Modification with SUMO: A role in transcriptional regulation (original) (raw)
Abstract
Small ubiquitin-related modifier (SUMO) is a protein moiety that is ligated to lysine residues in a variety of target proteins. The addition of SUMO can modulate the ability of proteins to interact with their partners, alter their patterns of subcellular localization and control their stability. It is clear that SUMO influences many different biological processes, but recent data suggest that it is particularly important in the regulation of transcription. Indeed, several transcription factors, such as Sp3, c-Jun, c-Myb and various nuclear receptors, have recently been shown to be subject to sumoylation and, although this modification can have a positive influence, a growing body of evidence highlights its role in the negative regulation of transcription. This review summarizes recent experiments focusing on sumoylation and transcriptional repression.
Introduction
Post-translational modifications of proteins, such as phosphorylation, acetylation, methylation, ubiquitination and sumoylation (the addition of the small ubiquitin-related modifier, or SUMO), have critical roles in many cellular processes owing to their ability to cause rapid changes in the functions of pre-existing proteins, multiprotein complexes and subcellular structures. Ubiquitination and sumoylation are unusual in that the modifier itself is a small polypeptide (Hochstrasser, 2000). The conjugation of ubiquitin to lysine residues has a well-established role in earmarking proteins for degradation by the 26S proteasome, but it is also involved in many other cellular functions including the regulation of translation and intracellular transport (Weissman, 2001). It has also been proposed that ubiquitination is essential for the function of acidic transcriptional activation domains (Salghetti et al., 2001), suggesting that this modification can be directly involved in gene activation.
SUMO is structurally related to ubiquitin, is of similar size (their molecular masses are 11 and 9 kDa, respectively) and is also ligated to lysine residues within its target proteins. In sumoylation, the target lysine generally falls within a recognizable consensus, namely ψ-Lys-X-Glu (where ψ is a large hydrophobic amino acid, most commonly isoleucine or valine, and X is any residue). Three SUMO family members, SUMO-1/Smt3C, SUMO-2/Smt3A and SUMO-3/Smt3B, are known to exist in mammals (Melchior, 2000; Muller et al., 2001). Like ubiquitin, SUMO-2 and SUMO-3 have been shown to have the ability to form polymeric chains, suggesting that modification by SUMO-1 and SUMO-2 or SUMO-3 might have distinct functional consequences (Saitoh & Hinchey, 2000; Tatham et al., 2001). However, because of the paucity of studies on SUMO-2 and SUMO-3, this review will focus on SUMO-1.
SUMO is emerging as a versatile modifier for a large number of proteins in many different pathways, and the consequences of this modification seem to be as diverse as its targets (Fig. 1A). Indeed, available data currently implicate SUMO in the regulation of protein–protein interactions (Matunis et al., 1998), subcellular nuclear localization (Seeler & Dejean, 2001; Pichler & Melchior, 2002), protein–DNA interactions (Goodson et al., 2001; Hong et al., 2001) and enzymatic activity (Hardeland et al., 2002), and there is also evidence that SUMO can act as an antagonist of ubiquitin (Desterro et al., 1998; Hoege et al., 2002).
Figure 1.
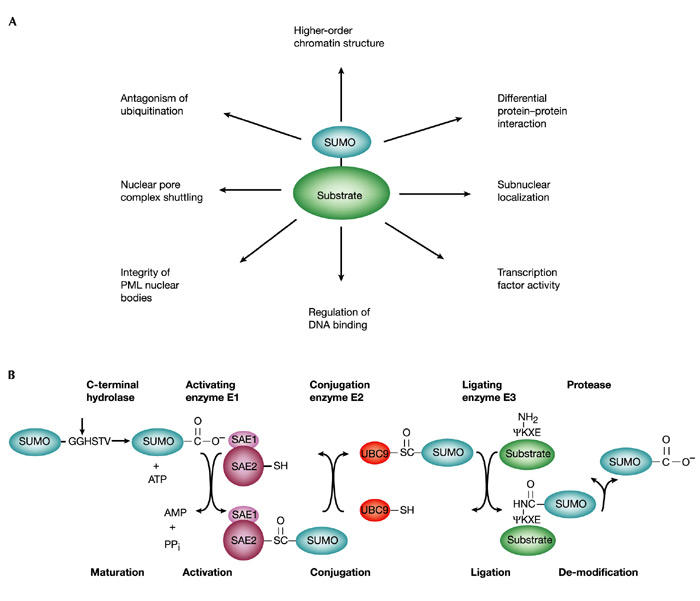
Specificity in SUMO signalling and conjugation. (A) Signalling functions of SUMO. Some of the known functions of sumoylation are indicated with respect to transcriptional regulation. (B) The SUMO conjugating pathway. SUMO is synthesized as a precursor and processed by hydrolases to make the carboxy-terminal double-glycine motif available for conjugation (vertical arrow). It is subsequently conjugated to proteins by means of E1 activating (SAE1(AOS1)/SAE2(UBA2)), E2 conjugating (Ubc9) and E3 ligating enzymes (PIAS family or RanBP2, not shown). The E3-like proteins might serve to increase the affinity between Ubc9 (E2) and the substrates by bringing them into close proximity in catalytically favourable orientations, allowing sumoylation to occur at a maximal rate. The resulting isopeptide bond is stable and its disruption requires a desumoylating enzyme. PML, promyelocytic leukaemia protein.
Despite the multiplicity of its cellular effects, the exact function of SUMO conjugation is unknown. This review will present basic features of the sumoylation process and will then focus on the negative effects of sumoylation on transcriptional regulation. Additional background on other effects of sumoylation can be found in several recent reviews (Melchior, 2000; Muller et al., 2001; Seeler & Dejean, 2001; Kim et al., 2002b; Pichler & Melchior, 2002).
The sumoylation of transcription factors has been shown to have a range of differing effects on their activity. For example, sumoylation of the heat shock factors HSF1 and HSF2 (Goodson et al., 2001; Hong et al., 2001) stimulates their DNA-binding activity. Moreover, conjugation of SUMO to the co-activator GRIP1 (glucocorticoid receptor-interacting protein-1) and to the viral protein IE2-p86 enhances their activity (Hofmann et al., 2000; Kotaja et al., 2002a). In contrast, the sumoylation of many diverse transcription factors, such as Sp3, c-Jun, c-Myb, AP2 (activating enhancer-binding protein-2) and nuclear receptors, has been shown to correlate with downregulation of their transcriptional activation potency (Muller et al., 2000; Poukka et al., 2000; Abdel-Hafiz et al., 2002; Bies et al., 2002; Eloranta & Hurst, 2002; Nishida & Yasuda, 2002; Ross et al., 2002; Sapetschnig et al., 2002; Tian et al., 2002). Interestingly, several negative regulatory domains, within these and other transcription factors, that were hitherto thought to be unrelated have now been found to contain motifs corresponding to the consensus for sumoylation (Fig. 2) (Iniguez-Lluhi & Pearce, 2000; Snowden et al., 2000; Yang et al., 2002). Most interestingly, perhaps, some of these sumoylation motifs themselves have been shown to be capable of mediating transcriptional repression, raising the possibility that one mechanistic element common to the dampening of transcriptional activity might be the attachment of a SUMO moiety.
Figure 2.
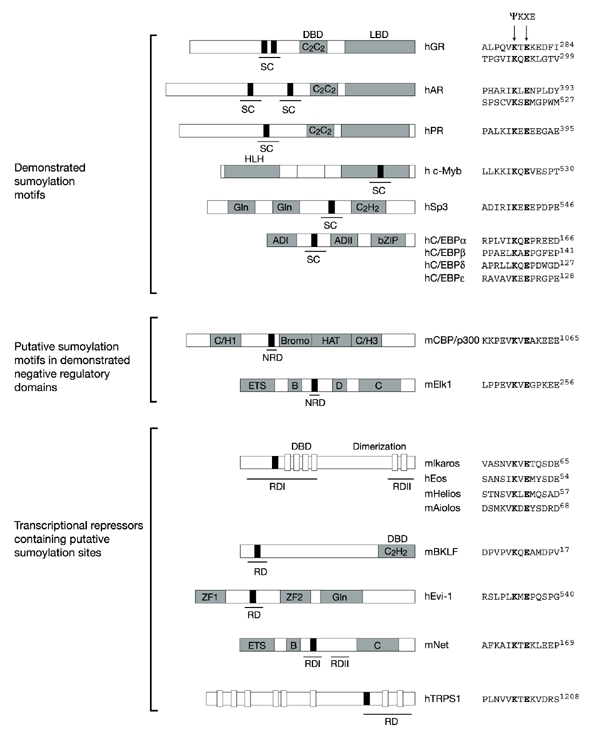
Sumoylation sites in various transcriptional regulators. Black boxes indicate sumoylation sites. The synergy control motifs (SC), negative regulatory domains (NRD) and repression domains (RD) are underlined. Other characterized domains are indicated with the following abbreviations: DBD, DNA-binding domain; LBD, ligand-binding domain; HLH, helix–loop–helix; Gln, rich in glutamine; AD, activation domain; bZIP, basic zipper; Bromo, bromodomain; HAT, histone acetyl transferase; ZF, zinc finger. Sequence alignments of sumoylation sites are indicated for each protein. h, human; m, mouse; GR, glucocorticoid receptor; AR, androgen receptor; PR, progesterone receptor; C/EBP, CCAAT/enhancer-binding protein; BKLF, basic krüppel-like factor; TRPS1, tricho-rhino-phalangeal syndrome type I.
The SUMO conjugation pathway
The pathway of sumoylation is analogous to that of ubiquitination, but SUMO conjugation involves a different set of enzymes (Fig. 1B). Specifically, SUMO is activated in an ATP-dependent manner by an E1-activating enzyme, which consists of an SAE1(AOS1)sAE2(UBA2) heterodimer. Activated SUMO is transferred to Ubc9, the E2-conjugating enzyme, and is subsequently attached to the ε-amino group of a specific lysine in the target protein (Melchior, 2000; Muller et al., 2001). Even though Ubc9 binds to the SUMO acceptor site and efficiently transfers SUMO to selected targets in vitro, a specific SUMO E3-ligating enzyme might be required for efficient and properly targeted modification in vivo. Two classes of SUMO E3 ligases have been identified recently. The first consists of Saccharomyces cerevisiae Siz1 and Siz2, and several related members of the metazoan protein inhibitor of activated STAT (PIAS) family (Johnson & Gupta, 2001; Kahyo et al., 2001; Sachdev et al., 2001). These proteins are characterized by an essential RING-like domain with similarities to the RING finger of ubiquitin E3 ligases. The second class currently has only one known member, RanBP2/Nup358, a component of the nuclear pore complex (Pichler et al., 2002). This protein contains domains that can bind to RanGTP and RanGDP, as well as repeats for nuclear transport receptor binding and a cyclophilin homology region, but has no obvious similarity to other E3 ligases.
Sumoylation is readily reversed by specific proteases (Fig. 1B). These enzymes seem to have a dual role, because they can also act as carboxy-terminal hydrolases in the processing of SUMO to its mature form.
Transcriptional activators can be restrained by sumoylation
The currently known SUMO target proteins fall into multiple categories, such as transcription factors, signal transducers, enzymes or viral proteins, making a simple explanation for SUMO's biological role unlikely (Supplementary Table 1). Nevertheless, the following examples illustrate the direct involvement of SUMO modifications in gene regulation.
Recently, the sumoylation of a number of transcriptional activators has been investigated and, in general, the mutation of their sumoylation sites has been found to increase their transcriptional activity (Muller et al., 2000; Poukka et al., 2000; Abdel-Hafiz et al., 2002; Bies et al., 2002; Eloranta & Hurst, 2002; Nishida & Yasuda, 2002; Ross et al., 2002; Sapetschnig et al., 2002; Tian et al., 2002). One interpretation of this is that SUMO is a negative regulator of transactivation domain potency. Consistent with this idea is the finding that overexpression of free SUMO-1 can suppress AP2 and AP2-mediated transcription specifically co-activated by the CREB-binding protein (CBP)/p300-interacting transactivator with ED-rich tail (CITED) protein (Eloranta & Hurst, 2002). Moreover, the E3 SUMO ligases protein inhibitor of activated STAT (PIAS1) and PIASxα repress androgen-receptor-mediated transcription in a manner dependent on the ectopic expression of SUMO-1 and the E3 RING-finger-like domain (Nishida & Yasuda, 2002). However, the generality of this interpretation is unclear because the repression of p53 and leukocyte enhancer factor-1 (LEF-1) activity, which is mediated by PIAS1 and PIASy, respectively, does not depend on the p53 or LEF-1 sumoylation sites (Sachdev et al., 2001; Schmidt & Muller, 2002). This result suggests that PIAS proteins might be recruited to, and act on, other associated targets that mediate repression. Indeed, a PIAS1 mutant lacking the RING-finger-like domain has been reported to activate p53-mediated gene expression as efficiently as the wild-type PIAS1 does (Megidish et al., 2002), implying that additional functions of PIAS proteins or PIAS-associated proteins are likely to be important (Kotaja et al., 2002b). Similarly, the SUMO E2 enzyme Ubc9, which interacts with the androgen receptor and with the transcriptional repressor TEL, seems to regulate their activities independently of its SUMO-E2 enzyme activity (Chakrabarti et al., 1999; Poukka et al., 1999). Thus, interpreting the effects of sumoylation has often been complicated by the possibility that the overexpression of components of the SUMO conjugation machinery might regulate transcription factor activity independently of the roles of these enzymes as providers of SUMO groups. Nevertheless, the most commonly observed effect of sumoylation is a negative effect on transcription.
Sumoylation sites in negative regulatory domains
Additional evidence for an association between the sumoylation machinery and transcriptional regulation has come from a study that involved mapping negative domains within transcription factors. It was found that several unrelated proteins such as nuclear receptors, c-Myb, CCAAT/enhancer-binding protein (C/EBP) and the Sp1-related protein Sp3 contain small domains known as synergy control motifs (Fig. 2; SC). These domains have been shown to downregulate the synergistic activation that occurs at compound, but not single, response elements (Iniguez-Lluhi & Pearce, 2000). Examination of the SC sequences has revealed that these seemingly distinct elements do share limited homology and, in particular, that each contains a potential sumoylation site, ψ-Lys-X-Glu (Fig. 2). Recent work on Sp3 supports the view that the sumoylation motif is directly involved in reducing transcriptional activity. Sp3 is typically found to be inactive or to act only as a weak activator. Remarkably, a single lysine residue within the SC motif of Sp3 is absolutely essential for the inhibition of its activity, and this residue has now been shown to be the target of sumoylation. Indeed, all mutations that prevented SUMO modifications strongly enhanced the transcriptional activity of Sp3, supporting a role for SUMO in transactivation silencing (Sapetschnig et al., 2002). It was originally proposed that the presence of multiple SC motifs (now known to encompass sumoylation sites) allows for the recruitment of a putative SC factor (SCF) that attenuates synergistic activation (Iniguez-Lluhi & Pearce, 2000). Recent results suggest that the negative factor is likely to be SUMO, or potentially the E2 ligase Ubc9, which is capable of binding SUMO consensus sites (Sampson et al., 2001). Evidence from Gal4–SUMO–Sp3 fusions is most consistent with the functional repressor being SUMO itself (Ross et al., 2002), although an involvement of Ubc9 cannot be excluded because SUMO can bind this enzyme (Gong et al., 1997).
Mapping of other negative regulatory domains has also led to the identification of putative sumoylation sites. It was recently shown that CBP/p300 and Elk-1 contain short motifs that function as repression domains (Fig. 2, NRD) when fused to the DNA-binding domain of Gal4. These short domains share little overall homology but, significantly, each contains ψ-Lys-X-Glu motifs. Most importantly, mutation of either the critical lysine or the consensus glutamate residue in the Elk-1 protein abolishes repression, suggesting that sumoylation (or Ubc9 binding) is mechanistically involved in repression (Snowden et al., 2000; Yang et al., 2002).
Can SUMO directly mediate gene repression?
It has been found that a Gal4–VP16 protein carrying a single SUMO-motif-containing inhibitory domain from C/EBPε, c-Jun (Kim et al., 2002a) or the glucocorticoid receptor (Iniguez-Lluhi & Pearce, 2000), possesses less than 10% of the activity of the parental protein and that the attachment of additional sumoylation motifs leads to further inhibition. Most strikingly, even when these elements alone are fused to Gal4, they are able to strongly repress the activity of other activator proteins. Inhibition is proportional to the number of sumoylation consensus sites, and these inhibitory domains are clearly highly potent and portable, indicating that SUMO modification does not merely modulate but actually provides inhibitory domain function (Iniguez-Lluhi & Pearce, 2000; Kim et al., 2002a; Yang et al., 2002). It has also been shown that a single SUMO moiety fused to Gal4 can repress transcription in reporter gene assays (Ross et al., 2002), which is again consistent with the view that SUMO itself can have a negative effect on transcription.
Although most of the experiments described so far have focused on well-characterized activators of transcription and on the attenuation of their function, the finding that Gal4–SUMO can repress transcription raises the possibility that SUMO might have a more direct role and that the repression domains of existing repressor proteins might operate through sumoylation. Indeed, it is notable that several well-characterized repressors contain Ψ-Lys-X-Glu SUMO consensus sites within their repression domains (Fig. 2, RD). However, the functional relevance of this must still be evaluated.
How might SUMO attenuate transcription?
There is currently no clear molecular basis for the mechanism by which SUMO addition regulates transcription factor activity. Obviously, modification with SUMO alters the surface of the target protein and might cause either general conformational changes or specific changes at critical interfaces, thereby influencing the ability of the protein to interact with binding partners. One model, therefore, is that modification with SUMO promotes or inhibits protein–protein interactions and thereby regulates the assembly of transcriptional complexes. Consistent with this idea is the fact that SUMO-1 interacts with the CHD3/ZFH zinc-finger-containing helicase that is present in histone deacetylase complexes (Minty et al., 2000). However, the precise interaction will vary in each instance depending on the exact sequence of the protein, and it is unlikely that it will be possible to develop any predictive model of action.
Sumoylation might also block alternative lysine-targeted modifications such as acetylation or ubiquitination. For example, sumoylation of the NF-κB inhibitor IκBα occurs on a lysine residue that is also a target for ubiquitination and thus stabilizes IκBα, preventing its ubiquitination and subsequent degradation and, consequently, the activation of NF-κB (Desterro et al., 1998). Modulation of the level of acetylation is another possible way in which conjugation of SUMO could repress transcriptional activity. Indeed, the sumoylated lysine of Sp3 is also subject to acetylation (Braun et al., 2001). Although the effect of this modification is still unclear, one could imagine a scenario in which the transcriptional activity of Sp3, and possibly other transcription factors, could be regulated by different modifications at the same lysine.
Several co-repressors of the histone deacetylase family have been shown to be subject to sumoylation, namely HDAC1, HDAC4 and HDAC6 (David et al., 2002; Kirsh et al., 2002). Indeed, sumoylation-deficient point mutants of HDAC1 and HDAC4 show a slightly impaired ability to repress transcription as well as a reduced histone deacetylase activity. This suggests that modifications with SUMO are required for the full repression activity of these proteins and further implicates this type of post-translational modification in gene regulatory events, although the precise mechanism again remains elusive.
Perhaps the best clue to how sumoylation modulates transcriptional activity has come from examinations of the effect that mutating sumoylation sites has on the subnuclear distribution of transcription factors. For example, modification of the promyelocytic leukemia protein (PML) with SUMO is involved in the formation of nuclear subdomains called nuclear PML oncogenic domains (PODs) or nuclear bodies, and in the recruitment of other nuclear-body-associated proteins such as Sp100, Daxx and SUMO-1 itself (Seeler & Dejean, 2001). Nuclear PODs are subnuclear structures that are associated with the nuclear matrix and it has been postulated that they have roles in transcription, cellular transformation, cell cycle regulation and viral infection. Interestingly, SUMO-1-modified, inactive or repressive forms of Sp3 and LEF-1 have been found to localize to the nuclear periphery and to nuclear dots, whereas the more transcriptionally active forms, which lack SUMO-1, have a more diffuse nuclear localization (Sachdev et al., 2001; Ross et al., 2002). These findings suggest that the sumoylation of transcription factors serves to organize specific protein complexes into nuclear matrix sites within nuclear bodies, thus regulating their activities either negatively or positively (Sachdev et al., 2001; Best et al., 2002). How transcription factor activity and subnuclear localization are functionally linked remains an intriguing question for further investigation.
Concluding remarks
Modifications of transcription factors with SUMO are being discovered at a rapid pace. Future efforts to understand in detail the many roles of SUMO in transcription will undoubtedly turn up additional, previously unknown E3 ligases, their protein targets and their mechanisms of action. Finally, because lysine residues can also be subject to acetylation, methylation and ubiquitination, and because phosphorylation can, in some instances, prevent the formation of SUMO-1 conjugates (Muller et al., 2000), competition between SUMO and other post-translational modifications is emerging as a common mechanism that permits the complex control of transcriptional activity.
Supplementary data are available at EMBO reports online (http://www.nature.com/embor/journal/vaop/ncurrent/extref/4-embor738-s1.pdf).
Supplementary Material
Supplementary Information



References
- Abdel-Hafiz H.A., Takimoto G.S., Tung L. & Horwitz K.B. (2002) The inhibitory function (IF) in human progesterone receptor N-termini binds small ubiquitin-like modifier (SUMO-1) protein to regulate autoinhibition and transrepression. J. Biol. Chem., 277, 33950–33956. [DOI] [PubMed] [Google Scholar]
- Best J.L. et al. (2002) SUMO-1 protease-1 regulates gene transcription through PML. Mol. Cell, 10, 843–855. [DOI] [PubMed] [Google Scholar]
- Bies J., Markus J. & Wolff L. (2002) Covalent attachment of the SUMO-1 protein to the negative regulatory domain of the c-Myb transcription factor modifies its stability and transactivation capacity. J. Biol. Chem., 277, 8999–9009. [DOI] [PubMed] [Google Scholar]
- Braun H., Koop R., Ertmer A., Nacht S. & Suske G. (2001) Transcription factor Sp3 is regulated by acetylation. Nucleic Acids Res., 29, 4994–5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti S.R. et al. (1999) Modulation of TEL transcription activity by interaction with the ubiquitin-conjugating enzyme UBC9. Proc. Natl Acad. Sci. USA, 96, 7467–7472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David G., Neptune M.A. & DePinho R.A. (2002) SUMO-1 modification of histone deacetylase 1 (HDAC1) modulates its biological activities. J. Biol. Chem., 277, 23658–23663. [DOI] [PubMed] [Google Scholar]
- Desterro J.M., Rodriguez M.S. & Hay R.T. (1998) SUMO-1 modification of IκBα inhibits NF-κB activation. Mol. Cell, 2, 233–239. [DOI] [PubMed] [Google Scholar]
- Eloranta J.J. & Hurst H.C. (2002) Transcription factor AP-2 interacts with the SUMO-conjugating enzyme UBC9 and is sumolated in vivo. J. Biol. Chem., 277, 30798–30804. [DOI] [PubMed] [Google Scholar]
- Gong L., Kamitani T., Fujise K., Caskey L.S. & Yeh E.T. (1997) Preferential interaction of sentrin with a ubiquitin-conjugating enzyme, Ubc9. J. Biol. Chem., 272, 28198–28201. [DOI] [PubMed] [Google Scholar]
- Goodson M.L. et al. (2001) Sumo-1 modification regulates the DNA binding activity of heat shock transcription factor 2, a promyelocytic leukemia nuclear body associated transcription factor. J. Biol. Chem., 276, 18513–18518. [DOI] [PubMed] [Google Scholar]
- Hardeland U., Steinacher R., Jiricny J. & Schar P. (2002) Modification of the human thymine-DNA glycosylase by ubiquitin-like proteins facilitates enzymatic turnover. EMBO J., 21, 1456–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstrasser M. (2000) Biochemistry. All in the ubiquitin family. Science, 289, 563–564. [DOI] [PubMed] [Google Scholar]
- Hoege C., Pfander B., Moldovan G.L., Pyrowolakis G. & Jentsch S. (2002) RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature, 419, 135–141. [DOI] [PubMed] [Google Scholar]
- Hofmann H., Floss S. & Stamminger T. (2000) Covalent modification of the transactivator protein IE2-p86 of human cytomegalovirus by conjugation to the ubiquitin-homologous proteins SUMO-1 and hSMT3b. J. Virol., 74, 2510–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y. et al. (2001) Regulation of heat shock transcription factor 1 by stress-induced SUMO-1 modification. J. Biol. Chem., 276, 40263–40267. [DOI] [PubMed] [Google Scholar]
- Iniguez-Lluhi J.A. & Pearce D. (2000) A common motif within the negative regulatory regions of multiple factors inhibits their transcriptional synergy. Mol. Cell. Biol., 20, 6040–6050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson E.S. & Gupta A.A. (2001) An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell, 106, 735–744. [DOI] [PubMed] [Google Scholar]
- Kahyo T., Nishida T. & Yasuda H. (2001) Involvement of PIAS1 in the sumoylation of tumor suppressor p53. Mol. Cell, 8, 713–718. [DOI] [PubMed] [Google Scholar]
- Kim J., Cantwell C.A., Johnson P.F., Pfarr C.M. & Williams S.C. (2002a) Transcriptional activity of CCAAT/enhancer binding proteins is controlled by a conserved inhibitory domain that is a target for sumoylation. J. Biol. Chem., 277, 38037–38044. [DOI] [PubMed] [Google Scholar]
- Kim K.I., Baek S.H. & Chung C.H. (2002b) Versatile protein tag, SUMO: its enzymology and biological function. J. Cell Physiol., 191, 257–268. [DOI] [PubMed] [Google Scholar]
- Kirsh O. et al. (2002) The SUMO E3 ligase RanBP2 promotes modification of the HDAC4 deacetylase. EMBO J., 21, 2682–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotaja N., Karvonen U., Janne O.A. & Palvimo J.J. (2002a) The nuclear receptor interaction domain of GRIP1 is modulated by covalent attachment of SUMO-1. J. Biol. Chem., 277, 30283–30288. [DOI] [PubMed] [Google Scholar]
- Kotaja N., Karvonen U., Janne O.A. & Palvimo J.J. (2002b) PIAS proteins modulate transcription factors by functioning as SUMO-1 ligases. Mol. Cell. Biol., 22, 5222–5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matunis M.J., Wu J. & Blobel G. (1998) SUMO-1 modification and its role in targeting the Ran GTPase-activating protein, RanGAP1, to the nuclear pore complex. J. Cell Biol., 140, 499–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megidish T., Xu J.H. & Xu C.W. (2002) Activation of p53 by protein inhibitor of activated Stat1 (PIAS1). J. Biol. Chem., 277, 8255–8259. [DOI] [PubMed] [Google Scholar]
- Melchior F. (2000) SUMO—nonclassical ubiquitin. Annu. Rev. Cell Dev. Biol., 16, 591–626. [DOI] [PubMed] [Google Scholar]
- Minty A., Dumont X., Kaghad M. & Caput D. (2000) Covalent modification of p73alpha by SUMO-1. Two-hybrid screening with p73 identifies novel SUMO-1-interacting proteins and a SUMO-1 interaction motif. J. Biol. Chem., 275, 36316–36323. [DOI] [PubMed] [Google Scholar]
- Muller S. et al. (2000) c-Jun and p53 activity is modulated by SUMO-1 modification. J. Biol. Chem., 275, 13321–13329. [DOI] [PubMed] [Google Scholar]
- Muller S., Hoege C., Pyrowolakis G. & Jentsch S. (2001) SUMO, ubiquitin's mysterious cousin. Nature Rev. Mol. Cell Biol., 2, 202–210. [DOI] [PubMed] [Google Scholar]
- Nishida T. & Yasuda H. (2002) PIAS1 and PIASxa function as SUMO-E3 ligases toward androgen receptor, and repress androgen receptor-dependent transcription. J. Biol. Chem., 277, 41311–41317. [DOI] [PubMed] [Google Scholar]
- Pichler A., Gast A., Seeler J.S., Dejean A. & Melchior F. (2002) The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell, 108, 109–120. [DOI] [PubMed] [Google Scholar]
- Pichler A. & Melchior F. (2002) Ubiquitin-related modifier SUMO1 and nucleocytoplasmic transport. Traffic, 3, 381–387. [DOI] [PubMed] [Google Scholar]
- Poukka H., Aarnisalo P., Karvonen U., Palvimo J.J. & Janne O.A. (1999) Ubc9 interacts with the androgen receptor and activates receptor-dependent transcription. J. Biol. Chem., 274, 19441–19446. [DOI] [PubMed] [Google Scholar]
- Poukka H., Karvonen U., Janne O.A. & Palvimo J.J. (2000) Covalent modification of the androgen receptor by small ubiquitin-like modifier 1 (SUMO-1). Proc. Natl Acad. Sci. USA, 97, 14145–14150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross S., Best J.L., Zon L.I. & Gill G. (2002) SUMO-1 modification represses Sp3 transcriptional activation and modulates its subnuclear localization. Mol. Cell, 10, 831–842. [DOI] [PubMed] [Google Scholar]
- Sachdev S. et al. (2001) PIASy, a nuclear matrix-associated SUMO E3 ligase, represses LEF1 activity by sequestration into nuclear bodies. Genes Dev., 15, 3088–3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh H. & Hinchey J. (2000) Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J. Biol. Chem., 275, 6252–6258. [DOI] [PubMed] [Google Scholar]
- Salghetti S.E., Caudy A.A., Chenoweth J.G. & Tansey W.P. (2001) Regulation of transcriptional activation domain function by ubiquitin. Science, 293, 1651–1653. [DOI] [PubMed] [Google Scholar]
- Sampson D.A., Wang M. & Matunis M.J. (2001) The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J. Biol. Chem., 276, 21664–21669. [DOI] [PubMed] [Google Scholar]
- Sapetschnig A. et al. (2002) Transcription factor Sp3 is silenced through SUMO modification by PIAS1. EMBO J., 21, 5206–5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D. & Muller S. (2002) Members of the PIAS family act as SUMO ligases for c-Jun and p53 and repress p53 activity. Proc. Natl Acad. Sci. USA, 99, 2872–2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeler J.S. & Dejean A. (2001) SUMO: of branched proteins and nuclear bodies. Oncogene, 20, 7243–7249. [DOI] [PubMed] [Google Scholar]
- Snowden A.W., Anderson L.A., Webster G.A. & Perkins N.D. (2000) A novel transcriptional repression domain mediates p21(WAF1/CIP1) induction of p300 transactivation. Mol. Cell. Biol., 20, 2676–2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatham M.H. et al. (2001) Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J. Biol. Chem., 276, 35368–35374. [DOI] [PubMed] [Google Scholar]
- Tian S., Poukka H., Palvimo J.J. & Janne O.A. (2002) SUMO-1 modification of the glucocorticoid receptor. Biochem. J., 367, 907–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman A.M. (2001) Themes and variations on ubiquitylation. Nature Rev. Mol. Cell. Biol., 2, 169–178. [DOI] [PubMed] [Google Scholar]
- Yang S.H., Bumpass D.C., Perkins N.D. & Sharrocks A.D. (2002) The ETS domain transcription factor Elk-1 contains a novel class of repression domain. Mol. Cell. Biol., 22, 5036–5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information