The SUMO E3 ligase RanBP2 promotes modification of the HDAC4 deacetylase (original) (raw)
Abstract
Transcriptional repression mediated through histone deacetylation is a critical component of eukaryotic gene regulation. Here we demonstrate that the class II histone deacetylase HDAC4 is covalently modified by the ubiquitin-related SUMO-1 modifier. A sumoylation-deficient point mutant (HDAC4-K559R) shows a slightly impaired ability to repress transcription as well as reduced histone deacetylase activity. The ability of HDAC4 to self-aggregate is a prerequisite for proper sumoylation in vivo. Calcium/calmodulin-dependent protein kinase (CaMK) signalling, which induces nuclear export, abrogates SUMO-1 modification of HDAC4. Moreover, the modification depends on the presence of an intact nuclear localization signal and is catalysed by the nuclear pore complex (NPC) RanBP2 protein, a factor newly identified as a SUMO E3 ligase. These findings suggest that sumoylation of HDAC4 takes place at the NPC and is coupled to its nuclear import. Finally, modification experiments indicate that the MEF2-interacting transcription repressor (MITR) as well as HDAC1 and -6 are similarly SUMO modified, indicating that sumoylation may be an important regulatory mechanism for the control of transcriptional repression mediated by both class I and II HDACs.
Keywords: E3 ligase/HDACs/nuclear pore complex/SUMO/ubiquitin-like modification
Introduction
Regulation of chromatin structure by acetylation of histone tails is a key mechanism in the control of gene expression. Histone deacetylases (HDACs), which serve to return chromatin to its condensed state, play a critical role in transcriptional repression (Kuo and Allis, 1998). Among the three defined classes of deacetylases, the class II HDAC4, -5, -6 and -7 exhibit the unique property of being regulated by nucleocytoplasmic shuttling (reviewed in Khochbin et al., 2001). Furthermore, class II HDACs, and the related MEF2-interacting transcription repressor (MITR) protein, have been shown to interact with members of the MEF2 family of transcription factors and with the 14-3-3 chaperone proteins in the control of myeloblast differentiation (Miska et al., 1999; Sparrow et al., 1999; Grozinger and Schreiber, 2000; McKinsey et al., 2000). Finally, like class I HDACs, members of the class II family are found in complexes with the SMRT/N-CoR corepressors (Huang et al., 2000; Kao et al., 2000); indeed, recent evidence suggests that the enzymatic activity of HDAC4/5 stems from the presence of the class I HDAC3 in the SMRT/N-CoR-containing deacetylase complexes (Guenther et al., 2001; Fischle et al., 2002).
A number of ubiquitin-like proteins have been discovered recently that are attached to lysine residues of target proteins in a way analogous to that of ubiquitylation. Among them, SUMO-1, which is highly conserved from yeast to humans, has been shown to modify a number of proteins such as RanGAP1 (Matunis et al., 1996; Mahajan et al., 1997), the two nuclear body PML and SP100 proteins (Sternsdorf et al., 1997; Kamitani et al., 1998; Muller et al., 1998), p53 (Gostissa et al., 1999; Rodriguez et al., 1999; Muller et al., 2000), IκBα (Desterro et al., 1998) and a growing number of other proteins including viral proteins (for reviews see Melchior, 2000; Muller et al., 2001; Seeler and Dejean, 2001). Unlike ubiquitylation, sumoylation does not appear to promote protein degradation but rather was shown to be involved in mediating protein–protein interactions, subcellular compartmentalization and protein stability.
Conjugation of SUMO-1 requires the E1-activating heterodimer Aos1/Uba2 and the single E2-conjugating Ubc9 enzyme. Very recently, two distinct families of SUMO E3 ligases have been identified. In yeast, the majority of SUMO conjugation requires the Siz1 and Siz2 proteins (Johnson and Gupta, 2001; Takahashi et al., 2001). The mammalian proteins to which Siz1 and Siz2 are most closely related are the PIAS (protein inhibitor of activated STAT) proteins, and PIAS1 and PIASy were shown to catalyse sumoylation of p53 and LEF-1, respectively (Kahyo et al., 2001; Sachdev et al., 2001; reviewed in Hochstrasser, 2001). A second type of SUMO E3, structurally unrelated to the Siz/PIAS members, is the nuclear pore complex (NPC) RanBP2 protein (Pichler et al., 2002). This protein, which strongly enhances SUMO-1 modification of the SP100 target, is believed to act at the cytoplasmic filaments of the NPC serving to modify susbtrates on their way to the nucleus.
Previous work by our laboratory and others (Seeler and Dejean, 2001 and references therein) prompted us to investigate further the possible role of sumoylation in transcriptional repression mechanisms. Here we show that the class II histone deacetylase HDAC4 undergoes SUMO-1 modification at a single lysine residue (K559) and that this process is regulated by CaMK signalling. We show that the NPC RanBP2 protein functions as an E3 ligase for HDAC4, suggesting that sumoylation and nuclear import are coordinated events. An HDAC4 mutant, defective for SUMO-1 conjugation, exhibits lower transcriptional repressing and histone deacetylase activities, indicating that sumoylation might be important for full biological activity of HDAC4-containing complexes. Finally, we provide in vitro evidence that other HDACs and HDAC-related proteins are modified similarly by SUMO.
Results
HDAC4 is modified by SUMO-1 in vivo
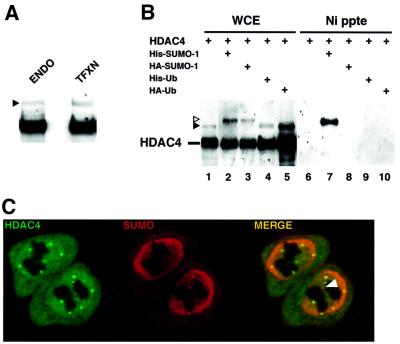
Western blotting of HeLa cell extracts with a rabbit polyclonal anti-HDAC4 antibody revealed a prominent anti-HDAC4-reactive band as well as a weaker more slowly migrating species (Figure 1A, lane ENDO, filled triangle). This band co-migrated with a higher molecular weight band also seen in extracts of cells transfected with an HDAC4 expression plasmid (Figure 1A, lane TFXN). To examine the possibility that these slower migrating species correspond to covalent adducts of ubiquitin (Ub) or the ubiquitin-related SUMO-1 modifiers, HeLa cells were co-transfected with vectors expressing either His-Ub or His-SUMO-1 together with HDAC4 (Figure 1B). As seen in Figure 1A, the same pattern of bands was detected in crude extracts (lanes 1–5). Upon purification of His-tagged complexes by nickel affinity chromatography (lanes 6–10), a major His-SUMO-1–HDAC4 conjugate was visible (lane 7), whereas no ubiquitylated HDAC4 could be detected (lane 9). When haemagglutinin (HA)-SUMO-1 was substituted for His-SUMO-1, the modified HDAC4 form was not retained on the beads (lane 8), thus confirming the specificity of the conjugates. The sumoylated form of HDAC4 was readily visible in crude extracts as either a His-SUMO-1 or an HA-SUMO-1 conjugate (open triangle in lanes 2 and 3, respectively) or as a conjugate with endogenous SUMO (filled triangle in lanes 1, 4 and 5). Finally, confocal microscopy of cells co-transfected with HDAC4- and SUMO-expressing plasmids showed the existence of subnuclear aggregates containing both HDAC4 and SUMO proteins (Figure 1C). Thus HDAC4 is a potent substrate for SUMO-1 modification in vivo.
Fig. 1. SUMO-1 is conjugated to HDAC4 in vivo. (A) Anti-HDAC4 western blot of HeLa whole-cell extract (WCE) from cells untransfected (ENDO) or transfected with HDAC4 expression plasmid (TFXN). The filled triangle shows the position of the higher molecular weight HDAC4 species. To yield equivalent signals, 20-fold less extract was loaded from transfected than from untransfected cells. (B) HeLa cells were co-transfected with the indicated plasmids (1 µg each) and empty expression vector up to 2 µg. WCE (lanes 1–5) and NTA precipitates (lanes 6–10) from transfected cells were analysed by western blotting using the anti-HDAC4 rabbit polyclonal antibody. The positions of unmodified and modified HDAC4 are indicated. The open and filled triangles mark the position of conjugates with exogenous His- (or HA-) SUMO-1 and endogenous SUMO, respectively. (C) Immunofluorescence labelling of HeLa cells co-transfected with expression plasmids encoding HDAC4 (labelled with anti-HDAC4 serum and FITC–anti-rabbit secondary antibodies, left panel) and HA-SUMO-2 (detected with anti-HA mAb 12CA5 and Texas Red-anti-mouse secondary antibody, middle panel). Co-localization appears yellow in the merged confocal image (right panel). The arrowhead indicates a SUMO-positive/HDAC4-negative nuclear aggregate probably corresponding to a PML nuclear body.
In vitro SUMO modification of HDACs
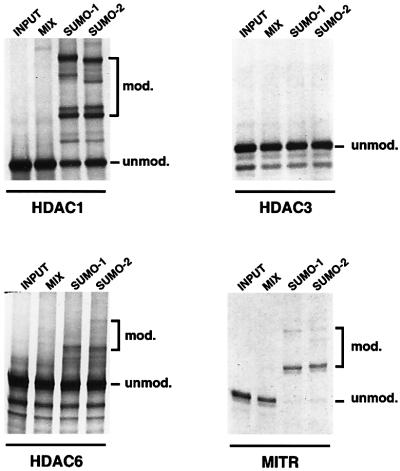
To see whether SUMO modification of HDAC proteins could be extended to other members of the HDAC family of proteins, we employed an in vitro modification system as described previously (Seeler et al., 2001). In this assay, 35S-radiolabelled, in vitro translated HDAC1, HDAC3, HDAC6 and MITR proteins were incubated with a HeLa fraction providing the E1 activity, together with recombinant Ubc9 in the presence or absence of recombinant SUMO-1 or SUMO-2. As shown in Figure 2, the addition of either SUMO-1 or SUMO-2 to the mix induces the formation of a number of conjugated forms of HDAC1, HDAC6 and MITR. HDAC3 failed to undergo modification in this assay. In contrast, MITR appeared as a potent SUMO substrate as nearly all 35S-labelled MITR protein was modified in this system. Together, these data demonstrate that SUMO modification is a relatively common process among HDAC and HDAC-related proteins and that both SUMO-1 and SUMO-2/-3 are efficient modifiers, at least in vitro.
Fig. 2. In vitro modification by SUMO-1 and SUMO-2 of HDACs and HDAC-related proteins. 35S-labelled in vitro translated HDAC1, HDAC3, HDAC6 and MITR were incubated in a sumoylation mix containing a fraction of HeLa cells providing the E1 activity together with recombinant Ubc9, in either the absence (MIX) or presence of recombinant SUMO-1 or SUMO-2. The positions of unmodified and modified substrate proteins are indicated.
SUMO-1 modification of HDAC4 occurs at Lys559
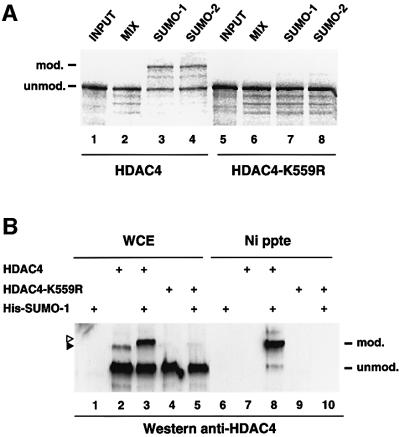
We concentrated further on HDAC4 and, to map the lysine residue(s) of HDAC4 serving as the SUMO-1 attachment site(s), we focused on lysines 133, 548, 559 and 697 because they reside in a motif that resembles the LKXE consensus sumoylation site (Duprez et al., 1999). Each of the four lysines was changed independently to arginine and the resulting HDAC4 point mutants were analysed for sumoylation in an in vitro modification assay as described in Figure 2. Mutation of K133, K548 and K697 was without consequence for SUMO modification (data not shown) whereas K559 mutation abrogated HDAC4 modification by both SUMO-1 and SUMO-2 (Figure 3A, lanes 7 and 8). To confirm these results in vivo, the same K559R mutant was co-expressed in cells together with His-SUMO-1 and the cell extracts subjected to nickel–agarose precipitation (Figure 3B). The wild-type HDAC4 protein served as a positive control in the same assay (lane 8). Consistent with the in vitro data, the HDAC4-K559R mutant was no longer able to undergo SUMO-1 modification (lane 10). Direct western blotting of the cell lysates indicated that the wild-type and mutant proteins were expressed at comparable levels (lanes 3 and 5, respectively). Thus Lys559, which lies within the N-terminal extension of class II HDACs, is required for sumoylation of HDAC4 in vitro and in vivo and is likely to be the predominant modification site for SUMO. Interestingly, mutation of this HDAC4 SUMO acceptor site had no visible consequence for the subcellular distribution of the protein compared with its native counterpart (data not shown).
Fig. 3. Lys559 is the major SUMO-1 modification site in HDAC4 in vitro and in vivo. (A) 35S-labelled in vitro translated wild-type HDAC4 (lanes 1–4) and HDAC4-K559R (lanes 5–8) were subjected to an in vitro modification reaction in the presence of a mix consisting of a HeLa fraction providing the E1 activity, recombinant Ubc9 and ATP, with or without recombinant SUMO-1 or SUMO-2. Reactions were revealed by autoradiography after separation by SDS–PAGE. (B) HeLa cells were co-transfected with the indicated plasmids (1 µg each) and empty expression vector up to 2 µg. WCE (lanes 1–5) and NTA precipitates (lanes 6–10) were separated by SDS–PAGE and probed with a rabbit anti-HDAC4 polyclonal antibody. HDAC4 conjugates are indicated by triangles as in Figure 1.
Contribution of sumoylation to HDAC4 functional properties
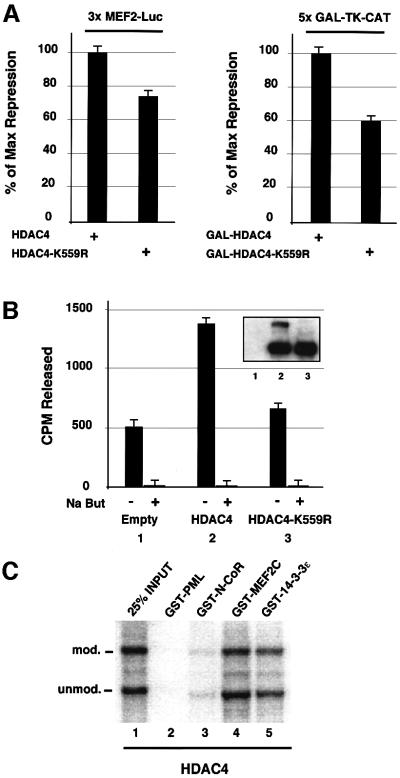
To gain insight into the role of SUMO-1 modification on HDAC4, we then assessed the impact of the modification on the transcription-repressing properties of HDAC4. The ability of the sumoylation-deficient HDAC4-K559R mutant to repress transcription was slightly impaired when compared with that of the wild-type HDAC4 protein (Figure 4A). The decrease in repression could be seen either with the HDAC4-K559R protein acting on an MEF2-responsive promoter (3×MEF2-Luc, 25% reduction, left panel) or with a GAL4 DNA-binding domain fusion of HDAC4-K559R on a GAL4 reporter gene (5×GAL-TK-CAT, 40% reduction, right panel). We then compared the relative histone deacetylase activity of the sumoylation-deficient HDAC4 mutant with that of its wild-type counterpart. COS-7 cells were transiently transfected with expression vectors for SUMO-1 together with HDAC4 or HDAC4-K559R followed by immunoprecipitation using the anti-HDAC4 antibodies and an assay for histone deacetylase activity (Figure 4B). Although the two proteins were expressed at a similar level (inset), the HDAC activity associated with HDAC4-K559R was significantly reduced, to a level barely above that of endogenous HDAC4 (compare lane 3 with lanes 1 and 2).
Fig. 4. K559R mutation reduces repressing and HDAC activities of HDAC4. (A) Left panel: HeLa cells were co-transfected with the 3×MEF2-Luc reporter (0.2 µg), a MEF2A expression vector (0.2 µg), HDAC4 or HDAC4-K559R expression vectors (0.5 µg) and empty expression vector up to 2 µg. Right panel: HeLa cells were co-transfected with the 5×GAL-TK-CAT reporter (1 µg), GAL-HDAC4 or GAL-HDAC4-K559R expression vectors (1 µg) and empty expression vector up to 4 µg. Values represent the average of three independent transfections. Luciferase and CAT reporter activities were normalized for transfection efficiency to an internal β-galactosidase control and expressed as a percentage of repression relative to that obtained with HDAC4 or GAL-HDAC4 alone. (B) Immunoprecipitation of histone deacetylase activity from COS-7 cells transfected with 2 µg of SUMO-1 expression vector and 2 µg of HDAC4, HDAC4-K559R or an empty vector as a control using anti-HDAC4 antibodies. HDAC activity was determined by release of [3H]acetate from an in vitro acetylated histone H4 peptide. The histone deacetylase inhibitor sodium butyrate abrogated HDAC activity in every immunoprecipitate. Immunoblot analysis with anti-HDAC4 antibodies confirmed the similar expression of wild-type and mutant HDAC4 proteins (inset). (C) [35S]methionine-labelled HDAC4 was modified in vitro by SUMO-1 as in Figure 3A and used in a pull-down assay with GST–N-CoR, GST–MEF2C, GST–14-3-3ε or GST–PML1–447 as a negative control.
To evaluate the relative affinity of the unmodified HDAC4 versus its modified form for known HDAC4 partners, we carried out GST pull-down assays. For this, in vitro translated HDAC4 was subjected to the in vitro sumoylation reaction and then incubated with GST–N-CoR, GST–MEF2C, GST–14-3-3ε and GST–PML as a negative control. As seen in Figure 4C, both the modified and unmodified HDAC4 species were retained efficiently by GST–N-CoR, GST–MEF2C and GST–14-3-3ε but not by the GST–PML control. Further, quantitative PhosphorImager analysis showed that in vitro modified and unmodified HDAC4 species bound HDAC4 partners with equal affinity since the relative proportion of the two forms in the mixture was unchanged after binding (data not shown).
Taken together, these results suggest that SUMO-1 modification of HDAC4 may be important for exerting the full repressing activity of HDAC4-containing complexes, but that direct interactions with the known HDAC4 binding partners are unlikely to be the mechanism by which sumoylation modulates this activity.
HDAC4 contains an N-terminal dimerization domain
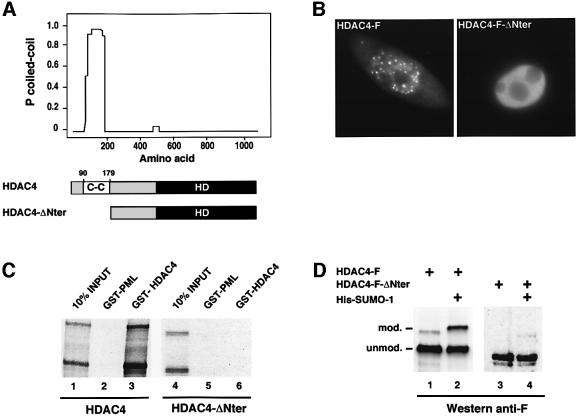
Previous work had shown that HDAC4 can form cytoplasmic and nuclear aggregates upon overexpression (Miska et al., 1999). This prompted us to investigate whether this protein contains a self-interaction domain. Analysis of the amino acid sequence with the COILS program (Lupas, 1997) revealed a domain encompassing amino acid residues 90–179 that possesses a high probability of forming a coiled-coil structure (Figure 5A). Expression in HeLa cells of an F-tagged HDAC4 derivative in which this domain had been deleted (HDAC4-F-ΔNter) revealed a diffuse, primarily nuclear distribution (Figure 5B, right panel). In contrast, full-length HDAC4-F exhibited numerous nuclear speckles and a granular appearance in the cytoplasm not seen with HDAC4-F-ΔNter (Figure 5B, left panel), indicating that this N-terminal coiled-coil domain is indispensable for HDAC4 aggregation. Next, we carried out in vitro binding experiments in which we compared the ability of the in vitro SUMO1-modified full-length HDAC4 protein to bind to a GST–HDAC4 fusion with that of the modifed HDAC4-ΔNter mutant (Figure 5C). While the unmodified and modified native protein each interacts strongly with itself (lane 3), the N-terminal deletion mutant, while competent for modification in vitro (lane 4), shows no binding at all (lane 6), indicating that the observed aggregation of HDAC4 is likely to be due to self-interaction through the coiled-coil motif.
Fig. 5. Self-aggregation of HDAC4 and its impact on sumoylation. (A) Computer analysis of the amino acid sequence of HDAC4 with the COILS program revealed a high probability for a coiled-coil structure between residues 90 and 179. (B) HeLa cells were transfected with expression plasmids encoding F-tagged versions of HDAC4 (left panel) or HDAC4-ΔNter (right panel) and immunolabelled with anti-F monoclonal antibody followed by fluorescein-conjugated secondary antibody. (C) GST interaction assay of 35S-radiolabelled, in vitro SUMO-1-modified HDAC4 and HDAC4-ΔNter proteins with GST–HDAC41–602. GST–PML1–447 served as a negative control. (D) HeLa cells were co-transfected with the indicated plasmids (1 µg each) and empty expression vector up to 2 µg. HDAC4 sumoylation was evaluated by direct western blotting of whole-cell extracts using the anti-F monoclonal antibody.
We next wished to study the effect of the aggregation property of the protein on its ability to undergo sumoylation in vivo (Figure 5D). When compared with its native counterpart, which is modified efficiently by either the endogenous SUMO (lane 1) or the transfected His-SUMO-1 (lane 2), the HDAC4-F-ΔNter mutant exhibits a highly reduced capacity to undergo modification (lanes 3 and 4), suggesting that the level of sumoylation in vivo is largely dependent on the ability of HDAC4 to form cellular aggregates.
Sumoylation of HDAC4 requires nuclear localization and is regulated in a signal-dependent manner
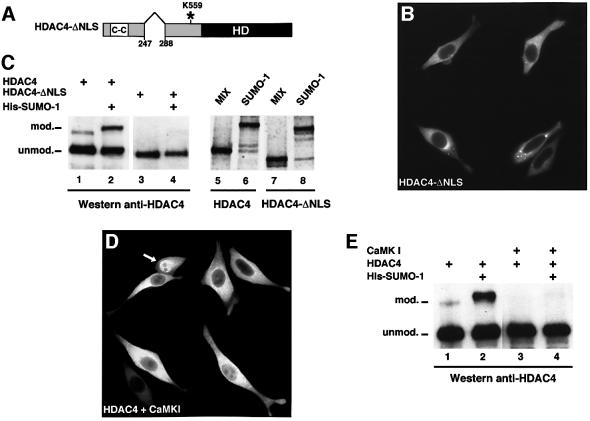
The analysis of numerous SUMO substrates (Rodriguez et al., 2001) has suggested SUMO modification to be a predominantly nuclear process. Given that HDAC4 shuttles between the nucleus and the cytoplasm, we wished to examine whether sumoylation of HDAC4 occurs in either or both of these cellular compartments. Based on the homology to HDAC5, we deduced the nuclear localization signal (NLS) of HDAC4 to lie between residues 247 and 288 (Figure 6A). As expected, expression of a construct (HDAC4-ΔNLS) lacking these residues rendered the protein entirely cytoplasmic, albeit without impairing the protein’s ability to aggregate (Figure 6B). Western blot analysis of cells expressing HDAC4-ΔNLS demonstrated this protein to be unable to undergo SUMO-1 modification (Figure 6C, lane 3), even upon co-expression of exogenous His-SUMO-1 (Figure 6C, lane 4). This inability, however, is not an intrinsic property of this protein since modification by SUMO-1 occurs as efficiently as for the full-length HDAC4 protein using the in vitro assay (Figure 6C, compare lane 8 with lane 6).
Fig. 6. Nuclear localization is a prerequisite for HDAC4 sumoylation. (A) Schematic representation of the HDAC4-ΔNLS deletion mutant. The NLS (amino acids 247–288), the coiled-coil (CC), the catalytic histone deacetylase (HD) domain and the sumoylation K559 site are indicated. (B) The cytoplasmic localization of HDAC4-ΔNLS overexpressed in HeLa cells was observed by indirect immunofluorescence using the anti-HDAC4 antibody and a fluorescein-conjugated secondary antibody. (C) Left panel: HDAC4-ΔNLS fails to undergo SUMO modification in vivo. WCEs of HeLa cells were transfected with the indicated plasmids (1 µg each) and empty expression vector up to 2 µg, and subjected to western blotting using the anti-HDAC4 antibody. Right panel: HDAC4-ΔNLS is sumoylated efficiently in vitro. Radiolabelled HDAC4 and HDAC4-ΔNLS proteins were subjected to the in vitro modification assay as described in Figure 3A. (D) Exclusion of HDAC4 from the nucleus by the activated form of CaMKI. HeLa cells were co-transfected with HDAC4 and CaMK1 (1 µg each) and stained with the anti-HDAC4 antibody. The white arrow indicates a cell showing retention of HDAC4 in the nucleus which most probably corresponds to a single HDAC4 transfectant. (E) Reduced sumoylation of HDAC4 upon CaMKI expression. WCEs of HeLa cells were transfected with the indicated plasmids and subjected to western blotting using the anti-HDAC4 antibody.
Previous work has shown the nuclear export of HDAC4 to depend on activation of calcium/calmodulin-dependent protein kinase (CaMK) signalling such that binding of the 14-3-3 protein to phosphorylated HDAC4 activates nuclear export of the protein (McKinsey et al., 2001; Wang and Yang, 2001). Consistent with previous observations, transfection of a constitutively active CaMKI vector in HeLa cells induced nuclear exclusion of HDAC4 in the vast majority of the cells (Figure 6D). Expression of CaMKI together with HDAC4 and SUMO-1 considerably reduced the pool of sumoylated protein (Figure 6E, compare lane 4 with lane 2). While it cannot be excluded that CaMKI affects sumoylation of HDAC4 by mechanisms distinct from its nuclear export-facilitating capacity, altogether these results further suggest the dependence of the modification process on the nuclear localization of HDAC4.
RanBP2 functions as a SUMO E3 ligase for HDAC4
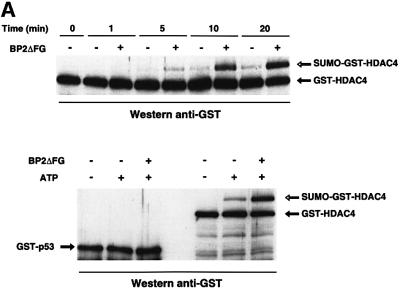
We have shown recently that RanBP2 strongly enhances SUMO-1 modification of the nuclear boby-associated SP100 protein in the presence of E1 and E2 enzymes (Pichler et al., 2002). The requirement for an intact NLS for sumoylation of HDAC4 suggested that the modification process could be, as for SP100, stimulated by RanBP2 en route to the nucleus. To test this hypothesis, we used a reconstituted in vitro modification assay with recombinant HDAC4, E1 (Aos1/Uba2), E2 (Ubc9) and SUMO-1 in the presence or absence of a 33 kDa fragment of RanBP2 (BP2ΔFG) which was shown previously to contain the E3 ligase activity (Pichler et al., 2002). Although the sumoylation of HDAC4 can occur marginally without SUMO ligase, the reaction was strongly stimulated by addition of BP2ΔFG (Figure 7A, upper panel). Moreover RanBP2 functions catalytically as 5 ng of BP2ΔFG were sufficient to promote modification of a 100-fold molar excess of HDAC4. Thus, RanBP2 plays an E3-like role in SUMO modification of HDAC4. To assess the specificity of the assay, we substituted recombinant p53 for HDAC4 in a similar in vitro modification experiment. Whereas sumoylation of HDAC4 was greatly enhanced by RanBP2, p53 failed to undergo SUMO modification in a parallel reaction (Figure 7A, lower panel). As a control, we ensured that p53 was competent for in vitro modification by PIASxβ, a member of the other SUMO E3 ligase family (Hochstrasser, 2001; Jackson, 2001) (data not shown).
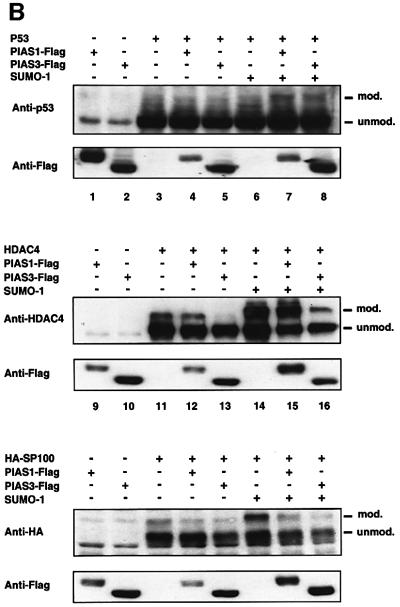
Fig. 7. RanBP2 catalyses SUMO-1 modification of HDAC4. (A) RanBP2 stimulates in vitro modification of HDAC4 (upper panel) but not p53 (lower panel). Reconstituted in vitro reactions containing 1 µg of GST–HDAC1–602 or GST–p53, 150 ng of E1, 10 ng of E2 and 1 µg of SUMO-1 in a 20 µl reaction volume were incubated for the indicated times (upper panel) or for 30 min (lower panel) at 30°C in the absence or presence of 5 mM ATP and in the absence or presence of 5 ng of RanBP2 (amino acids 2553–2838) (BP2ΔFG). Analysis was by immunoblotting with anti-GST antibodies. (B) PIAS stimulates in vivo modification of p53 (upper panel) but not HDAC4 (middle panel) and SP100 (lower panel). HeLa cells were co-transfected with expression vectors for the indicated substrates (0.1 µg), SUMO-1 (0.5 µg), PIAS1-Flag or PIAS3-Flag (0.6 µg) and empty expression vector up to 2 µg. The samples were subjected to SDS–PAGE, western blotted with anti-p53, anti-HDAC4 or anti-HA antibodies, and subsequently reprobed using anti-Flag antibodies to check the expression of the transfected PIAS proteins.
Next, we addressed the issue as to whether the sumoylation of HDAC4 could also be catalysed by the PIAS family of proteins. To validate the assay, we conducted an in vivo modification assay using p53 as a bona fide PIAS-responsive substrate (Figure 7B, upper panel). HeLa cells were co-transfected with p53 and PIAS1 or PIAS3 and the relative amount of sumoylated p53 was analysed by western blotting. As reported previously (Kahyo et al., 2001), sumoylation of p53 was found to be enhanced by PIAS1 in the presence of SUMO-1 (lane 7). A slightly less pronounced increase in p53 modification was also observed upon PIAS3 and SUMO-1 overexpression (lane 8). We then tested HDAC4 in a similar set of experiments. HeLa cells were co-transfected with HDAC4 and either PIAS1 or PIAS3 and then the sumoylation in the absence or presence of PIAS was checked by western blotting (Figure 7B, middle panel). The modified HDAC4 species was detected as a 175 kDa band (lanes 11–13) which was more abundant in cells co-transfected with SUMO-1 (lanes 14–16). Expression of either PIAS protein did not enhance SUMO modification of HDAC4 but rather decreased its efficiency. This reduction in sumoylation was particularly clear for PIAS3 (compare lane 13 with lane 11, and lane 16 with lane 14). These findings suggest that, under these conditions, PIAS1 and PIAS3 proteins do not function as bona fide SUMO E3 ligases for HDAC4 and that the observed decrease in sumoylation probably reflects competition for SUMO-1 in modifying PIAS-specific substrates. To check the possible sensitivity of SP100 to the E3-like activity of PIAS proteins, we performed similar in vivo experiments using SP100 as a substrate. As for HDAC4, the sumoylation of SP100 was not enhanced at all, even when SUMO-1 was co-expressed, and rather appeared to be reduced by the addition of either PIAS1 or PIAS3 (Figure 7B, lower panel, compare lanes 20 and 21 with lane 19 in the absence of SUMO-1, and lanes 23 and 24 with lane 22 in the presence of SUMO-1).
Altogether, the above data suggest that the two classes of structurally unrelated SUMO E3 ligases identified so far, the RanBP2 and PIAS proteins, probably act in a rather independent, though not necessarily mutually exclusive, way on distinct protein substrates, HDAC4 sharing with SP100 the property to be sumoylated in a RanBP2-dependent manner whereas p53 modification depends on PIAS proteins.
Discussion
In the present work, we have shown that the class II histone deacetylase HDAC4, as well as possibly other HDAC enzymes, belongs to a subfamily of transcriptional regulators that are subject to covalent modification by the ubiquitin-like SUMO protein. Previously described examples include p53, c-jun (Gostissa et al., 1999; Rodriguez et al., 1999; Muller et al., 2000; Kwek et al., 2001), androgen receptor (Poukka et al., 2000), Drosophila Dorsal (Bhaskar et al., 2000) and Tramtrack Ttk69 (Lehembre et al., 2000). The effects of this modification on the transcriptional regulatory properties of these proteins have largely proved to be subtle. Although it cannot be formally excluded that the observed effect on HDAC4 activity is not simply due to an overall structural alteration of the mutated protein, the present results suggest that the sumoylation of HDAC4 may be required for full activity of this factor, since both its deacetylase and repressing properties are impaired in the absence of a functional SUMO acceptor site.
The role of sumoylation
Two main hypotheses for the biological role of SUMO modification have been proposed to date. In the first, exemplified by IκBα, sumoylation antagonizes ubiquitylation by targeting a common acceptor lysine residue (Desterro et al., 1998). However, in contrast to HDAC1 which is modified efficiently by ubiquitin, we have not found HDAC4 to be ubiquitylated to a significant extent (our unpublished results), thus making this model an unlikely explanation for the role of sumoylation in HDAC4 function. The second model implicates SUMO in modifying the molecular interaction properties of its target proteins such that for the nuclear import factor RanGAP1 and the PML protein, sumoylation is a requirement for proper targeting to the NPC and the nuclear bodies, respectively (Mahajan et al., 1997; Matunis et al., 1998; Muller et al., 1998; Ishov et al., 1999). In the case of SP100, PML and LEF-1, recent evidence supports the idea that sumoylation enhances the stability of the subtrate protein’s interaction with a partner protein (HP1, Daxx and PIASy, respectively) (Ishov et al., 1999; Sachdev et al., 2001; Seeler et al., 2001). Our in vitro experiments, however, indicate that SUMO modification does not affect heteromeric interactions of HDAC4 with its known binding partners N-CoR, MEF2C or 14-3-3, suggesting that perhaps additional SUMO-dependent interactions of HDAC4 remain to be discovered. Moreover, the subcellular localization as well as the propensity of the sumoylation-deficient HDAC4-K559R to form conspicuous aggregates upon overexpression remain unaltered with respect to the native HDAC4 (our unpublished results).
SUMO and higher order chromatin complexes
Apart from the finding that sumoylation preferentially affects transcription factors and co-factors (Seeler and Dejean, 2001), there is significant evidence implicating SUMO in the functional regulation of higher order chromatin or chromosome structures. The budding yeast orthologue of SUMO, SMT3, was first isolated as a high-copy suppressor of a mutation affecting proper chromosome segregation, thus suggesting a role in centromere function (Meluh and Koshland, 1995). Furthermore, the Ulp2-type SUMO isopeptidase (SMT4) has been shown to be essential for the regulation of chromosome condensation (Strunnikov et al., 2001). Interestingly, this defect could be overcome by overexpression of Siz1, the yeast homologue of the mammalian PIAS proteins now known to perform a SUMO E3 ligase function (see below). More recently, the gene for the suppressor of position-effect variegation, Su(var)2-10/Zimp, which encodes the sole Drosophila homologue of the PIAS proteins, has been shown to be necessary for the maintenance of interphase chromosome structure and positioning (Hari et al., 2001). If this gene product, like other PIAS family members, here also performs an E3 ligase function, this finding would underscore further the evolutionarily conserved role of sumoylation in regulating higher order chromatin structure.
Sumoylation by PIAS versus RanBP2 E3 ligases
For ubiquitylation, the existence of numerous E2 and E3 enzymes provides for a high degree of combinatorial substrate specificity. In contrast, SUMO attachment to target proteins relies on only a single E2 enzyme, Ubc9, and the existence of E3-like factors has long remained doubtful. Recent work by several laboratories, however, has shown that PIAS protein family members perform an apparently evolutionarily conserved role as SUMO E3 ligases (Johnson and Gupta, 2001; Kahyo et al., 2001; Sachdev et al., 2001; Takahashi et al., 2001). More recently, we have shown that the nuclear import factor RanBP2 performs an E3 ligase function for the nuclear body protein SP100 (Pichler et al., 2002), and here we have extended these findings by showing that HDAC4 similarly requires RanBP2 for efficient modification. It thus appears that there are at least two modification pathways, the PIAS- and the RanBP2-dependent. It remains to be seen whether, as a rule, the two function in a mutually exclusive fashion or whether certain substrates undergo modification mediated by both types of proteins. In this context, preliminary in vitro experiments suggest that HDAC4 sumoylation could also be stimulated by PIASxβ, requiring, however, 1000-fold more PIASxβ than RanBP2 to achieve a similar yield of conjugate (data not shown). Furthermore, even within the PIAS protein family, it is not clear as yet whether, for example, the PIASy-dependent substrate LEF-1 (Sachdev et al., 2001) might not be modifed similarly in a PIAS1- or 3-dependent manner. We have found here that sumoylation of p53 may also be enhanced by PIAS3 and not only by PIAS1, as reported previously (Kahyo et al., 2001). Thus, the degree of substrate specificity conferred by these proteins remains an open question. Nevertheless, the finding that RanBP2 catalyses the modification of at least two proteins might suggest this E3-like factor to function in a more general fashion, coupling sumoylation with nuclear import. It is thus tempting to speculate that RanBP2-mediated sumoylation represents the constitutive ‘tagging’ of a substrate protein for a ‘nuclear’ mode of function, whereas PIAS-mediated sumoylation may occur in response to specific (stress-induced?) signals in the nucleus. In this context, it is noteworthy that PIASy preferentially augments modification of LEF-1 with SUMO-2 over SUMO-1, correlating well with the previously shown up-regulation of SUMO-2 modification in response to DNA-damaging agents and other stress-related stimuli (Saitoh and Hinchey, 2000).
Sumoylation and nucleo-cytoplasmic transport
Our findings link the shuttling of proteins between the cytoplasm and the nucleus to the sumoylation process. The histone deacetylases HDAC4 and HDAC5 have been shown to play a pivotal role in the control of the gene expression programme leading to muscle cell differentiation. Critical to this activity, not shared with class I HDACs, is their ability to translocate between the nuclear and cytoplasmic compartments in a phosphorylation-dependent manner. Current models thus envisage that MEF2/MyoD-dependent transcription leading to differentiation is de-repressed by phosphorylation that in turn leads to the 14-3-3-dependent nuclear export and/or cytoplasmic retention of HDAC4 (Grozinger and Schreiber, 2000; McKinsey et al., 2000; Miska et al., 2001; Wang and Yang, 2001). As shown here with HDAC4, the sumoylation of nuclear proteins, where tested, requires intact nuclear import capacity since proteins lacking a functional NLS remain unmodified (Rodriguez et al., 2001). A mechanistic rationale for this behaviour may lie in our finding that sumoylation of HDAC4 uses RanBP2 as an E3 ligase, supporting the notion that HDAC4 modification occurs at the NPC during its translocation into the nucleus and not thereafter. In this context, it would be interesting to know whether nuclear export of HDAC4 is coupled to its de-modification. Such has been inferred, for example, in the case of E1B-55 kDa protein in which sumoylation is necessary for nuclear retention, while its absence permits nuclear export, even in the presence of the nuclear export inhibitor leptomycin B (Endter et al., 2001). It might thus be hypothesized that the sumoylation of HDAC4 represents an additional nuclear retention signal, operating perhaps by inhibiting or retarding the re-export of the protein.
In conclusion, our demonstration here that HDAC4, like numerous chromatin-associated proteins, undergoes SUMO modification further strengthens the evidence implicating this type of post-translational modification in regulatory events occuring at the chromatin level. Furthermore, our work has permitted the delineation of two primary (the PIAS- and RanBP2-dependent) classes of sumoylation systems. Further work will undoubtedly show whether additional systems exist, and how they might contribute to the regulation of SUMO substrate specificity and function.
Materials and methods
Plasmid constructions
To prepare HDAC4-F, a full-length human HDAC4 cDNA was fused 3′ with the F-tag-encoding sequence (Ali et al., 1993) and cloned into expression vector pSG5 (Stratagene). Point mutant derivatives HDAC4 K133R, K548R, K559R and K697R were obtained using the QuikChange Kit (Stratagene) following the manufacturer’s protocol. Deletion derivatives HDAC4-ΔNter (amino acids 224–1084), HDAC4-F-ΔNter and HDAC4-ΔNLS (deletion of amino acids 247–288) were generated by PCR. GAL4-HDAC4 has been described previously (Miska et al., 1999), and GAL4-HDAC4-K559R was prepared from HDAC4-K559R using convenient restriction sites. Plasmids for in vitro translation of HDAC4, the above mutant derivatives and HDAC1 were obtained by cloning the appropriate cDNAs into vector PING14 (Brehm et al., 1998). HDAC3 and HDAC6 were subcloned in the pcDNA3.1/Myc-His vector. GST–HDAC41–602 was obtained by cloning a sequence encoding amino acids 1–601 into the _Bam_HI site of pGEX2T. His-HDAC41–602 was obtained by cloning the same fragment into vector pQE-30 (Qiagen). GST–PML1–447 was generated by cloning a sequence encoding amino acids 1–447 of PML into pGEX2T. A vector for GST–14-3-3ε was prepared by cloning a full-length RT–PCR-derived human cDNA into pGEX2T. BP2ΔFG, a fragment of human RanBP2 (amino acids 2553–2838), was prepared as described (Pichler et al., 2002). Expression of GST fusion proteins was carried out in BL21(DE3) pLysS cells, and native proteins were purified using standard protocols. A MITR cDNA in vector pKS was obtained from RIKEN (KIAA0744). Expression vectors for MEF2A, GST–MEF2C and the 3×MEF-Luc reporter (Lu et al., 2000), constitutively active CaMKI (Haribabu et al., 1995), expression vectors for HA- and His-tagged SUMO-1, -2 and ubiquitin (Muller et al., 2000), SP100 and 5×GAL-TK-CAT (Seeler et al., 1998), PIAS1-Flag (Liu and Shuai, 2001), PIAS3-Flag (Rodel et al., 2000), p53 and GST–p53 (Unger et al., 1999), and GST–N-CoR (amino acids 1122–1416) have been described previously. All constructions were verified by dideoxy sequencing, and detailed plasmid maps are available upon request.
Cell culture, transfections, reporter and deacetylase assays
HeLa and COS-7 cells were grown at 37°C in 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM, Life Technologies), supplemented with antibiotics, glutamate and 10% fetal calf serum. Cell transfections were performed with the LipofectAMINE Plus reagent (Life Technologies, Inc.) according to the manufacturer’s instructions. Luciferase and CAT reporter assays were carried out using standard protocols. Deacetylase assays were carried out on extracts from COS-7 cells transfected with HDAC4 and HDAC4-K559R as described previously (Magnaghi-Jaulin et al., 1998).
Antibodies and immunodetection
A rabbit polyclonal anti-HDAC4 antiserum was raised against GST–HDAC41–602 and affinity purified against His-HDAC41–602 protein immobilized on nickel–agarose (Qiagen). The anti-F-tag monoclonal antibody (mAb F3) was as described (Ali et al., 1993). Anti-GST antibodies were provided by Dr Ludger Hengst. HA- and Flag-tagged proteins were detected with mAb 12CA5 (Roche) and rabbit polyclonal F-7425 (Sigma), respectively. Anti-p53 antibody (CM1) was obtained from Novocastra Laboratory Ltd. Immunofluorescence staining was carried out as described (Seeler et al., 1998) using the indicated primary antibodies and fluorescein isothiocyanate (FITC)-coupled secondary antibodies (Vector Laboratories). Fluorescence microscopy was performed using a Leica DMRB microscope equipped with a Princeton CoolSnapFX CCD camera controlled by MetaVue software or with a Leica SM confocal microscope. Western blots were prepared on Hybond C-extra membranes (Amersham) and revealed using the CDP-Western Star kit (Perkin-Elmer) for chemiluminescent alkaline phosphatase detection.
Nickel pull-downs, sumoylation assays and GST pull-downs
In vivo sumoylation assays were carried out using the nickel pull-down technique, as described previously (Seeler et al., 2001). Sumoylation assays on [35S]methionine-labelled in vitro translated proteins, prepared using the T7 or Sp6 TNT coupled reticulocyte lysate kit (Promega), were performed using the protocol of Desterro et al. (1997). Recombinant in vitro sumoylation assays (20 µl total volume, 0–30 min incubation at 30°C) were carried out as described previously (Pichler et al., 2002) in buffer TB (20 mM HEPES, 110 mM K-acetate, 2 mM Mg-acetate, 0.5 mM EGTA) containing 1 µg of recombinant SUMO1, 150 ng of E1 (Aos1/Uba2), 10 ng of E2 (Ubc9) and 1 µg of GST–HDAC41–602 or GST–p53 substrates in the presence or absence of 5 mM ATP and in the presence or absence of 5 ng of BP2ΔFG. Reaction products were visualized by immunoblotting with appropriate antibodies.
GST pull-down assays were performed on in vitro translated, [35S]methionine-labelled and sumoylated proteins as described (Seeler et al., 2001).
Acknowledgments
Acknowledgements
We acknowledge Pierre Chambon for the anti-F antibodies and the 5×AL-TK-CAT reporter, Ludger Hengst for the anti-GST antibodies, Anthony Means for the CaMKI expression vector, Eric Olson for the 3×MEF2-Luc reporter, the MEF2A expression plasmid and the GST–MEF2C vector, Moshe Oren for the GST–p53 and p53 expression vectors, and Ke Shuai for the PIAS-Flag expression vectors. Our special thanks go to Enrico Garattini for helpful discussions. We also thank all members of the laboratory for sharing reagents and advice. This work was supported by grants from the European Economic Community (QLG1), the Association pour la Recherche contre le Cancer, the Ligue Nationale Contre le Cancer, the Fondation de France and the BMBF. J.S. was supported by the Pasteur–Negri–Weizmann Council, O.K. by the Ministère de la Recherche et la Technologie, and S.M. by the Association for International Cancer Research.
References
- Ali S., Metzger,D., Bornert,J.M. and Chambon,P. (1993) Modulation of transcriptional activation by ligand-dependent phosphorylation of the human oestrogen receptor A/B region. EMBO J., 12, 1153–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskar V., Valentine,S.A. and Courey,A.J. (2000) A functional interaction between Dorsal and components of the Smt3 conjugation machinery. J. Biol. Chem., 275, 4033–4040. [DOI] [PubMed] [Google Scholar]
- Brehm A., Miska,E.A., McCance,D.J., Reid,J.L., Bannister,A.J. and Kouzarides,T. (1998) Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature, 391, 597–601. [DOI] [PubMed] [Google Scholar]
- Desterro J.M., Thomson,J. and Hay,R.T. (1997) Ubch9 conjugates SUMO but not ubiquitin. FEBS Lett., 417, 297–300. [DOI] [PubMed] [Google Scholar]
- Desterro J.M., Rodriguez,M.S. and Hay,R.T. (1998) SUMO-1 modification of IκBα inhibits NF-κB activation. Mol. Cell, 2, 233–239. [DOI] [PubMed] [Google Scholar]
- Duprez E. et al. (1999) SUMO-1 modification of the acute promyelocytic leukaemia protein PML: implications for nuclear localisation. J. Cell Sci., 112, 381–393. [DOI] [PubMed] [Google Scholar]
- Endter C., Kzhyshkowska,J., Stauber,R. and Dobner,T. (2001) SUMO-1 modification required for transformation by adenovirus type 5 early region 1B 55-kDa oncoprotein. Proc. Natl Acad. Sci. USA, 98, 11312–11317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle W., Dequiedt,F., Hendzel,M.J., Guenther,M.G., Lazar,M.A., Voelter,W. and Verdin,E. (2002) Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol. Cell, 9, 45–57. [DOI] [PubMed] [Google Scholar]
- Gostissa M., Hengstermann,A., Fogal,V., Sandy,P., Schwarz,S.E., Scheffner,M. and Del Sal,G. (1999) Activation of p53 by conjugation to the ubiquitin-like protein SUMO-1. EMBO J., 18, 6462–6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grozinger C.M. and Schreiber,S.L. (2000) Regulation of histone deacetylase 4 and 5 and transcriptional activity by 14-3-3-dependent cellular localization. Proc. Natl Acad. Sci. USA, 97, 7835–7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther M.G., Barak,O. and Lazar,M.A. (2001) The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol. Cell. Biol., 21, 6091–6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hari K.L., Cook,K.R. and Karpen,G.H. (2001) The Drosophila Su(var)2-10 locus regulates chromosome structure and function and encodes a member of the PIAS protein family. Genes Dev., 15, 1334–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haribabu B., Hook,S.S., Selbert,M.A., Goldstein,E.G., Tomhave,E.D., Edelman,A.M., Snyderman,R. and Means,A.R. (1995) Human calcium–calmodulin dependent protein kinase I: cDNA cloning, domain structure and activation by phosphorylation at threonine-177 by calcium–calmodulin dependent protein kinase I kinase. EMBO J., 14, 3679–3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstrasser M. (2001) Sp-ring for sumo. new functions bloom for a ubiquitin-like protein. Cell, 107, 5–8. [DOI] [PubMed] [Google Scholar]
- Huang E.Y., Zhang,J., Miska,E.A., Guenther,M.G., Kouzarides,T. and Lazar,M.A. (2000) Nuclear receptor corepressors partner with class II histone deacetylases in a Sin3-independent repression pathway. Genes Dev., 14, 45–54. [PMC free article] [PubMed] [Google Scholar]
- Ishov A.M., Sotnikov,A.G., Negorev,D., Vladimirova,O.V., Neff,N., Kamitani,T., Yeh,E.T., Strauss,J.F.,3rd and Maul,G.G. (1999) PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J. Cell Biol., 147, 221–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson P.K. (2001) A new RING for SUMO: wrestling transcriptional responses into nuclear bodies with PIAS family E3 SUMO ligases. Genes Dev., 15, 3053–3058. [DOI] [PubMed] [Google Scholar]
- Johnson E.S. and Gupta,A.A. (2001) An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell, 106, 735–744. [DOI] [PubMed] [Google Scholar]
- Kahyo T., Nishida,T. and Yasuda,H. (2001) Involvement of PIAS1 in the sumoylation of tumor suppressor p53. Mol. Cell, 8, 713–718. [DOI] [PubMed] [Google Scholar]
- Kamitani T., Nguyen,H.P., Kito,K., Fukuda-Kamitani,T. and Yeh,E.T. (1998) Covalent modification of PML by the sentrin family of ubiquitin-like proteins. J. Biol. Chem., 273, 3117–3120. [DOI] [PubMed] [Google Scholar]
- Kao H.Y., Downes,M., Ordentlich,P. and Evans,R.M. (2000) Isolation of a novel histone deacetylase reveals that class I and class II deacetylases promote SMRT-mediated repression. Genes Dev., 14, 55–66. [PMC free article] [PubMed] [Google Scholar]
- Khochbin S., Verdel,A., Lemercier,C. and Seigneurin-Berny,D. (2001) Functional significance of histone deacetylase diversity. Curr. Opin. Genet. Dev., 11, 162–166. [DOI] [PubMed] [Google Scholar]
- Kuo M.H. and Allis,C.D. (1998) Roles of histone acetyltransferases and deacetylases in gene regulation. BioEssays, 20, 615–626. [DOI] [PubMed] [Google Scholar]
- Kwek S.S., Derry,J., Tyner,A.L., Shen,Z. and Gudkov,A.V. (2001) Functional analysis and intracellular localization of p53 modified by SUMO-1. Oncogene, 20, 2587–2599. [DOI] [PubMed] [Google Scholar]
- Lehembre F., Badenhorst,P., Muller,S., Travers,A., Schweisguth,F. and Dejean,A. (2000) Covalent modification of the transcriptional repressor tramtrack by the ubiquitin-related protein Smt3 in Drosophila flies. Mol. Cell. Biol., 20, 1072–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B. and Shuai,K. (2001) Induction of apoptosis by protein inhibitor of activated Stat1 through c-Jun NH2-terminal kinase activation. J. Biol. Chem., 276, 36624–36631. [DOI] [PubMed] [Google Scholar]
- Lu J., McKinsey,T.A., Nicol,R.L. and Olson,E.N. (2000) Signal-dependent activation of the MEF2 transcription factor by dissociation from histone deacetylases. Proc. Natl Acad. Sci. USA, 97, 4070–4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A. (1997) Predicting coiled-coil regions in proteins. Curr. Opin. Struct. Biol., 7, 388–393. [DOI] [PubMed] [Google Scholar]
- Magnaghi-Jaulin L., Groisman,R., Naguibneva,I., Robin,P., Lorain,S., Le Villain,J.P., Troalen,F., Trouche,D. and Harel-Bellan,A. (1998) Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature, 391, 601–605. [DOI] [PubMed] [Google Scholar]
- Mahajan R., Delphin,C., Guan,T., Gerace,L. and Melchior,F. (1997) A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell, 88, 97–107. [DOI] [PubMed] [Google Scholar]
- Matunis M.J., Coutavas,E. and Blobel,G. (1996) A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J. Cell Biol., 135, 1457–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matunis M.J., Wu,J. and Blobel,G. (1998) SUMO-1 modification and its role in targeting the Ran GTPase-activating protein, RanGAP1, to the nuclear pore complex. J. Cell Biol., 140, 499–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinsey T.A., Zhang,C.L., Lu,J. and Olson,E.N. (2000) Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature, 408, 106–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinsey T.A., Zhang,C.L. and Olson,E.N. (2001) Identification of a signal-responsive nuclear export sequence in class II histone deacetylases. Mol. Cell. Biol., 21, 6312–6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchior F. (2000) SUMO—nonclassical ubiquitin. Annu. Rev. Cell. Dev. Biol., 16, 591–626. [DOI] [PubMed] [Google Scholar]
- Meluh P.B. and Koshland,D. (1995) Evidence that the MIF2 gene of Saccharomyces cerevisiae encodes a centromere protein with homology to the mammalian centromere protein CENP-C. Mol. Biol. Cell, 6, 793–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miska E.A., Karlsson,C., Langley,E., Nielsen,S.J., Pines,J. and Kouzarides,T. (1999) HDAC4 deacetylase associates with and represses the MEF2 transcription factor. EMBO J., 18, 5099–5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miska E.A., Langley,E., Wolf,D., Karlsson,C., Pines,J. and Kouzarides,T. (2001) Differential localization of HDAC4 orchestrates muscle differentiation. Nucleic Acids Res., 29, 3439–3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller S., Matunis,M.J. and Dejean,A. (1998) Conjugation with the ubiquitin-related modifier SUMO-1 regulates the partitioning of PML within the nucleus. EMBO J., 17, 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller S., Berger,M., Lehembre,F., Seeler,J.S., Haupt,Y. and Dejean,A. (2000) c-Jun and p53 activity is modulated by SUMO-1 modification. J. Biol. Chem., 275, 13321–13329. [DOI] [PubMed] [Google Scholar]
- Muller S., Hoege,C., Pyrowolakis,G. and Jentsch,S. (2001) SUMO, ubiquitin’s mysterious cousin. Nature Rev. Mol. Cell Biol., 2, 202–210. [DOI] [PubMed] [Google Scholar]
- Pichler A., Gast,A., Seeler,J.S., Dejean,A. and Melchior,F. (2002) The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell, 108, 109–120. [DOI] [PubMed] [Google Scholar]
- Poukka H., Karvonen,U., Janne,O.A. and Palvimo,J.J. (2000) Covalent modification of the androgen receptor by small ubiquitin-like modifier 1 (SUMO-1). Proc. Natl Acad. Sci. USA, 97, 14145–14150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodel B. et al. (2000) The zinc finger protein Gfi-1 can enhance STAT3 signaling by interacting with the STAT3 inhibitor PIAS3. EMBO J., 19, 5845–5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez M.S., Desterro,J.M., Lain,S., Midgley,C.A., Lane,D.P. and Hay,R.T. (1999) SUMO-1 modification activates the transcriptional response of p53. EMBO J., 18, 6455–6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez M.S., Dargemont,C. and Hay,R.T. (2001) SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J. Biol. Chem., 276, 12654–12659. [DOI] [PubMed] [Google Scholar]
- Sachdev S., Bruhn,L., Sieber,H., Pichler,A., Melchior,F. and Grosschedl,R. (2001) PIASy, a nuclear matrix-associated SUMO E3 ligase, represses LEF1 activity by sequestration into nuclear bodies. Genes Dev., 15, 3088–3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh H. and Hinchey,J. (2000) Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J. Biol. Chem., 275, 6252–6258. [DOI] [PubMed] [Google Scholar]
- Seeler J.S. and Dejean,A. (2001) SUMO: of branched proteins and nuclear bodies. Oncogene, 20, 7243–7249. [DOI] [PubMed] [Google Scholar]
- Seeler J.S., Marchio,A., Sitterlin,D., Transy,C. and Dejean,A. (1998) Interaction of SP100 with HP1 proteins: a link between the promyelocytic leukemia-associated nuclear bodies and the chromatin compartment. Proc. Natl Acad. Sci. USA, 95, 7316–7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeler J.S., Marchio,A., Losson,R., Desterro,J.M., Hay,R.T., Chambon,P. and Dejean,A. (2001) Common properties of nuclear body protein SP100 and TIF1α chromatin factor: role of SUMO modification. Mol. Cell. Biol., 21, 3314–3324.11313457 [Google Scholar]
- Sparrow D.B., Miska,E.A., Langley,E., Reynaud-Deonauth,S., Kotecha,S., Towers,N., Spohr,G., Kouzarides,T. and Mohun,T.J. (1999) MEF-2 function is modified by a novel co-repressor, MITR. EMBO J., 18, 5085–5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternsdorf T., Jensen,K. and Will,H. (1997) Evidence for covalent modification of the nuclear dot-associated proteins PML and Sp100 by PIC1/SUMO-1. J. Cell Biol., 139, 1621–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunnikov A.V., Aravind,L. and Koonin,E.V. (2001) Saccharomyces cerevisiae SMT4 encodes an evolutionarily conserved protease with a role in chromosome condensation regulation. Genetics, 158, 95–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y., Kahyo,T., Toh,E.A., Yasuda,H. and Kikuchi,Y. (2001) Yeast Ull1/Siz1 is a novel SUMO1/Smt3-ligase for septin components and functions as an adaptor between conjugating enzyme and substrates. J. Biol. Chem., 27, 27. [DOI] [PubMed] [Google Scholar]
- Unger T., Juven-Gershon,T., Moallem,E., Berger,M., Vogt Sionov,R., Lozano,G., Oren,M. and Haupt,Y. (1999) Critical role for Ser20 of human p53 in the negative regulation of p53 by Mdm2. EMBO J., 18, 1805–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A.H. and Yang,X.J. (2001) Histone deacetylase 4 possesses intrinsic nuclear import and export signals. Mol. Cell. Biol., 21, 5992–6005. [DOI] [PMC free article] [PubMed] [Google Scholar]