found to exhibit an acceleration factor of at least 100-fold when using a substoichiometric CypA/CAN ratio of 1:12. Taken together, the interaction between CypA and CAN represents the first example of efficient CypA catalysis on a natively folded and biologically relevant protein substrate. It will be of great interest to further investigate the role of binding versus catalysis for the functions of other PPIases with respect to their natural protein substrates.
Conclusions.
The importance of the host protein CypA for HIV-1 virulence has been extensively stated and studied in the last few years. The crystal structure of CAN in complex with CypA provided a detailed atomic resolution picture of the interface. However, all present data do not explain the mode of action of CypA for virulence. Our results illustrating the catalytic action of CypA on CA call for a more serious consideration of CypA catalysis in HIV-1 replication. Solution NMR enabled not only the detection of catalysis, but also a characterization of both the cis- and trans-bound forms of CAN. The lack of information about catalysis and the cis-bound form provided by the CAN/CypA crystal structure is most likely due to trapping one of the existing conformers, a phenomenon often seen in crystallography. Furthermore, monitoring the active complex in solution identified residues in CAN outside the CypA binding loop that are affected by binding. The results shown in this paper, together with the atomic resolution structure of the CAN/CypA complex, demonstrate how x-ray crystallography and NMR spectroscopy complement each other. Ultimately, additional _in vivo_experiments are needed to elucidate the significance of catalysis and/or binding of the different conformers by CypA for HIV-1 replication.
Acknowledgments
We thank B. Chamberlin for technical assistance. This work was supported by National Institutes of Health Grants GM62117 (to D.K.) and AI40333 (to W.I.S.), and instrumentation grants awarded by the National Science Foundation and the Keck Foundation (to D.K.).
Abbreviations
CypA
cyclophilin A
CAN
capsid N-terminal domain
CsA
cyclosporin A
PPIase
peptidyl prolyl cis/trans isomerase
NOESY
nuclear Overhauser effect spectroscopy
HSQC
heteronuclear single quantum coherence
3D
three-dimensional
G89-P90
Gly-89–Pro-90
References
- 1.Braaten D, Luban J. EMBO J. 2001;20:1300–1309. doi: 10.1093/emboj/20.6.1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luban J, Bossolt K L, Franke E K, Kalpana G V, Goff S P. Cell. 1993;73:1067–1078. doi: 10.1016/0092-8674(93)90637-6. [DOI] [PubMed] [Google Scholar]
- 3.Franke E K, Yuan H E, Luban J. Nature (London) 1994;372:359–362. doi: 10.1038/372359a0. [DOI] [PubMed] [Google Scholar]
- 4.Thali M, Bukovsky A, Kondo E, Rosenwirth B, Walsh C T, Sodroski J, Gottlinger H G. Nature (London) 1994;372:363–365. doi: 10.1038/372363a0. [DOI] [PubMed] [Google Scholar]
- 5.Braaten D, Franke E K, Luban J. J Virol. 1996;70:3551–3560. doi: 10.1128/jvi.70.6.3551-3560.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fitzon T, Leschonsky B, Bieler K, Paulus C, Schroder J, Wolf H, Wagner R. Virology. 2000;268:294–307. doi: 10.1006/viro.1999.0178. [DOI] [PubMed] [Google Scholar]
- 7.Gothel S F, Marahiel M A. Cell Mol Life Sci. 1999;55:423–436. doi: 10.1007/s000180050299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fischer G, Bang H, Mech C. Biomed Biochim Acta. 1984;43:1101–1111. [PubMed] [Google Scholar]
- 9.Fischer G, Wittmann-Liebold B, Lang K, Kiefhaber T, Schmid F X. Nature (London) 1989;337:476–478. doi: 10.1038/337476a0. [DOI] [PubMed] [Google Scholar]
- 10.Harrison R K, Stein R L. Biochemistry. 1990;29:3813–3816. doi: 10.1021/bi00468a001. [DOI] [PubMed] [Google Scholar]
- 11.Kern D, Drakenberg T, Wikstrom M, Forsen S, Bang H, Fischer G. FEBS Lett. 1993;323:198–202. doi: 10.1016/0014-5793(93)81338-z. [DOI] [PubMed] [Google Scholar]
- 12.Kern D, Kern G, Scherer G, Fischer G, Drakenberg T. Biochemistry. 1995;34:13594–13602. doi: 10.1021/bi00041a039. [DOI] [PubMed] [Google Scholar]
- 13.Schreiber S L, Crabtree G R. Immunol Today. 1992;13:136–142. doi: 10.1016/0167-5699(92)90111-J. [DOI] [PubMed] [Google Scholar]
- 14.Ivery M T. Med Res Rev. 2000;20:452–484. doi: 10.1002/1098-1128(200011)20:6<452::aid-med2>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 15.Schiene C, Fischer G. Curr Opin Struct Biol. 2000;10:40–45. doi: 10.1016/s0959-440x(99)00046-9. [DOI] [PubMed] [Google Scholar]
- 16.Schmid F X. Curr Biol. 1995;5:993–994. doi: 10.1016/s0960-9822(95)00197-7. [DOI] [PubMed] [Google Scholar]
- 17.Stoller G, Rucknagel K P, Nierhaus K H, Schmid F X, Fischer G, Rahfeld J U. EMBO J. 1995;14:4939–4948. doi: 10.1002/j.1460-2075.1995.tb00177.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deuerling E, Schulze-Specking A, Tomoyasu T, Mogk A, Bukau B. Nature (London) 1999;400:693–696. doi: 10.1038/23301. [DOI] [PubMed] [Google Scholar]
- 19.Handschumacher R E, Harding M W, Rice J, Drugge R J, Speicher D W. Science. 1984;226:544–547. doi: 10.1126/science.6238408. [DOI] [PubMed] [Google Scholar]
- 20.Takahashi N, Hayano T, Suzuki M. Nature (London) 1989;337:473–475. doi: 10.1038/337473a0. [DOI] [PubMed] [Google Scholar]
- 21.Liu J, Farmer J D, Jr, Lane W S, Friedman J, Weissman I, Schreiber S L. Cell. 1991;66:807–815. doi: 10.1016/0092-8674(91)90124-h. [DOI] [PubMed] [Google Scholar]
- 22.Gamble T R, Vajdos F F, Yoo S, Worthylake D K, Houseweart M, Sundquist W I, Hill C P. Cell. 1996;87:1285–1294. doi: 10.1016/s0092-8674(00)81823-1. [DOI] [PubMed] [Google Scholar]
- 23.Gitti R K, Lee B M, Walker J, Summers M F, Yoo S, Sundquist W I. Science. 1996;273:231–235. doi: 10.1126/science.273.5272.231. [DOI] [PubMed] [Google Scholar]
- 24.Yoo S, Myszka D G, Yeh C, McMurray M, Hill C P, Sundquist W I. J Mol Biol. 1997;269:780–795. doi: 10.1006/jmbi.1997.1051. [DOI] [PubMed] [Google Scholar]
- 25.Braaten D, Franke E K, Luban J. J Virol. 1996;70:4220–4227. doi: 10.1128/jvi.70.7.4220-4227.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fischer G, Tradler T, Zarnt T. FEBS Lett. 1998;426:17–20. doi: 10.1016/s0014-5793(98)00242-7. [DOI] [PubMed] [Google Scholar]
- 27.Kordel J, Drakenberg T, Forsen S, Thulin E. FEBS Lett. 1990;263:27–30. doi: 10.1016/0014-5793(90)80697-h. [DOI] [PubMed] [Google Scholar]
- 28.Luban J. Cell. 1996;87:1157–1159. doi: 10.1016/s0092-8674(00)81811-5. [DOI] [PubMed] [Google Scholar]
- 29.Yin L, Braaten D, Luban J. J Virol. 1998;72:6430–6436. doi: 10.1128/jvi.72.8.6430-6436.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wiegers K, Rutter G, Schubert U, Grattinger M, Krausslich H G. Virology. 1999;257:261–274. doi: 10.1006/viro.1999.9669. [DOI] [PubMed] [Google Scholar]
- 31.Farrow N A, Zhang O, Forman-Kay J D, Kay L E. J Biomol NMR. 1994;4:727–734. doi: 10.1007/BF00404280. [DOI] [PubMed] [Google Scholar]
- 32.Sattler M, Schleucher J, Griesinger C. Prog NMR Spectrosc. 1999;34:93–158. [Google Scholar]
- 33.Pervushin K, Riek R, Wider G, Wuthrich K. Proc Natl Acad Sci USA. 1997;94:12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Delaglio F, Grzesiek S, Vuister G W, Zhu G, Pfeifer J, Bax A. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 35.Baine P. Magn Reson Chem. 1986;24:304–307. [Google Scholar]
- 36.Kraulis P J, Domaille P J, Campbell-Burk S L, Van Aken T, Laue E D. Biochemistry. 1994;33:3515–3531. doi: 10.1021/bi00178a008. [DOI] [PubMed] [Google Scholar]
- 37.Perrin C L, Dwyer T J. Chem Rev. 1990;90:935–967. [Google Scholar]
- 38.Wuthrich C G n K. Biopolymers. 1981;20:2623–2633. [Google Scholar]
- 39.Pervushin K. Q Rev Biophys. 2000;33:161–197. doi: 10.1017/s0033583500003619. [DOI] [PubMed] [Google Scholar]
- 40.Zydowsky L D, Etzkorn F A, Chang H Y, Ferguson S B, Stolz L A, Ho S I, Walsh C T. Protein Sci. 1992;1:1092–1099. doi: 10.1002/pro.5560010903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Braaten D, Ansari H, Luban J. J Virol. 1997;71:2107–2113. doi: 10.1128/jvi.71.3.2107-2113.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dorfman T, Weimann A, Borsetti A, Walsh C T, Gottlinger H G. J Virol. 1997;71:7110–7113. doi: 10.1128/jvi.71.9.7110-7113.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Endrich M M, Gehrig P, Gehring H. J Biol Chem. 1999;274:5326–5332. doi: 10.1074/jbc.274.9.5326. [DOI] [PubMed] [Google Scholar]
- 44.Pushkarsky T, Zybarth G, Dubrovsky L, Yurchenko V, Tang H, Guo H, Toole B, Sherry B, Bukrinsky M. Proc Natl Acad Sci USA. 2001;98:6360–6365. doi: 10.1073/pnas.111583198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sherry B, Zybarth G, Alfano M, Dubrovsky L, Mitchell R, Rich D, Ulrich P, Bucala R, Cerami A, Bukrinsky M. Proc Natl Acad Sci USA. 1998;95:1758–1763. doi: 10.1073/pnas.95.4.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baker E K, Colley N J, Zuker C S. EMBO J. 1994;13:4886–4895. doi: 10.1002/j.1460-2075.1994.tb06816.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gross I, Hohenberg H, Huckhagel C, Krausslich H G. J Virol. 1998;72:4798–4810. doi: 10.1128/jvi.72.6.4798-4810.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bukovsky A A, Weimann A, Accola M A, Gottlinger H G. Proc Natl Acad Sci USA. 1997;94:10943–10948. doi: 10.1073/pnas.94.20.10943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aberham C, Weber S, Phares W. J Virol. 1996;70:3536–3544. doi: 10.1128/jvi.70.6.3536-3544.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Braaten D, Aberham C, Franke E K, Yin L, Phares W, Luban J. J Virol. 1996;70:5170–5176. doi: 10.1128/jvi.70.8.5170-5176.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Campos-Olivas R, Summers M F. Biochemistry. 1999;38:10262–10271. doi: 10.1021/bi990991x. [DOI] [PubMed] [Google Scholar]
- 52.Zhou X Z, Kops O, Werner A, Lu P J, Shen M, Stoller G, Kullertz G, Stark M, Fischer G, Lu K P. Mol Cell. 2000;6:873–883. doi: 10.1016/s1097-2765(05)00083-3. [DOI] [PubMed] [Google Scholar]
- 53.Gaburjakova M, Gaburjakova J, Reiken S, Huang F, Marx S O, Rosemblit N, Marks A R. J Biol Chem. 2001;276:16931–16935. doi: 10.1074/jbc.M100856200. [DOI] [PubMed] [Google Scholar]
- 54.Lee J P, Palfrey H C, Bindokas V P, Ghadge G D, Ma L, Miller R J, Roos R P. Proc Natl Acad Sci USA. 1999;96:3251–3256. doi: 10.1073/pnas.96.6.3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jin Z G, Melaragno M G, Liao D F, Yan C, Haendeler J, Suh Y A, Lambeth J D, Berk B C. Circ Res. 2000;87:789–796. doi: 10.1161/01.res.87.9.789. [DOI] [PubMed] [Google Scholar]
- 56.Chazin W J, Kordel J, Drakenberg T, Thulin E, Brodin P, Grundstrom T, Forsen S. Proc Natl Acad Sci USA. 1989;86:2195–2198. doi: 10.1073/pnas.86.7.2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ng K K, Weis W I. Biochemistry. 1998;37:17977–17989. doi: 10.1021/bi9819733. [DOI] [PubMed] [Google Scholar]
- 58.Sheng X R, Zhang H J, Pan X M, Li X F, Zhou J M. FEBS Lett. 1997;413:429–432. doi: 10.1016/s0014-5793(97)00951-4. [DOI] [PubMed] [Google Scholar]
- 59.Lopez-Ilasaca M, Schiene C, Kullertz G, Tradler T, Fischer G, Wetzker R. J Biol Chem. 1998;273:9430–9434. doi: 10.1074/jbc.273.16.9430. [DOI] [PubMed] [Google Scholar]
- 60.Lu K P, Hanes S D, Hunter T. Nature (London) 1996;380:544–547. doi: 10.1038/380544a0. [DOI] [PubMed] [Google Scholar]
- 61.Yaffe M B, Schutkowski M, Shen M, Zhou X Z, Stukenberg P T, Rahfeld J U, Xu J, Kuang J, Kirschner M W, Fischer G, et al. Science. 1997;278:1957–1960. doi: 10.1126/science.278.5345.1957. [DOI] [PubMed] [Google Scholar]
- 62.Stukenberg P T, Kirschner M W. Mol Cell. 2001;7:1071–1083. doi: 10.1016/s1097-2765(01)00245-3. [DOI] [PubMed] [Google Scholar]
- 63.Lu P J, Wulf G, Zhou X Z, Davies P, Lu K P. Nature (London) 1999;399:784–788. doi: 10.1038/21650. [DOI] [PubMed] [Google Scholar]